Abstract
Context
Adrenocortical carcinomas (ACC) are a rare tumor type with a poor five-year survival rate and limited treatment options.
Objective
Understanding of the molecular pathogenesis of this disease has been aided by genomic analyses highlighting alterations in TP53, WNT, and IGF signaling pathways. Further elucidation is needed to reveal therapeutically actionable targets in ACC.
Design
In this study, global DNA methylation levels were assessed by the Infinium HumanMethylation450 BeadChip Array on 18 ACC tumors and 6 normal adrenal tissues. A new, non-linear correlation approach, the discretization method, assessed the relationship between DNA methylation/gene expression across ACC tumors.
Results
This correlation analysis revealed epigenetic regulation of genes known to modulate TP53, WNT, and IGF signaling, as well as silencing of the tumor suppressor MARCKS, previously unreported in ACC.
Conclusions
DNA methylation may regulate genes known to play a role in ACC pathogenesis as well as known tumor suppressors.
Introduction
Adrenocortical carcinomas (ACC) are rare neoplasms that account for up to 0.2% of cancer deaths. The estimated incidence of the disease is 0.7 to 2.0 cases per million persons per year [1, 2]. This disease usually strikes adults in their 40-50s, but may also be seen in children, typically with a Tumor Protein p53 (TP53) germline mutation [3]. The only curative treatment is surgical removal of a localized tumor; many patients have metastatic disease at the time of diagnosis, which further limits their therapeutic options [4, 5]. In a study looking at almost 4000 cases of ACC between 1985 and 2005, Bilimoria et al. reported 26.5% presented with nodal metastasis and 11.3% presented with distant metastasis, leading to a five-year survival rate of 11.5% compared to 55.1% without distant metastasis [4]. The response rate to the generally accepted first-line chemotherapy regimen consisting of etoposide, doxibucin, cisplatin and mitotane is only 23% [6], making it clear that there is an urgent need for new therapies.
Genomic analyses are elucidating the molecular pathogenesis of ACC and exposing potential therapeutic targets. The most well studied and consistently observed genomic aberrations involve the TP53 tumor suppressor gene, insulin-like growth factor type 2 (IGF2) signaling, and the Wingless-Type MMTV Integration Site Family (WNT) pathway. Several studies have clearly established a role for aberrant TP53 function, even though the reported mutation rate for the TP53 in sporadic adult ACC is only approximately 16–27% [7–9]. Overexpression of IGF2 is seen in at least 95% of ACC tumor samples studied [10], prompting clinical trials employing Insulin-Like Growth Factor 1 Receptor (IGF1R) inhibitors [11]. WNT pathway signaling is also aberrant, with activating somatic mutations of the β-catenin gene seen with a similar frequency of approximately 30% in both benign and malignant adrenal cortex tumors [12]. While genomic analyses have uncovered pathway perturbations in ACC, clinically actionable targets have remained elusive, leading one to conclude that there is a need for a deeper understanding of ACC tumorigenesis.
Investigation of epigenetic alterations in ACC is garnering interest. Global methylation analyses have demonstrated that distinct methylation patterns exist to distinguish ACC from benign tumors and/or normal adrenal tissue [13, 14]. Rechache et al. have demonstrated that metastatic ACC tumors showed a distinct methylation pattern compared to normal adrenal and primary ACC tumors, and malignant samples demonstrated global hypomethylation [14]. Barreau et al. identified a CpG island methylator phenotype (CIMP) in ACC associated with poor patient survival [15]. These global methylation analyses suggested hypermethylation of tumor suppressors such as cyclin-dependent kinase inhibitor 2A (CDKN2A), Deleted in Lung and Esophageal Cancer 1 (DLEC1), or N-Myc Downstream-Regulated Gene family member 2 (NDRG2) as a mechanism for reduced mRNA expression observed in ACC tumors compared to adenomas or normal adrenal tissue. More recently, an integrated genomic characterization identified two distinct molecular subgroups in ACC: C1A and C1B [7]. The C1A subgroup correlated with a poor patient prognosis, increased frequency of driver genetic mutations, distinct mRNA and miRNA clusters, and had a clear association with CIMP. Thus, methylation analyses have identified subgroups of ACC tumors with differential prognosis and suggested molecular alterations that might contribute to ACC pathogenesis; although, the full extent of the genes and pathways modulated by methylation in ACC remains unknown.
In this work, we sought to correlate DNA methylation changes with expression profiles in a set of ACC (n = 18 [17 tumors, 1 liver metastasis ACC_150]) and normal adrenal (n = 6) samples in order to better understand how methylation may result in the aberrant gene and pathway expression observed in ACC. Our study uses a non-linear correlation approach referred to as the discretization method [16] to assess DNA methylation/gene expression relationships within pathways, and reveals potential epigenetic regulation of genes involved in TP53, WNT, IGF2, and tumor suppressor gene signaling and/or stability. While other studies have reported DNA methylation changes in ACC in the past, our study is distinct in the manner in which we describe DNA methylation/gene expression associations to identify epigenetically regulated pathways of known importance to ACC.
Materials and Methods
Clinical Samples
The clinical samples used in this analysis represent a subset of samples previously described [8]. Briefly, a set of ACC flash frozen tumors and normal adrenal glands were collected at the Mayo Clinic (Rochester, Minnesota), the University Hospital Essen (Essen, Germany), the University of Calgary (Alberta, Canada), and Scottsdale Healthcare (Scottsdale, Arizona), as well as donated directly by patients through their community care settings; all samples were obtained under appropriate ethical procedures and written informed patient consent at the respective institutions. Normal adrenal glands were collected at the time of surgery for another indication, typically resection of a tumor of the kidney. The adrenal was taken en bloc, and the cortex was macrodissected from the medulla as best possible. Research materials for this study were obtained under protocols approved by the Western Institutional Review Board (WIRB # 20051769). The diagnosis of ACC was confirmed by review of the pathology report, and in most cases, by reexamination of the histopathology slides by an experienced endocrine pathologist.
