Abstract
B. xenovorans LB400 is a model bacterium for the study of the metabolism of aromatic compounds. The aim of this study was the genomic and functional characterization of a non-ribosomal peptide synthetase containing gene cluster that encodes a siderophore in B. xenovorans LB400. The mba gene cluster from strain LB400 encodes proteins involved in the biosynthesis and transport of a hydroxamate-type siderophore. Strain LB400 has a unique mba gene organization, although mba gene clusters have been observed in diverse Burkholderiales. Bioinformatic analysis revealed the presence of promoters in the mba gene cluster that strongly suggest regulation by the ferric uptake regulator protein (Fur) and by the alternative RNA polymerase extracytoplasmic function sigma factor MbaF. Reverse transcriptase PCR analyses showed the expression of iron-regulated transcriptional units mbaFGHIJKL, mbaN, mbaABCE, mbaO, mbaP and mbaD genes under iron limitation. Chrome azurol S (CAS) assay strongly suggests that strain LB400 synthesized a siderophore under iron limitation. Mass spectrometry ESI-MS and MALDI-TOF-MS analyses revealed that the siderophore is a non-ribosomal peptide, and forms an iron complex with a molecular mass of 676 Da. Based on bioinformatic prediction, CAS assay and MS analyses, we propose that the siderophore is L-Nδ-hydroxy-Nδ-formylOrn-D-β-hydroxyAsp-L-Ser-L-Nδ-hydroxy-Nδ-formylOrn-1,4-diaminobutane that is closely related to malleobactin-type siderophores reported in B. thailandensis.
Introduction
Iron is essential for diverse metabolic processes in microorganisms. This element is involved in metabolic processes such as cellular respiration, Krebs cycle, oxygen transport, DNA synthesis, nitrogen fixation, photosynthesis, methanogenesis and hydrogen metabolism. Iron forms part of heme cofactors, iron-sulfur clusters and di-iron centers in proteins. In the presence of oxygen and at physiological pH, ferric ion is poorly soluble and therefore presents a rather low bioavailability. Aerobic microorganisms have developed iron-uptake systems which are required under iron-restricted conditions. Siderophores are low molecular mass compounds (<1000 Da) containing hydroxamate, α-hydroxycarboxylate and catechol ligands that covalently bind iron (III) with high affinity [1,2].
Diverse siderophores are composed of peptides assembled via non-ribosomal peptide synthetases (NRPS). They are composed of proteinogenic and non-proteinogenic amino acids that may be subjected to modification such as epimerization, glycosylation, acylation, methylation, formylation, halogenation, hydroxylation and cyclization [3]. NRPS catalyze the formation of peptide bonds between amino acid substrates by using biosynthetic modules composed of adenylation (A), thiolation (T), condensation (C) and epimerization (E) domains. The A domain is responsible for the activation of amino acid monomer into aminoacyl-AMP using ATP and for the catalysis of its transfer to the adjacent T domain to which monomer and nascent chain are bound. The C domain catalyzes the formation of the peptide bond between two adjacent modules, which leads to the elongation of the peptide [4]. The E domain catalyzes the epimerization from L to D form of the thioester-bound amino acid. These multimodular enzymes are governed by the colinearity rule [5], which is the basis for in silico prediction of the substrates that will be joined to each module and the structure of the final product. This rule is useful when searching for novel bioactive compounds synthesized by NRPS clusters [6,7].
The Burkholderia genus comprises both non-pathogenic environmental species and human pathogens such as B. mallei and B. cepacia [8–10]. In Burkholderia, NRPS systems for siderophore synthesis have been reported [11,12]. The siderophore ornibactin is synthesized by B. vietnamiensis, B. cepacia, B. ambifaria and B. cenocepacia [13,14], whereas malleobactin is produced by B. pseudomallei, B. mallei and B. thailandensis [15–17]. In some Burkholderia strains an additional siderophore has been reported, such as cepabactin in B. cepacia [13], pyochelin in B. pseudomallei [15], and cepaciachelin in B. ambifaria [12].
B. xenovorans LB400 is a model bacterium for the study of the degradation of a wide range of aromatic compounds including polychlorobiphenyls [18–25]. Recently, a functional polyhydroxyalkanoate anabolic pathway in strain LB400 has been reported [26]. The aim of this study was the genomic and functional characterization of a non-ribosomal peptide synthetase containing gene cluster that encodes proteins for the synthesis and transport of a siderophore in B. xenovorans LB400. Experimental analyses demonstrated that under iron limitation B. xenovorans LB400 produces a hydroxamate-type siderophore, which structure was elucidated by mass spectrometry.
Materials and Methods
In silico search and analysis of NRPS genes
A search for conserved domains of NRPS genes was performed in LB400 genome using the RPS-BLAST algorithm. The pfam00668 condensation domains provided by the Conserved Domains Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd) were predicted. NRPS domain organization and binding pocket signatures of A domains were analyzed [5,27]. The Stachelhaus code is defined by 10 amino acid residues in the A domains that form the binding pocket of a specific substrate [27]. The substrate specificity prediction was carried out using the web-based program NRPS predictor2 (http://ab.inf.uni-tuebingen.de/toolbox/index.php?view=domainpred) [28]. Subsequently, this prediction was refined with available information in literature [16,17].
For the search of a siderophore gene cluster in other Burkholderiales strains, a BlastP search was accomplished using the translated NRPS sequence protein from the BxeB0525 gene (MbaA) for translated sequence proteins from complete bacterial genomes of the NCBI Genome database, including the genomes of 43 Burkholderiales strains. 836 proteins hits were manually analyzed to identify genomes that have malleobactin- or ornibactin-type gene cluster. For the identification of iron regulated promoters in the siderophore gene cluster of LB400 strain, extracytoplasmatic function (ECF) sigma factor and ferric uptake regulator (Fur) binding sequences in the non-coding regions of the LB400 minor chromosome (C2) were searched (between the nucleotides 2.774.645 and 2.808.477), using the Artemis software (www.sanger.ac.uk/resources/software/artemis/). Sequence alignments were performed using the CLUSTALW algorithm [29].
The 3D structure of the MbaO protein was built using Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/) to identify the structure homologous and obtain the alignment based on the secondary structure and Modeller (salilab.org/modeller/) to model the 3D structure. Ligand prediction was performed with 3Dligandsite (www.sbg.bio.ic.ac.uk/3dligandsite/). Transmembrane regions were analyzed by Sosui (harrier.nagahama-i-bio.ac.jp/sosui). As crystallized structures have not been reported, siderophore and ferrisiderophore 3D structures were predicted using Marvin Sketch (http://www.chemaxon.com). The binding site for siderophore and ferrisiderophore was predicted using a Docking strategy. Docking analyses between the 3D protein structure with the siderophore and ferrisiderophore were performed with Autodock 4 [30].
Bacterial strain and culture conditions
For iron limitation assays, B. xenovorans LB400 was cultivated in MM9 mineral medium with trace solution supplemented with glucose (5 mM) as sole carbon and energy source at 30°C [22]. For iron-rich condition, bacterial cells were grown in MM9 mineral medium with trace solution supplemented with glucose (5 mM) and FeSO4 (45 μM). To study the effect of L-ornithine (Orn) on bacterial growth and siderophore activity under iron limitation, MM9 mineral medium with trace solution was supplemented with glucose (5 mM) and Orn (5 mM). Growth was determined by measuring turbidity at 600 nm.
Siderophore activity
The presence of a siderophore was determined in LB400 culture supernatants by ferrisiderophore detection after incubation with FeCl3 (2 mM) for 30 min and measuring absorbance at 400 nm as well as by the chrome azurol S (CAS) colorimetric assay [31]. CAS activity was determined by the decrease in absorbance at 630 nm of the supernatant treated with CAS solution. Absorbance was measured after 2 h incubation at room temperature. Culture medium without cells treated with CAS solution was used as reference absorbance.
RNA isolation and RT-PCR
B. xenovorans LB400 was grown on glucose until a turbidity at 600 nm of 0.5 (exponential phase) in presence of Orn (5 mM) or FeSO4 (45 μM). Total RNA was isolated from LB400 cells using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer´s recommendations. To degrade any residual DNA, DNase I treatment was carried out using the RNase-Free DNase Set (Qiagen, Hilden, Germany). Amplification of the 16S rRNA gene was used as control for DNA contamination using the 27f (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492r (5’-TACGGYTAC CTTGTTACGACTT-3’) primers [22]. RNA concentration was quantified using a QubitTM fluorometer (Invitrogen, Carlsbad, CA, USA). Reverse transcription-PCR (RT-PCR) was carried out using the Thermo Scientific Verso cDNA Kit according to the manufacturer’s instructions. Primers spanning intergenic regions of the mba genes were designed to evaluate the co-transcription of genes using RT-PCR. Sequence-specific primer pairs are listed in S1 Table. The cycles of amplification (30 cycles) were carried out after an initial denaturation at 94°C for 5 min as follows: 94°C for 1 min, 59°C for 1 min, 72°C for 2 min, and a final elongation at 72°C for 7 min. All primer pairs amplified fragments of the expected size from LB400 genomic DNA.
