Summary
Approximately half of all deaths from liver cirrhosis, the 10th leading cause of mortality in the United States, are related to alcohol use. Chronic alcohol consumption is accompanied by intestinal dysbiosis and bacterial overgrowth, yet little is known about the factors that alter the microbial composition or their contribution to liver disease. We previously associated chronic alcohol consumption with lower intestinal levels of the antimicrobial-regenerating islet-derived (REG)-3 lectins. Here, we demonstrate that intestinal deficiency in REG3B or REG3G increases numbers of mucosa-associated bacteria and enhances bacterial translocation to the mesenteric lymph nodes and liver, promoting the progression of ethanol-induced fatty liver disease toward steatohepatitis. Overexpression of Reg3g in intestinal epithelial cells restricts bacterial colonization of mucosal surfaces, reduces bacterial translocation, and protects mice from alcohol-induced steatohepatitis. Thus, alcohol appears to impair control of the mucosa-associated microbiota, and subsequent breach of the mucosal barrier facilitates progression of alcoholic liver disease.
Introduction
Alcoholic liver disease can progress from simple hepatic steatosis and steatohepatitis to end-stage organ disease (cirrhosis). Liver cirrhosis is the 10th leading cause of mortality in the United States (Kim et al., 2002). Alcohol-attributable liver disease is a major factor in burden of disease - approximately 50% of all cirrhosis-associated deaths are related to alcohol (Rehm et al., 2013). Patients with chronic alcohol abuse and alcoholic liver disease undergo changes in the composition of intestinal microbial communities (Leclercq et al., 2014; Mutlu et al., 2012). Alcohol-associated dysbiosis is characterized not only by changes in the composition of the bacteria (Bull-Otterson et al., 2013), but also by bacterial overgrowth in the small intestine (Bode et al., 1984; Yan et al., 2011). These findings, from animal models and alcoholic patients, indicate that chronic alcohol administration is associated with impairments in mechanisms that maintain the homeostatic composition of the intestinal microbiota. Alcohol-associated changes in the enteric microbiota are required for development of liver disease, because intestinal decontamination with non-absorbable antibiotics can prevent (Adachi et al., 1995) and reduce features of alcoholic liver disease in animal models (Chen et al., 2015a).
Antimicrobial proteins are produced and secreted by intestinal epithelial and Paneth cells and serve as the first line of defense against pathogens; they maintain homeostasis of the commensal bacteria. The antimicrobial proteins REG3B and REG3G are secreted C-type lectins (Vaishnava et al., 2011) that are predominantly expressed in the gastrointestinal tract, but not in the liver, under homeostatic conditions (Nata et al., 2004). REG3G has bactericidal activity against Gram-positive bacteria and helps maintain the spatial segregation of luminal bacteria and the intestinal epithelial surface (Cash et al., 2006; Mukherjee et al., 2014; Vaishnava et al., 2011). REG3B has bactericidal activity against Gram-negative bacteria, and protects mice against intestinal infection and dissemination of Salmonella enteritidis (Miki et al., 2012; van Ampting et al., 2012).
We have previously shown that Reg3b and Reg3g gene and protein expression are suppressed after chronic intragastric feeding of ethanol to mice (Hartmann et al., 2013; Yan et al., 2011). This inhibition appears to be specific for Reg3b and Reg3g, as the expression of other intestinal antimicrobial molecules is not affected (Yan et al., 2011). Similarly, expression of Reg3g in the small intestine is suppressed in cirrhotic rats with bacterial translocation (Teltschik et al., 2012). However, it is not clear how reduced levels of REG3 lectins affect liver disease progression. We investigated whether absence or intestine-specific overexpression of REG3 lectins affects the progression of ethanol-induced liver disease via alterations to the composition of the microbiota and increased bacterial translocation.
Results
Loss of REG3B promotes progression of ethanol-induced steatohepatitis
We previously demonstrated that chronic alcohol feeding reduces intestinal levels of Reg3b and Reg3g mRNA and protein in mice (Hartmann et al., 2013; Yan et al., 2011). To investigate the role of REG3B in development of alcoholic liver disease, we compared the effects of feeding ethanol to Reg3b−/− mice (C57BL/6 background) vs littermates with a wild-type copy of the gene (WT). We created Reg3b−/− mice and documented loss of REG3B protein in the small intestine (Figures 1A and 1B).
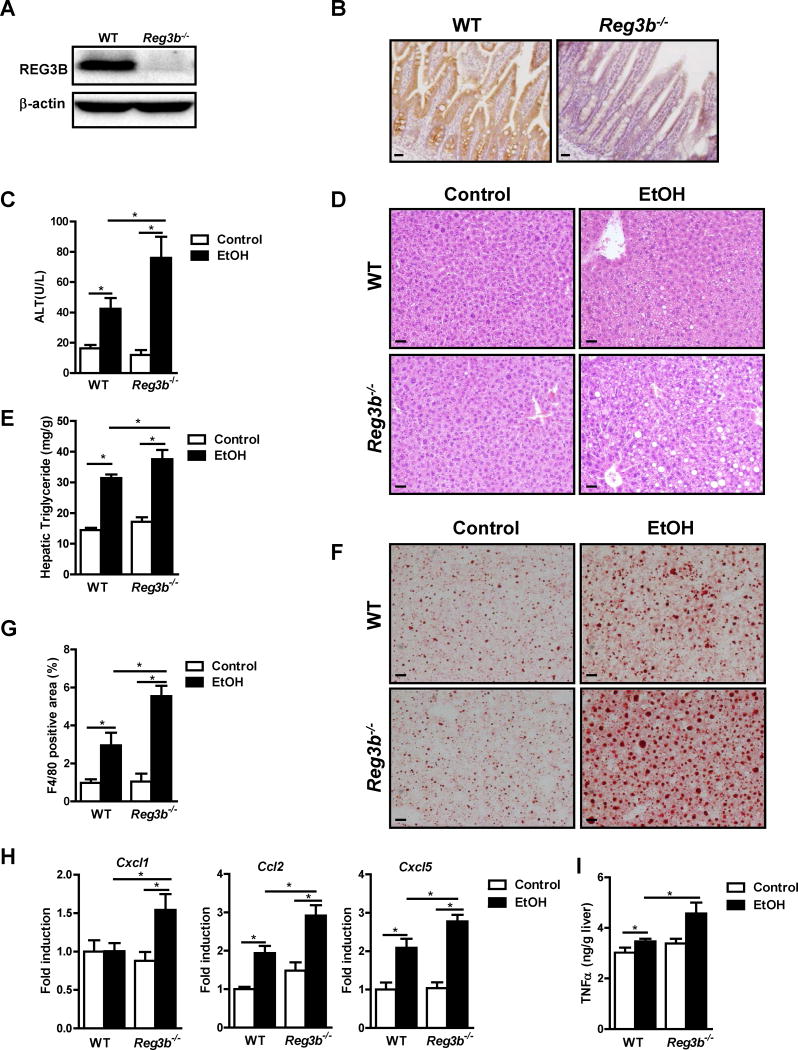
Figure 1. Exacerbated alcohol-induced liver disease in Reg3b−/− mice.
WT mice and their Reg3b−/− littermates were fed an oral control diet (n=4–7) or ethanol diet (n=9–19). (A, B) Immunoblot and immunohistochemical analysis of REG3B in the small intestine. (C) Plasma levels of ALT. (D) Representative liver sections after hematoxylin and eosin staining. (E) Hepatic triglyceride content. (F) Representative Oil Red O-stained liver sections. (G) Immunofluorescent analysis of F4/80 in the liver; positively stained area was quantitated by image analysis software. (H) Hepatic expression of mRNAs encoding chemokines. (I) Hepatic expression of TNFα protein. Scale bars, 50 μm. *P<.05. See also Figure S1.