Gene Expression Profiling
The mRNA expression and statistical analysis of ACC and normal tissues was previously described [8]. Briefly, RNA was extracted from 100 mg samples of ACC tumors and normal adrenal tissue, amplified and reverse transcribed utilizing the MessageAmp II Biotin Enhanced Kit (Ambion Life Technologies Corp, Carlsbad, CA). Biotin-labeled cRNA was synthesized according to their standard protocol, followed by purification through provided cRNA Filter Cartridges. Labeled cRNA was fragmented and hybridized to Affymetrix U133 Plus 2 human genome arrays following the standard Affymetrix protocol (Affymetrix Inc., Santa Clara, CA). Scanning and washing was completed on the Fluidic Stations FS450 and the GeneChip® Scanner 3000 with Workstation.
Array quality for ACC and the normal samples was assessed using the Affy QCReport package in Bioconductor and the R statistical language; all arrays passed the quality control metrics. All subsequent data normalization and statistical analysis was done using GenePattern (Broad Institute, www.broadinstitute.org) [17]. Expression array data was normalized by gcRMA with quantile normalization, and background subtraction after using the Expression File Creator [18]. Data were then floored at 5.5 using Preprocess Dataset, and filtered to remove 1) probes with more than 35 floored values and/or 2) probes where all values from one batch were floored while values from the other batch were not. Further batch effects were minimized using ComBat with the parametric option [19]. The expression data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE19776 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19776).
DNA Methylation Analysis
Global DNA methylation was evaluated using the Infinium® HumanMethylation450® BeadChip Array. (Illumina, San Diego, CA). Briefly, 1 μg of each DNA sample underwent bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer’s recommendation for the Illumina Infinium Assay. Bisulfite-treated DNA was then hybridized to arrays according to the manufacturer’s protocol. We used GenomeStudio V2011.1 (Illumina) for methylation data assembly and acquisition. Methylation levels for each CpG residue are presented as ß values, estimating the ratio of the methylated signal intensity over the sum of the methylated and unmethylated intensities at each locus. The average ß value reports a methylation signal ranging from 0 to 1, representing completely unmethylated to completely methylated values, respectively. Methylation data was preprocessed in R using the Illumina Methylation Analyzer (IMA) package [20]. Data preprocessing included background correction, probe scaling to balance Infinium I and II probes, quantile normalization, and logit transformation. A logit transformation converts otherwise heteroscedastic beta values (bounded by 0 and 1) to M values following a Gaussian distribution. Additionally, detection p-values >0.05 in 25% of samples, probes on X and Y chromosomes, and probes situated within 10 bp of putative SNPs were removed. Differential methylation analysis on logit-transformed values was performed to compare 18 ACC tumors to 6 normal adrenal samples in IMA. Wilcox rank test was conducted between ACC and normal samples and p-values were corrected by calculating the false discovery rate by the Benjamini-Hochberg method. Probes with adjusted p-values <0.05, and delta β values ≥0.2 or ≤-0.2 were considered statistically significant and differentially methylated. The methylation data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE77871 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77871).
Correlating DNA Methylation to Gene Expression by the Discretization Method
We used our newly reported non-linear discretization method by categorizing samples into different groups based on probe methylation levels to identify DNA methylation/expression correlations [16]. Gene expression data was available for 14 of the 18 samples analyzed for methylation changes. For each methylation probe, the 14 ACC samples were separated into hypermethylated (M) or hypomethylated (U) groups based on the degree of methylation differing from the average methylation levels of 6 normal adrenal samples. Samples with methylation levels > μ (mean methylation level of normal samples) were classified into the M group and samples with methylation levels < μ were classified as U for the given CpG locus interrogated by the probe. Every probe in the methylation array with a gene expression probe on the Affymetrix U133 Plus 2 human genome array was analyzed. Differential gene expression analysis, comparing samples that were categorized as M and U, was conducted with a t-test without assuming equal variance (Matlab software, www.mathworks.com). If a gene was differentially expressed (FDR-corrected p-value < 0.05) between samples in the M and U groups, CpG methylation was considered correlated to expression of that gene. A negative correlation was defined as the directionality of change for expression and methylation in opposite directions (e.g. hypermethylation and loss of expression, or vice versa). A positive correlation occurred when the directionality of change was the same between methylation and expression (e.g. hypermethylation and positive expression, or hypomethylation and reduced expression).
To validate our findings reported in S2 Table we used RNAseq and methylation data from 78 ACC samples generated by TCGA. Level 3 RNAseq data (TPM values) and methylation beta values were downloaded from TCGA Data Portal (https://tcga-data.nci.nih.gov). RNAseq data were log2 transformed: log2(TPM + 1), before further analysis. In level 3 TCGA methylation data, some values were measured as “NA” and those values were removed during differential analysis. We also did not include those methylation probes with < 2 samples in either hypo- or hyper-group in the analysis. The validation analysis was conducted by testing if the same methylation probes reported in S2 Table also had correlation with gene expression in the TCGA ACC dataset, when using the discretization method. In the current manuscript, we modified the discretization method first described in Jung et al. [16] to reflect the small number of samples. In this modified method, we discretized samples based on whether a sample was hypo or hypermethylated when compared to non-neoplastic adrenal tissue. For this, a delta beta value for each probe was calculated for each tumor’s methylation data when compared against the mean of the methylation data from the non-neoplastic adrenal tissue. In this binary discretization, a tumor sample was deemed hypomethylated or hypermethylated when its delta-beta value was negative or positive, respectively, compared to normal tissue. Since there were sufficient samples in the TCGA dataset, we also performed discretization according to the original method described (referred to here as ternary discretization) by Jung et al. [16]. Once the samples were grouped into U and M groups by either binary or ternary discretization, the expression values of genes with corresponding methylation probes were tested for differential expression using Welch’s t-test.
Pathway Analysis
The gene list of interest was uploaded into IPA (Ingenuity® Systems, Redwood City, CA) and the Core Analysis workflow was run with default parameters. The Core Analysis provides an assessment of significantly altered pathways, molecular networks, and biological processes represented in the samples' gene list.
