Abstract
Parameter estimation for high dimension complex dynamic system is a hot topic. However, the current statistical model and inference approach is known as a large p small n problem. How to reduce the dimension of the dynamic model and improve the accuracy of estimation is more important. To address this question, the authors take some known parameters and structure of system as priori knowledge and incorporate it into dynamic model. At the same time, they decompose the whole dynamic model into subset network modules, based on different modules, and then they apply different estimation approaches. This technique is called Rao‐Blackwellised particle filters decomposition methods. To evaluate the performance of this method, the authors apply it to synthetic data generated from repressilator model and experimental data of the JAK‐STAT pathway, but this method can be easily extended to large‐scale cases.
Inspec keywords: reduced order systems, biochemistry, parameter estimation, nonlinear dynamical systems
Other keywords: model reduction, parameter estimation, nonlinear dynamical biochemical reaction networks, subset network modules, Rao‐Blackwellised particle filters decomposition methods, repressilator model, JAK‐STAT pathway
1 Introduction
Biochemical reaction process is a complex and high‐dimension dynamics system, which includes a variety of feedback loop [1, 2] and possesses strongly non‐linear kinetic characteristics such as chaos, bifurcation, complex disturbance wave and so on [3–7]. The non‐linear complex and high‐dimension biochemical reaction can be decomposed into several sets of chemical substances, and then combined to consider the whole mechanism. On the basis of biochemical reaction, dynamic modelling is accordingly divided into many subset network modules. Both subset network modules and the whole dynamic characteristics should be analysed [8, 9]. This is defined as model reduction techniques. Model reduction techniques decompose a critical biochemical reactions and variables according to core dynamical characteristics of the system. There are two kinds of techniques mostly used to partition the state variables. One is fast and slow decompositions, another is linear and non‐linear decompositions. The former detailed model reduction approaches have singular perturbation techniques in papers [10, 11], hierarchical approach in paper [12], quasi‐steady‐states approximations in papers [13, 14], partial‐equilibriums in paper [15] and kernel‐based manifold learning techniques in paper [16]. The latter includes quasi‐steady and quasi‐equilibrium in paper [9], hierarchy of coarse grained model in paper [17], distribution state estimation in paper [18] and Rao‐Blackwellised particle filters (RBPFs) in paper [19]. In our work, we focus on linear and non‐linear decompositions by using RBPFs.
In the past, the dynamic model of the non‐linear biochemical reaction is generally based on black‐box framework to estimate the parameters and identify the structures of system. Since there exists large p small n problem (number of unknown parameters p is of much larger than sample size n, p ≫ n) in parameter estimation fields, we take some known parameters and structure of system as priori knowledge and incorporate it into dynamic model. In other words, we estimate the parameters and states in non‐linear biochemical reaction network based on grey‐box framework [20]. As we know, most of biochemical reaction networks are non‐linear and non‐Gaussian, however, in which, linear subsystem are still available. Pseudo‐monomolecular or monomolecular reaction is the simplest reaction described by a set of first‐order reactions. In papers [17, 21, 22], pseudo‐monomolecular or monomolecular reaction subsystems are considered as linear subset network modules, based on which, linear and non‐linear decompositions are easy to be realised. For linear kinetic models, appearing as pseudo‐monomolecular or monomolecular reaction subsystems, we propose to estimate the parameters using conventional Kalman filter algorithm. However, for the remaining non‐linear kinetic models, we develop an algorithm to estimate both states and parameters using the particle filter algorithm. It is known as RBPFs [19, 23–27]. Extended Kalman filter (EKF) [28] and unscented Kalman filter (UKF) [29] are the most widely used joint state and parameter estimation algorithm for the non‐linear state‐space model of biochemical network. In this paper, we will compare the three estimation methods that are RBPF, UKF and EKF in synthetic data generated from repressilator model and experimental data from the JAK‐STAT pathway. The results show that RBPF provides a way to handle high‐dimensional problems and bears very good accuracy with quite reasonable complexity.
2 Non‐linear state‐space models
Consider a general non‐linear dynamic system
| (1) |
where e is the individual and k is the time; is the state vector of the e individual at a k time; is the input vector of the e individual at a k time; is the observation vector of the e individual at a k time; f and h are non‐linear functions, θ is the vector of parameters; The initial state x 0 is a Gaussian vector with mean and covariance matrix ; w and v are vectors of white noises with zero mean and joint covariance matrix
Parameters in a non‐linear dynamic system (1) can be treated as additional states in the system. Thus, the state vector in (1) is augmented as
A state‐space equation treating parameters as states can be written as
| (2) |
where is a white Gaussian noise with mean zero and covariance matrix
Supposing a system is divided into two parts: a linear and a non‐linear and the noise is additive, then (2) can be expressed as follows
| (3) |
where and denote the non‐linear and linear states, respectively, and , is the process noise given by
where N(0, σ 2) denotes the normal distribution with 0 as the mean value and σ 2 the variance. Moreover, and v k , have arbitrary fixed probability density function (pdf).