Ratios of liver to body weight were slightly higher in Reg3b−/− mice fed ethanol for 8 weeks than WT mice (Figure S1A). Reg3b−/− mice developed more severe ethanol-associated liver disease; these mice had higher levels of liver injury (based on plasma level of alanine aminotransferase [ALT]) and hepatic steatosis than WT littermate mice (Figure 1C and 1D). This was confirmed by measurement of hepatic triglyceride levels (Figure 1E and 1F). In Reg3b−/− mice, liver disease progressed from simple steatosis to steatohepatitis. Expression of the macrophage marker, F4/80, as determined by immunofluorescence, was higher in the liver of ethanol-fed Reg3b−/− mice than ethanol-fed WT mice (Figure 1G; Figure S1B). Although livers of WT mice did not significantly increase expression of mRNA encoding the inflammatory chemokine CXCL1 with ethanol feeding, livers of Reg3b−/− mice showed significantly increased levels of Cxcl1 mRNA following ethanol administration. Livers of WT and Reg3b−/− mice each upregulated hepatic expression of Ccl2 and Cxcl5 mRNAs after ethanol feeding, but their levels increased to a significantly greater extent in Reg3b−/− mice (Figure 1H). These chemokines are mediating ethanol-induced liver disease in mice, and increased hepatic expression levels correlate with disease severity in patients with alcoholic hepatitis (Chang et al., 2015; Colmenero et al., 2007; Dominguez et al., 2009). Following ethanol administration, hepatic tumor necrosis factor (TNF)-α protein was higher in Reg3b−/− mice than in WT mice (Figure 1I). We confirmed that TNFα causes lipid accumulation in primary mouse hepatocytes (Ma et al., 2008) (Figure S1C). To determine whether REG3B affects intestinal absorption and hepatic metabolism of ethanol, we examined plasma levels of ethanol and hepatic ethanol metabolism. Plasma levels of ethanol were comparable between Reg3b−/− and WT mice following chronic ethanol administration (Figure S1D). Alcohol dehydrogenase (ADH) and cytochrome p450 enzyme 2E1 (CYP2E1) are main hepatic enzymes that metabolize ethanol and convert it to acetaldehyde (Hartmann et al., 2013). Levels of Adh1 mRNA level did not differ significantly between Reg3b−/− and WT after ethanol feeding (Figure S1E). Hepatic levels of CYP2E1 protein increased to a similar extent following ethanol administration in WT and Reg3b−/− mice (Figure S1F). REG3B deficiency therefore exacerbates alcoholic liver disease without affecting intestinal absorption or hepatic metabolism of ethanol.
REG3B restricts the mucosa-associated microbiota and prevents bacterial translocation
Development of alcoholic liver disease involves increased translocation of microbial products from the intestinal lumen to the liver, which is facilitated by a disruption of the intestinal barrier (Chen et al., 2015b). Following ethanol administration, intestinal permeability increased in WT mice, based on measurements of fecal albumin (Figure 2A) and plasma levels of endotoxin (Figure 2B). However, intestinal permeability (Figure 2A) and plasma levels of endotoxin (Figure 2B) did not differ significantly between WT and Reg3b−/− mice after ethanol feeding, indicating that REG3B does not affect paracellular intestinal permeability.
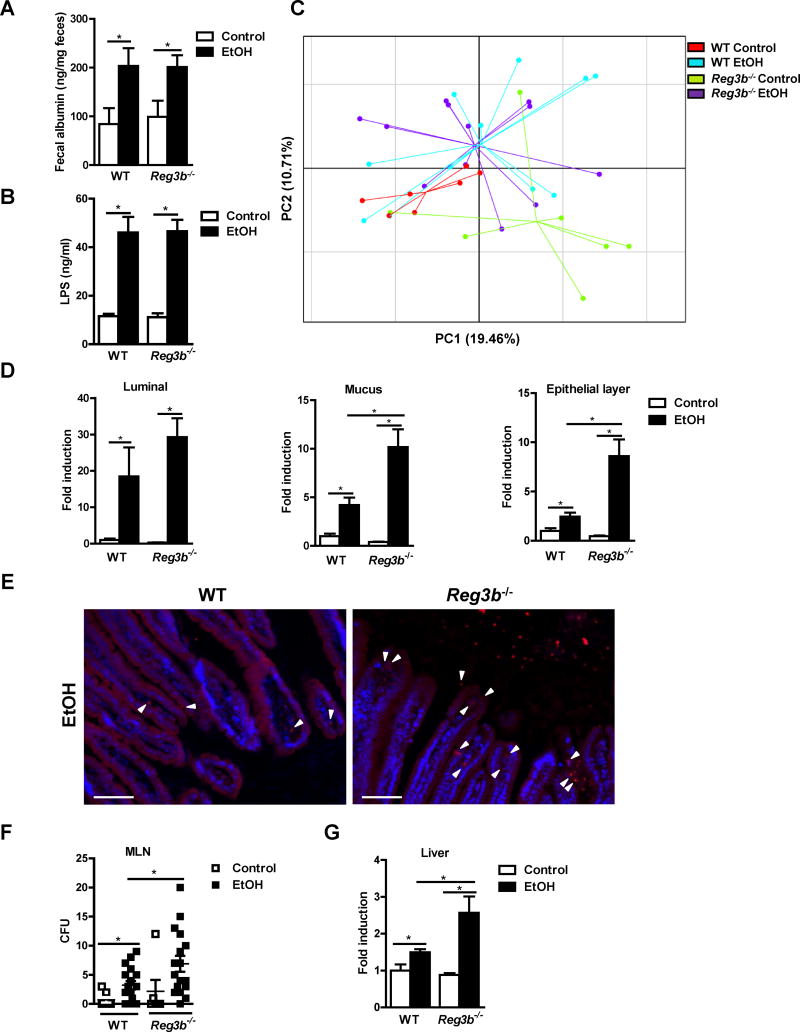
Figure 2. REG3B controls bacterial colonization of intestinal epithelial cells and reduces bacterial translocation after chronic alcohol feeding.
WT and Reg3b−/− mice were fed an oral control diet (n=4–7) or ethanol diet (n=7–17). (A) Fecal albumin content. (B) Plasma levels of LPS. (C) Principal component analysis (PCA) of microbiomes was performed. (D) Total bacteria in the lumen and mucus layer, and bacteria associated with epithelial cells, were assessed by qPCR. (E) FISH analysis using a universal probe for eubacteria. Arrowheads indicate bacteria present on epithelial surfaces and within the mucosa. (F) Colony-forming units (CFUs) were counted on anaerobic culture plates of mesenteric lymph nodes (MLN). (G) Total bacteria in the liver as assessed by qPCR. Scale bars, 50μm. *P<.05. See also Figure S1 and S2.
REG3B has antibacterial effects (van Ampting et al., 2012), so we analyzed changes in fecal microbiota, largely comprised of luminal microbes, by 16S ribosomal RNA (rRNA) gene sequencing. Comparisons of bacterial 16S rRNA data sets (Figure S1G) demonstrated that the microbiota of ethanol-fed WT mice clustered separately from that of control WT mice, based on principal component analysis (PCA) (Figure 2C) and Bray-Curtis dissimilarity index (Figure S1H, left panel); these findings are consistent with previous publications (Yan et al., 2011). The microbiota of ethanol-fed WT vs ethanol-fed Reg3b−/− mice did not cluster separately (Figure 2C) and did not differ significantly (Figure S1H, right panel). There are more Gram-negative bacteria in control mice deficient in REG3B when compared to WT control mice. This is in accordance with our prediction, since REG3B reduces the number of Gram-negative bacteria (Miki et al., 2012; van Ampting et al., 2012). Chronic ethanol administration makes this difference less prominent (Figure S1I). These findings indicate that REG3B does not affect the luminal composition of intestinal microbiota, which is largely comprised of luminal microbes, following chronic ethanol exposure.