Results
Global Methylation Patterns in ACC versus Normal Adrenal
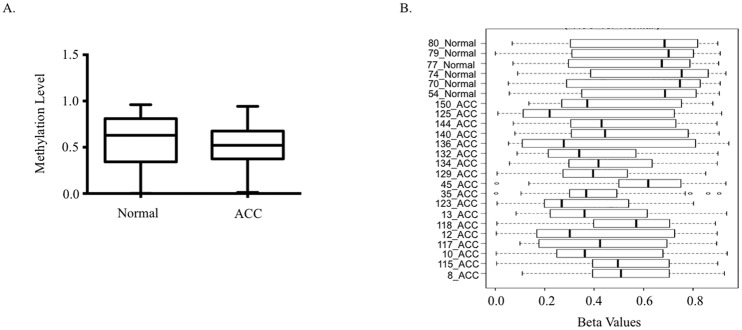
To assess global DNA methylation patterns in ACC tumors, we used the 450K-methylation platform to compare 18 ACC tumors with 6 normal adrenal gland samples. Overall, the analysis revealed 1291 differentially methylated CpG loci (DML) encompassing 629 unique genes (S1 Table). Beta values were used for generating box plots to represent overall methylation levels across DML for ACC and normal. Median overall methylation was slightly lower in ACC (β = 0.51) than in normal adrenal (β = 0.67) (Fig 1A), indicating hypomethylation in ACC (p-value < 0.0001). Individual sample box plots of DML demonstrated a more homogenous distribution of methylation values in normal adrenal samples compared to ACC which displayed varying degrees of methylation between samples (Fig 1B). Of all DML, 475 were hypermethylated and 816 were hypomethylated. Next we examined the distribution of DML across chromosomes, plotting the distribution of hypo- and hyper-DML after normalization to chromosome length (Table 1). According to the analysis chromosomes 5, 7, 12, and 19 had the most hypomethylated loci and chromosomes 11, 17, and 19 had the most hypermethylated loci, with chromosomes 7, 12, and 19 having the most overall DML.
Fig 1. Differential Methylation Analysis comparing ACC to normal adrenal.
A) Box plot demonstrating overall lower median β value methylation levels for 1291 differentially methylated probes. B) Box plots showing β values of 1291 differentially methylated loci for each individual sample (6 normal and 18 ACC samples).
Table 1. Frequency of DML by chromosome after normalization to chromosome length.
| Chromosome | Frequency Hypo | Frequency Hyper | Total Frequency |
|---|---|---|---|
| 1 | 694 | 520 | 1214 |
| 2 | 403 | 391 | 794 |
| 3 | 597 | 247 | 844 |
| 4 | 723 | 196 | 919 |
| 5 | 1354 | 653 | 2007 |
| 6 | 758 | 387 | 1145 |
| 7 | 1883 | 652 | 2535 |
| 8 | 965 | 335 | 1300 |
| 9 | 204 | 184 | 388 |
| 10 | 978 | 383 | 1361 |
| 11 | 811 | 854 | 1665 |
| 12 | 1937 | 581 | 2518 |
| 13 | 450 | 125 | 575 |
| 14 | 510 | 349 | 859 |
| 15 | 393 | 534 | 927 |
| 16 | 861 | 510 | 1371 |
| 17 | 568 | 1242 | 1810 |
| 18 | 332 | 184 | 516 |
| 19 | 1998 | 2193 | 4191 |
| 20 | 549 | 411 | 960 |
| 21 | 539 | 60 | 599 |
| 22 | 56 | 449 | 505 |
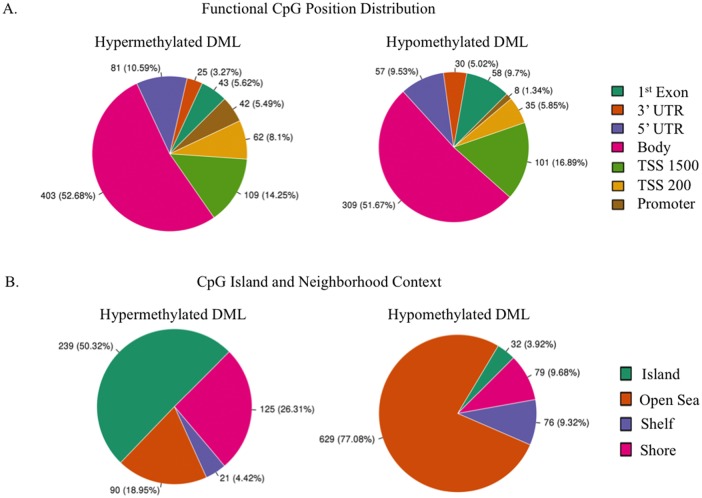
Next we examined the regional and functional CpG distribution of DML in ACC. Functional distribution relates CpG position to: transcription start sites (TSS −200 to −1500 bp), 5′ untranslated region (UTR), exon 1 for coding genes, and gene bodies. Overall, the majority of probes (>50%) were situated in gene bodies, followed by ~15% of probes situated within 1500 bp upstream of TSS, with a very similar breakdown between hyper and hypo DML (Fig 2A).
Fig 2. Frequency of differentially methylated regions according to functional and CpG Island contextual distribution.
A) Pie charts demonstrate the frequency by which hyper or hypomethylated loci are distributed according to their functional position, including distance from transcriptional start site (TSS). B) Frequency of DML to be situated in a CpG islands, shores, shelves, or open sea. Neighborhood context is an indication of proximity to a CpG island.