Isolation of the ferrisiderophore complex
B. xenovorans LB400 was grown until late stationary phase (48 h) in the presence of Orn (5 mM) or FeSO4 (45 μM). Cultures (2 L) were centrifuged at 4,000 × g for 10 min at 4°C. The supernatant was concentrated to 200 mL under vacuum. Ferrisiderophore chelates were detected, by mixing the supernatant with FeCl3 (2 mM) for 30 min at room temperature, until the color of the supernatant turned yellow. The iron treated supernatant was centrifuged for 10 min, the siderophore fraction was separated on a Sep-Pak C8 column (Waters, Milford, MA, USA) and eluted with methanol (100 mL) to eliminate salts and concentrate the yellow colored fractions. Colored crude extracts were fractionated by several passes through a preparative HPLC using a RP-C18 column. Fractions were eluted using a solvent gradient (solvent A: water containing 0.1% TFA, solvent B: acetonitrile containing TFA 0.1%): 100% solvent A for 5 min, decreasing to 60% solvent A and 40% solvent B in 25 min, maintained at 100% solvent B for 5 min, followed by 100% solvent A for 5 min. Flow rate was kept at 3 mL min-1 at room temperature. Absorbance was monitored at 400 nm. Ferrisiderophore complexes of yellow fractions were characterized by analytical HPLC. A Jasco HPLC system equipped with a Chromolith Performance RP-18e column (100 x 4.6 mm) (Alltech Grom GmbH, Rottenburg, Germany) and a diode array detector module MD-2015 was used. Data processing was carried out with Jasco ChromPass software 1.7 (http://www.jasco.de/; Jasco Corporation, Tokyo, Japan). Ferrisiderophores were eluted at room temperature using a solvent gradient (solvent A: water containing 0.1% TFA, solvent B: acetonitrile containing TFA 0.1%): 100% solvent A for 2 min, decreasing to 60% solvent A and 40% solvent B in 6 min, maintained with 100% solvent B for 2 min and 100% solvent A for 2 min. Flow rates were kept at 1 mL min-1. Absorbance was monitored at 400 nm.
Characterization of the ferrisiderophore complex by mass spectrometry
Ferrisiderophores complexes of yellow colored fractions were characterized by electrospray ionization (ESI-MS) and by matrix assisted laser desorption/ionization—time of flight (MALDI-TOF) mass spectrometry. For ESI-MS, samples prepared in a solvent containing acetonitrile 99.9% and TFA 0.1% were analyzed by Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA, USA) coupled to an Esquire 4000 electrospray ion trap mass spectrometer (Bruker Daltonik, Bremen, Germany) as previously described [32]. As stationary phase, a C18 Luna column (5 μm particle size 100Å; 150 x 4.6 mm; Phenomenex, Torrance, CA, USA) connected to a split cell that divided the flow into the UV detector and into the mass spectrometer was used. As a solvent system, distilled water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.085% formic acid (solvent B) were used. Ferrisiderophores were eluted using a solvent gradient from 3 to 53.3% solvent B within 30 min at room temperature. Flow rates were kept at 1 mL min-1. Absorbance was monitored at 400 nm. The ionization process by electrospray was performed at 4000 V, assisted by nitrogen as nebulizer gas, at a temperature of 325°C, pressure of 30 psi and a flow rate of 7.5 L/min. Mass spectra were acquired in positive polarity. The samples showed the corresponding [M(Fe)+H]+ ion as the ionized molecule of the ferrisiderophore complex.
For MALDI-TOF, samples containing ferrisiderophore were mixed with a α-cyano-4-hydroxycinnamic acid (CHCA 10 mg/mL in acetonitrile/formic acid 0.1% v/v in 1:1 ratio) matrix in a 1:1 ratio, and the mixture (2 μL) was deposited on a micro sample holder plate scout (Bruker Daltonics, Billerica, MA, USA). The peptide extracts were analyzed with a MALDI-TOF Microflex apparatus (Bruker Daltonics, Billerica, MA, USA) in positive ion mode using reflection detection (Centro de Estudio para el Desarrollo de la Química, Universidad de Chile, Santiago, Chile). The FlexControl 3.0 program (Bruker Daltonik, Bremen, Germany) was used to control the spectrometer. A calibration was performed with external standards: a mixture of peptides of 1000–3000 Da mass (Bruker Daltonics, Billerica, MA, USA). Spectra corresponds to the sum of 10 sweeps of 30 laser impacts applied at different points randomly taken from each sample deposited on the plate holder sample. The mMass program version 3.0 (http://www.mmass.org) was used to visualize the mass spectra.
Results
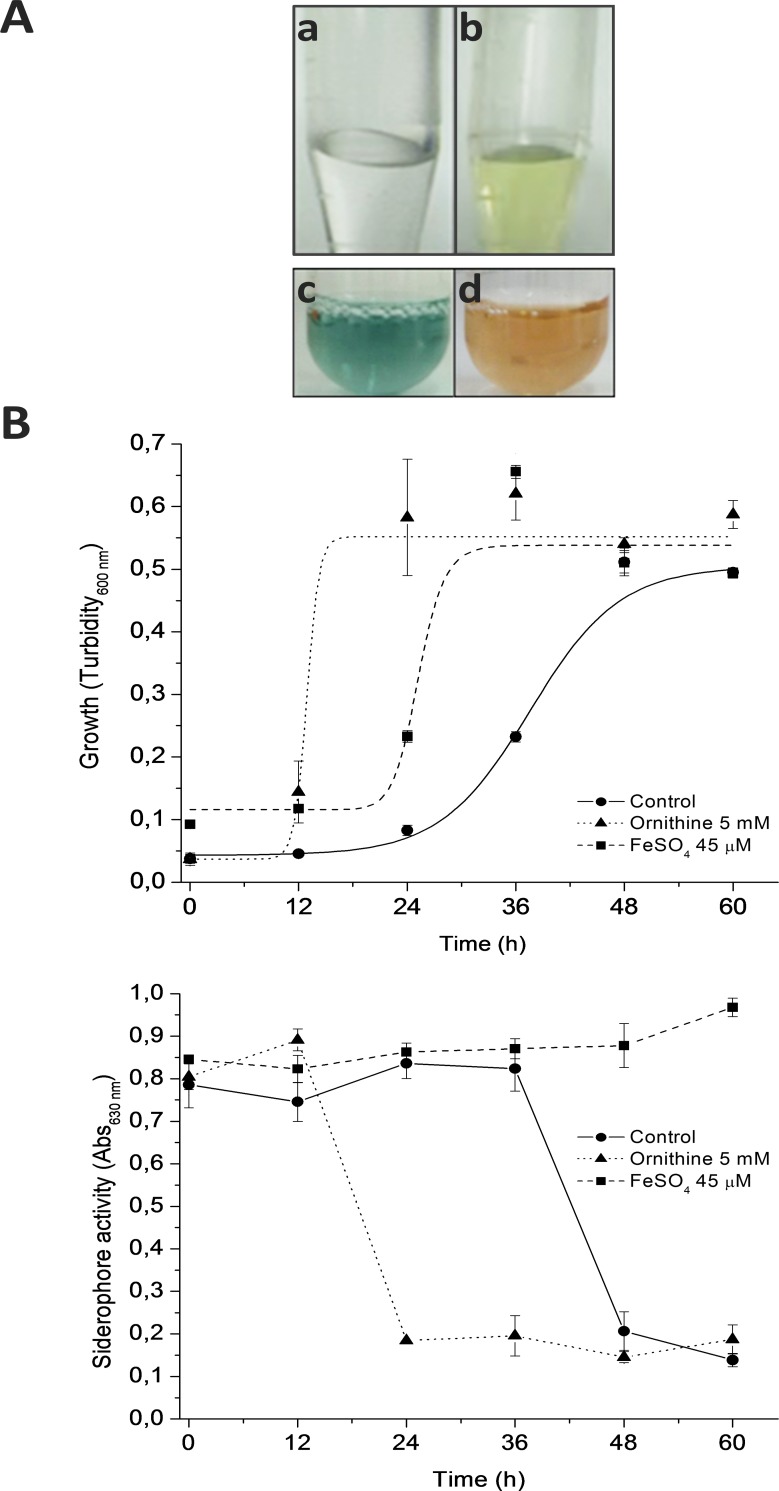
In this report, the production of a siderophore by B. xenovorans LB400 was studied. In a first approach for ferrisiderophore complex detection, the supernatants of LB400 cells grown under iron limitation and iron-rich condition were incubated with FeCl3. The supernatant of cells grown under iron limitation and incubated with FeCl3 turned yellow (Fig 1A), indicating the formation of a ferrisiderophore complex, whereas the supernatant of cells grown in iron-rich condition remained colorless. In a second approach, the supernatants of LB400 cells were analyzed by the CAS colorimetric assay. The color change from blue to orange was observed only in the supernatant of LB400 cells grown under iron-limitation (Fig 1A), suggesting the presence of a siderophore. In contrast, the LB400 supernatant from iron-rich condition maintained the blue color. When the iron-limited medium was supplemented with ornithine, which is a non-proteinogenic amino acid used for the synthesis of diverse peptidic siderophores [3], LB400 cells reached earlier the stationary phase and a siderophore activity was observed after a shorter incubation time (Fig 1B). Fig 1 illustrates that in stationary phase, LB400 supernatants showed a higher siderophore activity than in exponential phase.
Fig 1. Siderophore synthesis under iron limitation in B. xenovorans LB400.
A, Siderophore activity determined in supernatants of LB400 cells grown under iron-rich condition (a) and under iron limitation (b) and by the CAS assay of supernatants of LB400 cells grown under iron-rich condition (c) and under iron limitation (d). B, Effect of ornithine on LB400 growth and siderophore activity under iron limitation. LB400 cells were grown under iron-limitation in absence (control) or presence of ornithine (ornithine 5 mM), or under iron-rich condition (FeSO4 45 μM).