Alcohol-associated dysbiosis is characterized not only by compositional changes, but also by quantitative differences in microbes - particularly in the small intestine (Yan et al., 2011). Bacteria reside in different niches within the intestine; therefore, bacterial loads in the lumen, mucus layer, and epithelial cells of the small intestine were measured by quantitative PCR (qPCR). Wheat germ agglutinin (WGA) staining of intestinal sections confirmed that the mucus layer has been appropriately separated from the underlying epithelial cells (Figure S2A). After alcohol feeding for 8 weeks, WT mice showed bacterial overgrowth of the luminal and the mucosa-associated (mucus and epithelial layer) microbiota of the small intestine (Figure 2D). Although luminal increases in bacteria were similar between WT and Reg3b−/− mice with ethanol feeding, ethanol-fed Reg3b−/− mice had significantly higher numbers of mucosa-associated bacteria in the mucus and epithelial layer of the small intestine than ethanol-fed WT mice (Figure 2D).
Fluorescence in situ hybridization (FISH) analysis with a bacteria-specific 16S rDNA probe confirmed an increased number of bacteria on the surface of enterocytes and within the mucosa of ethanol-fed Reg3b−/− vs WT mice (Figure 2E). Use of a non-specific probe as negative control did not yield a fluorescent signal (Figure S2B). Increased colonization of the intestinal epithelial surface was accompanied by significant increases in numbers of bacteria translocated to the mesenteric lymph nodes and livers of ethanol-fed Reg3b−/− vs WT mice (Figure 2F and G). REG3B therefore appears to reduce ethanol-induced steatohepatitis by reducing mucosa-associated intestinal bacteria and preventing bacterial translocation.
Reg3g−/− mice are more susceptible to bacterial translocation and alcoholic liver disease
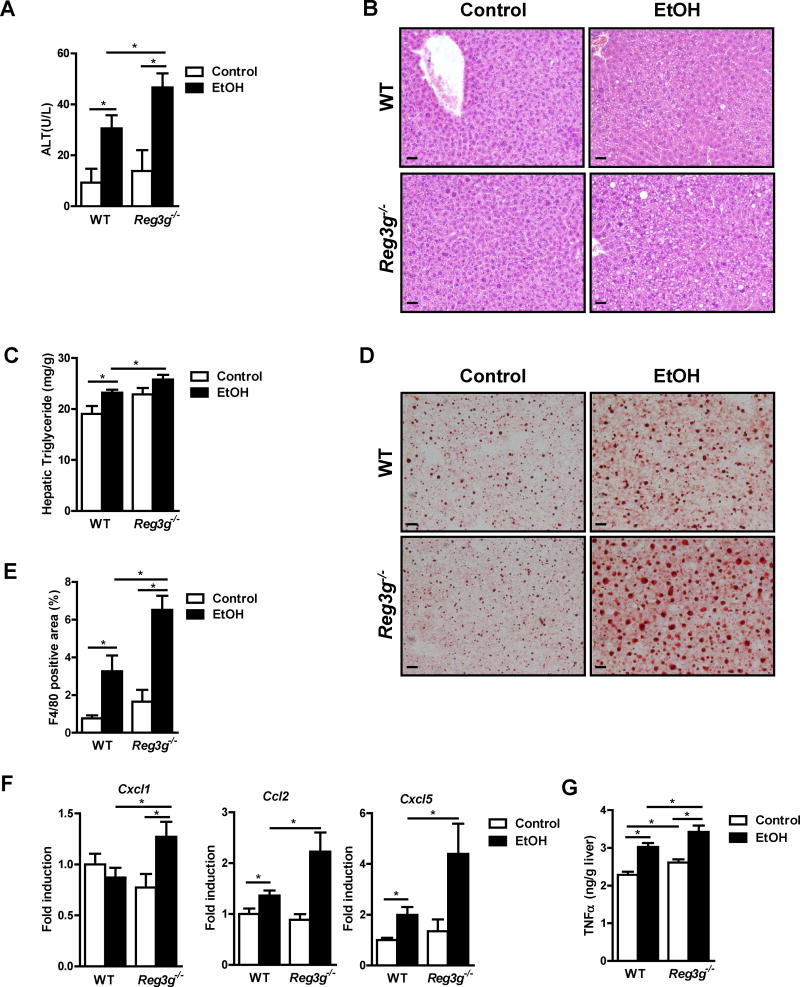
We investigated the role of REG3G in alcoholic liver disease using Reg3g−/− mice fed ethanol for 8 weeks. Ratios of liver to body weight were similar between ethanol-fed WT and littermate Reg3g−/− mice (Figure S3A). Similar to Reg3b−/− mice, Reg3g−/− mice developed more severe ethanol-induced liver injury (Figure 3A), steatosis (Figure 3B–D), and inflammation than WT mice (Figure 3E–G). In fact, livers of Reg3g−/− mice had more severe progression of ethanol-induced steatohepatitis. As in Reg3b−/− mice, Reg3g−/− mice showed more F4/80 positive cells, upregulated gene expression of chemokines Cxcl1, Ccl2 and Cxcl5, and higher TNFα protein levels in the liver following ethanol-feeding (Figure 3E–G; Figure S3B). Plasma levels of ethanol and hepatic ethanol metabolism did not differ significantly with ethanol feeding in Reg3g−/− mice vs WT mice (Figure S3C–S3E).
Figure 3. Chronic administration of ethanol exacerbates liver disease in Reg3g−/− mice.
WT and their Reg3g−/− littermates were fed an oral control diet (n=4–6) or ethanol diet (n=8–13). (A) Plasma level of ALT. (B) Representative liver sections after hematoxylin and eosin staining. (C) Hepatic triglyceride content. (D) Representative Oil Red O-stained liver sections. (E) Immunofluorescent analysis of F4/80 in the liver; positively stained area was quantitated by image analysis software. (F) Hepatic expression of mRNAs encoding chemokines. (G) Hepatic expression of TNFα protein. Scale bars, 50 μm. *P<.05. See also Figure S3 and S4.