Assume
where a = ((3δ − 1)/2δ), h 2 = 1 − a 2, δ is a discount factor (0, 1], typically around . is the Monte Carlo mean of the parameters and V k being the variance matrix of the parameters at time instant k.
We determine the unknown parameter θ by estimating the augmented state with ; The minimum mean‐squared errors estimation of given is
is approximated by particle filter, for each given parameter sample, is given by Kalman filter. This will result in each parameter particle being associated with one Kalman filter recursion.
3 RBPF algorithm for dual estimation
RBPF algorithms for dual estimation is summarised in Fig. 1.
Fig. 1.

RBPF algorithm for dual estimation
The following two applications of the algorithm (see Fig. 1) are from paper [28]; As the length of paper is limited, there is no explanation about the system equation. For detailed explanation, please see paper [28]. As far as signalling pathways, the topological structure of signalling pathways is known. In such case, the most important work is to estimate the parameters of non‐linear models based on Hill or mass action kinetics. The topological structure of network is used as prior knowledge and incorporated into the kinetic models and repressilator model.
4 Kinetic models for JAK2‐STAT5 signalling pathway
Measurement equation
Model reduction
For decomposing the system and dividing the state variables into linear state variable and non‐linear state variable, let
where denotes the state variable with conditional linear dynamics and denotes the non‐linear state variable. The system equation can be rewritten as the following
where
The measurement equation can be rewritten as the following
where
5 Repressilator
System and measurement equation [31]
where R i is the concentration of mRNA transcript from gene i and P i is the concentration of proteins translated from R i . Estimated parameters: V 1max, V 2max, V 3max, k 12, k 23, k 31.
Model reduction
For decomposing the system and dividing the state variables into linear state variable and non‐linear state variable, let
where denotes the state variable with conditional linear dynamics and denotes the non‐linear state variable. Then, the system equation can be rewritten as the following
where
The measurement equation can be rewritten as the following
where
6 Results
6.1 JAK‐STAT pathway
The initial values of the linear state variables of every particle are assumed as: and . The initial values of the non‐linear state variables of every particle are random number between 0 and 1. The initial values of parameters k 1, k 2, k 3 and k 4 are random number generated from the following intervals k 1 ∈ (−0.299, 0.421), k 2 ∈ (2.16, 2.76), k 3 ∈ (−0.2534, 0.3466) and k 4 ∈ (−0.14342, 0.30658). The estimates by the RBPF, the EKF and maximum likelihood (ML) method [32] are close, but significantly different from the estimates by UKF [29] as shown in Table 1. Under the given above initial values, using the concentration of EpoRA as input, Fig. 2 plot the predicted and observed concentrations of tyrosine phosphorylated STAT5 in the cytoplasm and total STAT5 in the cytoplasm (y 1 and y 2) by the RBPF method, the EKF method and the UKF method. The observed data were from experiment data 1. The results of experiment data 2–4 and the estimated parameters were listed in additional files AFigures 1, 2–3 and ATable 1.
Table 1.
Comparison of estimated parameters in the non‐linear state‐space model for the JAK‐STAT pathway using RBPF, EKF, ML approach and UKF
| Study | k 1 | k 2 | k 3 | k 4 | τ |
|---|---|---|---|---|---|
| RBPF (our study) | 0.022916 | 2.343347 | 0.117178 | 0.102687 | 6.1 |
| EKF | 0.0211 | 2.2788 | 0.1064 | 0.1057 | 6 min |
| ML | 0.0210 | 2.4600 | 0.1066 | 0.1066 | 6.4 min |
| UKF | 0.0515 | 3.3900 | 0.3500 |
Fig. 2.
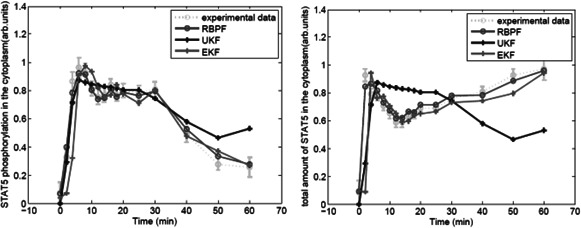
Predicted and observed concentrations of tyrosine phosphorylated STAT5 and total STAT5 in the cytoplasm for experiment 1
Fig. 3.
