Abstract
Although numerous linkage maps have been constructed in the genus Populus, they are typically sparse and thus have limited applications due to low throughput of traditional molecular markers. Restriction-site associated DNA sequencing (RADSeq) technology allows us to identify a large number of single nucleotide polymorphisms (SNP) across genomes of many individuals in a fast and cost-effective way, and makes it possible to construct high-density genetic linkage maps. We performed RADSeq for 299 progeny and their two parents in an F1 hybrid population generated by crossing the female Populus deltoides ‘I-69’ and male Populus simonii ‘L3’. A total of 2,545 high quality SNP markers were obtained and two parent-specific linkage maps were constructed. The female genetic map contained 1601 SNPs and 20 linkage groups, spanning 4,249.12 cM of the genome with an average distance of 2.69 cM between adjacent markers, while the male map consisted of 940 SNPs and also 20 linkage groups with a total length of 3,816.24 cM and an average marker interval distance of 4.15 cM. Finally, our analysis revealed that synteny and collinearity are highly conserved between the parental linkage maps and the reference genome of P. trichocarpa. We demonstrated that RAD sequencing is a powerful technique capable of rapidly generating a large number of SNPs for constructing genetic maps in outbred forest trees. The high-quality linkage maps constructed here provided reliable genetic resources to facilitate locating quantitative trait loci (QTLs) that control growth and wood quality traits in the hybrid population.
Introduction
The Populus genus not only has several attractive biological characteristics as a long-lived plant but also possesses tremendous economic and ecological importance. Because of its fast growth, asexual reproduction, small genome size (~480 Mbp) and easy genetic transformation, this genus was selected as a model system for forest trees [1, 2] and the species of P. trichocarpa and P. euphratica have had their genomes sequenced successively [3, 4]. Some species of Populus are cultivated worldwide to meet demand for pulp and paper, lumber, wood-based panels, and biofuels. Others are planted on a large scale to build windbreaks especially in Northwest China where sand-dust storms occur every year. The Populus genus comprises aspens, cottonwoods and poplars, and there are approximately 30 species that can be grouped into six separate sections [5]. The overwhelming majority of these species have an extensive distribution in the Northern Hemisphere, and harbour significant variability in adaptive traits such as growth rate, rooting ability, and drought or disease resistance. Nevertheless, there is an increasing interest in creating new varieties for superior adaptive and commercial traits through intraspecific or interspecific hybridization. High-quality genetic linkage maps provide valuable genomic information for achieving this goal by mapping quantitative trait loci (QTLs) and marker-assisted selection.
In the past two decades, great efforts have been made to develop numerous genetic linkage maps of different Populus species. Many of these linkage maps were constructed directly using the software MAPMAKER [6] with mapping strategies of inbred lines, including so-called ‘pseudo-testcross’ proposed by Grattapaglia and Sederoff [7–19]. Only a few studies have constructed Populus linkage maps with the software JoinMap [20], although this tool can incorporate different linkage phases and various segregation markers into linkage analyses for an outbred population [21–24]. All these maps were mainly constructed with molecular markers such as RAPD, RFLP, AFLP and SSR. However, these traditional molecular markers are of low throughput due to instability or time- and cost-consuming experiments, and hence cannot satisfy the needs of constructing high-density genetic linkage maps in forest trees. To date, the densest linkage map of P. trichocarpa contains more than 3,500 BeadArray SNP markers, which was reported in Slavov et al. [25] and again in Muchero et al. [26].
Restriction-site associated DNA sequencing (RADSeq), one of the next generation sequencing (NGS) technologies based at reduced genome complexity [27], allows for tens of thousands of single nucleotide polymorphisms (SNPs) to be identified in a fast and relatively cheap way across the genomes of many individuals from an experimental population. This powerful new sequencing method was first described by Baird et al. [28], where more than 13,000 SNPs were identified and used to map three traits in two model organisms, threespine stickleback and Neurospora crassa. Later on, Emerson et al. [29] used RADSeq technology to reveal an evolutionary mystery in the pitcher plant mosquito, Wyeomyia smithii. In the meantime, Hohenlohe et al. [30] applied RADSeq method to study parallel adaption in natural populations of threespine stickleback. Recently, a series of linkage maps constructed with RADSeq markers were reported in various organisms, including ryegrass, barley, moth, grape and gudgeon [31–35]. These RADSeq genetic linkage maps generally included more than 1,000 molecular markers with an average distance of less than 5 cM between adjacent markers, and hence provided reliable genetic resources for population genetics in related species.
The objective of this study was to construct high-density and high-quality genetic linkage maps of Populus with RADSeq technology for extensive QTL mapping studies in the future. An interspecific cross was conducted to generate an F1 mapping population by hybridizing Populus deltoides with P. simonii. RADSeq was performed using genomic DNA from the two parents and 299 progeny. A large number of SNPs that follow Mendel’s segregation ratio were identified and genotyped through mapping RAD reads of each individual to the reference genome of P. trichocarpa as well as some rigorous filtering procedures. Based on these SNP molecular markers, the maternal and paternal genetic linkage maps were constructed separately with linkage phase inferred between adjacent markers. The synteny and collinearity between the linkage maps and the reference genome were evaluated and were mostly consistent, indicating that the linkage maps are highly accurate in marker ordering. The dense and reliable linkage maps of the two parents are useful for identifying QTLs that control growth and timber traits in the permanent Populus population.
Materials and Methods
The Mapping Population and DNA Isolation
The mapping population was generated by hybridizing P. deltoides Marsh cv. ‘Lux’ (I-69/55) with P. simonii in 2011. P. deltoides has the superior characteristics of fast growth and resistance to Marssonina leaf spot disease, but has a low survival rate of field cutting propagation due to the poor rooting ability [17]. P. simonii, a native tree species widely distributed in northern China, displays excellent performance in cold, heat and drought, tolerance to alkali-salt and barren conditions, and regeneration ability [36]. These two parental Populus could produce hybrids with significant segregation of morphological and physiological traits. The female P. deltoides ‘I-69’ was chosen from Siyang Forest Farm (SFF) of Siyang County, Jiangsu Province, China, while the flowering branches of P. simonii ‘L-3’ were collected in a forest land managed by Luoning Forest Bureau of Henan Provicne, China. The interspecific cross was performed in the spring of 2011 in SFF. Approximately 500 seedlings of the F1 progeny were planted in Xiashu Forest Farm of Nanjing Forest University, Jurong County, Jiangsu Province, China. A total of 299 progeny were randomly chosen for constructing linkage maps of the two parents in this study. Young leaf tissue was collected from the two parents and each of the 299 individuals at the beginning of the vegetative period (late spring). The samples were immediately stored in a -80°C freezer. DNA was extracted from 150 individuals using the Plant Genomic DNA Kit (Tiangen, Tiangen Biotech Co. Ltd., Beijing) with the manufacturer's protocol, while the other 149 individuals DNA were extracted with the CTAB protocol [37].