Identification of a gene cluster involved in siderophore synthesis in B. xenovorans LB400
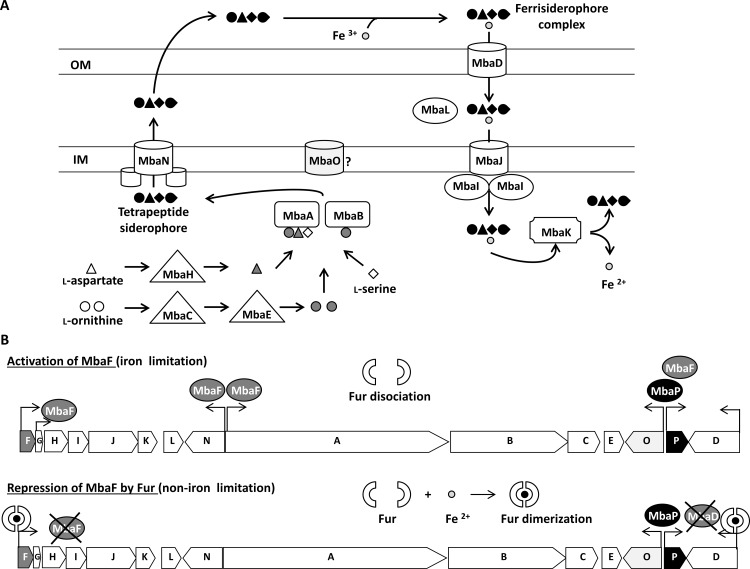
The pfam00668 protein domain that catalyzes the formation of a peptide bond through a condensation and is highly conserved in NRPS systems [33] was screened for in the LB400 genome. Five NRPS condensation domains were found to belong to two NRPS encoded by the mbaA (BxeB0525) and mbaB (BxeB0526) genes located on the LB400 chromosome 2 (Fig 2). A ~30 kb gene cluster with 15 coding sequences organized in operon-type structures that could be involved in the synthesis and transport of a hydroxamate-type siderophore was determined (Fig 2, Table 1). The cluster is flanked upstream by the BxeB0516 gene, which encodes a lipoprotein, and downstream by the BxeB0532 gene that encodes the NADP-dependent isocitrate dehydrogenase. The coding sequences involved in the siderophore synthesis are the BxeB0519 gene and the BxeB0525-BxeB0528 genes. The BxeB0525 (hereafter mbaA) and BxeB0526 (hereafter mbaB) genes encode two NRPS with high identity to MbaA (73%) and MbaB (72%) proteins that are involved in malleobactin synthesis in B. pseudomallei [15]. The predicted amino acid sequence of the BxeB0527 (hereafter mbaC) gene shows high identity (74%) to Orn monooxygenases (MOs) MbaC from B. pseudomallei and PvdA (73%) from B. cenocepacia that are involved in Nδ-hydroxylation of Orn [16,34]. The BxeB0528 (hereafter mbaE) gene encodes a Nδ-hydroxyornitine formyltransferase (FT) with 76% identity to PvdF from Pseudomonas aeruginosa PA01 [35]. Fig 3 illustrates a phylogenetic tree for MbaE FTsin that MbaE from strain LB400 clusters together with MbaE from B. phytofirmans PsJN and MbaE from Collimonas fungivorans Ter331. BxeB0519 gene (hereafter mbaH gene) encodes an α-ketoglutarate-dependent hydroxylase with high identity (84%) to OrbG that is involved in ornibactin synthesis in B. cenocepacia [14]. BxeB0518 gene (hereafter mbaG gene) encodes a MbtH-like protein that may participate as regulator in the siderophore synthesis. It has been proposed that MbtH-like proteins are regulators of NRPS, interacting with adenylating enzymes [36]. The coding sequences of the mba cluster probably involved in the transport and recognition of a hydroxamate ferrisiderophore complex are BxeB0520–BxeB0524 genes, BxeB0529 and BxeB0531 genes. BxeB0520 (hereafter mbaI), BxeB0521 (hereafter mbaJ) and BxeB0523 (hereafter mbaL) genes encode inner membrane, transmembrane and periplasmic transporter proteins from the ATP-binding cassette (ABC) family, which are involved in the uptake of hydroxamate-type ferrisiderophores [14,37,38]. The gene product of the BxeB0522 (hereafter mbaK) gene has 55% identity to protein OrbF from B. cenocepacia. This protein is probably involved in the reduction of intracellular ferric iron from the internalized ferrisiderophore complex [14]. The BxeB0524 (hereafter mbaN) gene encodes an ABC transporter protein with 82% identity to OrbE from B. cenocepacia [14]. MbaN is probably involved in the export of the siderophore through the cytoplasmic membrane. The BxeB0529 (hereafter mbaO) gene encodes a major facilitator superfamily (MFS) transporter [25] that may be involved in siderophore transport. MbaO protein shows high identity to MFS transporters from Burkholderia strains such as B. phytofirmans PsJN (90% identity), B. multivorans ATCC 17616 (70%), B. cenocepacia PC184 (67%), and B. cepacia GG4 (67%), and shows 67% identity to the MFS drug efflux transporter EmrB/QacA from Pseudomonas sp. GM102 [39]. Modelling of the MbaO 3D structure indicates that the MFS transporter possesses 14 transmembrane regions, and docking analyses indicate potential binding sites for malleobactin and ferrimalleobactin located close to the MbaO cytoplasmic side (S1 Fig). The BxeB0531 (hereafter mbaD) gene encodes an outer membrane TonB-dependent siderophore receptor that has 23% sequence identity to the ornibactin receptor OrbA from B. cenocepacia [40], and shows high conservation of TonB-dependent receptor domains. MbaD possess the TonB-dependent gated channel domain (cd01347) that permits the interaction with the ferrisiderophore complex, suggesting that MbaD is probably involved in the internalization of the ferrisiderophore. The BxeB0517 (hereafter mbaF) gene encodes a RNA polymerase ECF sigma factor that has high identity with OrbS (71%) from B. cenocepacia [14] and MbaS (79%) from B. pseudomallei [15]. The BxeB0530 (hereafter mbaP) gene encodes a transcriptional regulator of the LysR family that has 32% identity to the transcriptional regulator PtxR from the pyoverdin biosynthesis system in P. aeruginosa [41].
Fig 2. Organization of the mba gene cluster involved in the biosynthesis of siderophore in B. xenovorans LB400.
A, Genetic organization of the mba genes located at the minor chromosome (C2). Genes are represented with block arrows. Sizes of the genes and the intergenic regions are on scale. The transcriptional promoters are represented with black arrows oriented in the transcription sense. B, Genetic organization of the mba (orb) genes in Burkholderiales strains. The mba (orb) sequences (accession number) are B. xenovorans LB400 (BxeB0517–BxeB0531), B. phytofirmans PsJN (Bphyt_4063–Bphyt_4079), C. fungivorans Ter331 (CFU_2304–CFU_2292), B. pseudomallei K96243 (mbaF–mbaE), B. pseudomallei 668 (BURPS6681917–BURPS668 1931), B. pseudomallei MSHR346 (GBP346_A1956–GBP346_A1970), B. vietnamiensis G4 (Bcep1808 1577–Bcep1808 1590), Burkholderia sp. KJ006 (MYA_1458–MYA_1471), B. cenocepacia J2315 (orbS–orbL), B. cenocepacia PC184 (BCPG00175–BCPG00162) and B. phymatum STM815 (Bphy4047–Bphy4035). The gene organization of B. pseudomallei K96243 is shared by B. mallei SAVP1 (BMASAVP1A_1634–BMASAVP1A _1621), B. mallei NCTC 10229 (BMA10229_A0295–BMA10229_A0284), B. mallei NCTC 10247 (BMA10247_0864–BMA10247_0877), B. thailandesis E264 (BTH I2427–BTH I2414) and B. pseudomallei 1710b (BURPS1710b_2076–BURPS1710b_2092). The gene organization of B. pseudomallei 668 is shared by B. pseudomallei 1106a (BURPS1106A 1931–BURPS1106A 1917) and B. pseudomallei BPC006 (BPC006_I1984–BPC006_I1999). The gene organization of B. cenocepacia J2315 is shared by B. ambifaria MC40-6 (BamMC406_1550–BamMC406_1564), B. cepacia AMMD (Bamb 1529–Bamb 1543), B. cenocepacia AU1054 (Bcen 1152–Bcen 1166), B. cenocepacia HI2424 (Bcen2424 1632–Bcen2424 1646), Burkholderia sp. 383 (Bcep18194 A4778–Bcep18194 A4792), B. multivorans ATCC17616 (Bmul 1606–Bmul 1593) and B. cepacia GG4 (GEM_1784–GEM_1770).
Table 1. Predicted genes encoding the siderophore biosynthesis pathway in B. xenovorans LB400.