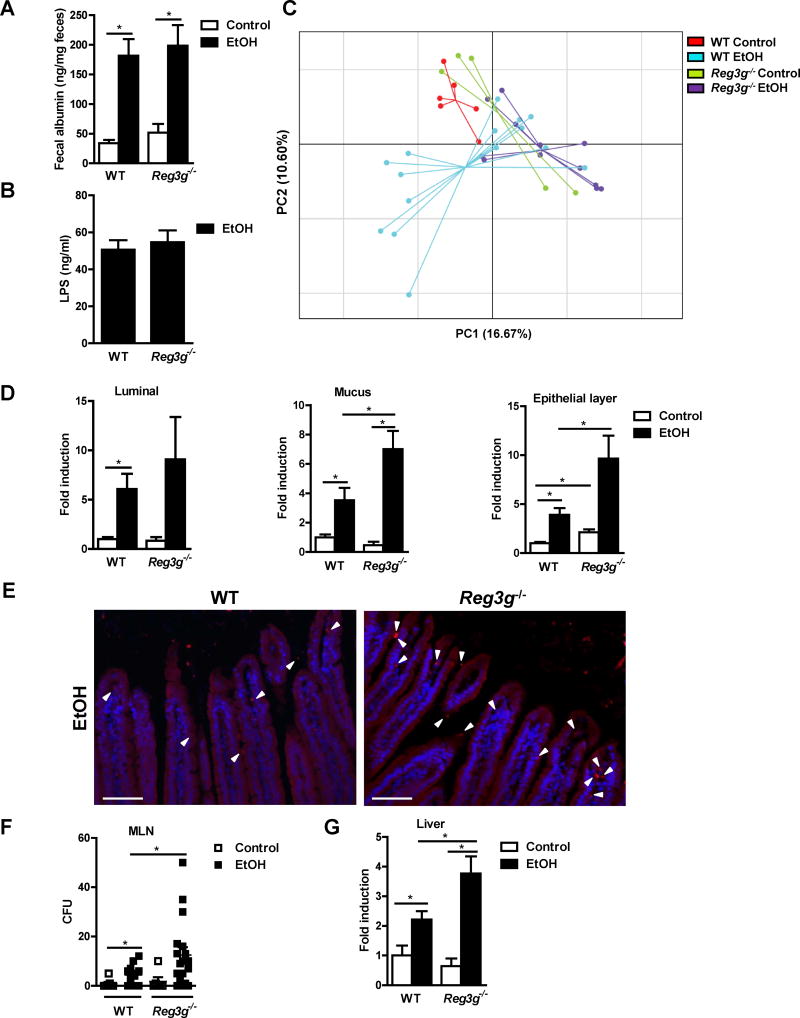
Ethanol feeding increased intestinal permeability and translocation of lipopolysaccharide (LPS) to a similar degree in Reg3g−/− vs WT littermate mice (Figure 4A and 4B). The microbiota of WT vs Reg3g−/− mice did not differ significantly in control mice or after chronic ethanol feeding (Figure 4C; Figure S3F), consistent with previously published data (Vaishnava et al., 2011). Similarly, ethanol feeding of Reg3g−/− mice did not alter the quantity of the luminal bacteria (Figure 4D). However, the number of mucosa-associated bacteria was significantly higher in Reg3g−/− than WT mice following ethanol administration (Figure 4D and 4E). This increase was associated with increased numbers of bacteria in mesenteric lymph nodes and livers of ethanol-fed, but not control, Reg3g−/− mice (Figure 4F and G). Similar to REG3B, intestinal REG3G limits mucosal colonization and translocation of commensal bacteria, and reduces ethanol-induced liver damage in mice.
Figure 4. Mucosa-associated bacterial colonization and bacterial translocation is increased in Reg3g−/− mice following chronic alcohol feeding.
WT mice and their Reg3g−/− littermates were fed an oral control diet (n=4–6) or ethanol diet (n=11–18). (A) Fecal albumin content. (B) Plasma level of LPS. (C) Principal component analysis (PCA) of microbiomes was performed. (D) Total bacteria in the lumen and mucus layer, and associated with epithelial cells, as assessed by qPCR. (E) FISH analysis using a universal probe for eubacteria. Arrowheads indicate bacteria present on epithelial surfaces and within the mucosa. (F) CFUs were counted on anaerobic culture plates of mesenteric lymph nodes (MLN). (G) Total bacteria in the liver, assessed by qPCR. Scale bars, 50μm. *P<.05. See also Figure S3 and S4.
CX3CR1hi cells are a sub-population of CD11c+ mononuclear phagocytes and have both dendritic cell- and macrophage-like characteristics. CX3CR1hi cells phagocytose commensal bacteria and migrate from the lamina propria to mesenteric lymph nodes in mice (Diehl et al., 2013; Medina-Contreras et al., 2011; Niess and Adler, 2010). To test the involvement of this pathway in bacterial translocation, CX3CR1/CD11c double positive cells were quantitated in mesenteric lymph nodes. The number of CX3CR1/CD11c double positive cells was similar in WT and Reg3g−/− mice after ethanol feeding (Figure S3G).
To demonstrate that bacterial translocation mediates increased susceptibility of Reg3g−/− mice to alcohol-induced liver disease, depletion of the commensal microbiota was performed using non-absorbable antibiotics. Increased numbers of luminal and mucosa-associated bacteria were reduced in ethanol-fed WT and Reg3g−/− mice receiving non-absorbable antibiotics (Figure S4A). Depletion of the intestinal microbiota reduced bacterial translocation (Figure S4B) and ameliorated alcoholic liver disease to a similar degree in WT and Reg3g−/− mice after chronic alcohol feeding (Figure S4C–E). These results support a link between bacterial translocation and ethanol-induced liver disease.
Intestinal overexpression of Reg3g reduces features of ethanol-induced liver disease in mice
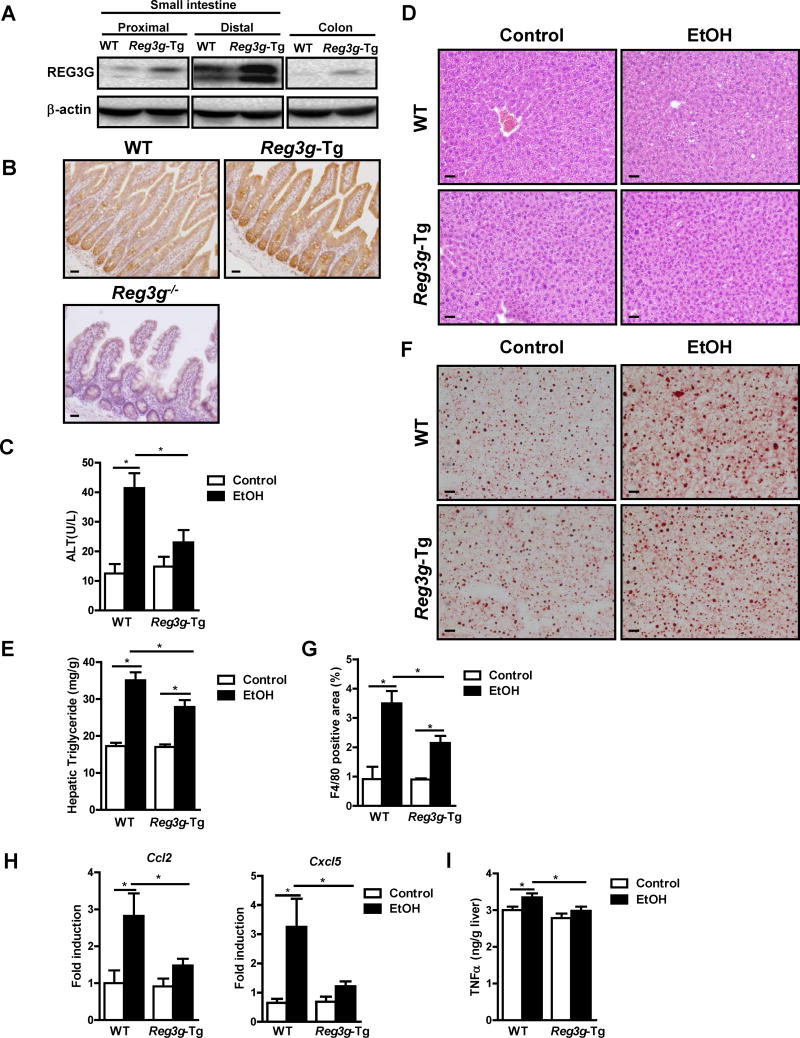
To determine whether restoring REG3G to the intestine is sufficient to reduce bacterial translocation and protect against ethanol-induced liver damage, we generated transgenic mice that overexpress Reg3g under the control of the intestinal epithelial cell-specific Villin promoter. We observed increased levels of Reg3g mRNA and protein in enterocytes and Paneth cells of Reg3g-transgenic (Reg3g-Tg) mice, compared to their C57BL/6 littermates without the transgene (WT) (Figure 5A and B; Figure S5A).