This study was permitted to conduct in Luoning County of Henan province, China, and Siyang County of Jiangsu province, China, by the two local Agriculture and Forestry Commissions, and in Xiashu Forest Farm of Nanjing Forestry University located in Jurong County, Jiangsu province, China, by the Academic Committee of the university. All the test fields belong to the local government or university and no endangered or protected species were involved in our study.
RAD Sequencing
The RAD library was constructed following the protocol described by Baird et al. [28]. Genomic DNA from each of the two parents (1.5 μg) and 299 progeny (300 ng) was digested for 4 h at 37°C in a 50-μL reaction with 20 units (U) of EcoRI (NEB, USA). The reactions were stopped by holding at 65°C for 20 min. 200 nM P1 adapter was ligated to the products of the restriction reaction, along with 5 μL of 10 mM ATP, 1 μL 10x NEB Buffer 2, 0.5 μL (1,000 U) T4 DNA Ligase (high concentration, NEB) and 5 μL H2O, and incubated at room temperature for 20 min. The P1 adapter, a modified Solexa adapter (top: 5’-CAAGCAGAAGACGGCATACGAGATXXXX XXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’, bottom: 3’-GTTCG TCTTCTGCCGTATGCTCTAXXXXXXCACTGACCTCAAGTCTGCACACGAG AAGGCTAGATTAA-p-5’, X indicated MID), contained a matching sticky-end to the fragments and a MID (Molecular Identifier), a short sequence that will uniquely identify an individual. Samples were again heat-inactivated for 20 min at 65°C, pooled and purified with a Qiagen PCR cleanup column and eluted in 30 μL of buffer EB. DNA was sheared using Covaris S220 into 300–700 bp. Fragment ends were repaired using the Quick Blunting kit Enzyme Mix (NEB) with 30 μL DNA, 5 μL 10x Blunting Buffer, 10 μL 1Mm dNTP and 9 μL H2O, incubated at 25°C for 30 min, purified with 1.6 times the volume of AMPure XP Beads and eluted in 21 μL water. dATP overhangs were added to the DNA using 20 μL of purified library template, dATP (1 μL 100 mM), 15U of Klenow exo- (NEB) and 3 μL 10x NEB Buffer 2. The reaction was incubated at 37°C for 30min, then purified with 1.6 times the volume of AMPure XP Beads column and eluted in 21 μL water.
Paired-end P2 adapter (top: 5’-AATGATACGGCGACCACCGAGATCTAC ACTCTTTCCCTACACGACGCTCTTCCGATCT-3’, bottom: 3’-TTACTATGCCG CTGGTGGCTCTAGATGTGAGAAAGGGATGTGCTGCGAGAAGGCTAG-p-5’), a divergent modified Solexa adapter (2006 Illumina, Inc.), was ligated to 20 μL sheared, size-selected, P1-ligated and pooled DNA template with 2 μL of 2 μM Adapter2, 3 μL of NEB Buffer 2 and 1 μL of 1,000 U T4 DNA Ligase in total reaction of 30 μL. The ligation was incubated overnight at 4°C, then DNA product purified with a same volume of AMPure XP Beads and eluted in 30 μL of water. PCR enrichment of the library was performed in a PCR amplification with 25 μL 2x Phusion PCR Master Mix (NEB), 1.5 μL of PCR Primer 1: 5'-AATGATA CGGCGACCACCGA-3', 1.5 μL of PCR Primer 2: 5'-CAAGCAGAAGACGGC ATACGAG-3' and 14 μL of H2O. Cycling conditions were: 98°C for 1 min, then 18 cycles of 98°C for 10 s, 60°C for 30 s, 72°C for 40 s, and a final extension at 72°C for 5 min. PCR amplicons were gel purified and fragments in the size range 300–700 bp were excised from the gel.
The RAD library was sequenced using an Illumina HiSeq 2000 sequencer. The two parents and one half of the progeny were sequenced in 8 lanes (paired-end, 100 bp) at Novogene Bioinformatics Institute (NBI), Beijing, China. In addition, the two parents and another half of the progeny were sequenced in 7 lanes (paired-end, 90 bp) at Beijing Genomics Institute (BGI), Shenzhen, China.
RAD Sequence Analysis, SNP Discovery and Genotyping
Standard quality control (QC) pipelines (NBI, Beijing, China; BGI, Shenzhen, China) were used to process the raw sequencing data. First, multiple sequences were segregated by the appropriate nucleotide MID assigned to each sample, and paired reads containing primer/adaptor sequence were removed. Second, when one single read contains more than 10% of its bases uncalled, the pair was discarded. Third, if the number of low-quality bases (Q score less than or equal to 5) was greater than 50% of the length in a single read, then both of the paired reads were also removed from the dataset. After these filtering procedures, the so-called clean data reads were generated for each sample. We further used NGS QC toolkit (v2.3.3, [38]) with the default cut-off values to filter these clean data and to obtain high-quality (HQ) reads that each has at least 70% bases with the Phred quality score more than or equal to 20.
The filtered HQ reads were mapped to the reference genome of P. trichocarpa [3] (V3.0, DOE-JGI, http://www.phytozome.net/poplar), which consists of approximately 434.1 Mb arranged into 19 primary scaffolds (corresponding to the 19 chromosomes, 394.5 Mb) and 1427 additional scaffolds. First, we used the BWA [39] mem command with default parameters to align the paired-end reads of each sample separately to the reference sequences. Subsequently, a sequence alignment/map (SAM) format file for each individual was produced [40]. Next, several steps were taken for SNP calling and genotyping with SAMtools (including bcftools, [40]) and Perl scripts (available upon request): (i) filtering out records in each SAM file, which have multiple mapping positions on the genome; (ii) producing BCF files with command samtools mpileup–g–f -I; (iii) generating the variant call format (VCF, [41]) files with command bcftools view–c–v for each parent; (iv) filtering SNPs with relatively relaxed conditions (quality more than 20 and DP more than or equal to 5) for each parent using Perl scripts, merging the SNP data sets and saving all parental SNP positions as a list site file used in the next step; (v) for all progeny, filtering SAM files as step (i), generating BCF files as step (ii), and creating VCF files with command bcftools view–l–c–g using the list site file; (vi) identifying genotype of each progeny according to the genotype likehihoods (GL) of all possible genotypes at each SNP in VCF files, and filtering with stringent conditions (DP more than or equal to 10 and GQ more than 50). Finally, we further filtered those SNPs for linkage mapping, which followed Mendelian segregation ratios by chi-square test across the whole population.
Linkage Map Construction
We performed chi-square tests with different degrees of freedom to check whether the genotyped SNPs follow the corresponding Mendelian segregation ratios, such as 1:1, 1:2:1 and 1:1:1:1. Those SNPs deviating seriously from the Mendelian ratios (p < 0.01) and having more than 10% missing genotypes in the population were removed from linkage analysis. Since the overwhelming majority of SNPs segregate in 1:1 in this study, the pseudo-testcross mapping strategy [7] was applied to construct two linkage maps, each for a different parent. The construction of both parental linkage maps was carried out with two softwares, JoinMap 4.1 [20] and FsLinkageMap 2.1 [42]. The maternal linkage map was constructed using the SNP markers that have the segregation type of abaa, where ab represents the heterozygous genotype from the mother and aa the homozygous genotype from the father, while the paternal linkage map was built with the markers of segregation type of aaab. First, two-point linkage analysis was conducted and linkage groups were clustered under the threshold of LOD score using FsLinkageMap. Second, markers in each linkage group were ordered three times using the maximum likelihood (ML) mapping algorithm with default parameters in JoinMap and one time using the ordering method in FsLinkageMap. The best order was chosen as the mapping order among the four results of the two programs, based on the minimum sum of adjacent recombination fractions (SARF; [43]). Third, the linkage maps were drawn in WMF format with FsLinkageMap and then edited in PDF or EPS format with the software of Mayura Draw (http://www.mayura.com).