Regional distribution of DML was assessed based on their proximity to the closest CpG island. In addition to islands, shores are 0–2 kb from CpG islands, shelves are 2–4 kb away, and open sea regions are isolated loci without a designation. When comparing the ACC samples to normal samples, we identified the majority of DML (55.7%) were in open sea, followed by islands (21%), shores (15.8%), and shelves (7.5%). In addition, we note that the majority of hypermethylated loci (50.3%) were located in CpG islands compared to the majority of hypomethylated loci being situated in open sea (77%) (Fig 2B). In contrast, of all hypomethylated loci, the smallest percentage (3.92%) was found in islands and the smallest percentage of hypermethylated probes (4.42%) were found in shelves.
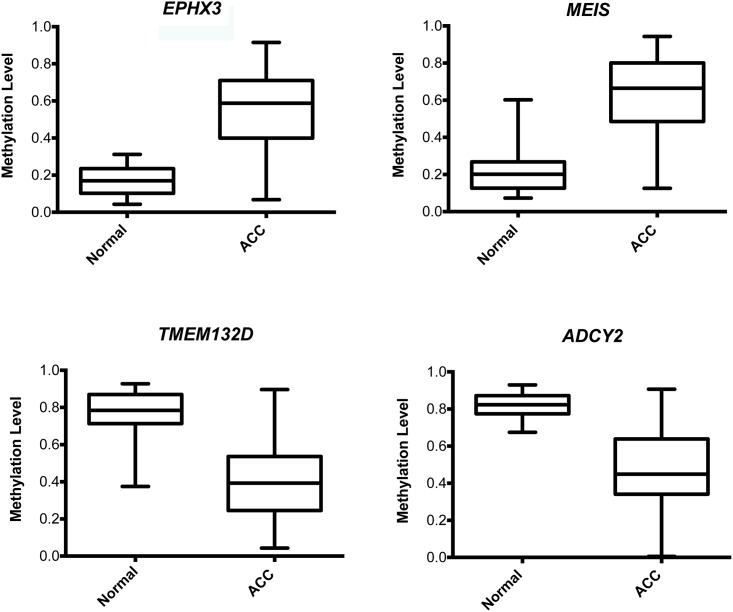
We next performed unsupervised clustering analysis (Euclidean distance, Complete hierarchical clustering methods) of DML and demonstrated a distinct separation of normal and ACC samples with evidence of gene clusters that are preferentially hyper or hypomethylated in ACC (Fig 3). Among the genes most hypermethylated in ACC were the EPHX3 and MEIS genes, with 20% and 9.2% of possible gene probes affected (Fig 4). Representative genes in the hypomethylated clusters are the ADCY2 and TMEM132D genes, which affected 16.2% and 18.5% of probes (Fig 4).
Fig 3. Unsupervised clustering analysis of normal and ACC samples using DML.
Clustering analysis revealed separation of ACC and normal samples. Samples are on the horizontal axis: normal samples are shown with a yellow bar and ACC samples are shown with a blue bar.
Fig 4. Box plots display examples of genes significantly hyper or hypomethylated in ACC.
EPHX3 and MEIS genes are significantly hypermethylated in ACC compared with TMEM132D and ADCY2 which are significantly hypomethylated.
Identification of Methylation-Expression Correlations Using the Discretization Method
In a separate analysis, we examined methylation-expression correlations using a non-linear approach: referred to as the discretization method. This method relies on the fact that cancer samples can be categorized into two groups according to their relative methylation levels for each probe on the methylation array, enabling a higher number of correlations to be detected than by comparisons using linear methods. Samples were either considered hypermethylated (M) or hypomethylated (U) compared to a reference (average methylation levels from normal samples in this study) and statistically significant gene expression differences between samples in the M and U groups would suggest a methylation-expression correlation at a given locus. By applying this binary discretization method, we identified 763 unique CpG loci (550 unique genes) with methylation-expression correlations. On average, there were 9 samples in the M group and 5 samples in the U group. There were 319 loci with positive correlations and 444 loci with negative correlations. All CpG methylation/expression correlations from this discovery cohort are shown in S2 Table.
In order to validate the findings of the current study in an independent cohort of samples, we downloaded RNA-seq expression data and matching DNA methylation data from TCGA portal for 78 ACC samples. This sample size not only allowed us to test correlations using our modified binary discretization method, but also provided the opportunity to utilize the ternary method as well. Due to absent/missing data in the TCGA set, 468 probes were available for binary testing of coordinated expression. Additionally, some probes lacked sufficient class size (M and U), thus leaving 433 probes for ternary grouping out of 763 unique probes reported in S2 Table. In binary grouping, 164/468 (35%) probes (S3 Table) showed significant correlation (p-value < 0.05) with corresponding gene expression values, and in ternary grouping 154/433 (35.6%) probes (S4 Table) showed significant correlation (p-value < 0.05).
Alterations in TP53 and WNT Signaling Pathways
In order to identify biological concepts enriched in S2 Table, we both visually inspected the list to identify obvious patterns and we submitted the gene list to Core Analysis workflow in IPA (Ingenuity® Systems). Significant findings were sorted based on p-values to identify the most distinguishing categories representing epigenetically regulated genes in ACC. In brief summary, the most significant Diseases and Disorders associated with the genes in S2 Table was Cancer with 471 genes represented from our list (S5 Table). The 5 top Molecular and Cellular Functions were Cellular Growth and Proliferation, Cell Morphology, Cellular Assembly and Organization, Cellular Function and Maintenance, and Cell Death and Survival (S6 Table).
Using the discretization method, we identified a number of genes with methylation-expression correlations known to be involved in cancer, or specifically ACC pathogenesis. All of the genes we point out below were enriched in the Ingenuity Pathway Analysis under one of the categories mentioned above. Their pathway association and whether or not they were validated is noted on each of the tables below. Among this list of correlated genes are regulators of the TP53 pathway, such as: SETD7, DYRK2, CCDC8, UBE2D1, RBM5, NDRG1 and DUSP7 (Table 2, Figs 5 and 6, S2–S4 Tables). In each example, samples with low methylation had statistically significant higher levels of gene expression and samples with high methylation had lower levels of gene expression. DUSP7, NDRG1, SETD7, and UBE2D1 were observed to be under-expressed in an independent data set comparing ACC mRNA expression to normal adrenal, with the under-expression agreeing with the observed hypermethylation/under-expression correlation [21].