Figure 5. Intestine-specific overexpression of REG3G protects mice from ethanol-induced liver disease.
WT mice and their Reg3g-Tg littermates were fed an oral control diet (n=5–9) or ethanol diet (n=10–18). (A, B) Immunoblot and immunohistochemical analyses of REG3G in the small intestine. (C) Plasma level of ALT. (D) Representative liver sections after hematoxylin and eosin staining. (E) Hepatic triglyceride content. (F) Representative Oil Red O-stained liver sections. (G) Immunofluorescent analysis of F4/80 in the liver; positively stained area was quantitated by image analysis software. (H) Hepatic expression of mRNAs encoding chemokines. (I) Hepatic expression of TNFα protein. Scale bars, 50μm. *P<.05. See also Figure S5 and S6.
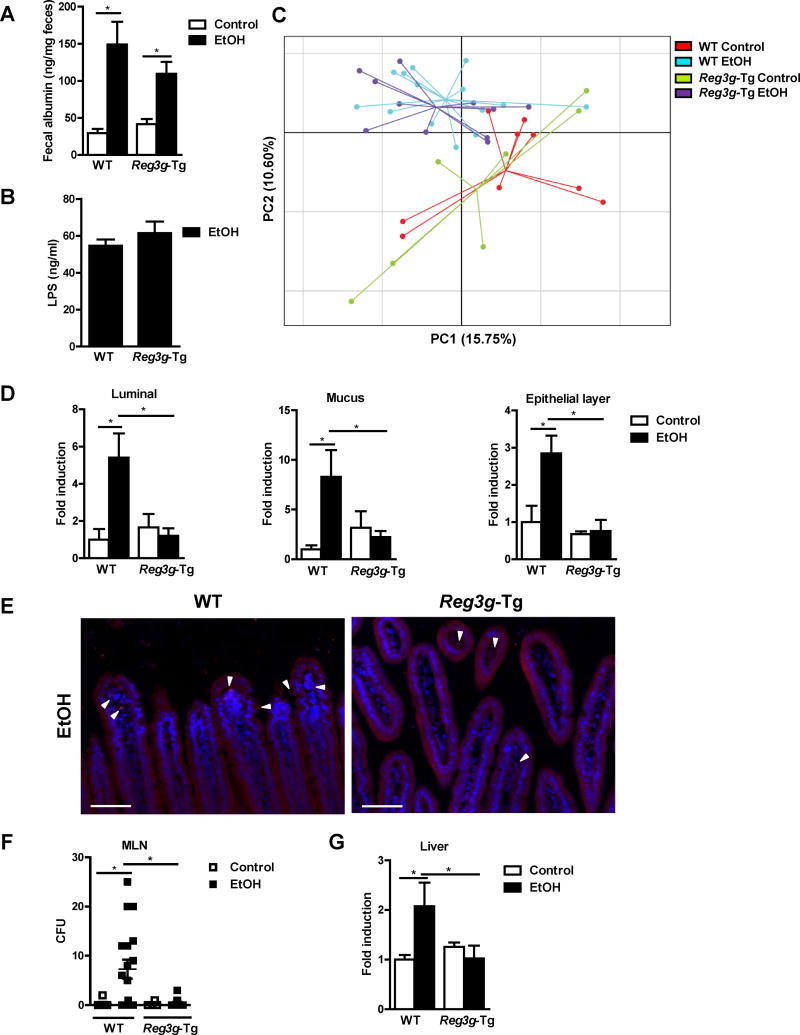
Consistent with findings from gene disruption experiments, Reg3g-Tg mice expressing physiologically relevant levels of REG3G protein, were protected from ethanol-induced liver injury, steatosis, and inflammation (Figure 5C–I; Figure S5B–C). Intestinal REG3G protein expression remained elevated following ethanol administration in Reg3g-Tg mice (Figure S5D). Ethanol absorption in the intestine and metabolism in the liver were not affected by the transgene (Figure S5E–G). Paracellular permeability and systemic levels of LPS did not differ significantly between Reg3g-Tg and WT mice after alcohol feeding (Figure 6A and 6B).
Figure 6. Reg3g-Tg mice have lower intestinal bacterial loads and bacterial translocation after ethanol feeding.
WT mice and their Reg3g-Tg littermates were fed an oral control diet (n=5–9) or ethanol diet (n=10–18). (A) Fecal albumin content. (B) Plasma levels of LPS. (C) Principal component analysis (PCA) of microbiomes was performed. (D) Total bacteria in the lumen and mucus layer, and associated with epithelial cells, as assessed by qPCR. (E) FISH analysis using a universal probe for eubacteria. Arrowheads indicate bacteria present on epithelial surfaces and within the mucosa. (F) CFUs were counted on anaerobic culture plates of mesenteric lymph nodes (MLN). (G) Total bacteria in the liver as assessed by qPCR. Scale bars, 50μm. *P<.05. See also Figure S5 and S6.
The luminal composition of the intestinal microbiota, largely comprised of microbes, differed between these strains after ethanol feeding (Figure 6C; Figure S5H). Similarly, ethanol-fed Reg3g-Tg mice had significantly lower numbers of luminal bacteria than WT mice (Figure 6D). Although REG3G activity is restricted to the inner mucus layer under homeostatic conditions (Vaishnava et al., 2011), overexpressed REG3G might penetrate through the mucus layer into the intestinal lumen, where it has antibacterial activity. Overexpression of intestinal REG3G also significantly reduced the numbers of mucosa-associated bacteria after ethanol feeding (Figure 6D and 6E). In Reg3g-Tg mice, significantly fewer bacteria translocated through the intestinal epithelial cell layer to mesenteric lymph nodes and liver after ethanol feeding, compared with WT mice (Figure 6F and G).
To investigate the role of the luminal microbiota for ethanol-induced liver disease, we performed co-housing studies. WT and Reg3g-Tg mice were co-housed after weaning and during the time of ethanol feeding, which allowed for a more homogeneous luminal microbiota composition and similar luminal numbers of bacteria in both strains (Figure S6A–B). Co-housed Reg3g-Tg mice still showed lower numbers of mucosa-associated bacteria (Figure S6B). Despite having a similar luminal microbiota, ethanol-fed Reg3g-Tg mice demonstrated less bacterial translocation and reduced alcohol-induced liver injury and steatosis (Figure S6C–G).
These findings support a model in which REG3G reduces the number of bacteria on mucosal surfaces of the intestine to limit bacterial translocation and liver disease following chronic alcohol administration.
Alcohol abuse increases the mucosa-associated microbiota in the small intestine of humans
We previously showed that duodenal biopsies from patients with alcohol dependency have lower levels of REG3G than healthy individuals (controls) (Yan et al., 2011). To determine how reduced expression of REG3G relates to the number of bacteria covering small intestinal mucosa surfaces in humans, we amplified the bacterial 16S rRNA gene by qPCR in whole duodenal biopsies from patients with alcohol dependency and individuals without alcohol dependency (controls). Mucosa-associated (mucus plus epithelial cells) bacteria were significantly increased in duodenal tissues from alcohol-dependent patients compared with controls (Figure 7).
Figure 7. Alcohol abuse increases the number of mucosa-associated bacteria in humans.