QTL Mapping
We measured each tree height and diameter at breast height (DBH) in the fall of 2014 and performed QTL mapping for the two growth traits based on the two parental linkage maps constructed here. The mapping method is a modified composite interval mapping (CIM; [44]) that can incorporate different segregation types of markers and various linkage phases. The log likelihood ratio (LR) at each map position was calculated with 15 background markers selected by forward regression method and a window size of 10 cM. The threshold value for asserting the existence of a QTL was determined by 1000 permutation tests [45]. The algorithm was implemented with in-house R scripts, which are available upon request.
Results
RAD Sequencing and SNP Genotype Calling
A total of 329.5 Gb raw RADSeq data containing 1,790,149,605 paired-end (PE) reads was generated on the Illumina HiSeq2000 (Table 1). Of these data, 142.0 Gb of 100-bp PE reads for the two parents and 150 progeny were produced from the sequencing experiment performed in NBI, while the rest of 187.5 Gb 90-bp PE reads for the same two parents and other 149 progeny was produced by sequencing in BGI. The RADSeq raw data are available under accession number SRP052929 at the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra). After a series of quality control and filtering procedures (described in Materials and Methods), we obtained 309.3 Gb of HQ reads. The female P. deltoides parent yielded 5.9 Gb of HQ reads (2.3 Gb from NBI, 3.6 Gb from BGI), whereas the male P. simonii parent yielded 12.2 Gb of HQ reads (2.5 Gb from NBI, 9.7 Gb from BGI). On average, 0.8 Gb of HQ 100-bp PE reads was obtained for the 150 progeny sequenced in NBI and 1.1 Gb of HQ PE reads for the additional 149 progeny sequenced in BGI. It is worthy of note that one sample with field ID, ‘C15-1’, as well as the two parents, were independently sequenced in both sequencing companies and can be used to confirm the accuracy of sequencing. The length of the HQ forward reads generated in BGI ranged from 82 to 86 bp due to discarding the original 4–8 bp MID barcode sequences that identify individual samples within a pooled library. All these RADSeq data of HQ PE reads allowed for calling SNP genotypes across the genomes of the two parents and 299 progeny in the full-sib family of P. deltoides × P. simonii.
Table 1. Summary of RADSeq Data from NBI and BGI with averages in brackets.
| Experiment | Sample | No. sample | No. raw reads | Raw reads data (Gb) | No. HQ reads | HQ reads data (Gb) |
|---|---|---|---|---|---|---|
| NBI | Male parent | 1 | 14,024,713 | 2.80 | 12,617,155 | 2.52 |
| Female parent | 1 | 12,949,974 | 2.59 | 11,500,364 | 2.30 | |
| Progeny | 150 | 683,250,607 (4,555,004) | 136.65 (0.91) | 612,650,551 (4,084,337) | 122.53 (0.82) | |
| BGI | Male parent | 1 | 57,159,139 | 9.91 | 55,789,694 | 9.68 |
| Female parent | 1 | 21,619,787 | 3.72 | 20,956,868 | 3.60 | |
| Progeny | 149 | 1,001,145,385 (6,719,097) | 173.87 (1.17) | 971,115,096 (6,517,551) | 168.66 (1.13) | |
| Total | Male parent | 1 | 71,183,852 | 12.72 | 68,406,849 | 12.20 |
| Female parent | 1 | 34,569,761 | 6.31 | 32,457,232 | 5.90 | |
| Progeny | 299 | 1,684,395,992 | 310.52 | 1,583,765,647 | 291.19 | |
| Total | 301 | 1,790,149,605 (5,947,341) | 329.55 (1.10) | 1,684,629,728 (5,596,777) | 309.29 (1.03) |
With the short read mapping program, BWA, 94.1% of the HQ reads from the female parent and the same ratio from the male parent were mapped to the Populus reference genome. However, only 23.6% of the maternal HQ reads each was aligned to the reference genome with the edit distance less than or equal to 8, the alignment score more than 60, and the alignment score less than 10 for second-best alignment, while for the male parent this fraction is nearly the same (23.1%) as the female parent. On average, these almost uniquely mapped HQ reads reached 31.24-fold and 61.66-fold coverage depth, and covered 8.98% and 9.14% of the reference genome for the female and male parents, respectively. For all the progeny, the average coverage depth of such reads ranged from the minimum of 3.54-fold to the maximum of 21-fold and the coverage of the reference genome from 4.96% to 8.83% (S1 Table). We used the almost uniquely mapped HQ reads from each parent for SNP calling. As a result, 836,895 SNPs with depth of more than or equal to 5 were revealed in the two parents, of which 475,965 SNPs were from the female, 475,591 SNPs from the male, and 114,661 SNPs from both. We further performed SNP genotype calling for the parents and progeny within those SNPs identified in the two parents. We filtered out those SNPs at which one parent has genotype, but the other does not due to no or low coverage of reads. Consequently, there were 385,470 SNPs genotyped in both parents; however, only 85,363 SNPs segregated in the progeny, with segregation patterns of abaa (51,817), aaab (32,654), abab (793), and abac (99) (Table 2). Finally, 1,603 and 942 SNPs with segregation types of abaa and aaab, respectively, were chosen for linkage mapping, which followed the Mendelian segregation ratio 1:1 with p ≥ 0.01 and at which at least 90% of the 299 progeny were genotyped. The distance of adjacent SNPs with same segregation patterns is more than 1 kb on the reference genome.
Table 2. Number of SNP loci genotyped in both parents and used for linkage mapping.
| Female genotypea | Male genotype | SNPs in parents | SNPs used for mapping |
|---|---|---|---|
| ab | aa | 51,817 | 1,603 |
| aa | ab | 32,654 | 942 |
| ab | ab | 793 | 0 |
| ab | ac | 99 | 0 |
| aa | bb | 300,107 | 0 |
| Total | 385,470 | 2,545 |
aThe notations of a, b, c and d denote up to four possible alleles from two parents at an SNP site.
To validate the accuracy of SNP genotypes, we used the two RADSeq data sets from NBI and BGI to independently call genotypes for the two parents, ‘I-69’ and ‘L-3’, and one progeny, ‘C15-1’. Of the 2545 SNPs for linkage mapping, over 97.0% were genotyped with the two data sets for each of the 3 samples. Among those SNPs genotyped with the two data sets for each sample, ~98% were confirmed with each other’s data set, resulting in totally 98.2% of the 7496 SNP genotypes confirmed for all the 3 samples (S2 Table).
