Abstract
Sorghum is an important C4 grain and grass crop used for food, feed, forage, sugar, and biofuels. In its native Africa, sorghum landraces often grow to approximately 3–4 meters in height. Following introduction into the U.S., shorter, early flowering varieties were identified and used for production of grain. Quinby and Karper identified allelic variation at four loci designated Dw1-Dw4 that regulated plant height by altering the length of stem internodes. The current study used a map-based cloning strategy to identify the gene corresponding to Dw1. Hegari (Dw1dw2Dw3dw4) and 80M (dw1dw2Dw3dw4) were crossed and F2 and HIF derived populations used for QTL mapping. Genetic analysis identified four QTL for internode length in this population, Dw1 on SBI-09, Dw2 on SBI-06, and QTL located on SBI-01 and SBI-07. The QTL on SBI-07 was ~3 Mbp upstream of Dw3 and interacted with Dw1. Dw1 was also found to contribute to the variation in stem weight in the population. Dw1 was fine mapped to an interval of ~33 kbp using HIFs segregating only for Dw1. A polymorphism in an exon of Sobic.009G229800 created a stop codon that truncated the encoded protein in 80M (dw1). This polymorphism was not present in Hegari (Dw1) and no other polymorphisms in the delimited Dw1 locus altered coding regions. The recessive dw1 allele found in 80M was traced to Dwarf Yellow Milo, the progenitor of grain sorghum genotypes identified as dw1. Dw1 encodes a putative membrane protein of unknown function that is highly conserved in plants.
Introduction
Sorghum is the fifth most widely cultivated cereal crop worldwide. This C4 grass is grown for grain, feed, forage, sugar, and biofuels. Sorghum diverged from a common ancestor with maize ~12 MYA and rice ~50 MYA [1]. It is native to Africa and parts of India and Australia with most African landraces growing to 3–4 meters in height before harvest. When grown in the U.S., many sorghum accessions from Africa produce tall, late flowering plants. However, after its initial introduction to the U.S., breeders found naturally occurring shorter genotypes that were subsequently used to breed short grain sorghum varieties to reduce stalk lodging. Sorghum genotypes with longer stems are grown for forage, sugar, and biomass to increase yield. Energy sorghum hybrids are 3–4 meters in height with long internodes and biomass yield ranging from 15–40 Mg/ha depending on genotype and environment [2–4]. Stem biomass of a first generation energy sorghum hybrid accounted for ~80% of harvested shoot biomass [5]. Therefore, a more complete understanding of the genetic and biochemical basis of stem growth could identify ways to increase the stem biomass yield of bioenergy sorghum.
Plant height is affected by the length of each internode, the rate of internode production, and the duration of vegetative growth. The latter influences height because production of internodes stops at floral induction even though internode elongation continues until anthesis. In the 1950s, Quinby and Karper [6] identified four loci, Dw1-Dw4, that control height by modifying internode length. Recessive alleles at the four loci reduce internode length [6]. Pleiotropic effects of Dw2 and Dw3 have been reported and include panicle length, seed weight, and leaf area for the former [7,8] and seed weight, panicle size, tiller number, and leaf angle for the latter [8–10]. However, pleiotropic effects have not been described for Dw1 or Dw4. Additionally, QTL for height, including Dw3 and a QTL on chromosome 9, have been found to co-localize with QTL for stem and total biomass [11].
The gene corresponding to Dw3 was cloned by Multani et al. [12] and determined to encode an ABCB1 auxin efflux transporter. Further analysis showed that the maize homolog, br2, transports auxin from intercalary meristems located at the base of a stem internode into the elongating internode [13]. QTL corresponding to Dw1 and Dw2 have been identified, but the underlying genes are unknown. Dw1 was mapped to the distal end of SBI-09 [14] and Dw2 to SBI-06 adjacent to Ma1 [15]. Recently, a QTL for stem length was identified on SBI-07 located near Dw3 in a RIL population from a cross of Tx430 and P898012 [16].
The Green Revolution dwarfing genes in rice and wheat reduce gibberellin induced stem elongation producing semi-dwarf varieties with reduced lodging. In rice, semi-dwarf genotypes were found to encode a less active version of gibberellin 20 oxidase, an enzyme involved in GA synthesis [17]. In wheat, dwarf varieties contain alleles of a gene encoding a DELLA protein that is involved in gibberellin (GA) signaling [18]. Because of this, several researchers have suggested that Dw1 encodes a gibberellin 2 oxidase that is located in the genomic region near SNPs associated with this height locus on SBI-09 [19–21]. However, recent work showed that gibberellin mutants in sorghum have bent stems, which are not observed in genotypes recessive for the sorghum dwarfing genes. Furthermore, there were no sequence variants in the GA2 oxidase coding region located on SBI-09 near Dw1 between genotypes that were Dw1 and dw1 [22].
In this study, the gene corresponding to Dw1 was map-based cloned using an F2 population and HIFs derived from Hegari and 80M. Dw1 encodes a protein of unknown function that is highly conserved in plants. In the process of identifying Dw1, a QTL that modulates internode length was identified on SBI-01 and a QTL on SBI-07 corresponding to one recently identified by Li et al. [16] was found to interact with Dw1.
Methods
QTL Mapping of Stem Traits in Hegari x 80M
A map-based cloning approach was used to identify the gene corresponding to Dw1. A population segregating for Dw1 was constructed by crossing Hegari (Dw1dw2Dw3dw4) and 80M (dw1dw2Dw3dw4) [23]. The F1 plants were selfed and the F2 population (n = 218) was planted in April 2011 and grown in a greenhouse in long days (14 hours light, 10 hours dark), three plants per 3.8 gallon pot in soil that was a mixture of vermiculite (Sun Gro Horticulture) and Belk Clay soil (2:1) obtained from the Texas A&M University Field Station west of College Station, Texas. Osmocote Classic 13-13-13 (Scotts) was mixed into the soil and plants were subsequently fertilized every two weeks with Peters General Purpose 20-20-20 (JR Peters, Inc.). Plants were phenotyped for days to flowering, total stem fresh and dry weight, total stem length, and length and diameter of each internode at grain maturity for early flowering plants and after 190 days of growth for late flowering genotypes. DNA was extracted from leaf tissue using the FastDNA Spin Kit (MP Biomedicals). Each plant was genotyped using Digital Genotyping [24], using the enzyme FseI for digesting the genomic DNA. The Illumina GAII was used for sequencing and the reads were mapped onto the Sorghum bicolor genome v1.0 (Phytozome v6).
