Abstract
Simian hemorrhagic fever (SHF) is an often lethal disease of Asian macaques. Simian hemorrhagic fever virus (SHFV) is one of at least three distinct simian arteriviruses that can cause SHF, but pathogenesis studies using modern methods have been scarce. Even seemingly straightforward studies, such as examining viral tissue and cell tropism in vivo, have been difficult to conduct due to the absence of standardized SHFV-specific reagents. Here we report the establishment of an in situ hybridization assay for the detection of SHFV and distantly related Kibale red colobus virus 1 (KRCV-1) RNA in cell culture. In addition, we detected SHFV RNA in formalin-fixed, paraffin-embedded tissues from an infected rhesus monkey (Macaca mulatta). The assay is easily performed and can clearly distinguish between SHFV and KRCV-1. Thus, if further developed, this assay may be useful during future studies evaluating the mechanisms by which a simian arterivirus with a restricted cell tropism can cause a lethal nonhuman primate disease similar in clinical presentation to human viral hemorrhagic fevers.
Introduction
Simian hemorrhagic fever (SHF) is an acute viral hemorrhagic fever characterized by high lethality. Thus far, SHF has only been observed in captive Asian macaques of several species [1]. Recent genomic sequencing studies revealed that past SHF outbreaks were caused by at least three distinct simian arteriviruses (Nidovirales: Arteriviridae: Arterivirus). Simian hemorrhagic encephalitis virus (SHEV) caused the 1964 Sukhumi, USSR, outbreak [2], simian hemorrhagic fever virus (SHFV) caused the Bethesda, USA, epizootic outbreak only a few months later [3], and Pebjah virus (PBJV) was detected in samples collected during an SHF outbreak in Alamogordo, USA, in 1989 [2]. Several additional simian arteriviruses, among them Kibale red colobus virus 1 (KRCV-1), were recently discovered in apparently healthy African nonhuman primates [4–7]. Among them, at least KRCV-1 is able to infect and cause disease in Asian macaques in experimental settings [8].
The pathogenesis of SHF unambiguously due to SHFV (rather than PBJV, SHEV or other simian arterivirus) infection has only been evaluated in three studies during which macaques were experimentally infected with different virus isolates [9–11]. SHF is characterized by sudden onset of fever, weight loss, facial edema and erythema, dyspnea, diarrhea, lymphadenopathy, and splenomegaly. Limited intestinal and lung hemorrhages are typical manifestations, and epistaxis, hematomas, hematuria, melena, periocular hemorrhages, and petechiae are frequent findings. Disseminated intravascular coagulation, focal necroses in the liver and adrenal glands, proteinuria, lymphocyte depletion in the absence of lymphocyte infection in all lymphoid organs, and the induction of proinflammatory cytokines contribute to the severity of the disease, which ultimately progresses to shock [9–11].
Macrophages appear to be initial targets of SHFV [12]. SHFV antigen has also been detected in astrocytes, vascular endothelial cells, glial cells, and neuronal cell bodies [10]. In vitro, SHFV has only been shown to grow in the embryonic grivet monkey kidney MA-104 cell line and its various subclones (e.g., MARC-145, CL2621) [3, 13] and in primary macrophages and myeloid dendritic cells [12], suggesting an overall narrow cell tropism. Of the various newly discovered simian arteriviruses [4–7], only KRCV-1 was isolated in tissue culture (Wahl Jensen and Johnson et al., submitted). KRCV-1 growth appears to be restricted to one cell line, the MARC-145 subclone of the MA-104 cell line.
A general method for the detection of simian arteriviruses in formalin-fixed, paraffin-embedded (FFPE) tissue, independent of specific antibodies, would be a valuable tool for further characterizing the distribution of these viruses during in vivo infection. Conventional and tyramide-enhanced in situ hybridization (ISH) methods have previously been used to study the SHFV/KRCV-1-related porcine reproductive and respiratory syndrome virus (PRRSV), to evaluate virus distribution in porcine tissues following experimental exposure to PRRSV [14, 15], to differentiate PRRSV genotypes [16], and to identify coinfection of PRRSV and porcine circovirus [17]. However, no standard ISH methods have been developed for the detection of simian arterivirus RNA.