Genetic Linkage Maps and QTL Mapping
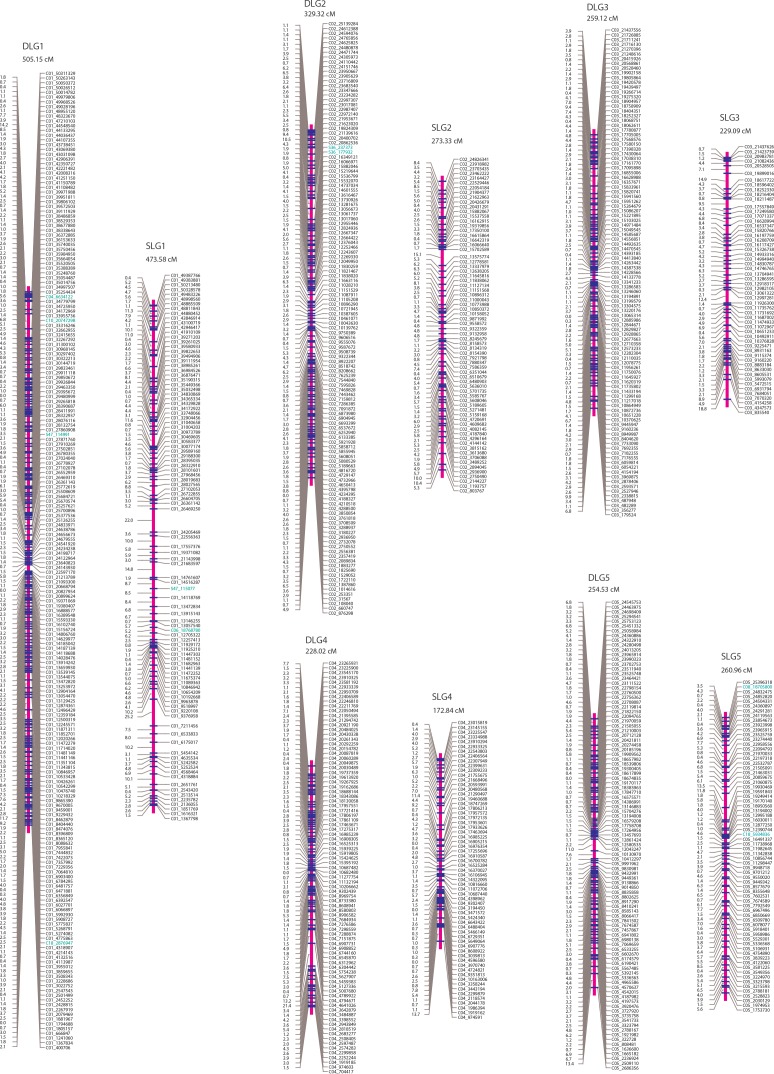
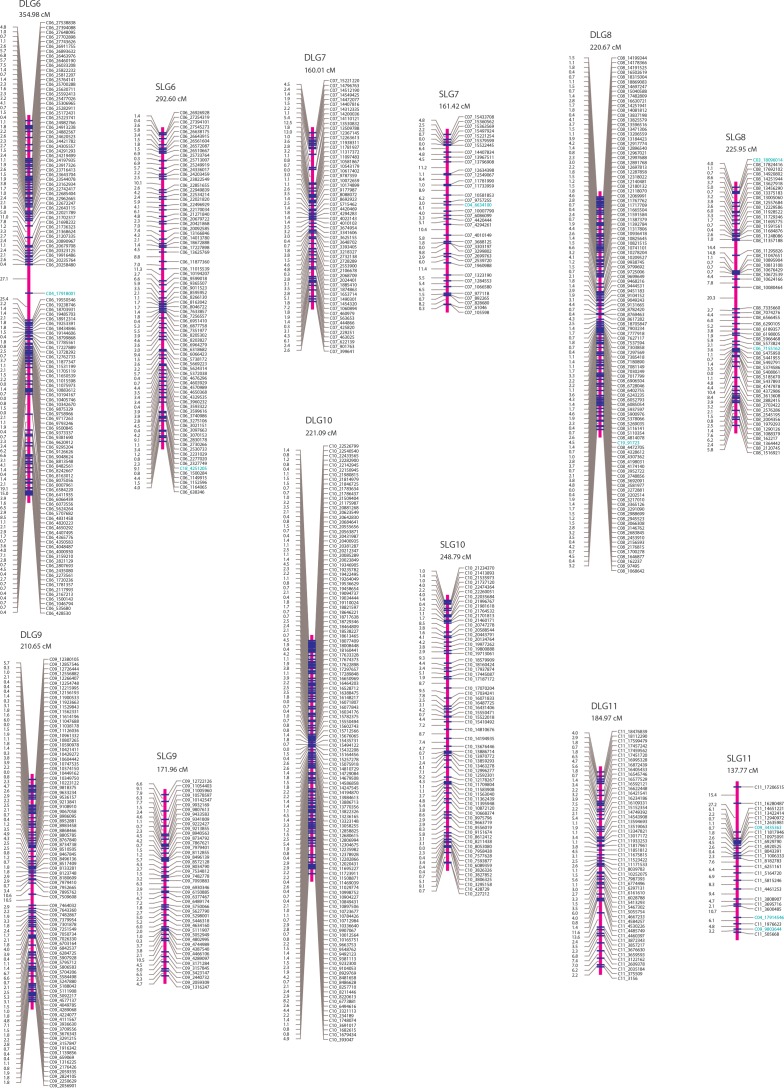
Two parent-specific linkage maps were constructed each with SNP markers that segregate in the ratio of 1:1 using the mapping strategy described in “Materials and Methods” (Figs 1–3). A total of 1,601 SNPs with the segregation type of abaa were assigned to 20 linkage groups (denoted as DLG1-20) at the LOD threshold of 15.0 and formed the genetic linkage map of the female P. deltoides ‘I-69’. On this maternal map, the lengths of linkage groups ranged from 84.93 to 505.15 cM, amounting to the total length of 4249.12 cM. On average, the distance between adjacent mapped markers was 2.69 cM, ranging from 0.00 to 27.97 cM. For the male P. simonii ‘L-3’, the linkage map was constructed with 940 SNP markers of segregation type aaab, which were also grouped into 20 linkage groups (denoted as SLG1-20) at the LOD threshold of 15.0. The total length of this parental map was 3816.24 cM, with an average length of 4.15 cM for marker intervals ranging from 0.00 to 28.07 cM and 190.81 cM for linkage groups ranging from 38.09 to 437.58 cM.
Fig 1. The genetic maps of linkage groups DLG1-DLG5 for the maternal P. deltoides ‘I-69’ and SLG1-SLG5 for the paternal P. simonii ‘L-3’.
The length of each linkage group is given below the linkage group name. SNP marker names are denoted by the names and positions of chromosomes or scaffolds of the reference genome P. trichocarpa. Markers from other chromosomes and additional scaffolds are in green.