| ORF | Gene | aa | Related gene products | ||
|---|---|---|---|---|---|
| Protein function | Organism | % Id (aa) | |||
| BxeB0517 | mbaF | 223 | extracytoplasmatic function sigma factor | B. pseudomallei K96243 | 79 (154/194) |
| BxeB0518 | mbaG | 105 | MbtH-like protein | B. cenocepacia J2315 | 79 (61/77) |
| BxeB0519 | mbaH | 340 | α-ketoglutarate dioxygenase | B. cenocepacia J2315 | 84 (275/329) |
| BxeB0520 | mbaI | 288 | Fe3+/hydroxamate transporter inner membrane subunit | B. cenocepacia J2315 | 86 (214(248) |
| BxeB0521 | mbaJ | 704 | Fe3+/hydroxamate transporter permease subunit | B. cenocepacia J2315 | 74 (512/695) |
| BxeB0522 | mbaK | 272 | Iron reductase | B. cenocepacia J2315 | 55 (148/270) |
| BxeB0523 | mbaL | 276 | Fe3+/hydroxamate transporter periplasmic component | B. cenocepacia J2315 | 67 (181/270) |
| BxeB0524 | mbaN | 562 | ABC peptide transporter | B. pseudomallei K96243 | 88 (492/561) |
| BxeB0525 | mbaA | 3180 | Non ribosomal peptide synthase NRPS I | B. pseudomallei K96243 | 73 (2378/3252) |
| BxeB0526 | mbaB | 1576 | Non ribosomal peptide synthase NRPS J | B. pseudomallei K96243 | 72 (1244/1731) |
| BxeB0527 | mbaC | 456 | Nδ-L-ornithine-monooxygenase | B. pseudomallei K96243 | 74 (347/468) |
| BxeB0528 | mbaE | 280 | Formyltransferase | B. pseudomallei K96243 | 81 (225/278) |
| BxeB0529 | mbaO | 543 | MFS transporter | Pseudomonas sp. GM102 | 67 (395/503) |
| BxeB0530 | mbaP | 312 | LysR-type transcriptional regulator | P. aeruginosa PAO1 | 32 (94/291) |
| BxeB0531 | mbaD | 727 | TonB-dependent siderophore receptor | B. pseudomallei K96243 | 27 (194/718) |
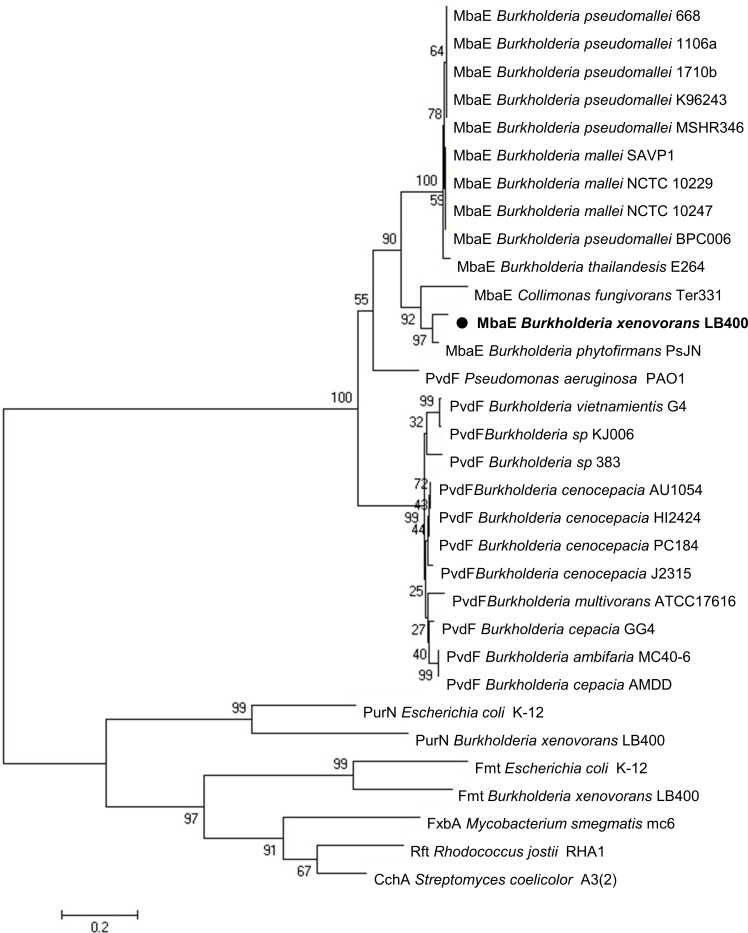
Fig 3. Phylogenetic analyses of MbaE formyltransferases.
The dendogram of MbaE formyltransferase from B. xenovorans LB400 and other formyltransferases was constructed by the neighbor-joining method using MEGA 6.0 based on multiple sequence alignment by Clustal W. Sequence of deduced protein from mbaE gene from B. xenovorans LB400 is highlighted (black circle). One clade includes the L-Nδ-hydroxy- ornithine formyltransferases (accession number) that may produce L-Nδ-hydroxy-Nδ-formylornithine and L-Nδ-hydroxy-Nα-formylornithine: MbaE B. xenovorans LB400 (YP554769.1), MbaE B. phytofirmans PsJN (YP001887830.1), MbaE C. fungivorans Ter331 (YP004752942.1), MbaE B. pseudomallei 668 (YP001058966.1), MbaE B. pseudomallei 1106a (YP001066211.1), MbaE B. pseudomallei 1710b (YP333487.1), MbaE B. pseudomallei K96243 (YP108374.1), MbaE B. pseudomallei MSHR345 (YP002896675.1), MbaE B. mallei SAVP1 (YP992947.1), MbaE B. mallei NCTC 10229 (YP001026286.1), MbaE B. mallei NCTC 10247 (YP001080440.1), MbaE B. pseudomallei BPC006 (YP006652779.1) and MbaE B. thailandesis E264 (YP442935.1). The other clade includes L-Nδ-hydroxy-ornithine formyltransferases (accession number) that produce L-Nδ-hydroxy-Nδ-formylornithine: PvdF P. aeruginosa PA01 (NP251086.1), PvdF B. vietnamientis G4 (YP001119431.1), PvdF Burkholderia sp. KJ006 (YP006332563.1), PvdF Burkholderia sp. 383 (YP369030.1), PvdF B. cenocepacia AU1054 (YP621045.1), PvdF B. cenocepacia HI2424 (YP835290.1), PvdF B. cenocepacia PC184 (YP002091473.1), PvdF B. cenocepacia J2315 (YP002230827.1), PvdF B. multivorans ATCC17616 (YP001579778.1), PvdF B. cepacia GG4 (YP006615890.1), PvdF B. ambifaria MC40-6 (YP001808266.1), PvdF B. cepacia AMDD (YP773434.1). The methionyl-tRNAfMet formyltransferases (accession number) are Fmt B. xenovorans LB400 (YP556637.1) and Fmt E. coli K-12 (YP492145.1) and the glycinamide ribonucleotide formyltransferases (accession number) are PurN B. xenovorans LB400 (YP560177.1) and PurN E. coli K-12 (YP490728.1).
A bioinformatic search of additional NRPS-dependent and NRPS-independent biosynthetic genes involved in siderophore production in LB400 genome was performed. In the LB400 genome, genes from the pyochelin, cepabactin and cepaciachelin biosynthetic pathways were not found. Catalytic domains of enzymes involved in the biosynthesis of non-peptidic siderophores, such as aerobactin, desferroxyamin, alcalygin, antrachelin, acromobactin, estafilobactin, rhizobactin 1021 and vibrioferrin were also not detected in LB400 genome. In this study, the mba cluster was the only gene cluster involved in siderophore biosynthesis found in strain LB400 genome.
Gene clusters involved in siderophore biosynthesis in Burkholderiales
The LB400 NRPS gene cluster shows high similarity with gene clusters that synthesize hydroxamate-type siderophores from other Burkholderia strains. To compare NRP siderophore gene cluster among Burkholderia genomes, a search of these genes in other Burkholderiales strains was performed. Eleven different gene clusters were identified in 24 Burkholderia strains and one Collimonas strain (Fig 2). These gene clusters differ in the presence and distribution of specific genes. In general, there are two types of gene clusters for the synthesis of siderophores belonging either to malleobactin or ornibactin-type compounds. The mba and orb gene clusters differ in the first A domain (A1) from NRPS that activates L-Nδ-hydroxy-Nδ-formylornithine (hfOrn) and L-Nδ-hydroxy-Nα-formylornithine (hαfOrn) in malleobactins and L-Nδ-hydroxy-Nδ-acylornithine (haOrn) in ornibactins. Another difference is the presence or absence of the orbK and orbL genes, which encode N-acetyltransferases probably involved in the acylation of L-Nδ-hydroxyornithine (hOrn) in ornibactins [17,42–44]. On the one hand, B. xenovorans LB400, B, vietnamensis G4, B. phytofirmans PsJN, C. fungivorans Ter331 and B. pseudomallei strains K96243, 668 and MSHR346 mba gene clusters are involved in the synthesis of malleobactin-type siderophores that contain the first Orn of the tetrapeptide as hfOrn or hαfOrn. On the other hand, Burkholderia sp. KJ006, B. cenocepacia strains J2315 and PC184, and B. phymatum STM815 orb gene clusters are associated with the synthesis of ornibactin-type compounds that possess the first tetrapeptide Orn as haOrn. Interestingly, two novel coding sequences (mbaO and mbaP) located downstream of the biosynthesis genes were identified in the LB400 gene cluster (Fig 2). These genes are present in the gene cluster identified in this study in B. phytofirmans PsJN (Fig 2), but are not present in the functional mba and orb gene clusters from B. cenocepacia and B. pseudomallei [14,15].
In silico analyses of promoter regions in strain LB400
To search for promoters, the non-coding regions upstream of each mba gene from LB400 strain were analyzed in silico. Bioinformatic analyses suggest the presence of different operons in the mba gene cluster of strain LB400. Six potential iron-regulated promoters were identified. Promoter regions of mbaG, mbaN, mbaA and mbaP genes contain sequences with high identity to the consensus sequence GCGGCTAAAATAATATN(GC)9NNNCGTC that includes the -10 and -35 elements for the promoter recognition of the RNA polymerase sigma factor OrbS for ornibactin synthesis in B. cenocepacia [14]. The mbaG, mbaN, mbaA and mbaP gene promoter sequences show a match of 25/28, 26/28, 25/28 and 20/28 with the consensus OrbS binding sequence, respectively. The RNA polymerase ECF sigma factor OrbS in B. cenocepacia and PvdS in P. aeruginosa (MbaF in B. xenovorans LB400) activates the expression of genes for the ornibactin and pyoverdin synthesis, respectively, when the ferric uptake regulator protein Fur is not exercising its repressor activity [14,45]. Strain LB400 possesses a fur gene (BxeA0571) that probably encodes a ferric uptake regulator protein. LB400 Fur protein shows 56% sequence identity with the Fur regulator from P. aeruginosa [46]. The promoter regions of the mbaF and mbaD genes were found to contain sequences with high identity to the Fur box (GTAAACGCAAATCATTCTC) in B. cenocepacia [14]. The Fur box of mbaF and mbaD genes promoters show a match of 17/19 and 12/19 with the fur sequence of the orbS gene from B. cenocepacia. It has been reported that a match of 13/19 with the Fur binding sequence (GATAATGATAATCATTATC) from E. coli [45] may constitute a functional Fur box in the orbS gene promoter [14]. The presence of Fur-binding elements in the promoters of mbaF and mbaD genes suggests the regulation of their expression by the Fur protein. Two potential rho-independent terminators transcription sequences were identified in the mba gene cluster from strain LB400. One terminator is located 2343 nt upstream from the predictive initiation codon of the mbaO gene and the other is located 3417 nt upstream from the predictive initiation codon of the mbaP gene. The mbaP gene promoter possesses a probably Fur box and also a potential LysR-type transcriptional regulator binding site (5’-TCGGCGTCCCTA-3´ sequence that is at -57 from the mbaP gene) for its autoregulation. The divergent orientation of transcription of the mbaP and mbaO genes, the presence of a LysR-type transcriptional regulator binding site (5’-TCCGATCGCACGGA-3´ sequence at -57 from the mbaO gene) and the absence of Fur and MbaF promoter binding elements upstream of the mbaO gene suggest that its transcription is activated by the MbaP regulator.