A genetic map for this population was constructed using MapMaker [25], with the Kosambi function. QTL analysis was performed in QTL Cartographer [26] using Composite Interval Mapping with a walk speed of 1.0cM and forward and backward model selection. The threshold was set using 1000 permutations and α = 0.05. QTL mapping was performed with the entire population, early flowering plants only (n = 85), and late flowering plants only (n = 118). To look for possible gene interactions multiple-QTL analysis was used. A single QTL analysis using the EM algorithm initially identified four primary additive QTL which were used to seed model selection. The method of Manichaikul et al. [27] was employed for model selection as implemented in R/qtl [28] for multiple-QTL analysis. Computational resources on the WSGI cluster at Texas A&M were used to calculate the penalties for main effects, heavy interactions, and light interactions. These penalties were calculated from 24,000 permutations for the average internode length to find a significance level of 5% in the context of a two-dimensional, two-genome scan.
Fine Mapping of Dw1
To refine the location of Dw1, plants were selected from early flowering lines that were segregating for Dw1, but fixed for the other loci controlling internode length. These plants (n = 6) were selfed to create Heterogeneous Inbred Families (HIFs) [29]. For each family, the F3 plants (n = 75 for each HIF) were planted in December 2011 and grown in the greenhouse as with the F2 population, phenotyped as described above, and genotyped using Digital Genotyping. The phenotypes were used to classify plants as dominant, heterozygous, or recessive at Dw1. The phenotype data were then correlated with genotype data spanning Dw1. The region encoding Dw1 was further refined using F4 HIFs derived from F3 plants that were heterozygous at Dw1. The plants were planted in June 2013 and grown in the greenhouse as with the previous generations, except in Sunshine MVP soil (Sun Gro Horticulture). At grain maturity the plants were phenotyped for stem and internode length (n = 78 for each HIF). The population was screened for individuals with breakpoints in the delimited Dw1 region using two CAPS (Cleaved Amplified Polymorphic Sequence) markers, except for Family 2 which was genotyped using Digital Genotyping because one of the CAPS markers was fixed in that family. The CAPS markers are described in S1 Table. Restriction enzyme digests were performed using the manufacturer’s recommended temperature for each enzyme (New England Biolabs) and incubations of at least 2 hours. All PCR amplification was done with Phusion (New England Biolabs). The breakpoints were refined using SNPs that were genotyped through Sanger sequencing using Big-Dye Terminator cycle sequencing kit v3.1 (Invitrogen) (S1 Table).
Sequencing of Candidate Genes
All of the genes in the region encoding Dw1 delimited by fine mapping were sequenced in the parental genotypes used for Dw1 mapping as well as Standard Yellow Milo (Dw1Dw2Dw3dw4) and Dwarf Yellow Milo (dw1Dw2Dw3dw4) by Sanger sequencing. The yellow milos are nearly isogenic except at Dw1. The primers used to amplify and sequence genes in the delimited Dw1 region are listed in S2 Table. A polymorphism in Sobic.009G229800 that distinguished 80M and Hegari created a stop codon and truncated protein in 80M (dw1).
cDNA Sequencing and qRT-PCR
RNA was collected from stem tissue for cDNA sequencing and to characterize the expression of Dw1 (Sobic.009G229800). The two parents (n = 3 for each) were planted in the greenhouse in August 2013, and after 42 days of growth, stem tissue was collected from plants in the mid-morning. Plants were cut at soil level and leaves and leaf sheaths were quickly stripped from the stem. Internodes that were in the process of elongating were located and divided into an upper portion of the internode that had stopped elongating, a mid-lower region containing cells that are in the process of elongation, and the base of the internode containing the intercalary meristem. A fully expanded internode was also harvested. The tissue was ground in liquid nitrogen and the RNA extracted using a Direct-zol RNA kit (Zymo Research) with TRI-Reagent (Molecular Research Center). The RNA was quantified on the Nanodrop spectrophotometer. RNA quality was confirmed by visualizing final samples with the BioAnalyzer (Agilent Technologies). Two technical replicates of cDNA and a no reverse transcriptase control were made using SuperScript III primed with both random hexamers and oligo (dT) at a ratio of 9:1 from 1μg of RNA.
Sobic.009G229800 cDNA from elongating stem tissue from each parental genotype was Sanger sequenced. The primers used to sequence the cDNA are listed in S3 Table. Gene expression was analyzed using qRT-PCR on the 7900HT Fast Real-Time PCR System (Applied Biosystems) running SDS v2.3 software. Dw1 was amplified in the presence of SYBR green using the following conditions: hold at 95°C for 10 mins, 40 cycles of 95°C 15 sec. and 60°C for 1 min. Primer efficiencies were determined based on a standard curve from a serial dilution of five 10-fold dilutions of PCR product for each parent. Primer specificity was checked using a dissociation curve and running PCR products on a gel. The primers used for Dw1 amplification were: 5’-TACGCTAAAGATGGCACAAGTC-3’ and 5’-TCCTTTGAACACGTCCAAGC-3’. The data was analyzed according to the comparative Ct (ΔΔCt) method [30] using the 18S ribosomal RNA to normalize the expression values and the sample from the 80M mature tissue as the calibrator. 18S ribosomal RNA reactions were performed with the TaqMan rRNA primers and probe (Applied Biosystems) and TaqMan MasterMix. Three technical replicates of qPCR were performed for each sample. The three biological replicates were averaged and the standard error of the mean calculated.
Protein Sequence Analysis
To gain insight into the function of Dw1, the protein sequence translated from the Hegari cDNA sequence was compared to other plants, using BLAST in Phytozome v.10 and to the NCBI database using NCBI BLAST. A sequence comparison of the protein’s homologs in maize, rice, and Arabidopsis was generated in Jalview [31] using T-Coffee [32] with default settings. A phylogenetic tree of several protein homologs was constructed with MEGA6 [33] using MUSCLE [34,35] to align the sequences and Maximum Likelihood to construct the tree. Protein function and structure was examined using several web-based programs: PSIPRED-MEMSAT-SVM [36,37], PSIPRED-DISOPRED[38], PONDR [39], and FoldIndex [40] using default settings for each program.