The RNAscope® chromogenic assay (Advanced Cell Diagnostics, Hayward, CA) is an ISH technique that detects RNA more rapidly and with greater sensitivity than conventional ISH methods. This assay has not been previously used for the detection and quantification of RNA of any arterivirus, but has been established for several other viruses (e.g., fox circovirus [18], human immunodeficiency virus-1 [19], human papillomavirus [20], human respiratory syncytial virus [21], raccoon polyomavirus [22]). In general, RNAscope® utilizes a unique probe design strategy that simultaneously amplifies signal and suppresses background to achieve single RNA molecule visualization while preserving tissue morphology [23]. A multiple set of proprietary RNAscope® probe pairs (also called the “target probe”) is designed by the company on request of a client to target distinct areas of a target RNA stretch. Each probe pair consists of two oligonucleotides that readily diffuse across a variety of sample types, including cryopreserved (fresh frozen) tissue, perfused and frozen tissue, or cultured cells. Each oligonucleotide contains a region complementary and highly specific to a specific region of the RNA target, a short linker, and one half of a so-called PreAmplifier (PreAMP) sequence. The successful hybridization of both oligonucleotides of the target probe on a target RNA joins the two halves of PreAmp. So-called double z probe pairs then bind to the complete PreAMP sequence, leading to a cascade of signal amplification events that mediate the binding of label molecules that catalyze the deposition of chromogens such as diaminobenzidine (DAB). The signal detected by microscopy is thus both highly specific and highly sensitive [24]. Here, we report on the results of an RNAscope® assay that can detect and differentiate between SHFV and KRCV-1, which are highly divergent simian arteriviruses [7].
Materials and Methods
Cells
Embryonic grivet monkey kidney MA-104 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA; #CCL-2378) and were seeded at 1 × 107 cells/T175 flask and grown overnight in Eagle’s minimal essential medium (EMEM, Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO) at 37°C in a humidified 5% CO2 atmosphere. MARC-145 cells, which are derived from MA-104 cells, were obtained from Kay Faaberg (US Department of Agriculture National Animal Disease Center, Ames, IA) and grown, infected, and processed in the same way as MA-104 cells.
Virus infection in vitro
We first aimed at establishment of a proof-of-principle assay, i.e., detection of SHFV in vitro with an SHFV-targeting RNAscope® target probe. MA-104 cell growth medium was removed, and cells were exposed to EMEM or EMEM containing SHFV prototype isolate LVR42-0/M6941 [ATCC #VR-533; GenBank AF180391.2 [25, 26]], prepared as described previously [13], at a multiplicity of infection (MOI) of 0.1. Flasks were returned to the incubator and rocked every 15 min for 1 h. Inoculates were removed, and cells were washed with EMEM before adding EMEM with 10% FBS to the flasks. Cells were incubated for 24–48 h until a cytopathic effect (CPE; cell death) was observed in approximately 50–60% of cells exposed to SHFV compared to an uninfected control flask.
KRCV-1 was recently isolated in tissue culture at the Wisconsin National Primate Research Center (WNPRC) in Madison, WI [8]. KRCV-1 was amplified on MARC-145 cells prior to use as described previously [8].
Experimental infections
Experimental infection of rhesus monkeys (Macaca mulatta) by intramuscular administration of SHFV LVR42-0/M6941 [GenBank KM371111 [26]] was reported previously by our laboratory [10]. We obtained >5-year-old formalin-fixed, paraffin-embedded (FFPE) brain-, liver-, and spleen-tissue sections that were derived from rhesus monkeys that had died on day 9 post-inoculation with a target dose of 5 × 103 pfu of SHFV or were mock-infected.
Slide preparation
For in vitro studies, supernatant was removed, 5 ml of cold phosphate-buffered saline (PBS, pH7.4, Life Technologies, Grand Island, NY) were added to each flask, and cells were harvested by scraping. Cells were transferred into labeled 50-ml conical tubes and centrifuged at 224 × g in a table-top centrifuge for 5 min. Cell pellets were then washed with cold PBS, resuspended in 2 ml of 10% neutral buffered formalin (Life Technologies), and fixed overnight at 4°C. Next, cells were pelleted by centrifugation, and formalin was aspirated prior to washing the cells twice with cold PBS. HistoGel (Life Technologies) was liquefied by heating at 55°C. After removal of the PBS from the sample tubes, cells were resuspended in 500 μl of HistoGel and immediately re-pelleted at 224 × g for 1 min in a table-top centrifuge. Supernatant-containing HistoGel was removed, and the cell pellets were solidified on ice. The resulting cell pellet-HistoGel blocks were dehydrated, embedded in paraffin following standard histology procedures, and then cut in sections of 5-μm thickness.