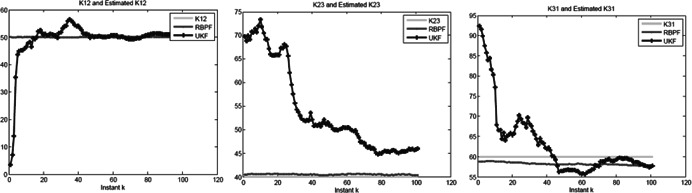
Estimated parameters of K12, K23 and K31 as a function of the time in the non‐linear state‐space model for the synthetic data generated from the repressilator model
6.2 Synthetic data generated from repressilator model
Let k 1m = 1, k 2m = 1, k 3m = 1, k 1p = 1, k 2p = 1, k 3p = 1, K 1 = 1, K 2 = 2, K 3 = 3, n = 3 and V 1max = 150, V 2max = 80, V 3max = 100, K 12 = 50, K 23 = 40, K 31 = 60, the initial values of linear and non‐linear state variables are random numbers between 0 and 1. The initial values of parameters are random number generated from the following intervals: V 1max ∈ (140, 160), V 2max ∈ (70, 90), V 3max ∈ (90, 110), K 12 ∈ (40, 60), K 23 ∈ (20, 40), K 31 ∈ (30, 50). The estimated parameters as a function of time k are shown in Figs. 3 and 4. From Figs. 3 and 4, we can see that at the beginning the estimated parameters quickly converge to the true parameters. This example demonstrates that although the parameters are treated as the states of the systems and hence may change over time, they can reach stable values. The estimated parameters over time k are summarised in ATable 2 in additional files, which demonstrated that the estimates of the parameters were very close to set the value of parameters. In this example, EKF does not converge, high non‐linearity of the repressilator model makes EKF a failure to converge to an optimum. Therefore, we only compare the two methods of RBPF and UKF.
Fig. 4.
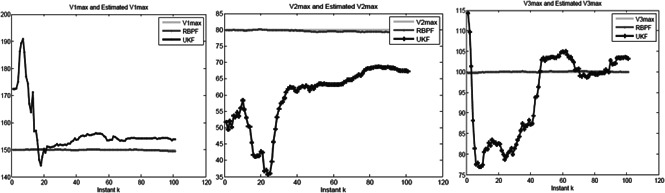
Estimated parameters of V1max, V2max and V3max as a function of the time in the non‐linear state‐space model for the synthetic data generated from the repressilator model
7 Conclusions
To evaluate the performance of our new methods, we have applied it to both synthetic data generated from repressilator model and experimental data of the JAK‐STAT pathway [31, 32]. The structure of both the above examples is known in modelling literature [29, 33]. Therefore, we use structure information and partly known parameters as priori knowledge and then conduct the identification of biochemical reaction networks based on grey box [34–37]. We consider the pseudo‐monomolecular or monomolecular reaction subsystems as linear subset network modules, then the whole dynamic model are decomposed into linear and non‐linear subset modules dynamic model. For linear subset modules dynamic model, we use Kalman filter algorithm to estimate both states and parameters, however, for non‐linear subset modules dynamic model, we adopt the particle filter algorithm to estimate both states and parameters. This model reduction technique is called RBPF. The results show that RBPF method perform well as a new model reduction techniques for high dimension non‐linear dynamic model. As future work, we will apply our algorithms to a high‐dimensional biochemical network in order to improve and validate it.
6 Acknowledgments
Xiaodian Sun and Mario Medvedovic are funded by the NIH Data Coordination and Integration Center for LINCS‐BD2 K grant U54HG008230.