Results
QTL Mapping of Stem Traits
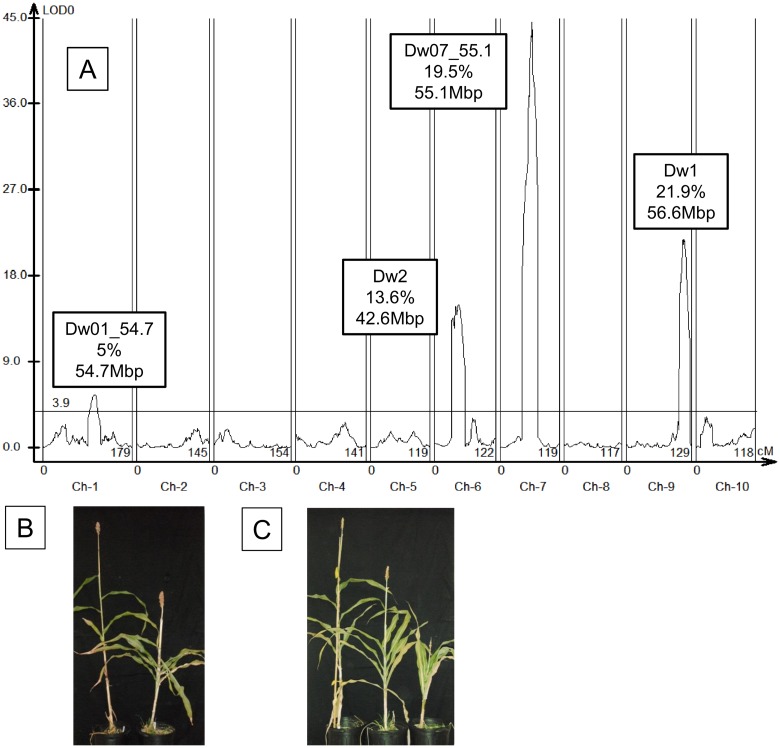
An F2 population for mapping Dw1 was constructed by crossing Hegari (Dw1) and 80M (dw1). The F2 population segregated for flowering time and height. The length of expanded internodes was measured for all plants in the population with the first expanded internode being labeled as number 5. Four QTL were identified that modulate the average length of internodes 5–10 (Fig 1, Table 1). A QTL corresponding to Dw1 was identified on SBI-09 with a peak at ~56.6 Mbp on Sorghum bicolor genome v2 (Phytozome v10). This QTL explained ~22% of the trait variance observed. The Dw1 allele in Hegari increased the lengths of all expanded internodes compared to plants containing the dw1 allele present in 80M (S1 Fig). A second QTL for internode length was located on SBI-06 at ~42.6 Mbp that aligned with Dw2 [15]. A previously reported QTL for internode length was identified on SBI-01 at ~54.7 Mbp (Dw01_54.7) that explained ~5% of the variance [41,42]. A QTL on SBI-07 at ~55.1 Mbp (Dw07_55.1) that was recently described by Li et al [16] explained 19% of the variance. The QTL on SBI-07 (Dw07_55.1) was 3 Mbp from the ABCB1 gene corresponding to Dw3 (58.6 Mbp). No QTL aligned with ABCB1 as expected because both parental genotypes are Dw3.
Fig 1. Stem internode length QTL identified in a population from Hegari x 80M.
F2 plants from a cross of Hegari and 80M (n = 218) were grown in the greenhouse and the length of each internode was measured. The average internode length was used to map QTL. (A) The resulting graph shows four QTL, including Dw1 and Dw2. The x-axis is the genetic map and the y-axis is the LOD score. The boxes above each trait identify the Dw loci, if any, the percentage of the variation explained by the QTL, and the location of the peak LOD value. (B) Photograph of Hegari (left) and 80M. (C) Photograph of F5 plants that are Dw1Dw1 (left), Dw1dw1 (center), and dw1dw1 (right) in otherwise uniform genetic backgrounds at the other loci that affect internode length.
Table 1. QTL for Average Internode Length Identified in the Entire Population of Hegari x 80M F2.
| QTL | Chr | Peak (cM) | Peak LOD | Peak (Mbp) | Additive* | Dominance* | R2 | Dw locus |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 104.2 | 5.53 | 54.7 | 12.5848 | -5.5165 | 0.0503 | Dw01_54.7 |
| 2 | 6 | 46.5 | 15 | 42.6 | -22.8162 | 4.1926 | 0.1358 | Dw2 |
| 3 | 7 | 62.4 | 44.37 | 55.1 | 39.2763 | 22.2605 | 0.1945 | Dw07_55.1 |
| 4 | 9 | 112.2 | 21.8 | 56.6 | -27.3763 | 6.4375 | 0.2186 | Dw1 |
* Positive means the allele from 80M increases length; negative is Hegari.
QTL mapping was also performed using data on fresh and dry weight per internode, fresh or dry weight per unit stem length, and diameter of internode 7 (S4 Table). Alleles of Dw1 contributed to variation for internode fresh weight and dry weight.
Analysis of Epistasis
Potential interactions among the four QTL modulating internode length were investigated using multiple-QTL mapping in R/qtl [27]. The best model (y ~ Dw01_54.7 + Dw2 + Dw07_55.1+ Dw1 + Dw10_3.2+ Dw07_55.1:Dw1) had a pLOD of 50.1 and included five QTL and an interaction between two of the QTL (Dw1 and Dw07_55.1, S5 Table). The analysis showed an interaction between Dw1 and Dw07_55.1 such that allelic variation in Dw1 has minimal impact on internode length in the presence of the 80M allele at Dw07_55.1 which increased internode length (S2 Fig). In addition, the 80M allele of Dw07_55.1 increased internode length in Dw1Dw1, Dw1dw1, and dw1dw1 backgrounds, although to a greater extent in genotypes that were dw1dw1. These results indicate that Dw1 and Dw07_55.1 independently activate the same downstream regulator of internode elongation, or act through different pathways to stimulate internode growth.
Fine Mapping Dw1
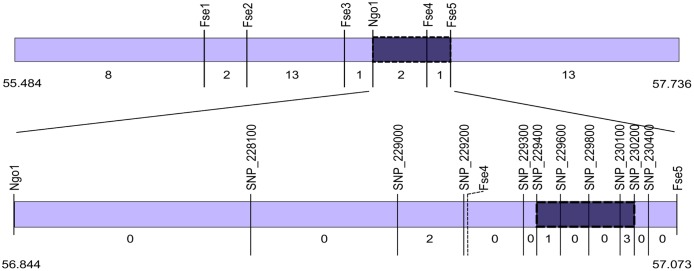
Dw1 was fine mapped by constructing HIFs from seed of F2 plants of the QTL mapping population that were heterozygous for Dw1 and homozygous at the other QTL that affect internode length. HIFs derived from F2 plants homozygous for the Hegari allele at Dw07_55.1 were most useful for fine mapping Dw1. F3 individuals from each HIF family were grown in the greenhouse until grain maturity, and then stems were phenotyped for internode length. Histograms of the average internode length for each HIF are shown in S3 Fig. Breakpoint analysis of the first set of HIFs narrowed the region encoding Dw1 to 313kb. The location of breakpoints in a few key lines was further refined using Digital Genotyping based on the restriction enzyme NgoMIV [24]. This information delimited the Dw1 locus to 230kb, a region encoding 35 genes as annotated in the v1.4 gene set (Phytozome v.9). A further round of fine mapping was carried out using five HIFs derived from F3 plants heterozygous for Dw1dw1. These plants were screened for recombinants with CAPS markers and six plants were identified with recombination breakpoints in the delimited Dw1 region. Phenotyping and identification of breakpoints by sequencing SNPs delimited Dw1 to a region that spanned 33kb and encoded seven genes as annotated in v2.1 (Phytozome v.10) (Table 2). Markers used for fine mapping and the location of the delimited Dw1 locus are shown in Fig 2. Information about the seven putative genes in the delimited Dw1 locus is provided in Table 2. Four of the genes were annotated with a function: an E3-ubiquitin ligase involved in syntaxin degradation, Photosystem I reaction center subunit VI, PRONE-Rop nucleoide exchanger, and a serine/threonine kinase. There were also three genes annotated as having unknown functions.