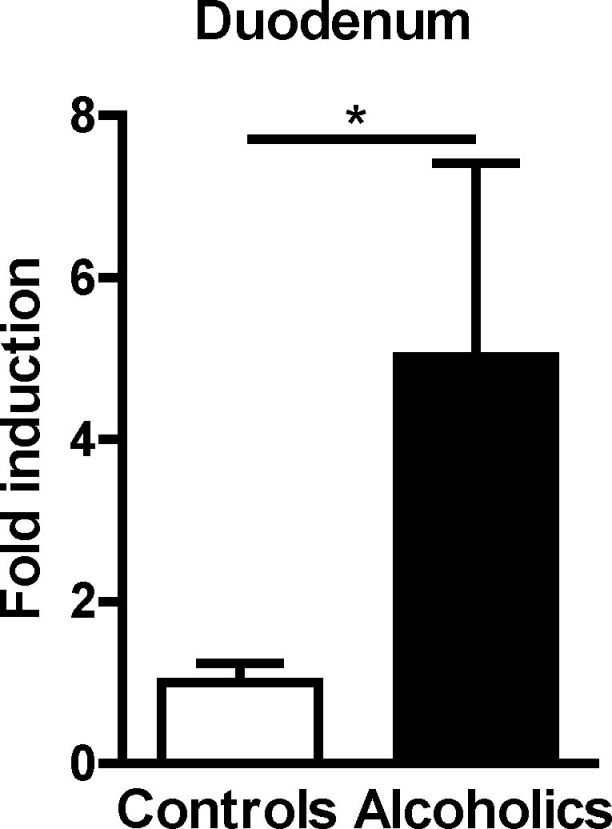
Total bacteria in duodenal biopsies from controls without alcohol dependency (n=5) and alcohol dependent patients (n=8). *P<0.05. See also Table S1.
Discussion
Antimicrobial molecules are required to maintain intestinal homeostasis. Impaired function of intestinal antimicrobial molecules can contribute to disease (Mukherjee and Hooper, 2015). Mice with genetic modifications that reduce secretion of antimicrobial molecules can develop chronic intestinal inflammation. Variants in the human homologues of these genes (ATG16L1 and XBP1) increase risk for Crohn’s disease (Cadwell et al., 2008; Kaser et al., 2008). Conversely, antimicrobial molecules protect against intestinal pathogens. Transgenic overexpression of human α-defensin-5 in Paneth cells protects mice from enteric salmonellosis (Salzman et al., 2003). REG3G protects mice against intestinal infection with Listeria monocytogenes and vancomycin-resistant Enterococcus (VRE) (Brandl et al., 2008; Brandl et al., 2007; Loonen et al., 2014).
Our study links the function of intestinal antimicrobial molecules to extra-intestinal disease. REG3 lectins restrict the access of bacteria to host epithelial cell surfaces and reduce the number of commensal bacteria that translocate to extra-intestinal tissues following chronic ethanol administration. Preventing bacterial translocation protects mice from alcoholic-induced liver disease. This process occurs independently from intestinal permeability, which is increased in alcoholic liver disease.
Our findings suggest that translocation of bacteria to the mesenteric lymph nodes is not linked to the bacterial load that is present in the intestinal lumen or to luminal dysbiosis. Rather, the determining variant is the number of bacteria that penetrate the intestinal mucus layer and colonize epithelial cell surfaces. REG3B and REG3G target different bacteria, but deficiency in either lectin exacerbates alcoholic liver disease, while overexpression of the Gram-positive bacteria targeting protein REG3G is sufficient to decrease disease. It is therefore conceivable that the number of mucosa-associated bacteria rather than the simple bacterial phylotype determines bacterial translocation and alcoholic liver disease. The genomic function of bacteria might be also an important determinant (Turnbaugh et al., 2009). It is also possible that either Gram-negative or Gram-positive bacteria can elicit the required immune responses in the liver that produce inflammation, and not a single species of bacteria. TLR2 and TLR4 deficient mice are resistant to alcoholic liver disease (Roh et al., 2015; Uesugi et al., 2001).
Chronic administration of ethanol to mice does not produce obvious morphologic damage to the lining epithelial cells (such as ulcerations or mucosal injury) or a significant increase in apoptosis of enterocytes (Chen et al., 2015b), which would allow bacteria to escape through intestinal surface openings. Instead, ethanol feeding increases expression of cytokines that promote inflammation in the lamina propria, which results in disruption of tight junction complexes and an increase in intestinal permeability (Chen et al., 2015a). The intercellular space between enterocytes with disrupted tight junctions is too small to allow the passage of viable bacteria (Turner, 2009). The most likely route for viable bacteria to reach mesenteric lymph nodes is therefore the transcellular route (Wiest et al., 2014). CX3CR1hi mononuclear cells phagocytose commensal bacteria from intestinal lumen and migrate from lamina propria to mesenteric lymph nodes in mice (Diehl et al., 2013; Medina-Contreras et al., 2011; Niess and Adler, 2010). Although the number of CX3CR1/CD11c double positive cells was not different in mesenteric lymph nodes of WT and Reg3g−/− mice after ethanol administration, future studies are required to specifically determine phagocytic capacity of these cells.
Deficiency in REG3G does not directly affect adaptive immunity (Vaishnava et al., 2011). Similarly, REG3G deficiency does not affect the immune response in the mesenteric lymph nodes (Loonen et al., 2014), indicating that changes in bacterial translocation do not result from impaired intestinal immunity. This is supported by findings from Reg3g-Tg mice (which overexpress Reg3g in only intestinal epithelial cells and Paneth cells). The exact mechanism that allows commensal bacteria to transcytose through intestinal epithelial cells is poorly understood and deserves further investigation.
How does increased bacterial translocation exacerbate alcoholic liver disease? Microbes that escape immune surveillance in mesenteric lymph nodes enter the blood stream and reach the liver, where they are phagocytosed by resident hepatic macrophages (Kupffer cells) (Balmer et al., 2014). Activated Kupffer cells are mediators of alcoholic liver disease. Elimination of resident liver macrophages prevents ethanol-induced liver disease in rodents (Niemela et al., 2002). These findings are consistent with our results, which demonstrate increased numbers of bacteria and an activated inflammatory response in the liver of Reg3b−/− and Reg3g−/− mice. In fact, an absence of REG3 lectins causes progression of simple alcoholic hepatic fatty liver disease to steatohepatitis.
Antimicrobial molecules serve as endogenous antibiotics to rapidly kill or inactivate microorganisms (Mukherjee and Hooper, 2015). Since expression of REG3 is reduced by chronic alcohol consumption in mice and humans (Hartmann et al., 2013; Yan et al., 2011), a strategy designed to increase intestinal concentrations of REG3 lectins or their production by intestinal epithelial cells might be developed to prevent alcohol-induced liver disease. Future studies are required to determine whether REG3 lectins are useful as intervention approach to revert alcohol-induced liver disease.
Experimental Procedures
Mice
The mouse strain used for this research project, B6;129S5-Reg3btm1Lex/Mmcd, identification number 032538-UCD, was obtained from the Mutant Mouse Regional Resource Center, a NCRR-NIH funded strain repository, and was donated to the MMRRC by Genentech, Inc. Rederived Reg3b−/− mice were backcrossed onto a pure C57BL/6 genetic background for 6 generations. Reg3g−/− mice on a C57BL/6 genetic background have been described (Vaishnava et al., 2011). Heterozygous mice were used for breeding, and WT and knockout littermate mice were used in all experiments.