7 References
- 1. Sobieszczanski‐Sobieski J.: ‘Sensitivity of complex, internally coupled systems’, AIAA J., 1990, 28, (1), pp. 153–160 (doi: 10.2514/3.10366) [DOI] [Google Scholar]
- 2. Hurty W.C.: ‘Dynamic analysis of structural systems using component modes’, AIAA J., 1965, 3, (4), pp. 678–685 (doi: 10.2514/3.2947) [DOI] [Google Scholar]
- 3. Roesky P.W., Doumbouya S.I., Schneider F.W.: ‘Chaos induced by delayed feedback’, J. Phys. Chem., 1993, 97, (2), pp. 398–402 (doi: 10.1021/j100104a022) [DOI] [Google Scholar]
- 4. Nitzan A., Ortoleva P., Deutch J., Ross J.: ‘Fluctuations and transitions at chemical instabilities: the analogy to phase transitions’, J. Chem. Phys., 1974, 61, (3), pp. 1056–1074 (doi: 10.1063/1.1681974) [DOI] [Google Scholar]
- 5. Matheson I., Walls D., Gardiner C.: ‘Stochastic models of firstorder nonequilibrium phase transitions in chemical reactions’, J. Stat. Phys., 1975, 12, (1), pp. 21–34 (doi: 10.1007/BF01024182) [DOI] [Google Scholar]
- 6. Lee K.J., McCormick W., Ouyang Q., Swinney H.L.: ‘Pattern formation by interacting chemical fronts’, Science, 1993, 261, (5118), pp. 192–194 (doi: 10.1126/science.261.5118.192) [DOI] [PubMed] [Google Scholar]
- 7. McNeil K., Walls D.: ‘Nonequilibrium phase transitions in chemical reactions’, J. Stat. Phys., 1974, 10, (6), pp. 439–448 (doi: 10.1007/BF01020400) [DOI] [Google Scholar]
- 8. Matthews M.L., Williams C.: ‘Region of attraction estimation of biological continuous Boolean models’, in (Eds.): ‘Book region of attraction estimation of biological continuous Boolean models’ (IEEE, 2012), pp. 1700–1705 [Google Scholar]
- 9. Radulescu O., Gorban A.N., Zinovyev A., Noel V.: ‘Reduction of dynamical biochemical reaction networks in computational biology’, arXiv preprint arXiv:1205.2851, 2012. [DOI] [PMC free article] [PubMed]
- 10. Prescott T.P., Papachristodoulou A.: ‘Layered decomposition for the model order reduction of timescale separated biochemical reaction networks’, J. Theor. Biol., 2014, 356, pp. 113–122 (doi: 10.1016/j.jtbi.2014.04.007) [DOI] [PubMed] [Google Scholar]
- 11. Kourdis P.D., Palasantza A.G., Goussis D.A.: ‘Algorithmic asymptotic analysis of the NF‐κB signaling system’, Comput. Math. Appl., 2013, 65, (10), pp. 1516–1534 (doi: 10.1016/j.camwa.2012.11.004) [DOI] [Google Scholar]
- 12. Radulescu O., Gorban A.N., Vakulenko S., Zinovyev A.: ‘Hierarchies and modules in complex biological systems’. Proc. ECCS'06, 2006.
- 13. Schneider K.R., Wilhelm T.: ‘Model reduction by extended quasi‐steady‐state approximation’, J. Math. Biol., 2000, 40, (5), pp. 443–450 (doi: 10.1007/s002850000026) [DOI] [PubMed] [Google Scholar]
- 14. Segel L.A., Slemrod M.: ‘The quasi‐steady‐state assumption: a case study in perturbation’, SIAM Rev., 1989, 31, (3), pp. 446–477 (doi: 10.1137/1031091) [DOI] [Google Scholar]
- 15. Gallagher P.M., Athayde A.L., Ivory C.F.: ‘The combined flux technique for diffusion – reaction problems in partial equilibrium: application to the facilitated transport of carbon dioxide in aqueous bicarbonate solutions’, Chem. Eng. Sci., 1986, 41, (3), pp. 567–578 (doi: 10.1016/0009‐2509(86)87039‐7) [Google Scholar]
- 16. Dsilva C.J., Talmon R., Gear C.W., Coifman R.R., Kevrekidis I.G.: ‘Data‐driven reduction for multiscale stochastic dynamical systems’, arXiv preprint arXiv:1501.05195, 2015.