Table 2. Genes in the Delimited Dw1 Locus.
| Gene Name | Probable Function | Location |
|---|---|---|
| Sobic.009G229500 | Unknown | 57,026,900–57,027,289 |
| Sobic.009G229600 | E3 ubiquitin ligase/syntaxin degradation | 57,027,335–57,036,566 |
| Sobic.009G229700 | Photosystem I reaction center, subunit VI | 57,036,793–57,037,995 |
| Sobic.009G229800 | Unknown | 57,042,620–57,045,133 |
| Sobic.009G229900 | PRONE-Rop nucleotide (guanine) exchanger | 57,046,394–57,049,526 |
| Sobic.009G230000 | Unknown | 57,050,065–57,051,463 |
| Sobic.009G230100 | Serine/threonine kinase | 57,051,814–57,055,008 |
Fig 2. A schematic of the region of SBI-09 encoding Dw1.
The top bar shows the Dw1 locus delimited by QTL mapping in the F2. The region was refined in the F3 population (n = 75 for each of six families) using the DG markers labeled in the diagram. The numbers below the bar are the number of recombinants (both bars). Note that all members of one of the families (237) had a breakpoint in between Fse5 and the end of the region shown. The lower bar represents the delimited Dw1 locus defined by mapping in the F3 generation with SNP markers labeled. Dark purple shows the location of Dw1 based on fine mapping. SNP markers are named with the last six digits of the gene name of the gene the SNP is in or near. Fse4 is included for perspective though it was not scored in the F4.
Identification of Polymorphisms in the Delimited Dw1 locus
All seven genes located in the fine mapped Dw1 locus were sequenced in Hegari and 80M (Table 3). No sequence variants were found in Sobic.009G229700 or Sobic.009G229900. The only sequence variants in Sobic.009G229600 and Sobic.009G230100 were located in introns and/or the 5’UTR. Of the genes annotated with an unknown function, Sobic.009G229500 had no sequence variants while Sobic.009G230000 had two INDELs in the 5’UTR and a SNP in the first exon that resulted in a synonymous mutation. Sobic.009G229800 was the only gene in the delimited Dw1 locus that had a polymorphism distinguishing the parental genotypes that resulted in a change in amino acid sequence (Table 3). Hegari (Dw1) encoded a full-length protein, whereas the sequence in 80M (dw1) (and BTx623 (dw1)) contained an A > T mutation that caused a Lys199 > stop codon change in the second exon of Sobic.009G229800 (Fig 3B).
Table 3. Polymorphisms Distinguishing 80M and Hegari in Genes in the Delimited Dw1 Locus.
| Gene | Number | Type | Polymorphism | Location | Region |
|---|---|---|---|---|---|
| Sobic.009G229500 | None | ||||
| Sobic.009G229600 | 1 | SNP | C > T | 2660 | Intron |
| 2 | INDEL | - > A | 6597 | Intron | |
| Sobic.009G229700 | None | ||||
| Sobic.009G229800 | 1 | INDEL | A > - | -707 | 5' UTR |
| 2 | SNP | A > T; K > Stop | 1350 | Exon | |
| Sobic.009G229900 | None | ||||
| Sobic.009G230000 | 1 | INDEL | - > CAGGCAGG | -64 | 5'UTR |
| 2 | INDEL | - > ACGACG | -25 | 5'UTR | |
| 3 | SNP | G > T; L > L | 126 | Exon | |
| Sobic.009G230100 | 1 | INDEL | T > - | -397 | 5' UTR |
| 2 | SNP | A > T | 537 | Intron | |
| 3 | INDEL | A > - | 1841 | Intron |
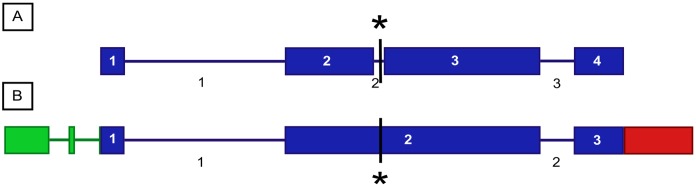
Fig 3. Gene annotation models of Dw1 (Sobic.009G229800).
(A) Gene model from Sorghum bicolor Genome v2.1 (Phytozome). (B) Gene model based on cDNA sequence analysis. Boxes (blue) represent exons and lines are introns. Regions colored green represent the 5’ UTR and those colored red the 3’ UTR. Exons are numbered within boxes and introns are numbered in black. The asterisk/vertical line marks the location of the Lys199 > stop codon mutation that distinguishes Dw1 from dw1.
All seven of the genes in the delimited region were also sequenced in Standard Yellow Milo (Dw1) and Dwarf Yellow Milo (dw1). Quinby [23] noted that dw1 was originally identified in the Standard Yellow Milo (Dw1, Dw2, Dw3) background [43]. The shorter version of Yellow Milo containing dw1 was named Dwarf Yellow Milo. Therefore, the sequences of Standard Yellow Milo and Dwarf Yellow Milo are expected to vary only at Dw1. Sequence analysis revealed only one polymorphism in the delimited Dw1 region that distinguished the two milo lines: the A > T SNP in Sobic.009G229800 that caused a premature stop codon. For all the other polymorphisms found between Hegari and 80M in the region, Standard Yellow Milo and Dwarf Yellow Milo had the same allele as 80M.