Fig 3. The genetic maps of linkage groups DLG12-DLG20 for the maternal P. deltoides ‘I-69’ and SLG12-SLG20 for the paternal P. simonii ‘L-3’.
The length of each linkage group is given below the linkage group name. SNP marker names are denoted by the names and positions of chromosomes or scaffolds of the reference genome P. trichocarpa. Markers from other chromosomes and additional scaffolds are in green.
Fig 2. The genetic maps of linkage groups DLG6-DLG11 for the maternal P. deltoides ‘I-69’ and SLG6-SLG11 for the paternal P. simonii ‘L-3’.
The length of each linkage group is given below the linkage group name. SNP marker names are denoted by the names and positions of chromosomes or scaffolds of the reference genome P. trichocarpa. Markers from other chromosomes and additional scaffolds are in green.
More detailed information on the two parental linkage maps is listed in Table 3 and S3 and S4 Tables. Except for DLG20 and SLG20, each linkage group of the two maps corresponds to the reference chromosome of P. trichocarpa. As we expected, there remained strong positive correlations among the SNP number, the genetic distance, and the physical size (Table 4). The number of SNP markers was significantly correlated with the length for each linkage group, with a high correlation coefficient of 0.9364 for the female map and 0.9376 for the male map. The correlations between the genetic and physical size for the two parents were also relatively high both at coefficients of over 0.90. Furthermore, the relationship between the SNP number and chromosome size was consistent for the two linkage maps, both with an absolutely high but relatively low correlation coefficient of ~0.85. In addition to these high consensuses for the genetic maps, we also predicted linkage phases between any two adjacent SNP markers, which were listed in the fifth column of S3 and S4 Tables. The predicted linkage phases would provide valuable information for deriving conditional probabilities of QTL genotypes on marker genotypes when performing QTL mapping in the hybrid population of P. deltoides × P. simonii.
Table 3. SNP number and length of linkage groups in two parental genetic maps of P. deltoides ‘I-69’ and P. simonii ‘L-3’.
| P. deltoides ‘I-69’ | P. simonii ‘L-3’ | Chromosome size (Mb)c | ||||
|---|---|---|---|---|---|---|
| Groupa | SNP number | Length (cM) | Groupb | SNP number | Length (cM) | |
| DLG1 | 206 | 505.15 | SLG1 | 100 | 437.58 | 50.50 |
| DLG2 | 125 | 329.32 | SLG2 | 70 | 273.33 | 25.26 |
| DLG3 | 103 | 259.12 | SLG3 | 55 | 229.09 | 21.82 |
| DLG4 | 87 | 228.02 | SLG4 | 63 | 172.84 | 24.27 |
| DLG5 | 99 | 254.53 | SLG5 | 70 | 260.96 | 25.89 |
| DLG6 | 120 | 354.98 | SLG6 | 79 | 292.60 | 27.91 |
| DLG7 | 58 | 160.01 | SLG7 | 36 | 161.42 | 15.61 |
| DLG8 | 109 | 220.67 | SLG8 | 57 | 225.95 | 19.47 |
| DLG9 | 90 | 210.65 | SLG9 | 44 | 171.96 | 12.95 |
| DLG10 | 126 | 221.09 | SLG10 | 66 | 248.79 | 22.58 |
| DLG11 | 53 | 184.97 | SLG11 | 25 | 137.77 | 18.50 |
| DLG12 | 56 | 159.27 | SLG12 | 37 | 133.87 | 15.76 |
| DLG13 | 51 | 199.87 | SLG13 | 47 | 155.71 | 16.32 |
| DLG14 | 92 | 236.46 | SLG14 | 34 | 161.91 | 18.92 |
| DLG15 | 64 | 151.74 | SLG15 | 40 | 156.39 | 15.28 |
| DLG16 | 37 | 115.32 | SLG16 | 11 | 91.50 | 14.49 |
| DLG17 | 50 | 128.72 | SLG17 | 37 | 135.49 | 16.08 |
| DLG18 | 37 | 122.30 | SLG18 | 34 | 164.57 | 16.96 |
| DLG19 | 28 | 122.00 | SLG19 | 26 | 130.42 | 15.94 |
| DLG20 | 10 | 84.93 | SLG20 | 9 | 38.09 | |
| Total | 1601 | 4249.12 | 940 | 3816.24 | 394.51 | |
aDLG indicates the linkage group of P. deltoides 'I-69';
bSLG indicates the linkage group of P. simonii 'L-3';
cThe genome size refers to the reference genome of P. trichocarpa (Tuskan et al. 2006).
Table 4. Correlations among the SNP number, genetic length and chromosome size for the linkage groups of the two parental maps.
| SNP number in DLG | DLG Length | SNP number in SLG | SLG length | |
|---|---|---|---|---|
| DLG Length | 0.9364 | |||
| SNP number in SLG | 0.9010 | 0.8905 | ||
| SLG length | 0.9421 | 0.9394 | 0.9376 | |
| Chromosome size | 0.8673 | 0.9149 | 0.8437 | 0.9216 |
To demonstrate the potential applications of the linkage maps constructed above in QTL mapping, we preliminarily performed composite interval mapping of the tree height and DBH. S1–S4 Figs showed the profiles of LRs against the map positions and the thresholds for existing QTLs determined by 1000 permutation tests. As a result, 8 QTLs for tree height and 7 for DBH were significantly identified and they explained 81.1% and 70.4% of the phenotypic variances, respectively (S5 and S6 Tables).