Table 2. Significantly regulated genes in the TP53 pathway.
| Gene | Methylation Discretization (high/low) | BH-corrected p-value | Direction of correlation | TCGA Validated (Binary/Ternary) | Alternate Validation Cohort (mRNA) | TP53 relationship |
|---|---|---|---|---|---|---|
| CCDC8 | 9/5 | 0.00003 | -1 | Yes/Yes | No | Tumor suppressor, co-factor in TP53-mediated apoptosis |
| DUSP7 | 12/2 | 0.0005 | -1 | NA | Yes (0.00001) | MAPK inactivator, TP53 target gene53 target gene |
| DYRK2 | 12/2 | 0.0004 | -1 | NA | No | Regulates TP53 stability |
| RBM5 | 12/2 | 0.0010 | -1 | NA | No | Regulates TP53 activity |
| SETD7 | 12/2 | 0.0005 | -1 | NA | Yes (0.00001) | Stabilizes TP53 |
| NDRG1 | 11/3 | 0.0200 | -1 | NA | Yes (0.03) | Tumor suppressor, necessary for TP53-mediated apoptosis |
| UBE2D1 | 12/2 | 0.0300 | -1 | NA | Yes (0.000005) | TP53 ubiquitination and turnover |
(NA = data not available for gene in TCGA dataset)
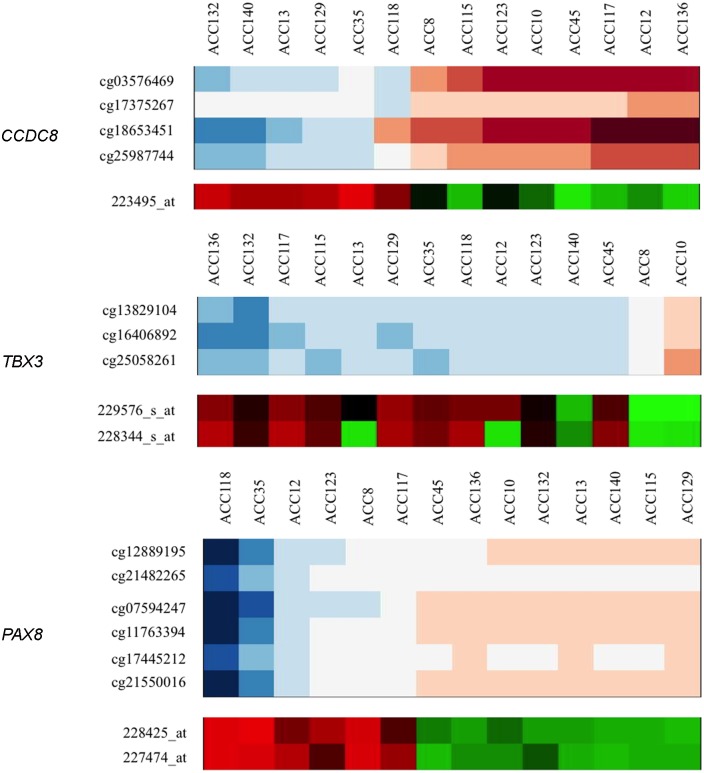
Fig 5. Methylation/expression correlations according to discretization method.
For each gene, the upper heatmap represents the log2 methylation values for 14 ACC samples each normalized to the average of 6 normal adrenal samples. Log2 methylation ratios >0 represent hypermethylation and <0 represent hypomethylation. The lower heat map shows expression of z-transformed expression levels, where a value 0 indicates no expression change compared to average expression level of 14 ACC samples. Three genes with negative correlations were selected for visualization: CCDC8 (TP53 pathway), TBX3 (WNT pathway), and PAX8 (other known cancer gene, involved in invasion and migration). Samples with higher methylation (peach-maroon) had lower expression (green). Samples with lower methylation (blue) had higher expression (red). Only the correlated methylation loci and expression probes are shown, and samples are organized by their discretization classification of M or U for each gene.
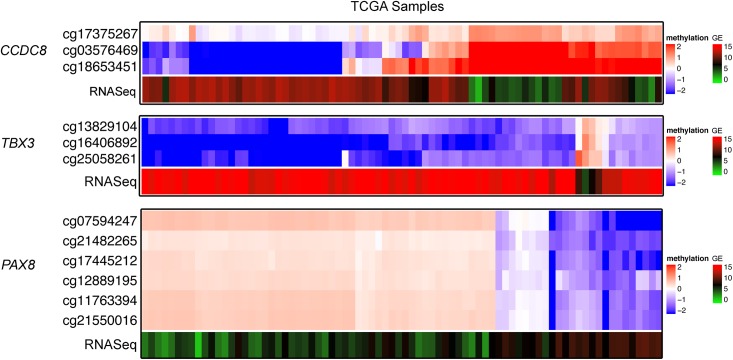
Fig 6. TCGA Validation: Methylation/expression correlations according to discretization method.
For each gene, the upper heat map represents the log2 methylation values (beta) for 78 TCGA ACC samples each normalized to the average of 6 TGen normal adrenal samples. Log2 methylation ratios >0 (red) represent hypermethylation and <0 (blue) represent hypomethylation. The lower heatmap shows gene expression of corresponding TCGA ACC samples, log2 (TPM+1). The same three genes used for TGen ACC study (discovery) were used for visualization: CCDC8 (TP53 pathway), TBX3 (WNT pathway), and PAX8 (other known cancer gene, involved in invasion and migration). Samples with higher methylation (red) had lower expression (green). Samples with lower methylation (blue) had higher expression (red). Only the correlated methylation loci and expression probes are shown, and samples are organized by similarity of methylation patterns between samples for each gene. As shown, all three genes show the same negative correlation between methylation status and gene expression as TGen ACC samples.