The gene-model for Sobic.009G229800 in v2.1 (Phytozome v10) included a very short intron (intron 2) (Fig 3A). However, cDNA sequence analysis of Sobic.009G229800, and RNA-seq analysis (see below), failed to provide evidence for intron 2. Instead, cDNA sequences from Hegari (Dw1) contain a continuous coding region that spanned intron 2 of the v2.1 gene-model. Gene-models of homologs of Sobic.009G229800 in other plant species (e.g. maize, rice, and Arabidopsis) also lack intron 2 and show continuous reading frames across this region. The cDNA sequence also clarified splicing in the 5’UTR (Fig 3, regions in green). Based on this analysis, we propose the revised annotation of Sobic.009G229800 shown in Fig 3B that contains three exons and conclude that the polymorphism that distinguishes Hegari and 80M generates a truncated protein lacking most of exon 2 and all of exon 3 (mutation marked by an asterisk in Fig 3) presumably resulting in a loss of function.
The intron/exon structures of the other genes in the delimited Dw1 locus were identical to homologs in maize and/or rice (S6 Table). Furthermore, the RNA-seq data for v3.1 (Phytozome v11) is consistent with the annotations of the other genes in the delimited Dw1 locus and the updated annotation of Sobic.009G229800 that lacks intron 2 (Fig 3B).
Sobic.009G229800 was sequenced in other genotypes of sorghum previously identified as Dw1 or dw1 (Tables 4 and 5; S7 Table). Genotypes previously designated as Dw1 encoded full-length proteins similar to Hegari. Numerous grain sorghum-breeding lines with shorter internodes were generated from the Dwarf Yellow Milo source of dw1. Therefore, it is not surprising that all of the lines designated dw1 have the same recessive allele as Dwarf Yellow Milo. Sobic.009G229800 sequences from Rio and Early White Milo (both Dw1) contain several additional polymorphisms (Table 5). SIFT [44] analysis of a non-synonymous coding mutation found in Rio and Early White Milo (A425S) predicted that this change in Dw1 would not affect function.
Table 4. Sequence Variants in Exons of Sobic.009G229800 in Diverse Sorghum Genotypes.
| Number | 2 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 22 |
|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | A > T | C > T | G > A | G > A | C > A | T > C | T > A | T > G | T > C |
| Location (bp)* | 1350 | 1127 | 1259 | 1583 | 1586 | 1667 | 1733 | 2028 | 2316 |
| Region | Exon 2 | Exon 2 | Exon 2 | Exon 2 | Exon 2 | Exon 2 | Exon 2 | Exon 2 | Exon 3 |
| Type | Syn | Syn | Syn | Syn | Syn | Syn | Nonsyn | Syn | |
| Change in Protein | K > Stop | F > F | P > P | S > S | P > P | T > T | P > P | S > A | N > N |
| SIFT | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 0.36 = tolerated | N/A |
*From the start codon
Table 5. Distribution of Dw1 Coding Sequence Variants in Sorghum Genotypes.
The polymorphism number corresponds to the number in Table 4.
| Line | Dw1 Genotype | Polymorphism Number | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 22 | ||
| Hegari | Dw1 | A | T | A | A | A | C | A | G | C |
| 80M | dw1 | T | T | A | A | A | C | A | G | C |
| Standard Yellow Milo | Dw1 | A | T | A | A | A | C | A | G | C |
| Dwarf Yellow Milo | dw1 | T | T | A | A | A | C | A | G | C |
| Double Dwarf Yellow Milo | dw1 | T | T | A | A | A | C | A | G | C |
| BTx623 | dw1 | T | T | A | A | A | C | A | G | C |
| BTx406 | dw1 | T | T | A | A | A | C | A | G | C |
| SC170 | dw1 | T | T | A | A | A | C | A | G | C |
| R.07007 | dw1 | T | T | A | A | A | C | A | G | C |
| IS3620c | dw1 | T | T | A | A | A | C | A | G | C |
| Rio | Dw1 | A | C | G | G | C | T | T | T | T |
| M35-1 | Dw1 | A | T | A | A | A | C | A | G | C |
| Texas Blackhull Kafir | Dw1 | A | T | A | A | A | C | A | G | C |
| Spur Feterita | Dw1 | A | T | A | A | A | C | A | G | C |
| Early White Milo | Dw1 | A | C | G | G | C | T | A | T | T |
Expression of Dw1 in Stem Tissue
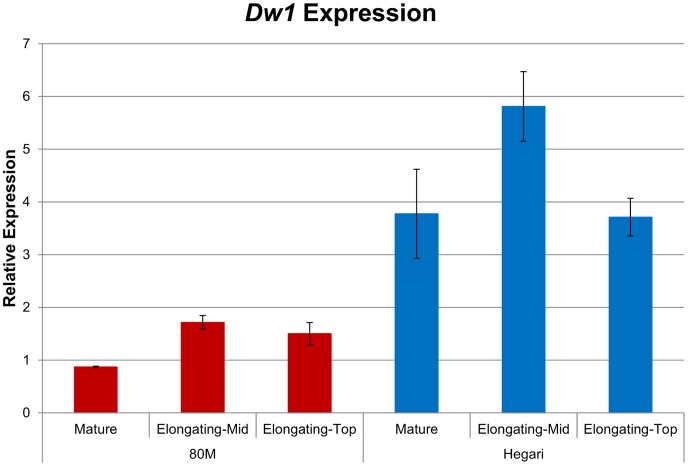
Sobic.009G229800 was expressed in fully elongated internodes and elongating internodes (Fig 4). The highest levels of expression were observed in the lower portion of the elongating internode. Dw1 mRNA levels were ~3-fold higher in stems of Hegari compared to 80M.
Fig 4. Relative expression of Dw1 in stem internodes.
RNA was extracted from a full length internode (Mature), the lower half of an elongating internode, and the upper half of an elongating internode for each parental genotype (n = 3 each). Relative expression was determined by qRT-PCR using the ΔΔCt method with 18S rRNA as the normalizer and the sample from 80M mature tissue as the calibrator.
Protein Sequence Analysis
Sobic.009G229800 is currently annotated as having an unknown function. BLAST analysis showed that homologous genes/proteins are present in maize, rice, and Arabidopsis among other plants (S8 and S9 Tables). Fig 5 shows the sequence alignment of Sobic.009G229800 and maize, rice, and Arabidopsis homologs. A phylogenetic tree of select homologs has two distinct groups corresponding to the monocots and dicots (S4 Fig). The Arabidopsis homolog of Dw1 is annotated as associated with the plasma membrane based on experimental evidence [45] and located in the nucleus based on prediction (TAIR). PSIPRED-MEMSAT-SVM predicts that the sorghum Dw1 protein contains a single transmembrane/pore-lining domain from residues 263–278. Interestingly, these residues are missing in the Arabidopsis homolog (Fig 5). PSIPRED-DISOPRED, PONDR, and FoldIndex all predicted a high degree of disorder in the protein (S10 Table).