Synteny and Collinearity between Genetic and Physical Maps
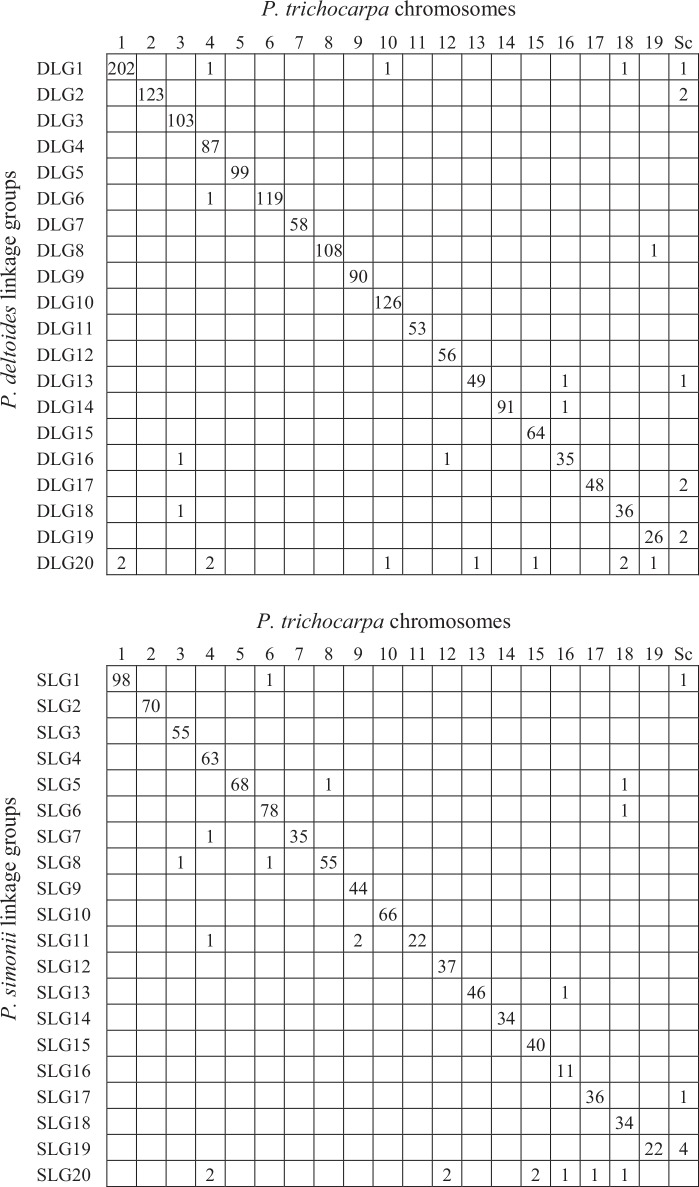
Oxford grid comparison of the genetic and physical maps showed that high levels of synteny were conserved between the parental genomes and the reference genome of P. trichocarpa (Fig 4). It can be found that all the SNPs on each of the 9 female linkage groups (DLGs 3, 4, 5, 7, 9, 10, 11, 12, 15) and 10 male linkage groups (SLGs 2, 3, 4, 9, 10, 12, 14, 15, 16, 18) were located in the corresponding chromosomes of P. trichocarpa (Fig 4). For the other linkage groups, except for DLG 20 and SLG 20, only a few (1–4) SNPs were identified in discordant reference chromosomes. However, synteny between DLG 20 (or SLG 20) and the physical map was inconclusive because all the 10 (9) SNPs were found in 7 (6) different reference chromosomes. Of all the 1,601 SNPs on the female genetic map of P. deltoides, 98.3% had a one-to-one relationship between the genetic linkage groups and the reference chromosomes, indicating highly conserved synteny between P. deltoides and P. trichocarpa. A similar high level of synteny between P. simonii and P. trichocarpa was also retained because 97.2% SNP markers on the male genetic map had a one-to-one relationship with the physical map.
Fig 4. Oxford grid comparison of the genetic and physical maps.
Each number in a cell denotes the number of homologous pair of SNP markers in each linkage group or genome. The last columns correspond to all the additional scaffolds of P. trichocarpa.
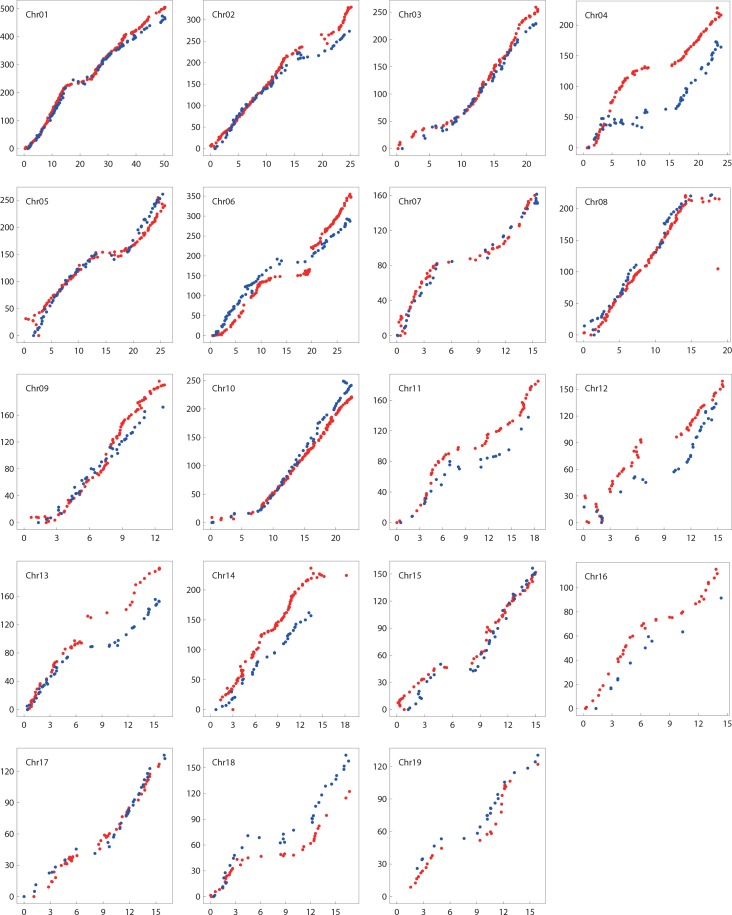
Further comparison of the linkage groups with the physical map revealed extensive conserved marker order except for a few of inversions in each chromosome (Fig 5). All the 19 syntenic pairs of linkage groups and chromosomes showed apparent collinearity between the female parent P. deltoides and P. trichocarpa, and between the male parent P. simonii and P. trichocarpa. However, there existed some linkage groups exhibiting rearrangements in local regions. For DLGs 2, 4, 8, 14, 15 and SLGs 2, 4, 6, 10, 11, 12, 17, a few short dot lines in Fig 5 were perpendicular to the 45-degree line, suggesting inverse orders of some SNPs relative to the reference genome positions.
Fig 5. Collinear comparison of the genetic and physical maps.
The x-axis indicates the reference sequence position with the unit of Mbp; the y-axis indicates the genetic map position with the unit of cM. The red and blue points, respectively, indicate the SNP position on the female and male genetic maps against the reference genome position.
Discussion
Here, we reported the high-quality SNP linkage maps of P. deltoides and P. simonii with the most recent RAD sequencing technology. The two parent-specific linkage maps will serve as important genetic resources for identifying QTLs that contribute to growth and timber traits as well as resistance with different QTL mapping approaches implemented in such an F1 hybrid population of P. deltoides and P. simonii [46,47]. The two linkage maps will also allow for comparative genomics among different species of Populus, and provide additional information on evolution in poplar. The less number of SNPs on the male linkage map suggests that the male parent P. simonii ‘L-3’ is less heterozygous than the female parent P. deltoides ‘I-69’. Each parental linkage map with a total of 20 linkage groups closely matches the karyotype of Populus (2n = 38). The last group DLG20 of the female map would be merged into group DLG4 with a large interval distance of 71.4 cM if the LOD threshold for linkage grouping decreased to a lower value of 8.0. Similarly, group SLD20 of the male map would be incorporated into group SLD8 with a large interval distance of 39.8 cM if the LOD threshold were chosen as 6.0. These suggested that the last small linkage group of each parental map might collapse with another linkage group when additional markers are provided. Although a few SNP markers come from other chromosomes and there exist a few apparent local discrepancies in marker order on some linkage groups (Figs 4 and 5), there are no apparent rearrangements and thus highly conserved synteny and collinearity can be inferred between the parental maps and the reference genome. In addition, on the parental maps there are 14 SNPs from the additional scaffolds of the reference genome, which could link unmapped scaffolds 47, 36, 34, 41, and 20 to chromosomes 1, 2, 13, 17, and 19 in poplar, respectively (Fig 4, S7 Table). Apparently, the result of synteny and collinearity was based on SNPs from conserved genome regions of different Populus species. However, there would likely be much less synteny and collinearity if divergent regions of the genomes were considered.