Transcriptional analysis of the mba gene cluster
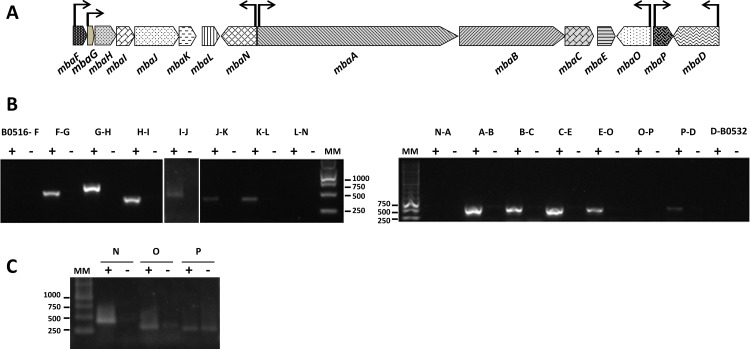
To evaluate if the mba genes from LB400 are functional, expression of the mba gene cluster in strain LB400 was evaluated under iron limitation and iron-rich condition. Under iron limitation, transcription products were observed for intergenic regions mbaFGHIJKL and mbaABCE, and for the mbaN, mbaO and mbaP genes. In contrast, cells grown in a medium supplemented with iron only expressed mbaO and mbaP genes, suggesting that the mba cluster transcription in strain LB400 is regulated by iron (Fig 4). Intergenic regions between BxeB0516-mbaF, mbaN-mbaA, mbaO-mbaP and mbaD-BxeB0532 were not transcribed since adjacent genes are orientated in opposite directions. Although both mbaE-mbaO and mbaP-mbaD genes are orientated in opposite directions, these intergenic regions were expressed under iron limitation. For both cases, rho-independent terminator transcription sequences were identified. Co-expression of intergenic regions and the bioinformatic analysis of the promoters, strongly suggests the presence of the transcriptional units mbaFGHIJKL, mbaN, mbaABCE, mbaO, mbaP and mbaD genes in the mba cluster from B. xenovorans LB400 (Fig 4).
Fig 4. Expression of the mba genes from B. xenovorans LB400.
A, Gene organization of the mba operon. Genes are represented with block arrows indicating gene orientation. Proposed biosynthetic genes are shown in dark gray, transport and utilization genes in light gray, and regulation genes in black. Transcriptional units predicted from RT-PCR analyses are shown by horizontal line arrows, with the promoter regions depicted by the start of the arrows. B, RT-PCR analysis of transcripts from the intergenic regions of the mba gene cluster. C, RT-PCR analysis of the expression of the mbaN, mbaO and mbaP genes. Amplification products were subjected to electrophoresis in an agarose (1%) gel. MM, molecular size markers (1 kb and Express DNA ladder). Expression of each region was analyzed under iron-limitation (+) and without iron limitation (-).
Prediction of the siderophore structure
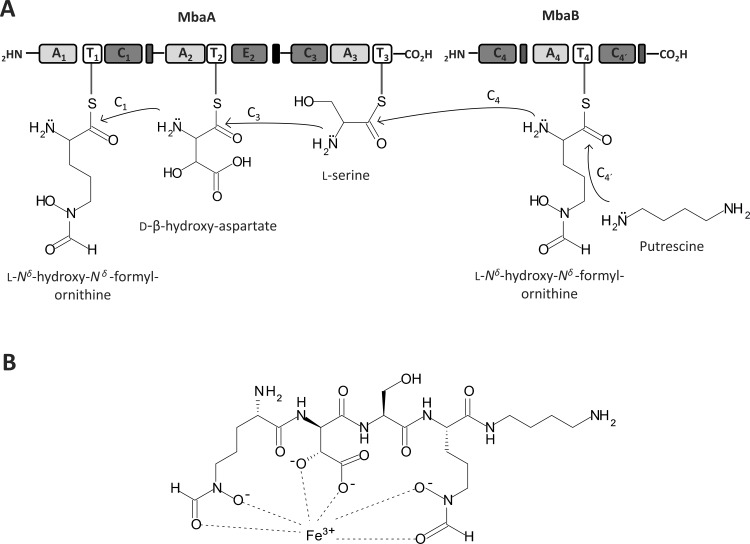
The substrate prediction for the NRPS enzymes encoded by genes mbaA and mbaB from B. xenovorans LB400 and the comparison of bacterial mba and orb gene clusters allowed the in silico prediction of the siderophore structure. Proteins encoded by mbaA and mbaB genes from strain LB400 have conserved NRPS domains that show high identity to NRPS domains for malleobactin and ornibactin synthesis in B. pseudomallei and B. cenocepacia, respectively [14–16,42]. MbaA has A, T, C and E domains organized in three biosynthetic modules (modules 1, 2 and 3). MbaB presents A, T and C domains organized in one module (module 4) as well as a second C domain (Fig 5). A prediction of the specific substrates for each module was carried out based on amino acid code of Stachelhaus of the A domains and information available in literature (Table 1). MbaA module 1 is predicted to use as substrate hOrn, hfOrn, hαfOrn or Phe based on previous reports [16,17]. For MbaA module 2 the use of L-β-hydroxy-aspartic acid (hAsp) as substrate is proposed [16]. Three conserved residues (Thr239, Lys278 and His322) that interact with hAsp are present in the second A domain [27,47]. Substrate prediction for MbaA module 3 indicates the use of L-serine (Ser). The catalytic dyad His278 and Ser301 that interacts with a Ser is present in the third A domain [27,47]. MbaB module 4 shows specificity for hfOrn (Table 2). The substrates Orn and Asp probably undergo tailoring reactions before assembling to its corresponding module. Asp is probably hydroxylated at the β-carbon by the α-ketoglutarate-dependent hydroxylase MbaH and epimerized to D-β-hydroxy-aspartic acid by the E2 domain of MbaA module 2. Orn for modules 1 and 4 is probably first hydroxylated by MO MbaC and subsequently formylated by FT MbaE. The fourth Orn attached to MbaB may be subjected to an additional modification that is an amidation at the C-terminus with the addition of 1,4-diaminobutane (putrescine) by the C4´ domain from MbaB module 4 (Fig 5). The Stachelhaus-code of the predicted Orn activation domains of modules 1 and 4 in strain LB400 show the universally conserved residues Asp235 and Lys517 (in all amino acid activating domains), as well as the Orn-specific hydrophobic residues Gly301 and Ile330 [47]. However, the presence in all Orn activation domains in Burkholderiales strains of Glu239 instead of Glu278 that is present in gramicidin, tyrocidine, fengycin and bacitracin synthetase Orn A domains, suggests its interaction with oxidized forms of Orn. This Glu position change, which also has been observed in the first A domain of exochelin synthetase FxbC (A1_FxbC) involved in the activation of hOrn [47–49], is probably a feature of domains that activate an oxidized Orn. Hydroxylation occurs at nitrogen δ in both Orn 1 and Orn 4 of malleobactin, producing hOrn. However, contrasting with ornibactin, the malleobactin molecule can undergo formylation at nitrogen δ and α of hOrn 1 [17]. On the other hand, phylogenetic analysis of FTs MbaE in Burkholderiales strains suggests that MbaE of strain LB400 is closely related to MbaE from B. thailandensis that is able to formylate nitrogen δ and α from the first hOrn and nitrogen δ from the hOrn 4 in malleobactins [17], whereas FT PvdF is able to formylate only nitrogen δ of both hOrn in ornibactin and pyoverdin. FTs MbaE and PvdF belong to markedly different clades in the dendogram (Fig 3). Incubation with ferric iron and CAS assay indicate that strain LB400 produces a malleobactin siderophore that forms a stable ferrisiderophore complex, suggesting that the first hOrn of the LB400 malleobactin is formylated at nitrogen δ generating a hydroxamate group that provides one of the predicted Fe-binding sites. Franke et al. proposed that malleobactin E is the only malleobactin of B. thailandensis E264 that possesses siderophore activity due to its three Fe-binding sites including the hydroxamate group at Orn 1 [17]. Therefore, the predicted chemical structure of the active siderophore synthesized by the Mba NRPS system of strain LB400 is the linear tetrapeptide hfOrn-D-β-hydroxyAsp-Ser-hfOrn-1,4-diaminobutane (Fig 5). The molecule has three functional hydroxamate groups that coordinate Fe3+ with three bidentate bonds, forming the ferrisiderophore complex. The predicted molecular mass for the ferritetrapeptide complex matches 675.2037 Da, with a siderophore: iron stoichiometry of 1:1 (Fig 5).
Fig 5. Siderophore synthesis by the non-ribosomal peptide synthetases MbaA and MbaB in B. xenovorans LB400 and the ferrisiderophore complex structure.
A, Modular organization of the non-ribosomal peptide synthetases encoded by the mbaA and mbaB genes. Domains are represented as blocks. Adenylation (A), thiolation (T), condensation (C) and epimerization (E) domains of MbaA are organized in three modules. Domains of MbaB are organized in one module. The synthesis of the siderophore starts with specific substrates assembly to each NRPS module. Substrate of modules 1 and 4 is L-Nδ-hydroxy-Nδ-formylornithine (hfOrn). Substrates of module 2 and 3 are D-β-hydroxy-aspartic acid (OH-Asp) and L-serine (Ser), respectively. Condensation domains of each module allow the attachment of the four substrates to form the tetrapeptide. C1 condenses hfOrn with OH-Asp, C3 condenses OH-Asp with Ser and C4 condenses Ser with hfOrn. Chemical modifications of the amino acids are catalyzed by the protein products of mbaH, mbaC and mbaE genes before the assembly to each NRPS module. The α-ketoglutarate-dependent hydroxylase MbaH hydroxylates the β-carbon of Asp. The L-ornithine-5-monooxygenase encoded by the mbaC gene hydroxylates the nitrogen δ of both Orn. The formyltranserase encoded by the mbaE gene formylates the nitrogen δ of Orn 1 and 4. C4´ domain of module 4 adds a diaminobutane (putrescine) molecule in the N-terminus of the modified tetrapeptide. B, Ferrisiderophore complex structure synthesized by B. xenovorans LB400. The ferrisiderophore structure was predicted by bioinformatic analyses and confirmed by ESI-MS and MALDI-TOF MS of the complex purified by HPLC.