To generate C57BL/6 mice that expressed Reg3g from a transgene, the mouse REG3G coding sequence was amplified by PCR from small intestinal cDNA and subcloned into a plasmid containing the 12.4-kb Villin promoter (Taniguchi et al., 2015) using SmaI and Xhol restriction enzyme sites. The Reg3g expression cassette was excised by PmeI digestion, purified, and injected into fertilized C57BL/6 oocytes to obtain founder mice which transmitted the Reg3g transgene (Transgenic Core Facility, UC San Diego). Littermate WT and transgenic mice were used for experiments. Mice not demonstrating increased intestinal Reg3g expression postmortem, compared with WT littermates, were excluded from the study (about 10% of transgenic mice). Littermate WT, knockout, and transgenic mice were separated at the time of weaning (age 3 weeks).
Animal model
Age-matched female mice (2 mice per cage) were subjected to the Lieber DeCarli diet model of chronic alcohol feeding for 8 weeks as described by us (Chen et al., 2015a). Pair-fed control mice received a diet with an isocaloric substitution of dextrose. Antibiotics treatment of Reg3g−/− and their WT littermates was started at 4 weeks after liquid diet feeding, and mice were gavaged three times per week until harvesting. The composition of antibiotics mixture has been described (Polymyxin B (150 mg/kg BW/day) and Neomycin (200 mg/kg BW/day)) (Chen et al., 2015a). Control mice were gavaged with an equal volume of vehicle (water). For co-housing studies, weaned WT and Reg3g-Tg littermate mice were housed together in a cage (2 mice per cage), and ethanol feeding was started at 8 weeks of age. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
Human samples
Patients fulfilling the DSM IV criteria (Ball et al., 1997) for alcohol dependence and with active alcohol consumption were compared to individuals without alcohol dependency (controls). Duodenal biopsies were taken from patients with endoscopically normal duodenum and were snap frozen. Written informed consent was obtained from all patients and controls. Patient characteristics are shown in Table S1. Patients did not take antibiotics or immunosuppressive medication during the two months preceding enrollment. Other exclusion criteria were diabetes, inflammatory bowel disease, known liver disease of any other etiology, and clinically significant cardio-vascular, pulmonary or renal co-morbidities. The study protocol was approved by the Ethics Committee of the Université Catholique de Louvain, in Brussels, Belgium, by the Human Research Protections Program of the University of California San Diego and of the VA San Diego Healthcare System. Genomic DNA was extracted from the entire biopsy and 16S qPCR performed as described below. The qPCR value of the 16S rRNA gene was normalized to the weight of the biopsy.
Bacterial cultures
At the time of collection, mesenteric lymph nodes were collected in a sterile fashion, homogenized using a beads beater, and plated onto on BBL Brucella Agar Plates (containing 5% horse blood; BD) for culture of anaerobic bacteria. The plates were incubated for 72 hrs at 37°C under anaerobic conditions followed by the counting of colony-forming units (CFUs).
Bacterial DNA isolation and qPCR for 16S
Genomic DNA was isolated from a 2 cm piece of the proximal small intestine (jejunum). The exact length, width, and weight of this piece was measured. Luminal contents were collected by flushing with 1 ml sterile phosphate-buffered saline (PBS). The remaining intestine was cut longitudinally and washed vigorously in 1 ml PBS to collect the mucus and its associated bacteria. The remaining part of the intestine was homogenized and used for isolating genomic DNA from the epithelial cell layer. DNA from intestinal contents, mucus layer, epithelial cell layer, and cecum was extracted as previously described (Fouts et al., 2012b; Yan et al., 2011). The qPCR value of the 16S rRNA gene (Hartmann et al., 2013) of luminal contents was normalized to weight, whereas the qPCR value of the 16S rRNA gene of the mucus and epithelial layer was normalized to the area of the intestine. Hepatic 16S rRNA gene expression was normalized to host 18S.
Real-time qPCR
RNA was extracted from mouse tissues, and cDNAs were generated (Hartmann et al., 2012). Primer sequences for mouse genes were obtained from the NIH qPrimerDepot. The qPCR value was normalized to 18S.
16S rRNA sequencing
Deep DNA pyrosequencing of the hypervariable V1–V3 region of prokaryotic 16S rRNA loci was performed to generate microbial community profiles using species-level (97% similarity) operational taxonomic unit-based classification and analysis as described in our previous publication (Chen et al., 2015b), except that sequences were generated on the Illumina MiSeq platform using a dual indexing strategy (Kozich et al., 2013), and our open source automated 16S pipeline, YAP, was modified accordingly (https://github.com/andreyto/YAP). Principal component analysis (PCA) was performed using the ade4 R package (Dray and Dufour, 2007) as described previously (Fouts et al., 2012a). Beta diversity was determined by computing Bray-Curtis distances using MOTHUR. Individual pairwise distances were extracted from the resulting lower triangle Phylip-style distance matrix using R and only relevant pairwise distances were plotted using boxplot and stripchart with jitter in R. Sequence data was registered at NCBI under BioProject PRJNA289260. Sequence reads are available at NCBI under the following consecutive BioSample ids: Reg3b−/− (SAMN03853805 - SAMN03853841), Reg3g−/− (SAMN03853842 - SAMN03853879), and Reg3g-Tg (SAMN03853880 - SAMN03853920).
FISH analysis
Samples fixed in Carnoy’s fixative and embedded in paraffin were deparaffinized and hybridized to universal eubacterial or control probes labeled with Alexa594 as described (Grivennikov et al., 2012).
Biochemical assays
Plasma levels of ethanol were measured using the Ethanol Assay Kit (BioVision). Levels of ALT were determined using Infinity ALT kit (Thermo Scientific). Plasma LPS level was measured by a commercial ELISA kit (Cloud-Clone Corp). Hepatic triglyceride levels were measured using the Triglyceride Liquid Reagents Kit (Pointe Scientific).
Staining procedures
Formalin-fixed tissue samples were embedded in paraffin and stained with hematoxylin-eosin (Surgipath), or with anti-REG3B (R&D Systems) and anti-REG3G (Vaishnava et al., 2011) antibodies using standard staining protocols. To preserve the mucus layer, intestinal samples were fixed in Carnoy’s fixative consisting of 60% ethanol, 30% chloroform, and 10% glacial acetic acid for 1h. To visualize the intestinal mucus layer, we used WGA (Invitrogen) staining that detects N-acetyl-D-glucosamine and N-acetyl-D-neuraminic acid residues (Wright, 1984) from the mucins. WGA was purchased as fluorescent coupled reagent. Frozen sections were used for Oil Red O staining. F4/80 (eBioscience) immunofluorescence staining was performed as described (Chen et al., 2015b). Ten random fields per slide were chosen for quantification. To detect CX3CR1/CD11c double positive cells, mesenteric lymph nodes were stained with primary antibodies against CX3CR1 (Novus Biologicals) and CD11c (Novus Biologicals), and positive reactions were visualized using fluorescent labeled secondary antibodies. 10 random high power fields per slide were chosen for analysis, and double positive cells were expressed as double-positive cells per high power field. Nuclei are stained with DAPI (blue).
Immunoblot analyses and ELISA
Immunoblot analysis was performed using anti-CYP2E1 (Millipore Corporation), anti-Reg3b (R&D Systems), and anti-Reg3g (Vaishnava et al., 2011) antibodies. Anti-β-actin (Sigma-Aldrich) antibody was used as loading control. Fecal albumin was determined by ELISA (Bethyl Labs) as described (Hartmann et al., 2013). Whole liver cell lysates were extracted using a buffer containing 100 mM Tris, pH 7.4; 150 mM NaCl and protease inhibitors (Roche). Liver TNFα was measured using enzyme-linked immunosorbent assay (eBioscience).
Hepatocyte isolation and culture
Primary mouse hepatocytes were isolated as described (Iwaisako et al., 2012). Hepatocytes were cultured in Medium 199 (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS) for 4 hours. Following an overnight starvation in medium without FBS, hepatocytes were stimulated with TNFα (5 ng/ml; R&D Systems) for 24 hours and stained with Oil Red O.