It is desirable to construct an integrated linkage map in an F1 population generated by hybridizing two individual trees [11, 21]. However, like most linkage mapping studies in forest trees [7, 12, 17], we had to build two linkage maps each specially for one parent since there were no enough markers heterozygous in both parents with segregation type abab or abcd as bridges, and any two markers one with segregation type abaa and the other aaab cannot provide any linkage information. The main reason why the overwhelming number of SNPs were not selected in our linkage analysis is that many individuals were not genotyped at most SNPs owning to low or no read coverage (S5 Fig). Another reason is that there were a lot of distorted SNPs not following Mendelian segregation ratio (p < 0.05), which accounted for 41.3% of the SNPs with segregation type abaa and 47.3% aaab that each has at least 50 individuals genotyped (S6 Fig). Segregation distortion was previously thought to be attributed to factors such as chromosome loss, genetic isolating mechanisms, viability genes and even genotyping errors [48], but one recent NGS study revealed that gene conversion events were unexpectedly abundant during meiosis in Arabidopsis [49], which skew segregation rates of alleles and were typically ignored in linkage mapping. Of all the SNPs genotyped in both parents, 77.9% (300,107) were homozygous and could not segregate in the progeny (Table 2), which suggested that a considerable number of SNP markers would segregate in a ratio of 1:2:1 in an F2 hybrid population.
The two parental maps may be in an excess of genetic map size compared with most previous poplar maps. Since obtaining the true order of a large number of markers in a linkage group is challenging, we chose the optimal orders based on SARF among several ordering results from JoinMap and FsLinkageMap (see Materials and Methods). We used the ML method in JoinMap because it can build a more accurate genetic map in a full-sib family of an outbreeding species than the previous regression method [50]. The genetic distances between adjacent markers were directly calculated from the results of two-point linkage analysis. This could expand the size of linkage maps because the double recombinants were not considered properly, but the more accurate marker orders and genetic distances between adjacent markers are most valuable in comparative genomics and in identifying QTLs [44,51]. The total sizes of our two linkage maps matched the results with the ML mapping algorithm in JoinMap 4.1, but disagreed substantially with the results from the regression algorithm in the same software (S8 Table). Interestingly, the results of the regression algorithm almost matched the estimates using the method of Hulbert et al. [52], which resulted in 2,085.83 cM for the female map and 2,434.59 cM for the male map. Investigations into the previous studies of genetic mapping in poplar showed that the observed genome length ranged from 1,600 to 3,800 cM [16, 53]. These discrepancies between different studies or between different algorithms even with the same software and data may reflect the difficulty of obtaining a perfect linkage map with high-density and high-quality in Populus.
Conclusions
We have constructed high-density genetic linkage maps of the maternal Populus deltoides ‘I-69’ and paternal P. simonii ‘L3’ in the F1 hybrid population. We demonstrate that RAD sequencing technology is capable of generating a large number of SNPs in a fast and cost-effective way for constructing genetic maps in outbred forest trees. Further analysis reveals that the synteny and collinearity are highly conserved between the parental linkage maps and the reference genome of P. trichocarpa. The linkage maps provide useful genetic resources for detecting QTLs that control growth and timber traits, especially that underlie the developmental trajectories in the permanent Populus population.
Supporting Information
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The median and 75% quantile are 54 and 135 for abaa, and 54 and 134 for aaab.
(PDF)
Those SNPs with less than 50 individuals genotyped were excluded. The Medians are 0.1317 and 0.0699 for abaa and aaab. The p-value of 0.05 corresponds to 41.3% quantile for abaa and 47.3% for aaab.
(PDF)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Huzhi Xu in Luoning Forest Bureau of Henan Provicne, China, and Jiangtao Zhang in Forest Science Research Institute of Henan Province, China, for collecting flowering branches of P. simonii. We also thank Xiangjin Yan in Siyang Agroforestry Center of Jiangsu Province, China, and Jinhai Yang in Nanjing Qiaolin Forestry Science and Technology Co. Ltd., Nanjing, China, for their great help in our crossing experiments and seedling cultivation.
Data Availability
The RADSeq raw data is available under accession number SRP052929 at the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra). Other relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding for this research was provided by the National Natural Science Foundation of China (No. 31270706) and the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bradshaw HD, Ceulemans R, Davis J, Stettler R. Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J Plant Growth Regul. 2000;19:306–313. [Google Scholar]
- 2.Taylor G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann Bot. 2002; 90: 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006; 313:1596–1604. [DOI] [PubMed] [Google Scholar]
- 4.Ma T, Wang J, Zhou G, Yue Z, Hu Q, Chen Y, et al. Genomic insights into salt adaptation in a desert poplar. Nature Communications. 2013; 4:2797 10.1038/ncomms3797 [DOI] [PubMed] [Google Scholar]
- 5.Eckenwalder JE. Systematics and evolution of Populus In: Stettler RF, Bradshaw HD, Heilman PE, Hinckley TM, editors. Biology of Populus and its implications for management and conservation. Ottawa: NRC Research Press, National Council of Canada; 1996. pp. 7–32. [Google Scholar]
- 6.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. [DOI] [PubMed] [Google Scholar]
- 7.Grattapaglia D, Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137:1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradshaw HD, Villar M, Watson BD, Otto KG, Stewart S, Stettler RF. Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor Appl Genet. 1994;89:167–178. 10.1007/BF00225137 [DOI] [PubMed] [Google Scholar]
- 9.Su XH, Zhang QW, Zheng XW, Zhang XH, Harrish S. Construction of Populus deltoides Marsh × P. cathayana Rehd. molecular linkage map. Science Silvae Sinicae. 1998; 34(6):29–37. [Google Scholar]
- 10.Frewen BE, Chen TH, Howe GT, Davis J, Rohde A, Boerjan W, et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics. 2000;154(2):837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu RL, Han YF, Hu JJ, Fang JJ, Li L, Li ML, et al. An integrated genetic map of Populus deltoides based on amplified fragment length polymorphisms. Theor Appl Genet. 2000;100:1249–1256. [Google Scholar]
- 12.Yin T, Huang M, Wang M, Zhu LH, Zeng ZB, Wu RL. Preliminary interspecific genetic maps of the Populus genome constructed from RAPD markers. Genome. 2001; 44:602–609. [PubMed] [Google Scholar]
- 13.Yin T, Zhang X, Huang M, Wang M, Zhuge Q, Tu S, et al. Molecular linkage maps of the Populus genome. Genome. 2002; 45:541–555. [DOI] [PubMed] [Google Scholar]