Table 2. Prediction of MbaA and MbaB adenylation domains specificities in B. xenovorans LB400 based on literature [16,17,27].
| Stachelhaus-code residues | Predicted substrate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A-Domain | 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | |
| A1 | D | V | E | T | L | G | G | I | S | K | hfOrn/hαfOrn/hOrn/Phe |
| A4 | D | G | E | Y | T | G | G | I | T | K | hfOrn |
| A2 | D | L | T | K | V | G | H | V | G | K | β-OH-Asp |
| A3 | D | V | W | H | V | S | L | I | D | K | Ser |
Structure analyses of the siderophore
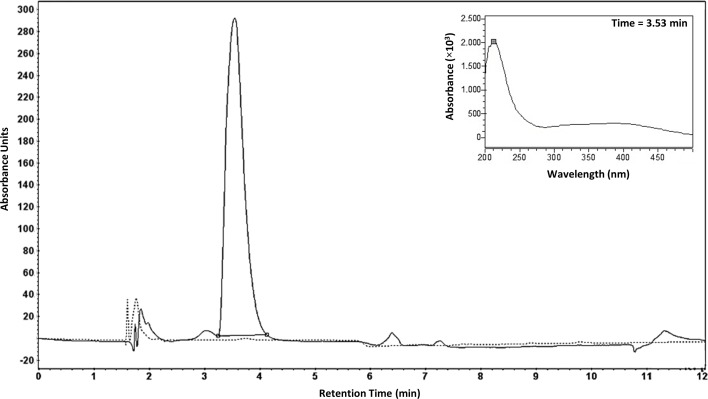
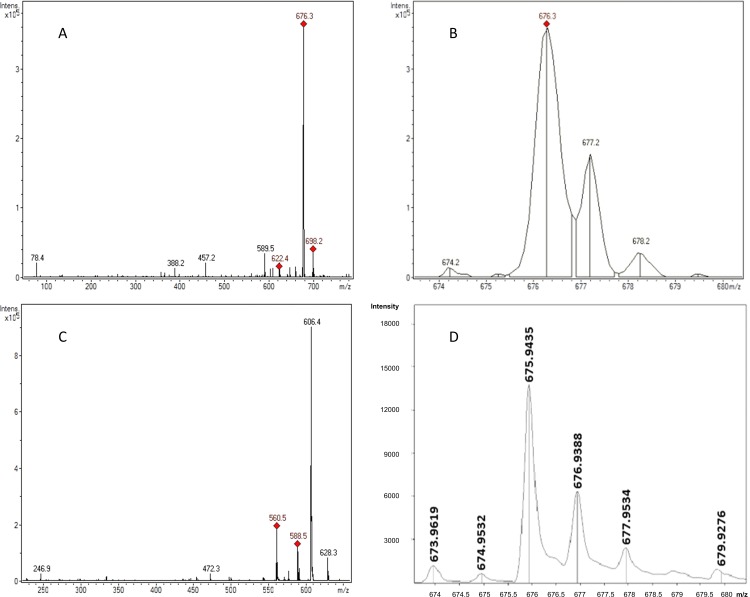
To determine the product synthesized by the mba gene cluster, the synthesis of a siderophore by strain LB400 was evaluated. Cells were grown in iron-limited medium supplemented with glucose and Orn. To study potential ferrisiderophore complexes, supernatants were treated with iron and subsequently collected, concentrated and subjected to HPLC analysis. HPLC chromatogram of a supernatant from LB400 cells grown under iron-limiting conditions revealed a peak with a retention time of 3.5 min (Fig 6). To determine the structure of the siderophore, HPLC fractions containing the ferrisiderophore complex were collected and subjected to electrospray ionization and mass spectrum analysis (ESI-MS). A quasimolecular ion of 676.3 Da [M(Fe)+H]+ with the isotope iron56 was identified in the ESI mass spectrum (Fig 7A). The in silico analysis predicted that the mass for the non-protonated form of the malleobactin is 622.5526 Da [M] and for the non-protonated form of the ferrimalleobactin complex is 675.2037 Da [M(Fe)]. The presence of iron in this compound was confirmed by the isotopic iron distribution pattern of the quasimolecular ion. The molecular mass of 674.2 Da is the isotopic peak from the siderophore complex with the isotope iron54 (Fig 7B). The peptide nature of the compound was confirmed by obtaining peptide fragments when the molecular ion of 676.3 Da was subjected to soft ionization by electrospray. The peak of 589.5 Da is the fragmentation product [M(Fe)+H-putrescine]+. The elimination of putrescine (-NHCH2CH2CH2CH2NH2) from malleobactin in mass spectra has been reported [16]. The fragments of the tetrapeptide bound to iron 606.4 [M(Fe)-70]+, 588.5 Da [M(Fe)-88]+ and 560.5 Da [M(Fe)-116]+ were determined (Fig 7C). The fragments 588.5 Da and 560.5 Da were probably formed by elimination of NHCH2CH2CH2CH2NH and CONHCH2CH2CH2CH2NH, respectively. The fragment 606.4 Da may be formed mainly by the elimination through a McLafferty rearrangement of CH2CHCH2CH2NH2. The presence of iron in the ferrisiderophore complex was confirmed by the isotopic distribution pattern in the 675.9435 Da peak by MALDI-TOF mass spectrometry (Fig 7D). These results confirm the formation of an iron siderophore complex by B. xenovorans LB400 cultures grown in iron-limited medium and treated with FeCl3. The molecular mass of the ferrisiderophore complex matches the theoretically predicted mass for the tetrapeptide bound to iron.
Fig 6. Purification of the ferrisiderophore complex from B. xenovorans LB400.
HPLC chromatogram of the supernatant of LB400 cells in an iron-depleted (black line) and an iron-supplemented (dotted line) culture. The peak with retention time of 3.53 min contains the ferrisiderophore complex. The absorbance spectrum of the ferrisiderophore complex is depicted in the insert.
Fig 7. Mass spectrometry of the ferrisiderophore complex.
A-C, Siderophore iron complex determined by ESI-MS. A, The molecular peak of the ferrisiderophore complex has a mass of 676.3 Da [M(Fe)+H]+. The peak of the siderophore without iron has a molecular mass of 622.4 Da [M)+H]+. The peak of 589.5 Da is the fragmentation product [M(Fe)+H-putrescine]+. The peak of 698.2 Da is a Na adduct [M(Fe)+Na]+. B, Isotopic distribution pattern of the quasimolecular ion of 676.3 Da. The peak of 676.3 Da is the ferrisiderophore complex with most abundant isotopes. The peaks of 674.2 Da, 677.2 Da and 678.2 Da are isotope peaks. C, Fragments of the siderophore iron complex. The peak of 606.4 Da is the fragmentation product formed mainly by the elimination through a McLafferty rearrangement of CH2CHCH2CH2NH2. The peaks of 588.5 Da and 560.5 Da are other fragmentation products bound to iron. D, Isotopic distribution pattern of the ferrisiderophore determined by MALDI-TOF mass spectrometry. The peak of 675.9435 Da corresponds to a population of ferrsiderophore complex that contains most abundant carbon12, hydrogen1, nitrogen14, oxygen16 and iron56 isotopes. The peaks of 676.9388 Da, 677.9534 and 679.9276 Da correspond to populations of complexes that contain lower abundance isotopes in their structure. The peak of 673.9619 Da and 674.9532 Da corresponds to a population of complexes that contain the isotope iron54.
Discussion
This is the first report that characterized a siderophore and the NRPS gene cluster associated to its biosynthesis in B. xenovorans LB400. This NRPS gene cluster comprises 15 coding sequences that are involved in the synthesis and transport of a hydroxamate-type siderophore and shows an unique gene organization. Based mainly on bioinformatic analyses, Fig 8 illustrates a model for the synthesis, transport and regulation of the malleobactin sideophore in B. xenovorans LB400. The presence of NRPS gene clusters that synthesize hydroxamate-type siderophores have been described in Burkholderia. The orb gene cluster encoding proteins of the siderophore ornibactin pathway from B. cenocepacia has been characterized [14]. The mba gene clusters from B. pseudomallei and B. thailandensis encode enzymes for the synthesis of malleobactins [15–17]. Two novel coding sequences (mbaO and mbaP) are present in LB400 mba gene cluster, which are absent in B. pseudomallei and B. cenocepacia. In this study, we also identified by bioinformatic analysis the NRPS mba gene cluster from the plant-promoting strain B. phytofirmans PsJN that contains the mbaO and mbaP genes. The mba gene clusters from strains LB400 and PsJN are highly similar, suggesting a closely related evolution. However, the mba gene cluster from strain PsJN possess two additional genes: the mbaM gene encoding a protein of unknown function that is also present in the mba gene cluster from B. pseudomallei K96243 [16], and a second mbaD gene. B. phytofirmans PsJN is the only Burkholderia strain that possess two mbaD genes (Fig 2).
Fig 8. Model for the biosynthesis, transport and regulation of the non-ribosomal peptide siderophore in B. xenovorans LB400 based on bioinformatic analyses.