Statistics
Results are expressed as mean±SEM. Significance was evaluated using the unpaired Student t test. The non-parametric Wilcoxon rank sum test (wilcox.test in R) was used for statistical analysis of Bray-Curtis distances and the Mann-Whitney test for human samples. A p-value less than 0.05 was considered to be a statistically significant difference.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants K08 DK081830, R01 AA020703, U01 AA021856 and by Award Number I01BX002213 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to BS). This study utilized Neuroscience microscopy equipment, which is funded by the UCSD School of Medicine Microscopy Core Grant P30 NS047101. We thank Karen Beeri for technical assistance, and Andrey Tovchigrechko for updating YAP for processing MiSeq data.
Abbreviations
- Adh
alcohol dehydrogenase
- ALT
alanine aminotransferase
- CFU
colony-forming units
- Cyp
cytochrome p450 enzyme
- MLN
mesenteric lymph nodes
- PCA
principal component analysis
- Reg3
regenerating islet derived-3
- Tg
transgenic
- TNF
tumor necrosis factor
- WGA
wheat germ agglutinin
- WT
wild-type
Footnotes
Author contribution
L.W. was responsible for acquisition of data, analysis and interpretation of data, and drafting of the manuscript; D.E.F., J.D. and K.M. were responsible for 16S rDNA sequencing and analysis; P.S. was responsible for collection of human samples; P.H., P.C. and C.L. assisted with experiments and data analysis; S.B.H., D.A.B. and L.V.H. provided experimental resources and edited the manuscript; B.S. was responsible for the study concept and design, writing the manuscript, and study supervision.
None of the authors has a financial, personal or professional conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology. 1995;108:218–224. doi: 10.1016/0016-5085(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Ball SA, Tennen H, Poling JC, Kranzler HR, Rounsaville BJ. Personality, temperament, and character dimensions and the DSM-IV personality disorders in substance abusers. J Abnorm Psychol. 1997;106:545–553. doi: 10.1037//0021-843x.106.4.545. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N, et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Science translational medicine. 2014;6:237ra266. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- Bode JC, Bode C, Heidelbach R, Durr HK, Martini GA. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31:30–34. [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. The Journal of experimental medicine. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Xu MJ, Zhou Z, Cai Y, Li M, Wang W, Feng D, Bertola A, Wang H, Kunos G, et al. Short- or long-term high-fat diet feeding plus acute ethanol binge synergistically induce acute liver injury in mice: An important role for CXCL1. Hepatology. 2015;62:1070–1085. doi: 10.1002/hep.27921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Stärkel P, Turner JR, Ho SB, Schnabl B. Dysbiosis-induced intestinal inflammation activates TNFRI and mediates alcoholic liver disease in mice. Hepatology. 2015a;61:883–894. doi: 10.1002/hep.27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015b;148:203–214. doi: 10.1053/j.gastro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenero J, Bataller R, Sancho-Bru P, Bellot P, Miquel R, Moreno M, Jares P, Bosch J, Arroyo V, Caballeria J, et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology. 2007;132:687–697. doi: 10.1053/j.gastro.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, Miquel R, Colmenero J, Moreno M, Garcia-Pagan JC, Bosch J, Arroyo V, Gines P, Caballeria J, Bataller R. Hepatic expression of CXC chemokines predicts portal hypertension and survival in patients with alcoholic hepatitis. Gastroenterology. 2009;136:1639–1650. doi: 10.1053/j.gastro.2009.01.056. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour AB. The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20. [Google Scholar]
- Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012a;10:174. doi: 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Torralba M, Nelson KE, Brenner DA, Schnabl B. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol. 2012b;56:1283–1292. doi: 10.1016/j.jhep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann P, Chen P, Wang HJ, Wang L, McCole DF, Brandl K, Starkel P, Belzer C, Hellerbrand C, Tsukamoto H, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58:108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann P, Haimerl M, Mazagova M, Brenner DA, Schnabl B. Toll-like receptor 2-mediated intestinal injury and enteric tumor necrosis factor receptor I contribute to liver fibrosis in mice. Gastroenterology. 2012;143:1330–1340. doi: 10.1053/j.gastro.2012.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaisako K, Haimerl M, Paik YH, Taura K, Kodama Y, Sirlin C, Yu E, Yu RT, Downes M, Evans RM, et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor delta agonist. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1369–1376. doi: 10.1073/pnas.1202464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WR, Brown RS, Jr, Terrault NA, El-Serag H. Burden of liver disease in the United States: summary of a workshop. Hepatology. 2002;36:227–242. doi: 10.1053/jhep.2002.34734. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, Windey K, Tremaroli V, Backhed F, Verbeke K, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4485–4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, van Baarlen P, Wells JM. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- Ma KL, Ruan XZ, Powis SH, Chen Y, Moorhead JF, Varghese Z. Inflammatory stress exacerbates lipid accumulation in hepatic cells and fatty livers of apolipoprotein E knockout mice. Hepatology. 2008;48:770–781. doi: 10.1002/hep.22423. [DOI] [PubMed] [Google Scholar]
- Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. The Journal of clinical investigation. 2011;121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Holst O, Hardt WD. The bactericidal activity of the C-type lectin RegIIIbeta against Gram-negative bacteria involves binding to lipid A. The Journal of biological chemistry. 2012;287:34844–34855. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang QX, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103–107. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966–978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nata K, Liu Y, Xu L, Ikeda T, Akiyama T, Noguchi N, Kawaguchi S, Yamauchi A, Takahashi I, Shervani NJ, et al. Molecular cloning, expression and chromosomal localization of a novel human REG family gene, REG III. Gene. 2004;340:161–170. doi: 10.1016/j.gene.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Niemela O, Parkkila S, Bradford B, Iimuro Y, Pasanen M, Thurman RG. Effect of Kupffer cell inactivation on ethanol-induced protein adducts in the liver. Free radical biology & medicine. 2002;33:350–355. doi: 10.1016/s0891-5849(02)00894-8. [DOI] [PubMed] [Google Scholar]
- Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J Hepatol. 2013;59:160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Roh YS, Zhang B, Loomba R, Seki E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am J Physiol Gastrointest Liver Physiol. 2015;309:G30–41. doi: 10.1152/ajpgi.00031.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teltschik Z, Wiest R, Beisner J, Nuding S, Hofmann C, Schoelmerich J, Bevins CL, Stange EF, Wehkamp J. Intestinal bacterial translocation in rats with cirrhosis is related to compromised Paneth cell antimicrobial host defense. Hepatology. 2012;55:1154–1163. doi: 10.1002/hep.24789. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Uesugi T, Froh M, Arteel GE, Bradford BU, Thurman RG. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology. 2001;34:101–108. doi: 10.1053/jhep.2001.25350. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ampting MT, Loonen LM, Schonewille AJ, Konings I, Vink C, Iovanna J, Chamaillard M, Dekker J, van der Meer R, Wells JM, et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect Immun. 2012;80:1115–1120. doi: 10.1128/IAI.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol. 2014;60:197–209. doi: 10.1016/j.jhep.2013.07.044. [DOI] [PubMed] [Google Scholar]
- Wright CS. Structural comparison of the two distinct sugar binding sites in wheat germ agglutinin isolectin II. J Mol Biol. 1984;178:91–104. doi: 10.1016/0022-2836(84)90232-8. [DOI] [PubMed] [Google Scholar]
- Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric Dysbiosis Associated with a Mouse Model of Alcoholic Liver Disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.