In situ hybridization
In situ hybridization was performed using the RNAscope® 2.0 HD Brown Chromogenic Reagent Kit according to the manufacturer’s instructions (Advanced Cell Diagnostics, Hayward, CA). Target probes with proprietary sequences were designed using custom software as described previously [23] to target the SHFV and KRCV-1 nucleocapsid (N) genes. GenBank accession numbers, target regions, and catalog order numbers for the proprietary target probes are: SHFV (GenBank AF180391.2; nucleotides 15,315–15,594; Advanced Cell Diagnostics #402481) and KRCV-1 (GenBank HQ845737; nucleotides 15,042–15,401; Advanced Cell Diagnostics #406651). Briefly, prepared slides were baked for 1 h at 60°C prior to use. After deparaffinization and hydration, tissues and cells were air-dried and treated with a peroxidase blocker before heating in a target retrieval solution (pretreatment 2 solution as part of the RNAscope® kit; Advanced Cell Diagnostics #320043) for 20 min at 95–100°C. Protease (pretreatment 3 solution of the RNAscope® kit; Advanced Cell Diagnostics #320045) was then applied for 30 min at 40°C. Target probes were hybridized for 2 h at 40°C, followed by a series of signal amplification and washing steps. Hybridization signals were detected by chromogenic reactions using DAB chromogen followed by 1:1 (vol/vol)-diluted hematoxylin (Fisher Scientific, Pittsburg, PA) counterstaining. Only in vitro samples with an average of at least 1 positive (brown) dot per cell were included for analysis. Slides were examined by microscopy (Leica Microsystems, Buffalo Grove, IL). At least 4 fields (200X-magnified images) were captured for each section using Leica Application Suite (LAS) v3.8 (Leica Microsystems). For in situ studies, formalin-fixed, paraffin-embedded (FFPE) tissues from SHFV-infected or mock-infected rhesus monkeys were processed as the in vitro slides, starting from the baking step. For statistical analysis of in situ samples, positive foci (most likely resembling single positive cells) were counted manually for each field, and standard deviation was calculated using GraphPad Prism 6 software (La Jolla, CA, USA).
Results and Discussion
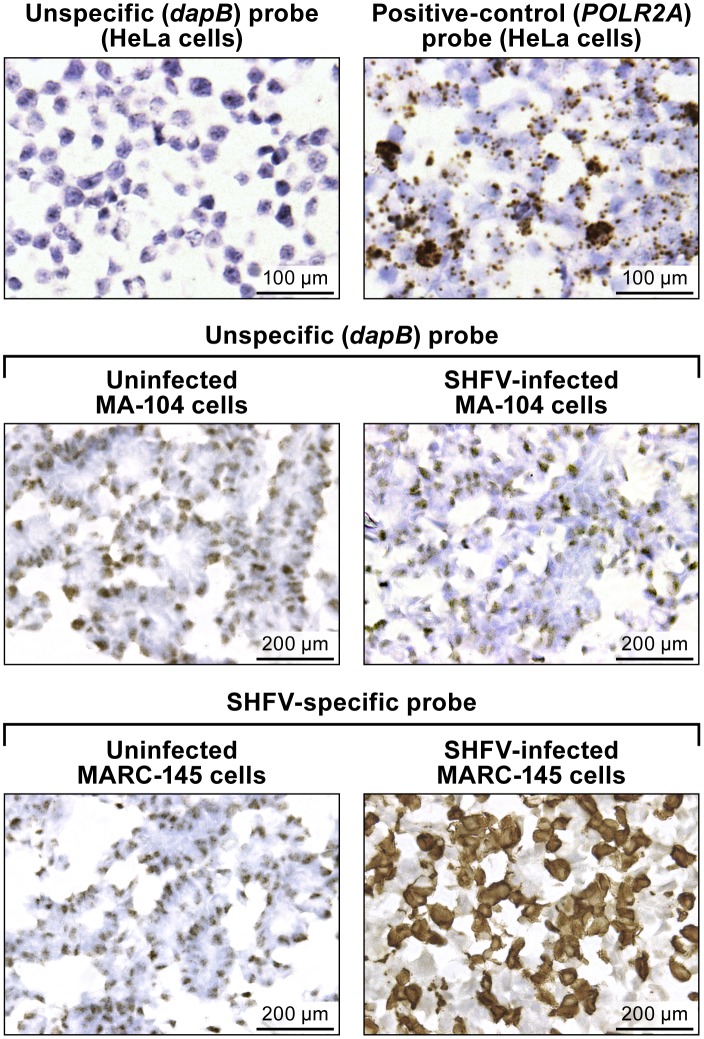
Staining of a control HeLa cell slide, provided by the company, with the RNAScope® bacterial dihydrodipicolinate reductase (dapB; Advanced Cell Diagnostics #310043) negative control target probe resulted in light background staining, probably due to unspecific binding without amplification to cellular nucleic acids (Fig 1, top left). Amplification was clearly visible as brown staining on the control Hela cells slide stained with the RNAscope® DNA-directed RNA polymerase II subunit RPB1 (POLR2A) (Advanced Cell Diagnostics #310451) positive control target probe (Fig 1, top right). Unspecific staining was also observed with the unspecific (dapB) control target probe on uninfected and SHFV-infected MA-104 cells (Fig 1, center) and on uninfected MARC-145 cells. Positive (brown) DAB staining was detected only in SHFV-infected MA-104 cells with the SHFV-target probe (Fig 1, bottom right).