A, The amino acids are modified by MbaH, MbaC and MbaE enzymes and assembled by MbaA and MbaB NRPS that synthetize the siderophore. The siderophore is exported through MbaN transporter localized in the inner membrane (IM). The siderophore binds iron(III) in the extracellular space, forming the ferrisiderophore complex. The complex is recognized by MbaD in the outer membrane (OM) and imported through the inner membrane by the MbaIJL ABC transporter. From the cytoplasmic ferrisiderophore complex the iron is released by MbaK. MbaO MFS transporter may be involved in siderophore transport, but its function is unknown. B, The transcriptional expression of the mba gene cluster from B. xenovorans LB400 is repressed by iron. In presence of iron, dimerized Fur binds to the Fur box present in the mbaF and mbaD genes and prevents their transcription. In absence of iron, Fur inactivation released ECF sigma factor binding site of mbaF gene, which allows the transcription of the mbaF gene. MbaF factor binds to the MbaF boxes in the mbaG, mbaN, mbaA and mbaP genes promoters for their expression. The mbaP gene transcription is also expressed in iron-rich condition. The LysR-type transcriptional regulator MbaP binds to the LysR binding site present in the mbaP and mbaO genes allowing their expression.
The NRPS gene cluster from strain LB400 allowed the prediction of the siderophore, the linear tetrapeptide hfOrn-D-β-OH-Asp-Ser-hfOrn-1,4-diaminobutane. The supernatant of LB400 cells grown under iron limitation showed a yellow color after incubation with FeCl3 and an orange color by the CAS assay, indicating the presence of a siderophore. ESI-MS and MALDI-TOF-MS analyses confirmed the siderophore structure. The presence of iron in the complex and the molecular mass of the iron-siderophore complex (676 Da) strongly support the bioinformaticpredicted siderophore structure. Closely related siderophore structures have been recently reported as malleobactin E and malleobactin B in B. mallei and B. thailandensis [16,17]. In strain LB400 the synthesis of a malleobactin isomer that has the formyl group in the nitrogen α of the first Orn could not be excluded. Interestingly, eight different malleobactins were detected in B. thailandensis [17]. The phylogenetic analyses of specific genes in the NRPS clusters from Burkholderia strains differentiate malleobactin-type and ornibactin-type gene clusters. The main structural difference between these two tetrapeptides is the first Orn: hfOrn or hαfOrn in malleobactins and haOrn in ornibactins. Other bacterial siderophores that exhibit hfOrn in their structure are pyoverdin, coelichelin, exochelin and rhodochelin, which are synthesized by P. aeruginosa, S. coelicolor [50], Mycobacterium smegmatis [51] and Rhodococcus jostii [52], respectively. In our study it has been found that malleobactin-type siderophores may be synthesized by several Burkholderia strains and a C. fungivorans strain. A second difference of the malleobactin- and ornibactin-type gene clusters of Burkholderiales is the phylogenetic clade distribution of FTs MbaE and PvdF. MbaE-type FTs involved in the production of both hαfOrn and hfOrn for malleobactin-type siderophores, are grouped in a different clade than FTs from the ornibactin (PvdF), pyoverdin (PvdF), coelichelin (CchA), exochelin (FxbA) and rhodochelin (Rft) systems that only produce hfOrn. FTs involved in formylation of primary or secondary amines are clustered in different phylogenetic branches. FTs involved in secondary metabolism such as the FTs mentioned above, possess the catalytic triad Asn106, His108 and Asp144 [53], however, lack the N-terminus for substrate binding and the SLLP motif that participates in the binding of tetrahydrofolate co-substrate present in FTs involved in primary metabolism such as glycinamide ribonucleotide transformylase and methionyl-tRNAfMet FT [54]. Based on the phylogenetic analyses of A domains and FTs, as well as the detection of a malleobactin-type compound in B. xenovorans LB400, a model for the siderophore synthesis is proposed.
Diverse Burkholderia strains have multiple iron acquisition mechanisms. Some Burkholderia strains (e.g., B. cepacia) produce more than one siderophore [12,13,15]. Alternative strategies for iron uptake have also been reported [12,55]. B. cenocepacia and B. pseudomallei use haemin and ferritin for iron acquisition [56,57]. Recently, a novel siderophore-independent ferric iron-uptake system has been identified in Burkholderia strains. The locus FtrBccABCD (BxeA3149–BxeA3152) from strain LB400 encodes a high affinity ferrous iron transporter [58]. Nevertheless, the mba gene cluster is the only LB400 gene cluster that encodes proteins involved in the synthesis of a siderophore.
Iron limitation and the presence of Orn increased LB400 siderophore activity, suggesting a regulated expression of the mba genes. RT-PCR results showed that the expression of the mba genes from strain LB400 is regulated by iron. Based mainly on bioinformatic analyses and previous studies, we propose a model for regulation of the mba gene cluster from strain LB400 (Fig 8). The presence of the mbaP gene that encodes a LysR-type transcriptional regulator in the LB400 mba cluster and the presence of LysR-type transcriptional regulator binding sites in the promotor regions of the mbaP and mbaO genes suggest an additional transcriptional regulation. RT-PCR assays showed that the mbaP and mbaO genes are transcribed in presence of iron-limitation, suggesting that their expression is not repressed by Fur. The transcriptional activation of siderophore biosynthesis genes by LysR-type transcriptional regulators has also been reported in other bacteria. In P. aeruginosa, under iron limitation the LysR-type regulator PtxR activates through the sigma factor PvdS the transcription of genes associated to pyoverdin biosynthesis. The regulation of ptxR gene expression by PvdS is Fur-independent under aerobic conditions [46,59]. Based on bioinformatics analyses we propose that the expression of most of the siderophore biosynthesis, transport and regulation genes is regulated by Fur and/or MbaF (Fig 8), whereas the mbaP gene is also autoregulated and mbaO gene may be regulated by the transcriptional regulator MbaP.
B. xenovorans strains have been associated with the rhizosphere [25]. The biosynthesis of a siderophore in B. xenovorans LB400 suggests that the capture of iron in its rhizosphere ecosystem is relevant for bacterial fitness. Iron could be scarce in soils and, therefore, a limiting nutrient for their bacterial inhabitants. The malleobactin synthesized by B. xenovorans LB400 may improve its fitness in the rhizosphere and could potentially be used also by plants for iron acquisition [60–62]. Siderophore-producing bacteria may participate also in control of phytopathogens [63–65]. The presence of orthologous mba genes in diverse Burkholderia species strongly suggests that the production of non-ribosomal peptide siderophores are relevant for Burkholderia living in diverse niches such as rhizosphere, soil and (immunocompromised) mammalian organisms. In conclusion, this study describes in the model bacterium B. xenovorans LB400 a functional and regulated NRP siderophore pathway that synthesized a malleobactin siderophore under iron limitation.
Supporting Information
Malleobactin (A) and ferrimalleobactin complex (D) 3D structures were predicted. The binding site for malleobactin 3D structure (B) and ferrimalleobactin 3D structure (E) was predicted using a Docking strategy. The frontal view of the best model of the protein-ligand complexes (C,F) are shown as ribbon representations, where the periplasmic side is located up and the malleobactin and ferrimalleobactin ligands surfaces are highlighted in blue. The cytoplasmic (G) and periplasmic (H) sides, and the volume (I) of the protein ligand complex are shown.
(TIF)
(DOC)
Acknowledgments
The authors acknowledge Fanny Guzmán and Andrés Barriga for support with mass spectrometry analyses.
Funding: The authors acknowledge Conicyt (MJV, FMS), Mecesup FSM0710 (FC), Mecesup CD FSM1204 (FMS), UTFSM (FC), RIABIN (FMS) and UTFSM-PIIC (FMS) fellowships. MS and BC acknowledge financial support by FONDECYT (1110992 & 1151174) (http://www.fondecyt.cl), Mecesup (FSM710 & CD FSM1204) (www.mineduc.cl), FDI Mecesup (2012, 2013 & 2014) (www.mineduc.cl) and USM (131562, 131342 & 131109) (http://www.usm.cl) grants.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors acknowledge Conicyt (MJV, FMS), Mecesup FSM0710 (FC), Mecesup CD FSM1204 (FMS), UTFSM (FC), RIABIN (FMS), and UTFSM-PIIC (FMS) fellowships. MS and BC acknowledge financial support by FONDECYT (1110992, 11121571 & 1151174) (http://www.fondecyt.cl), Mecesup (FSM710 & CD FSM1204) (www.mineduc.cl), FDI Mecesup (2012, 2013 &2014) (www.mineduc.cl), and USM (131562, 131342 & 131109) (http://www.usm.cl) grants.