We also identified methylation/expression correlations in 4 WNT signaling-related genes; this is another pathway known to be associated with ACC tumorigenesis (Table 3, S2–S4 Tables). Samples displaying higher methylation of PRDM5 and DKK3, which are both antagonists of WNT signaling, were accompanied by loss of mRNA expression and vice versa. Conversely, TBX3 (a WNT target gene) was hypomethylated and overexpressed; these correlations were observed in both the original and TCGA datasets (Figs 5 and 6). WNT3 (WNT ligand) was methylated and underexpressed in 11 samples and unmethylated and overexpressed in 3 samples. Loss of DKK3 mRNA expression was validated in an independent, publicly available data set comparing ACC to normal adrenal tissue [21].
Table 3. Significantly regulated genes in the WNT pathway.
| Gene | Methylation Discretization (high/low) | BH-corrected p-value | Direction of correlation | TCGA Validated (Binary/Ternary) | Independent mRNA validation (p-value) | WNT relationship |
|---|---|---|---|---|---|---|
| TBX3 | 2/12 | 0.0001 | -1 | Yes/Yes | No | WNT target gene |
| PRDM5 | 12/2 | 0.01 | -1 | Yes/Yes | No | Tumor suppressor, regulates WNT signaling |
| DKK3 | 11/3 | 0.04 | -1 | Yes/Yes | Yes (1.76E-10) | WNT antagonist |
| WNT3 | 11/3 | 0.02 | -1 | No/No | No | WNT ligand |
Other Cancer-Related Gene Alterations
The transcription factor PAX8, shown to be hypermethylated in other tumor types, largely demonstrated a negative correlation to gene expression, where hypermethylation correlated with reduced expression (Table 4, Figs 5 and 6, S2–S4 Tables). Negative methylation/expression correlations were also found for HDAC4 (a chromatin modifier), ERBB3 (a known oncogene), as well as MARCKS and CXCR4; genes known to induce tumor cell invasion and therapeutic resistance in other tumor types (Table 4, S2–S4 Tables). The role of IGF2 and IGF signaling as an oncogenic driver in ACC is also well described. Our data reveal a significant methylation/expression correlation for the IGF2 gene. One probe (cg04057455) situated in the gene body correlated positively with two gene expression probes (S2 Table). The other probe (cg08986368) located in a CpG island correlated negatively with one gene expression probe. The end result was increased expression for samples with CpG island hypomethylation or gene body hypermethylation, which is consistent with the expected effects of methylation on gene expression.
Table 4. Significantly regulated genes associated with cancer.
| Gene | Methylation Discretization (high/low) | BH-correctedp-value | Direction of correlation | TCGA Validated (Binary/Ternary) | IndependentmRNA validation (p-value) | Proposed Function |
|---|---|---|---|---|---|---|
| PAX8 | 9/5 | 0.05 | -1 | Yes/Yes | Yes (0.0002) | Transcription factor |
| HDAC4 | 12/2 | 0.009 | -1 | No | Yes (0.00001) | Chromatin modifier |
| MARCKS | 2/12 | 0.03 | -1 | No | Yes (0.00002) | Cell invasion and therapy resistance |
| CXCR4 | 3/11 | 0.03 | -1 | Yes/Yes | No | Cell invasion and metastasis |
| ERBB3 | 12/2 | 0.0002 | -1 | Yes/Yes | Yes (0.006) | Oncogene |
Discussion
The purpose of the current study was to analyze global DNA methylation patterns in ACC tumors compared to normal adrenal tissue and to correlate DNA methylation changes with mRNA expression. Our study demonstrated global hypomethylation in ACC compared to normal adrenal tissue. Global genomic hypomethylation has been observed across numerous cancer cell types [22, 23], including malignant adrenal tumors when compared to benign or normal adrenal tissue [14]. Consistent with previous global methylation analyses, including ACC, hypomethylation was most frequent in ‘open seas’, away from the CpG islands, and hypermethylation events occurred most commonly in CpG islands [15].
To identify significant correlations between methylation and gene expression in ACC, a discretization method was employed. This non-linear method for identifying correlations is more sensitive than linear methods because it relies on subsets of samples having opposite methylation trends [16]. With this method we identified 550 genes with significant negative or positive correlation of methylation and gene expression. The genes identified using this method implicate pathways known to be perturbed in ACC such as TP53, WNT, and IGF2 signaling. In addition, classical tumor suppressor, other cancer related, and novel genes not previously implicated in ACC were also identified. The TP53 tumor suppressor is mutated in more than 50% of all tumors. Genetic alterations in TP53, including mutations (16%) [7] and epigenetic silencing (rarely seen) [24], occur in a lower percentage of ACC tumors. Our recent work in ACC demonstrated significant differential expression of genes involved in TP53 canonical signaling [8]. Gene relationships to TP53 were determined by their inclusion in the Gene Set Enrichment Analysis (GSEA) gene sets PID_P53DOWNSTREAMPATHWAY, PID_P53REGULATIONPATHWAY, and/or other reported evidence of TP53 binding or modulation of activity. In the current study, we observed several genes including SETD7 [25], DYRK2 [26], CCDC8 [27], and UBE2D1 [28] which are known to control TP53 protein stability and turnover through post-translational modifications and were epigenetically regulated in ACC tumors. Thus, while TP53 is not differentially methylated or frequently highly mutated in ACC tumors, the methylation status of genes known to modulate TP53 stability and activity may provide an explanation for the observed TP53 deregulation.
Another pathway associated with ACC tumorigenesis is the WNT signaling pathway. WNT signaling has been aberrantly detected in ACC tumors evidenced in part by frequent mutations in the β-catenin gene [7, 29]. Discretization analysis also revealed epigenetic regulation of WNT signaling-related genes in ACC. PRDM5 is a putative tumor suppressor through repression of WNT signaling, and known to be frequently silenced due to methylation across a number of other tumor types [30, 31]. We observed both promoter and gene body methylation, which correlated with altered mRNA expression. The known WNT antagonist DKK3, normally expressed in the human adrenal cortex [32], is under-expressed in childhood adrenocortical tumors [33]. DKK3 affects apoptosis and cell proliferation [34], and our study now points to CpG methylation of DKK3 as a possible mechanism for gene deregulation in ACC tumors [21].