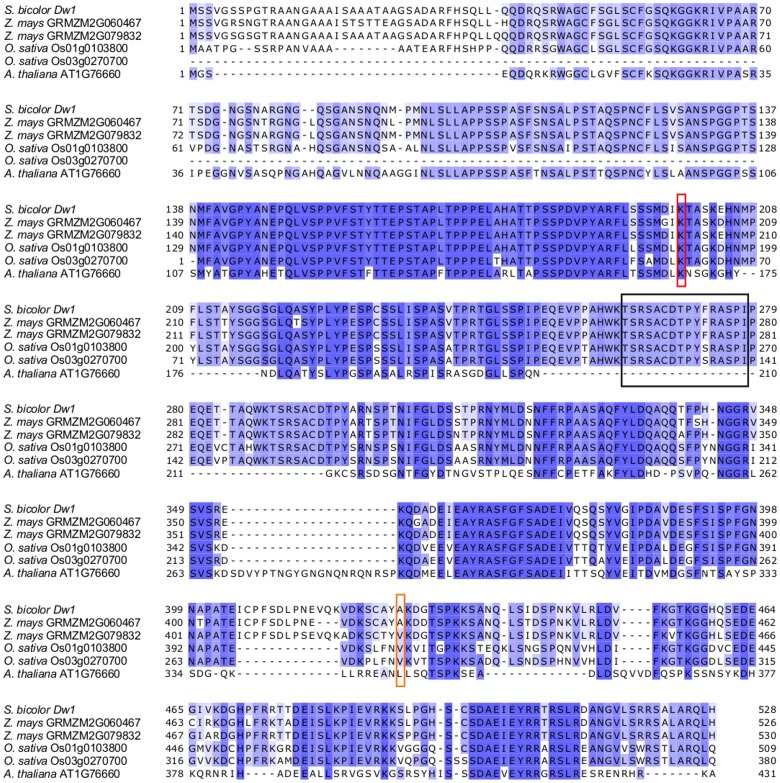
Fig 5. Protein alignment of Dw1 and select homologs.
Alignment of Dw1 with the two maize homologs, the two rice homologs, and the Arabidopsis homolog compiled in Jalview using the T-Coffee function (dark blue color indicates higher percent identity). The red rectangle marks the functional polymorphism that distinguishes Hegari (Dw1) and 80M (dw1). The orange rectangle marks a polymorphism present in Rio and Early White Milo not found in the other sequenced lines. The black box is the possible transmembrane domain predicted by PSIPRED-MEMSAT-SVM.
Maize homologs of Sobic.009G229800 located on chromosomes 6 and 8 are syntenic to sorghum chromosome 9. Genes flanking ZmDw1 on maize chromosome 8 show collinearity with the region on SBI-09 encoding Dw1. On the other hand, the OsDw1 homologs are located on rice chromosomes 1 and 3 while sorghum chromosome 9 is syntenic to rice chromosome 5. This suggests that Dw1 moved to its position on SBI-09 after separation from rice and before separation from maize.
Discussion
In this study, Dw1 was identified using a F2 population and HIFs derived from Hegari (Dw1) and 80M (dw1). Dw1 was identified as Sobic.009G229800 a gene of unknown function that is highly conserved in plants. The recessive dw1 allele corresponds to a loss of function mutation that creates a stop codon in the middle of the protein encoded by Sobic.009G229800. The recessive dw1 allele identified in 80M was present in Dwarf Yellow Milo (dw1) and Double Dwarf Yellow Milo (dw1,dw2) but not in Standard Yellow Milo (Dw1) consistent with reports that short plants containing dw1 originated as a spontaneous mutation in Standard Yellow Milo [6,23]. 80M and the other maturity standards (i.e., 100M, 90M, 80M, 60M) were derived from a cross of Early White Milo (Dw1) and Double Dwarf Yellow Milo (dw1, dw2) and progeny recessive for dw1 and dw2 were selected so that the maturity standards have similar internode lengths (dw1dw2Dw3dw4) [23].
The Dwarf Yellow Milo dw1 allele is present in BTx623, an elite seed parent, and in other genotypes used for grain sorghum breeding in the U.S. (i.e., BTx406, SC170, R07007). The dw1 allele described in this study is present in many grain sorghum lines because BTx406 (dw1) was used to convert tall late flowering sorghum accessions to short early flowering genotypes useful for grain sorghum breeding in the U.S. [15]. This also explains why Brown et al. [14] mapped a QTL for height (Sb_HT9.1) corresponding to allelic variation at the Dw1 locus in a panel of grain genotypes many of which included BTx406 in their pedigrees. Markers most tightly linked to Sb_HT9.1 identified a region of SBI-09 from 57.14–57.21, the same region we found that encodes Dw1. This region includes Sobic.009G229800; however, this gene was initially annotated in Phytozome as two genes (v1.4 gene set). Subsequently, Sobic.009G229800 was annotated with an intron spanning the portion of the coding region that contains the causative mutation (v2.1). Two additional mapping studies identified the same region of SBI-09 as encoding Dw1 [20,21]. Both studies suggested that mutations in a GA2 oxidase (GA2ox5) could be responsible for variation in height caused by Dw1. However, subsequent sequence analysis of GA2ox5 from genotypes that were Dw1Dw1 and dw1dw1 did not show sequence variants consistent with the identification of this gene as Dw1 [22]. Moreover, mutations causing reduced GA levels in sorghum result in short internodes but also abnormal culm bending, a phenotype not observed in dw1dw1 sorghum genotypes [22].
Dw1 (Sobic.009G229800) is present in maize, rice, other grasses, and dicots such as Arabidopsis. Several large INDELS distinguish the proteins in grasses and Arabidopsis. Homologs of Sobic.009G229800 in maize are collinear with Dw1 in sorghum; however, homologs in rice are not located on the homeologous chromosome suggesting that this gene moved to its current location in sorghum after separation of these grasses. The closest homolog in Arabidopsis is annotated as a plasma membrane protein, a localization that was verified experimentally [45]. The Arabidopsis protein was also annotated with a nuclear location. Analysis of the sorghum protein identified a stretch of amino acids (263–278) that could be associated with the lining of a transmembrane pore. The protein was also predicted to have highly disordered protein domains. Research clarifying the localization and biochemical function of the protein encoded by Sobic.009G229800 will be needed to understand how Dw1 regulates the length of stem internodes.