- 14.Yin T, DiFazio SP, Gunter LE, Riemenschneider D, Tuskan GA. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor Appl Genet. 2004;109:451–463. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D, Zhang Z, Yang K, Li B. Genetic mapping in (Populus tomentosa × Populus bolleana) and P. tomentosa Carr. using AFLP markers. Theor Appl Genet. 2004;108:657–662. [DOI] [PubMed] [Google Scholar]
- 16.Woolbright SA, DiFazio S, Yin T, Martinsen GD, Zhang X, Allan GJ, et al. A dense linkage map of hybrid cottonwood (Populus fremontii × P. angustifolia) contributes to long-term ecological research and comparison mapping in a model forest tree. Heredity. 2008;100:59–70. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Tong CF, Yin T, Zhang X, Zhuge Q, Huang M, et al. Detection of quantitative trait loci influencing growth trajectories of adventitious roots in Populus using functional mapping. Tree Genet Genomes. 2009;5:539–552. [Google Scholar]
- 18.Paolucci I, Gaudet M, Jorge V, Beritognolo I, Terzoli S, Kuzminsky E, et al. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet Genomes. 2010; 6: 863–875. [Google Scholar]
- 19.Wang Y, Zhang B, Sun X, Tan B, Xu L, Huang M, et al. Comparative genome mapping among Populus adenopoda, P. alba, P. deltoides, P. euramericana and P. trichocarpa. Genes Genet Syst. 2011;86:257–268. [DOI] [PubMed] [Google Scholar]
- 20.Van Ooijen JW. JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations Kyazma B V, Wageningen, Netherlands: 2006. Available: https://www.kyazma.nl/index.php/mc.JoinMap. [Google Scholar]
- 21.Yin T, DiFazio S, Gunter LE, Zhang X, Sewell MM, Woolbright SA, et al. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 2008;18:422–430. 10.1101/gr.7076308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pakull B, Groppe K, Meyer M, Markussen T, Fladung M. Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet Genomes. 2009;5:505–515. [Google Scholar]
- 23.Pakull B, Groppe K, Mecucci F, Gaudet M, Sabatti M, Fladung M. Genetic mapping of linkage group XIX and identification of sex-linked SSR markers in a Populus tremula × Populus tremuloides cross. Can J For Res. 2011;41: 245–253. [Google Scholar]
- 24.Wang Y, Sun X, Tan B, Zhang B, Xu L, Huang M, et al. A genetic linkage map of Populus adenopoda Maxim × P. alba L. hybrid based on SSR and SRAP markers. Euphytica. 2010;173:193–205. [Google Scholar]
- 25.Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, Rodgers-Melnick E, et al. Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytologist. 2012;196:713–725. 10.1111/j.1469-8137.2012.04258.x [DOI] [PubMed] [Google Scholar]
- 26.Muchero W, Guo J, DiFazio SP, J-G C, Ranjan P, Slavov GT, et al. High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC genomics. 2015;16:24 10.1186/s12864-015-1215-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davey JW, Blaxter ML. RADSeq: next-generation population genetics. Briefings in Functional Genomics. 2011; 9(5): 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerson KJ, Merz CR, Catchen JM, Hohenlohe PA, Cresko WA, Bradshaw WE, et al. Resolving postglacial phylogeography using high-throughput sequencing. Proc Natl Acad Sci USA. 2010;107:16196–16200. 10.1073/pnas.1006538107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6: e1000862 10.1371/journal.pgen.1000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfender WF, Saha MC, Johnson EA, Slabaugh MB. Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor Appl Genet. 2011;122: 1467–1480. 10.1007/s00122-011-1546-3 [DOI] [PubMed] [Google Scholar]
- 32.Chutimanitsakun Y, Nipper RW, Guesta-Marcos A, Cistue L, Corey A, Filichkina T, et al. Construction and application for QTL analysis of a restriction site associated DNA (RAD) linkage map in barley. BMC Genomics. 2011;12:4 10.1186/1471-2164-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baxter SW, Davey JW, Johnston JS, Shelton AM, Heckel DG, Jiggins CD, et al. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS ONE. 2011;6(4):e19315 10.1371/journal.pone.0019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N, Fang L, Xin H, Wang L, Li S. Construction of a high-density genetic map for grape using next generation restriction-site associated DNA sequencing. BMC Plant Biology. 2012; 12:148 10.1186/1471-2229-12-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakioka R, Kokita T, Kumada H, Watanabe K, Okuda N. A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae). BMC Genomics. 2013;14:32 10.1186/1471-2164-14-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Z, Zhao X, Pan W, Zhang J, Li B, Zhang D. Phenotypic variation among five provenances of Populus simonii in northern China. For Stud China. 2011;13(2):97–103. [Google Scholar]
- 37.Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytologist Bulletin. 1987;19:11–15. [Google Scholar]
- 38.Patel RK, Jain M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong CF, Zhang B, Shi JS. A hidden Markov model approach to multilocus linkage analysis in a full-sib family. Tree Genet Genomes. 2010; 6(5): 651–662. [Google Scholar]
- 43.Falk CT. A simple scheme for preliminary ordering of multiple loci: application to 45 CF families. In: Elston RC, Spence MA, Hodge SE, MacCluer JW, editors. Multipoint mapping and linkage based upon affected pedigree members, Genetic Workshop 6. Liss, New York; 1989. pp. 17–22.
- 44.Zeng Z-B. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996; 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong CF, Zhang B, Wang Z, Xu M, Pang XM, Si JN, et al. Multiallelic epistatic model for an outbred cross and mapping algorithm of interactive quantitative trait loci. BMC Plant Biology. 2011;11:148 10.1186/1471-2229-11-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong CF, Zhang B, Li HG, Shi JS. Model selection for quantitative trait loci mapping in a full-sib family. Genetics and Molecular Biology. 2012;35(3):622–631. 10.1590/S1415-47572012005000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuang H, Richardson T, Carson S, Wilcox P, Bongarten B. Genetic analysis of inbreeding depression in plus tree 850.55 of Pinus radiata D. Don. I. Genetic map with distorted markers. Theor Appl Genet. 1999;98:697–703. [Google Scholar]
- 49.Yang S, Yuan Y, Wang L, Li J, Wang W, Liu H, et al. Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci USA. 2012; 109(51): 20992–20997. 10.1073/pnas.1211827110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Ooijen JW. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genetics Research. 2011;93(5):343–349. 10.1017/S0016672311000279 [DOI] [PubMed] [Google Scholar]
- 51.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hulbert SH, Ilott TW, Legg EJ, Lincoln SE, Lander ES, Michelmore RW. Genetic analysis of the fungus, Bremia lactucae, using restriction fragment length polymorphisms. Genetics. 1988; 120:947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cervera M, Storme V, Ivens B, Gusmao J, Liu BH, Hostyn V, et al. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The threshold value for asserting the existence of a QTL at the significant level p = 0.05 is indicated as horizontal dashed lines, which was determined by 1000 permutation tests.
(PDF)
The median and 75% quantile are 54 and 135 for abaa, and 54 and 134 for aaab.
(PDF)
Those SNPs with less than 50 individuals genotyped were excluded. The Medians are 0.1317 and 0.0699 for abaa and aaab. The p-value of 0.05 corresponds to 41.3% quantile for abaa and 47.3% for aaab.
(PDF)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The RADSeq raw data is available under accession number SRP052929 at the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra). Other relevant data are within the paper and its Supporting Information files.