Using the discretization method, we also identified a number of putative tumor suppressor genes whose methylation correlated with decreased gene expression. MARCKS, a substrate for protein kinase C phosphorylation, displays tumor suppressive roles in multiple cancer types[35]; our study is the first report of MARCKS epigenetic silencing in ACC. Similarly, hypermethylation of PAX8 correlated with downregulation and low methylation correlated with increased expression. Elevated levels of PAX8 have been seen in several tumor types and epigenetic silencing has been observed in squamous cell lung cancer [36]. The prognostic and biological implication of PAX8 expression is likely tissue and organ specific.
In conclusion, we have demonstrated that differential DNA methylation and expression correlation of ACC tumors and normal adrenal tissue identified potential genes and pathways with relevance to underlying biology and potentially patient outcome. Although this study had a limitation in small sample size that precluded a statistical evaluation of patient outcome data, the results were validated by analyzing the TCGA ACC dataset. Future work will focus on molecular and biological subclassification of ACC tumors with DNA methylation and making correlations to patient outcomes. Lastly, correlations of methylation and gene expression by the discretization method identified, for the first time, epigenetic modulation of genes involved in TP53 stability and function, WNT signaling, and tumor suppressor genes not previously associated with ACC.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PPTX)
(XLSX)
Acknowledgments
The authors would like to acknowledge support provided by the ATAC Research Fund and the Kirsten’s Legacy Fund. Research reported in this publication was supported by a generous philanthropic contribution from Mr. Ray Thurston. Funding support was also provided by the Virginia G. Piper Charitable Trust (TGW and BS).
Data Availability
The expression data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE19776 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19776). The methylation data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE77871 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77871).
Funding Statement
The authors would like to acknowledge support provided by the ATAC Research Fund and the Kirsten’s Legacy Fund. Research reported in this publication was supported by a generous philanthropic contribution from Mr. Ray Thurston. Funding support was also provided by the Virginia G. Piper Charitable Trust (TGW and BS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Golden SH, Robinson KA, Saldanha I, Anton B, Ladenson PW. Clinical review: Prevalence and incidence of endocrine and metabolic disorders in the United States: a comprehensive review. The Journal of clinical endocrinology and metabolism. 2009;94(6):1853–78. 10.1210/jc.2008-2291 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kebebew E, Reiff E, Duh QY, Clark OH, McMillan A. Extent of disease at presentation and outcome for adrenocortical carcinoma: have we made progress? World journal of surgery. 2006;30(5):872–8. 10.1007/s00268-005-0329-x . [DOI] [PubMed] [Google Scholar]
- 3.Allolio B, Hahner S, Weismann D, Fassnacht M. Management of adrenocortical carcinoma. Clinical endocrinology. 2004;60(3):273–87. . [DOI] [PubMed] [Google Scholar]
- 4.Bilimoria KY, Shen WT, Elaraj D, Bentrem DJ, Winchester DJ, Kebebew E, et al. Adrenocortical carcinoma in the United States: treatment utilization and prognostic factors. Cancer. 2008;113(11):3130–6. 10.1002/cncr.23886 . [DOI] [PubMed] [Google Scholar]
- 5.Demeure MJ, Somberg LB. Functioning and nonfunctioning adrenocortical carcinoma: clinical presentation and therapeutic strategies. Surgical oncology clinics of North America. 1998;7(4):791–805. . [PubMed] [Google Scholar]
- 6.Fassnacht M, Terzolo M, Allolio B, Baudin E, Haak H, Berruti A, et al. Combination chemotherapy in advanced adrenocortical carcinoma. The New England journal of medicine. 2012;366(23):2189–97. 10.1056/NEJMoa1200966 . [DOI] [PubMed] [Google Scholar]
- 7.Assie G, Letouze E, Fassnacht M, Jouinot A, Luscap W, Barreau O, et al. Integrated genomic characterization of adrenocortical carcinoma. Nature genetics. 2014. 10.1038/ng.2953 . [DOI] [PubMed] [Google Scholar]
- 8.Demeure MJ, Coan KE, Grant CS, Komorowski RA, Stephan E, Sinari S, et al. PTTG1 overexpression in adrenocortical cancer is associated with poor survival and represents a potential therapeutic target. Surgery. 2013;154(6):1405–16; discussion 16. 10.1016/j.surg.2013.06.058 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reincke M, Karl M, Travis WH, Mastorakos G, Allolio B, Linehan HM, et al. p53 mutations in human adrenocortical neoplasms: immunohistochemical and molecular studies. The Journal of clinical endocrinology and metabolism. 1994;78(3):790–4. 10.1210/jcem.78.3.8126158 . [DOI] [PubMed] [Google Scholar]
- 10.Libe R, Fratticci A, Bertherat J. Adrenocortical cancer: pathophysiology and clinical management. Endocrine-related cancer. 2007;14(1):13–28. 10.1677/erc.1.01130 . [DOI] [PubMed] [Google Scholar]
- 11.Demeure MJ, Bussey KJ, Kirschner LS. Targeted therapies for adrenocortical carcinoma: IGF and beyond. Hormones & cancer. 2011;2(6):385–92. 10.1007/s12672-011-0090-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tissier F, Cavard C, Groussin L, Perlemoine K, Fumey G, Hagnere AM, et al. Mutations of beta-catenin in adrenocortical tumors: activation of the Wnt signaling pathway is a frequent event in both benign and malignant adrenocortical tumors. Cancer research. 2005;65(17):7622–7. 10.1158/0008-5472.CAN-05-0593 . [DOI] [PubMed] [Google Scholar]
- 13.Fonseca AL, Kugelberg J, Starker LF, Scholl U, Choi M, Hellman P, et al. Comprehensive DNA methylation analysis of benign and malignant adrenocortical tumors. Genes, chromosomes & cancer. 2012;51(10):949–60. 10.1002/gcc.21978 . [DOI] [PubMed] [Google Scholar]