Quinby and Karper [46] showed that alleles of Dw1 do not affect leaf size, only internode lengths. The restriction of Dw1 action to stems is useful because dw1dw1 can be used to reduce internode length without affecting leaf morphology or canopy development. Furthermore, a QTL corresponding to Dw1 was also found to modulate the weight of the stem but not weight per unit length of stem. Thus, Dw1 increases length and weight of internodes. Heterozygous Dw1dw1 progeny derived from Hegari x 80M had internode lengths that were intermediate compared to plants that were dw1dw1 and Dw1Dw1 (S2 Fig), indicating gene dosage alters the gene’s action on internode growth. Dw1 was expressed in stem internodes, with ~3-fold higher expression in Hegari (Dw1) compared to 80M (dw1). Higher expression in Hegari could be due to feedback from Dw1 resulting from greater growth of the internode, or due to differences in Hegari/80M genetic background.
This research was undertaken to further our understanding of genetic factors influencing internode elongation and stem length in sorghum with a focus on Dw1. QTL analysis of an F2 population derived from Hegari and 80M used for fine mapping Dw1 identified QTL that modulate stem internode length aligned with Dw1, Dw2, a minor QTL on SBI-01 (Dw01_54.7) and a QTL on SBI-07 approximately 3Mbp from Dw3 (Dw07_55.1) [16]. Interactions between Dw07_55.1 and Dw1 were detected and plants homozygous for the Dw07_55.1 allele from 80M had long internodes and showed attenuated influence of Dw1 alleles in this background. Dw3 is an ABCB1 efflux auxin transporter that has homologs in many other plants. However, the phenotypic effect of mutation of ABCB1 is attenuated in dicots like Arabidopsis where auxin is exported from apical meristems via two different ABCB transporters: ABCB1 and ABCB19 [47]. In grasses, auxin is exported from the apical meristem and intercalary meristems of the stem. ABCB1 in maize is the only ABCB transporter in the intercalary meristem leading to more severe stem internode length phenotypes when this gene is mutated. Interestingly, in maize the ABCB1 mutant causes severe shortening of the lower internodes while the upper internodes are essentially normal in length [13]. In contrast, dw1dw1 caused a reduction in the length of all internodes (S1 Fig). The current study and prior studies showed that recessive dw1 alleles decrease internode length/plant height in Dw3 backgrounds (Standard Yellow Milo, Dwarf Yellow Milo) as well as in plants that are homozygous for dw3 (Texas Blackhull Kafir (Dw1Dw2dw3) vs Martin (dw1Dw2dw3) [6]. This result suggests that Dw1 action is not dependent on Dw3, although Dw3 alleles may modulate the extent of Dw1 action on internode elongation. As noted above, Dw1 is not a GA2 oxidase as previously suggested and recessive alleles do not result in stem bending. However, it is possible that Dw1 mediates signaling by hormones (GA, auxin, brassinosteroids, strigolactone, ethylene), photoreceptors (phytochromes, PIFs), or other factors that modulate internode growth. Ongoing research is focused on characterizing the molecular basis of Dw1 action.
Supporting Information
The average internode length for each internode was calculated for each genotype at Dw1 for one of the F3 HIFs (n = 75). In (A) the internodes are numbered from the bottom of the stem, whereas in (B) they are numbered from the peduncle.
(TIFF)
The interaction plots show the interaction between Dw1 and the locus on chromosome 7 (Dw07_55.1) in the Hegari x 80M F2. The A allele is 80M and the B allele is Hegari. Phenotypes distinguishing Dw1 from dw1 are greater when the Dw07_55.1 locus on LG-07 is BB (fixed Hegari).
(TIFF)
For each HIF, the lines that had recombination break points in the region of Dw1 were removed and the remainder of the plants grouped into Dw1Dw1 (blue), Dw1dw1 (red), and dw1dw1 (green) and plotted in a histogram. Note that HIFs 74 and 237 have the 80M allele at Dw7_55.1 while the others have the Hegari allele.
(TIFF)
Tree was constructed in MEGA6 using Maximum Likelihood. Sorghum Dw1 is in bold letters.
(TIFF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The number in the polymorphism table is the same as in the genotypes scored table. The numbers also correspond to the numbers in Tables 4 & 5.
(XLSX)
Accessions included in the phylogenetic tree are bolded.
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
The authors would like to thank Patricia Klein and Ryan McCormick for their assistance in analyzing genotype data used in this study and Susan Hall for assistance in phenotyping.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Perry Adkisson Chair in Agricultural Biology, the Feedstock Genomics Program (NIFA No. 2010-67009-21507), and Ceres, Inc.
References
- 1.Swigoňová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14: 1916–1923. 10.1101/gr.2332504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullet J, Morishige D, McCormick R, Truong S, Hilley J, McKinley B, et al. Energy Sorghum-a genetic model for the design of C4 grass bioenergy crops. J Exp Bot. 2014;65: 3479–3489. 10.1093/jxb/eru229 [DOI] [PubMed] [Google Scholar]
- 3.Rooney WL, Blumenthal J, Bean B, Mullet JE. Designing sorghum as a dedicated bioenergy feedstock. Biofuels, Bioprod Biorefining. 2007;1: 147–157. 10.1002/bbb.15 [DOI] [Google Scholar]
- 4.Gill JR, Burks PS, Staggenborg S a., Odvody GN, Heiniger RW, Macoon B, et al. Yield results and stability analysis from the sorghum regional biomass feedstock trial. BioEnergy Res. 2014;7: 1026–1034. 10.1007/s12155-014-9445-5 [DOI] [Google Scholar]
- 5.Olson SN, Ritter K, Rooney W, Kemanian A, McCarl BA, Zhang Y, et al. High biomass yield energy sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels, Bioprod Biorefining. 2012; 640–655. 10.1002/bbb.1357 [DOI] [Google Scholar]
- 6.Quinby JR, Karper RE. Inheritance of Height in Sorghum. Agron J. 1954;46: 211–216. [Google Scholar]
- 7.Graham D, Lessman KJ. Effect of Height on Yield and Yield Components of Two Isogenic Lines of Sorghum vulgare Pers. Crop Sci. 1966;6: 372–374. 10.2135/cropsci1966.0011183X000600040024x [DOI] [Google Scholar]
- 8.Pereira MG, Lee M. Identification of genomic regions affecting plant height in sorghum and maize. Theor Appl Genet. 1995;90: 380–388. 10.1007/BF00221980 [DOI] [PubMed] [Google Scholar]
- 9.Casady AJ. Effect of a Single Height (Dw3) Gene of Sorghum on Grain Yield, Grain Yield Components, and Test Weight. Crop Sci. 1965;5: 385–388. 10.2135/cropsci1965.0011183X000500050002x [DOI] [Google Scholar]
- 10.Truong SK, McCormick RF, Rooney WL, Mullet JE. Harnessing genetic variation in leaf angle to increase productivity of sorghum bicolor. Genetics. 2015;201: 1229–1238. 10.1534/genetics.115.178608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray SC, Sharma A, Rooney WL, Klein PE, Mullet JE, Mitchell SE, et al. Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci. 2008;48: 2165–2179. 10.2135/cropsci2008.01.0016 [DOI] [Google Scholar]