References
- 1.Neilands JB (1995) Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270: 26723–26726. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Robinson AK, Rodríguez-Quiñones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106: 3468–3496. [DOI] [PubMed] [Google Scholar]
- 4.Konz D, Marahiel MA (1999) How do peptide synthetases generate structural diversity?. Chem Biol 6: R39–R48. [DOI] [PubMed] [Google Scholar]
- 5.Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97: 2651–2674. [DOI] [PubMed] [Google Scholar]
- 6.Yadav G, Gokhale RS, Mohanty D (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol 328: 335–363. [DOI] [PubMed] [Google Scholar]
- 7.Challis GL, Ravel J (2000) Coelichelin a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett 187: 111–114. [DOI] [PubMed] [Google Scholar]
- 8.Coenye T, Vandamme P (2003) Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5: 719–729. [DOI] [PubMed] [Google Scholar]
- 9.Mahenthiralingam E, Urban TA, Goldberg JB (2005) The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3: 144–156. [DOI] [PubMed] [Google Scholar]
- 10.Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V (2012) Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63: 249–266. 10.1007/s00248-011-9929-1 [DOI] [PubMed] [Google Scholar]
- 11.Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24: 1073–1109. [DOI] [PubMed] [Google Scholar]
- 12.Thomas MS (2007) Iron acquisition mechanisms of the Burkholderia cepacia complex. BioMetals 20: 431–452. [DOI] [PubMed] [Google Scholar]
- 13.Meyer JM, Van VT, Stintzi A, Berge O, Winkelmann G (1995) Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia. BioMetals 8: 309–317. [DOI] [PubMed] [Google Scholar]
- 14.Agnoli K, Lowe CA, Husnain SI, Thomas MS (2006) The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the Fur regulon. J Bacteriol 188: 3631–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alice AF, López CF, Lowe CA, Ledesma MA, Crosa JH (2006) Genetic and transcriptional analysis of the siderophore malleobactin bio-synthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol 188: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke J, Ishida K, Ishida-Ito M, Hertweck C (2013) Nitro versus hydroxamate in siderophores of pathogenic bacteria: effect of missing hydroxylamine protection in malleobactin biosynthesis. Angew Chemie 52: 8271–8275. [DOI] [PubMed] [Google Scholar]
- 17.Franke J, Ishida K, Hertweck C (2015) Plasticity of the malleobactin pathway and its impact on siderophore action in human pathogenic bacteria. Chem Eur J 21: 8010–8014. 10.1002/chem.201500757 [DOI] [PubMed] [Google Scholar]
- 18.Seeger M, Timmis KN, Hofer B (1995) Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl Environ Microbiol 61: 2654–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeger M, Zielinski M, Timmis KN, Hofer B (1999) Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl Environ Microbiol 65: 3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cámara B, Herrera C, González M, Couve E, Hofer B, Seeger M (2004) From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol 6: 842–850. [DOI] [PubMed] [Google Scholar]
- 21.Denef VJ, Park J, Tsoi TV., Rouillard JM, Zhang H, Wibbenmeyer JA, et al. (2004) Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl Environ Microbiol 70: 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, et al. (2006) Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci 103: 15280–15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Méndez V, Agulló L, González M, Seeger M (2011) The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS ONE 6: e17583 10.1371/journal.pone.0017583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero-Silva MJ, Méndez V, Agulló L, Seeger M (2013) Genomic and functional analyses of the gentisate and protocatechuate ring-cleavage pathways and related 3-hydroxybenzoate and 4-hydroxybenzoate peripheral pathways in Burkholderia xenovorans LB400. PLoS ONE 8: e56038 10.1371/journal.pone.0056038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chirino B, Strahsburger E, Agulló L, González M, Seeger M (2013) Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS ONE 8: e75746 10.1371/journal.pone.0075746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urtuvia V, Villegas P, González M, Seeger M (2014) Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int J Biol Macromol 70: 208–213. 10.1016/j.ijbiomac.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 27.Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6: 493–505. [DOI] [PubMed] [Google Scholar]
- 28.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH (2005) Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines. Nucleic Acids Res 33: 5799–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160: 47–56. [DOI] [PubMed] [Google Scholar]
- 32.Almodóvar I, Ramírez-Rodríguez O, Barriga A, Rezende MC, Araya-Maturana R (2011) Electrospray ionization mass spectrometric fragmentation of hydroquinone derivatives. Rapid Commun Mass Spectrom 25: 370–378. 10.1002/rcm.4868 [DOI] [PubMed] [Google Scholar]
- 33.Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA (1998) Peptide bond formation in nonribosomal peptide biosynthesis: catalytic role of the condensation domain. J Biol Chem 273: 22773–22781. [DOI] [PubMed] [Google Scholar]
- 34.Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C (1999) Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding L-ornithine N5-oxygenase. Infect Immun 67: 4443–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMorran BJ, Shantha Kumara HMC, Sullivan K, Lamont IL (2001) Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiol 147: 1517–1524. [DOI] [PubMed] [Google Scholar]
- 36.Herbst DA, Boll B, Zocher G, Stehle T, Heide L (2013) Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem 288: 1991–2003. 10.1074/jbc.M112.420182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMorran BJ, Merriman ME, Rombel IT, Lamont IL (1996) Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene 176: 55–59. [DOI] [PubMed] [Google Scholar]
- 38.Saurin W, Hofnung M, Dassa E (1999) Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48: 22–41. [DOI] [PubMed] [Google Scholar]
- 39.Brown SD, Utturkar SM, Klingeman DM, Johnson CM, Martin SL, Land ML, et al. (2012) Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J Bacteriol 194: 5991–5993. 10.1128/JB.01243-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol PA, Darling P, Lewenza S, Corbett C, Kooi C (2000) Identification of a siderophore receptor required for ferric-ornibactin uptake in Burkholderia cepacia. Infect Immun 68: 6554–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamood AN, Colmer JA, Ochsner UA, Vasil ML (1996) Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol 21: 97–110. [DOI] [PubMed] [Google Scholar]
- 42.Franke J, Ishida K, Hertweck C (2014) Evolution of siderophore pathways in human pathogenic bacteria. J Am Chem Soc 136: 5599–5602. 10.1021/ja501597w [DOI] [PubMed] [Google Scholar]
- 43.Stephan H, Freund S, Beck W, Jung G, Meyer JM, Winkelmann G (1993) Ornibactins–a new family of siderophores from Pseudomonas. BioMetals 6: 93–100. [DOI] [PubMed] [Google Scholar]
- 44.Stephan H, Freund S, Meyer JM, Winkelmann G, Jung G (1993) Structure elucidation of the gallium–ornibactin complex by 2D-NMR spectroscopy. Liebigs Ann Chemie 1993: 43–48. [Google Scholar]
- 45.Ochsner UA, Vasil ML (1996) Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci 93: 4409–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasil ML, Ochsner UA, Johnson Z, Colmer JA, Hamood AN (1998) The Fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator PtxR in Pseudomonas aeruginosa. J Bacteriol 180: 6784–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Challis GL, Ravel J, Townsend CA (2000) Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol 7: 211–224. [DOI] [PubMed] [Google Scholar]
- 48.Marahiel MA, Krause M, Skarpeid HJ (1985) Cloning of the tyrocidine synthetase 1 gene from Bacillus brevis and its expression in Escherichia coli. Mol Gen Genet 201: 231–236. [DOI] [PubMed] [Google Scholar]
- 49.Yu S, Fiss E, Jacobs WR (1998) Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J Bacteriol 180: 4676–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lautru S, Deeth RJ, Bailey LM, Challis GL (2005) Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1: 265–269. [DOI] [PubMed] [Google Scholar]
- 51.Sharman GJ, Williams DH, Ewing DF, Ratledge C (1995) Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem J 305: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bosello M, Mielcarek A, Giessen TW, Marahiel MA (2012) An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for rhodochelin iron coordination. Biochemistry 51: 3059–3066. 10.1021/bi201837f [DOI] [PubMed] [Google Scholar]
- 53.Shim JH, Benkovic SJ (1999) Catalytic mechanism of Escherichia coli glycinamide ribonucleotide transformylase probed by site-directed mutagenesis and pH-dependent studies. Biochemistry 38: 10024–10031. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt E, Panvert M, Blanquet S, Mechulam Y (1998) Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J 17: 6819–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornelis P (2010) Iron uptake and metabolism in Pseudomonads. Appl Microbiol Biotechnol 86: 1637–1645. 10.1007/s00253-010-2550-2 [DOI] [PubMed] [Google Scholar]
- 56.Whitby PW, VanWagoner TM, Springer JM, Morton DJ, Seale TW, Stull TL (2006) Burkholderia cenocepacia utilizes ferritin as an iron source. J Med Microbiol 55: 661–668. [DOI] [PubMed] [Google Scholar]
- 57.Kvitko BH, Goodyear A, Propst KL, Dow SW, Schweizer HP (2012) Burkholderia pseudomallei known siderophores and hemin uptake are dispensable for lethal murine melioidosis. PLoS Negl Trop Dis 6: e1715 10.1371/journal.pntd.0001715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathew A, Eberl L, Carlier AL (2014) A novel siderophore-independent strategy of iron uptake in the genus Burkholderia. Mol Microbiol 91: 805–820. 10.1111/mmi.12499 [DOI] [PubMed] [Google Scholar]
- 59.Gaines JM, Carty NL, Tiburzi F, Davinic M, Visca P, Colmer-Hamood JA, et al. (2007) Regulation of the Pseudomonas aeruginosa toxA, regA and ptxR genes by the iron-starvation sigma factor PvdS under reduced levels of oxygen. Microbiol 153: 4219–4233. [DOI] [PubMed] [Google Scholar]
- 60.Loper JE, Henkels MD (1999) Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Appl Environ Microbiol 65: 5357–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carrillo-Castañeda G, Juárez Muños J, Peralta-Videa J, Gómez E, Tiemann K, Duarte-Gardea M, et al. (2002) Alfalfa growth promotion by bacteria grown under iron limiting conditions. Adv Environ Res 6: 391–399. [Google Scholar]
- 62.Jin CW, Li GX, Yu XH, Zheng SJ (2010) Plant Fe status affects the composition of siderophore-secreting microbes in the rhizosphere. Ann Bot 105: 835–841. 10.1093/aob/mcq071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Penyalver R, Oger P, López MM, Farrand SK (2001) Iron-Binding compounds from Agrobacterium spp.: biological control strain Agrobacterium rhizogenes K84 produces a hydroxamate siderophore. Appl Environ Microbiol 67: 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De los Santos-Villalobos S, Barrera-Galicia GC, Miranda-Salcedo MA, Peña-Cabriales JJ (2012) Burkholderia cepacia XXVI siderophore with biocontrol capacity against Colletotrichum gloeosporioides. World J Microbiol Biotechnol 28: 2615–2623. 10.1007/s11274-012-1071-9 [DOI] [PubMed] [Google Scholar]
- 65.Chaiharn M, Chunhaleuchanon S, Lumyong S (2009) Screening siderophore producing bacteria as potential biological control agent for fungal rice pathogens in Thailand. World J Microbiol Biotechnol 25: 1919–1928. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Malleobactin (A) and ferrimalleobactin complex (D) 3D structures were predicted. The binding site for malleobactin 3D structure (B) and ferrimalleobactin 3D structure (E) was predicted using a Docking strategy. The frontal view of the best model of the protein-ligand complexes (C,F) are shown as ribbon representations, where the periplasmic side is located up and the malleobactin and ferrimalleobactin ligands surfaces are highlighted in blue. The cytoplasmic (G) and periplasmic (H) sides, and the volume (I) of the protein ligand complex are shown.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.