- 17. Radulescu O., Gorban A.N., Zinovyev A., Lilienbaum A.: ‘Robust simplifications of multiscale biochemical networks’, BMC Syst. Biol., 2008, 2, (1), p. 86 (doi: 10.1186/1752-0509-2-86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo Y., Wu W., Zhang B., Sun H.: ‘A distributed state estimation method for power systems incorporating linear and nonlinear models’, Int. J. Electr. Power Energy Syst., 2015, 64, pp. 608–616 (doi: 10.1016/j.ijepes.2014.07.053) [DOI] [Google Scholar]
- 19. Schon T., Gustafsson F., Nordlund P.‐J.: ‘Marginalized particle filters for mixed linear/nonlinear state‐space models’, IEEE Trans. Signal Process., 2005, 53, (7), pp. 2279–2289 (doi: 10.1109/TSP.2005.849151) [DOI] [Google Scholar]
- 20. Kristensen N.R., Madsen H., Jørgensen S.B.: ‘Parameter estimation in stochastic grey‐box models’, Automatica, 2004, 40, (2), pp. 225–237 (doi: 10.1016/j.automatica.2003.10.001) [DOI] [Google Scholar]
- 21. Gorban A.N., Radulescu O.: ‘Dynamic and static limitation in multiscale reaction networks, revisited’, Adv. Chem. Eng., 2008, 34, pp. 103–173 (doi: 10.1016/S0065‐2377(08)00003‐3) [Google Scholar]
- 22. Gorban A., Radulescu O., Zinovyev A.Y.: ‘Asymptotology of chemical reaction networks’, Chem. Eng. Sci., 2010, 65, (7), pp. 2310–2324 (doi: 10.1016/j.ces.2009.09.005) [DOI] [Google Scholar]
- 23. Schön T., Gustafsson F.: ‘Particle filters for system identification of state‐space models linear in either parameters or states’, 2003.
- 24. Li P., Goodall R., Kadirkamanathan V.: ‘Parameter estimation of railway vehicle dynamic model using Rao‐Blackwellised particle filter’, in (Eds.): ‘Book parameter estimation of railway vehicle dynamic model using Rao‐Blackwellised particle filter’ (2003) [Google Scholar]
- 25. Daly M.J., Reilly J.P., Morelande M.R.: ‘Rao‐Blackwellised particle filtering for blind system identification’, in (Eds.): ‘Book Rao‐Blackwellised particle filtering for blind system identification’ (IEEE, 2005), vol. 324, pp. iv/321–iv/324 [Google Scholar]
- 26. Karlsson R., Schön T., Gustafsson F.: ‘Complexity analysis of the marginalized particle filter’, 2004.
- 27. Schön T., Karlsson R., Gustafsson F.: ‘The marginalized particle filter in practice’, 2005.
- 28. Sun X., Jin L., Xiong M.: ‘Extended Kalman filter for estimation of parameters in nonlinear state‐space models of biochemical networks’, PloS One, 2008, 3, (11), p. e3758 (doi: 10.1371/journal.pone.0003758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quach M., Brunel N., d'Alché‐Buc F.: ‘Estimating parameters and hidden variables in non‐linear state‐space models based on ODEs for biological networks inference’, Bioinformatics, 2007, 23, (23), pp. 3209–3216 (doi: 10.1093/bioinformatics/btm510) [DOI] [PubMed] [Google Scholar]
- 30. Kisseleva T., Bhattacharya S., Braunstein J., Schindler C.: ‘Signaling through the JAK/STAT pathway, recent advances and future challenges’, Gene, 2002, 285, (1), pp. 1–24 (doi: 10.1016/S0378‐1119(02)00398‐0) [DOI] [PubMed] [Google Scholar]
- 31. Elowitz M.B., Leibler S.: ‘A synthetic oscillatory network of transcriptional regulators’, Nature, 2000, 403, (6767), pp. 335–338 (doi: 10.1038/35002125) [DOI] [PubMed] [Google Scholar]
- 32. Swameye I., Müller T., Timmer J., Sandra O., Klingmüller U.: ‘Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling’, Proc. Natl. Acad. Sci., 2003, 100, (3), pp. 1028–1033 (doi: 10.1073/pnas.0237333100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klipp E., Liebermeister W.: ‘Mathematical modeling of intracellular signaling pathways’, BMC Neurosci., 2006, 7, (Suppl 1), p. S10 (doi: 10.1186/1471-2202-7-S1-S10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holst J., Holst U., Madsen H., Melgaard H.: ‘Validation of grey box models’, in (Eds.): ‘Book validation of grey box models’ (Elsevier, 2014), p. 53 [Google Scholar]
- 35. Liu Z.‐P.: ‘Reverse engineering of genome‐wide gene regulatory networks from gene expression data’, Curr. Genomics, 2015, 16, (1), pp. 3–22 (doi: 10.2174/1389202915666141110210634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Z.‐P., Wu H., Zhu J., Miao H.: ‘Systematic identification of transcriptional and post‐transcriptional regulations in human respiratory epithelial cells during influenza A virus infection’, BMC Bioinf., 2014, 15, (1), p. 336 (doi: 10.1186/1471-2105-15-336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Z.‐P., Zhang W., Horimoto K., Chen L.: ‘Gaussian graphical model for identifying significantly responsive regulatory networks from time course high‐throughput data’, IET Syst. Biol., 2013, 7, (5), pp. 143–152 (doi: 10.1049/iet-syb.2012.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]