- 12.Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302: 81–84. 10.1126/science.1086072 [DOI] [PubMed] [Google Scholar]
- 13.Knöller AS, Blakeslee JJ, Richards EL, Peer WA, Murphy AS. Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J Exp Bot. 2010;61: 3689–96. 10.1093/jxb/erq180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown PJ, Rooney WL, Franks C, Kresovich S. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics. 2008;180: 629–37. 10.1534/genetics.108.092239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein RR, Mullet JE, Jordan DR, Miller FR, Rooney WL, Menz MA, et al. The Effect of Tropical Sorghum Conversion and Inbred Development on Genome Diversity as Revealed by High-Resolution Genotyping. Crop Sci. Crop Science Society of America; 2008;48: S12–S26. 10.2135/cropsci2007.06.0319tpg [DOI] [Google Scholar]
- 16.Li X, Li X, Fridman E, Tesso TT, Yu J. Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc Natl Acad Sci. 2015;112: 11823–11828. 10.1073/pnas.1509229112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, et al. Positional Cloning of Rice Semidwarfing Gene, sd-1: Rice “Green Revolution Gene” Encodes a Mutant Enzyme Involved in Gibberellin Synthesis DNA Res. Oxford University Press; 2002;9: 11–17. 10.1093/dnares/9.1.11 [DOI] [PubMed] [Google Scholar]
- 18.Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, et al. “Green revolution” genes encode mutant gibberellin response modulators. Nature. 1999;400: 256–261. 10.1038/22307 [DOI] [PubMed] [Google Scholar]
- 19.Thurber CS, Ma JM, Higgins RH, Brown PJ. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 2013;14: R68 10.1186/gb-2013-14-6-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci U S A. 2013;110: 453–8. 10.1073/pnas.1215985110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins RH, Thurber CS, Assaranurak I, Brown PJ. Multiparental mapping of plant height and flowering time QTL in partially isogenic sorghum families. G3 (Bethesda). 2014;4: 1593–602. 10.1534/g3.114.013318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ordonio RL, Ito Y, Hatakeyama A, Ohmae-Shinohara K, Kasuga S, Tokunaga T, et al. Gibberellin deficiency pleiotropically induces culm bending in sorghum: an insight into sorghum semi-dwarf breeding. Sci Rep. Nature Publishing Group; 2014;4: 5287 10.1038/srep05287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinby J. Sorghum Improvement amd the Genetics of Growth. College Station, TX: Texas A&M University Press; 1974. [Google Scholar]
- 24.Morishige DT, Klein PE, Hilley JL, Sahraeian SME, Sharma A, Mullet JE. Digital genotyping of sorghum—a diverse plant species with a large repeat-rich genome. BMC Genomics. 2013;14: 448 10.1186/1471-2164-14-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1: 174–181. 10.1016/0888-7543(87)90010-3 [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Basten C, Zeng Z. Windows QTL Cartographer 2.5 [Internet]. Raleigh, NC: Department of Statistics, North Carolina State University; 2012. Available: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm [Google Scholar]
- 27.Manichaikul A, Moon JY, Sen Ś, Yandell BS, Broman KW. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics. 2009;181: 1077–1086. 10.1534/genetics.108.094565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broman KW, Wu H, Sen Ś, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- 29.Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. TAG Theor Appl Genet. 1997;95: 1005–1011. 10.1007/s001220050654 [DOI] [Google Scholar]
- 30.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal. 2003;1: e012 10.1621/nrs.01012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302: 205–17. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5: 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41: 349–357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10: 159 10.1186/1471-2105-10-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J Mol Biol. 2004;337: 635–645. 10.1016/j.jmb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 39.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins Struct Funct Genet. 2001;42: 38–48. [DOI] [PubMed] [Google Scholar]
- 40.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, et al. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21: 3435–3438. 10.1093/bioinformatics/bti537 [DOI] [PubMed] [Google Scholar]
- 41.Ritter KB, Jordan DR, Chapman SC, Godwin ID, Mace ES, Lynne McIntyre C. Identification of QTL for sugar-related traits in a sweet x grain sorghum (Sorghum bicolor L. Moench) recombinant inbred population. Mol Breed. 2008;22: 367–384. 10.1007/s11032-008-9182-6 [DOI] [Google Scholar]
- 42.Mocoeur A, Zhang YM, Liu ZQ, Shen X, Zhang LM, Rasmussen SK, et al. Stability and genetic control of morphological, biomass and biofuel traits under temperate maritime and continental conditions in sweet sorghum (Sorghum bicolour). Theor Appl Genet. Springer Berlin Heidelberg; 2015;128: 1685–1701. 10.1007/s00122-015-2538-5 [DOI] [PubMed] [Google Scholar]
- 43.Karper RE. A Dominant Mutation of Frequent Recurrence in Sorghum. Am Nat. 1932;66: 511–529. [Google Scholar]
- 44.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11: 863–74. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6: 1198–1214. 10.1074/mcp.M600429-MCP200 [DOI] [PubMed] [Google Scholar]
- 46.Quinby JR, Karper RE. Inheritance of Duration of Growth in the Milo Group of Sorghum1. Crop Sci. 1961;1: 8–10. 10.2135/cropsci1961.0011183X000100010004x [DOI] [Google Scholar]
- 47.Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44: 179–94. 10.1111/j.1365-313X.2005.02519.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The average internode length for each internode was calculated for each genotype at Dw1 for one of the F3 HIFs (n = 75). In (A) the internodes are numbered from the bottom of the stem, whereas in (B) they are numbered from the peduncle.
(TIFF)
The interaction plots show the interaction between Dw1 and the locus on chromosome 7 (Dw07_55.1) in the Hegari x 80M F2. The A allele is 80M and the B allele is Hegari. Phenotypes distinguishing Dw1 from dw1 are greater when the Dw07_55.1 locus on LG-07 is BB (fixed Hegari).
(TIFF)
For each HIF, the lines that had recombination break points in the region of Dw1 were removed and the remainder of the plants grouped into Dw1Dw1 (blue), Dw1dw1 (red), and dw1dw1 (green) and plotted in a histogram. Note that HIFs 74 and 237 have the 80M allele at Dw7_55.1 while the others have the Hegari allele.
(TIFF)
Tree was constructed in MEGA6 using Maximum Likelihood. Sorghum Dw1 is in bold letters.
(TIFF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The number in the polymorphism table is the same as in the genotypes scored table. The numbers also correspond to the numbers in Tables 4 & 5.
(XLSX)
Accessions included in the phylogenetic tree are bolded.
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.