- 14.Rechache NS, Wang Y, Stevenson HS, Killian JK, Edelman DC, Merino M, et al. DNA methylation profiling identifies global methylation differences and markers of adrenocortical tumors. The Journal of clinical endocrinology and metabolism. 2012;97(6):E1004–13. 10.1210/jc.2011-3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barreau O, Assie G, Wilmot-Roussel H, Ragazzon B, Baudry C, Perlemoine K, et al. Identification of a CpG island methylator phenotype in adrenocortical carcinomas. The Journal of clinical endocrinology and metabolism. 2013;98(1):E174–84. 10.1210/jc.2012-2993 . [DOI] [PubMed] [Google Scholar]
- 16.Jung S, Kim S, Gale M, Cherni I, Fonseca R, Carpten J, et al. DNA methylation in multiple myeloma is weakly associated with gene transcription. PloS one. 2012;7(12):e52626 10.1371/journal.pone.0052626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nature genetics. 2006;38(5):500–1. 10.1038/ng0506-500 . [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. 10.1093/biostatistics/4.2.249 . [DOI] [PubMed] [Google Scholar]
- 19.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. 10.1093/biostatistics/kxj037 . [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Yan L, Hu Q, Sucheston LE, Higgins MJ, Ambrosone CB, et al. IMA: an R package for high-throughput analysis of Illumina's 450K Infinium methylation data. Bioinformatics. 2012;28(5):729–30. 10.1093/bioinformatics/bts013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giordano TJ, Kuick R, Else T, Gauger PG, Vinco M, Bauersfeld J, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(2):668–76. 10.1158/1078-0432.CCR-08-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annual review of biochemistry. 2012;81:97–117. 10.1146/annurev-biochem-052610-091920 . [DOI] [PubMed] [Google Scholar]
- 23.Fotouhi O, Adel Fahmideh M, Kjellman M, Sulaiman L, Hoog A, Zedenius J, et al. Global hypomethylation and promoter methylation in small intestinal neuroendocrine tumors: An in vivo and in vitro study. Epigenetics: official journal of the DNA Methylation Society. 2014;9(7). . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidhu S, Martin E, Gicquel C, Melki J, Clark SJ, Campbell P, et al. Mutation and methylation analysis of TP53 in adrenal carcinogenesis. European journal of surgical oncology: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2005;31(5):549–54. 10.1016/j.ejso.2005.01.013 . [DOI] [PubMed] [Google Scholar]
- 25.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Molecular cell. 2008;29(3):392–400. 10.1016/j.molcel.2007.12.025 . [DOI] [PubMed] [Google Scholar]
- 26.Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Molecular cell. 2007;25(5):725–38. 10.1016/j.molcel.2007.02.007 . [DOI] [PubMed] [Google Scholar]
- 27.Clayton PE, Hanson D, Magee L, Murray PG, Saunders E, Abu-Amero SN, et al. Exploring the spectrum of 3-M syndrome, a primordial short stature disorder of disrupted ubiquitination. Clinical endocrinology. 2012;77(3):335–42. 10.1111/j.1365-2265.2012.04428.x . [DOI] [PubMed] [Google Scholar]
- 28.Tokumoto M, Fujiwara Y, Shimada A, Hasegawa T, Seko Y, Nagase H, et al. Cadmium toxicity is caused by accumulation of p53 through the down-regulation of Ube2d family genes in vitro and in vivo. The Journal of toxicological sciences. 2011;36(2):191–200. . [DOI] [PubMed] [Google Scholar]
- 29.Gaujoux S, Grabar S, Fassnacht M, Ragazzon B, Launay P, Libe R, et al. beta-catenin activation is associated with specific clinical and pathologic characteristics and a poor outcome in adrenocortical carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(2):328–36. 10.1158/1078-0432.CCR-10-2006 . [DOI] [PubMed] [Google Scholar]
- 30.Meani N, Pezzimenti F, Deflorian G, Mione M, Alcalay M. The tumor suppressor PRDM5 regulates Wnt signaling at early stages of zebrafish development. PloS one. 2009;4(1):e4273 10.1371/journal.pone.0004273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu XS, Geng H, Li L, Ying J, Ma C, Wang Y, et al. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/beta-catenin signaling and is frequently silenced in multiple tumors. PloS one. 2011;6(11):e27346 10.1371/journal.pone.0027346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suwa T, Chen M, Hawks CL, Hornsby PJ. Zonal expression of dickkopf-3 and components of the Wnt signalling pathways in the human adrenal cortex. The Journal of endocrinology. 2003;178(1):149–58. . [DOI] [PubMed] [Google Scholar]
- 33.Leal LF, Mermejo LM, Ramalho LZ, Martinelli CE Jr., Yunes JA, Seidinger AL, et al. Wnt/beta-catenin pathway deregulation in childhood adrenocortical tumors. The Journal of clinical endocrinology and metabolism. 2011;96(10):3106–14. 10.1210/jc.2011-0363 . [DOI] [PubMed] [Google Scholar]
- 34.Veeck J, Dahl E. Targeting the Wnt pathway in cancer: the emerging role of Dickkopf-3. Biochimica et biophysica acta. 2012;1825(1):18–28. 10.1016/j.bbcan.2011.09.003 . [DOI] [PubMed] [Google Scholar]
- 35.Bickeboller M, Tagscherer KE, Kloor M, Jansen L, Chang-Claude J, Brenner H, et al. Functional characterization of the tumor-suppressor MARCKS in colorectal cancer and its association with survival. Oncogene. 2015;34(9):1150–9. 10.1038/onc.2014.40 . [DOI] [PubMed] [Google Scholar]
- 36.Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, Campan M, et al. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Molecular cancer. 2008;7:62 10.1186/1476-4598-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PPTX)
(XLSX)
Data Availability Statement
The expression data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE19776 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19776). The methylation data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE77871 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77871).