Fig 1. In vitro detection of SHFV RNA in infected cells using RNAscope® in situ hybridization.
(top): uninfected control HeLa cell slides. (top left) Result RNAscope® in situ hybridization with a negative-control target probe targeting the bacterial dapB gene. (top right) Result with a positive-control target probe targeting the human POLR2A gene. (center and bottom) Uninfected- or SHFV-infected MA-104 cells treated with unspecific (dapB) or SHFV-specific target probes. Positive results manifest as brown staining after amplification (top right, bottom left). All images were originally taken at 400X magnification.
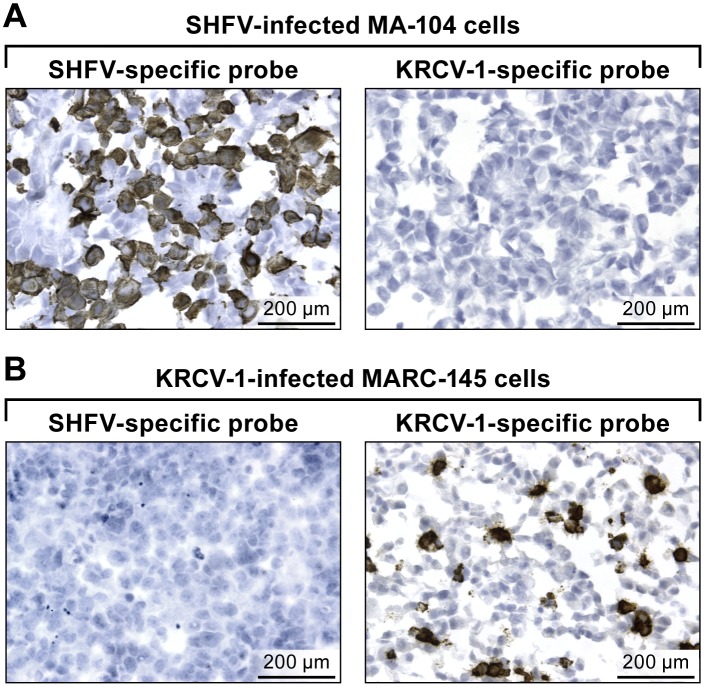
Using either the SHFV or the KRCV-1 target probe on SHFV-infected MA-104 cells, we show that the KRCV-1 target probe does not react with SHFV RNA (Fig 2A). On the other hand, by using MARC-145 cells infected with KRCV-1, we show that the KRCV-1 target probe detects KRCV-1 RNA, whereas the SHFV probe does not detect KRCV-1 RNA In contrast to the experiment shown in Fig 1, very little background staining was observed with the KRCV-1-specific target probe on MA-104 cells or the SHFV-specific target probe on MARC-145 cells (Fig 2B).
Fig 2. In vitro detection of KRCV-1 RNA in infected cells using RNAscope® in situ hybridization.
(A) SHFV-infected MA-104 cells labeled with SHFV- (left) or KRCV-1-specific (right) probes. (B) KRCV-1-infected MARC-145 cells labeled with SHFV- (left) or KRCV-1-specific (right) probes. Positive results manifest as brown staining after amplification. All images were originally taken at 400X magnification.
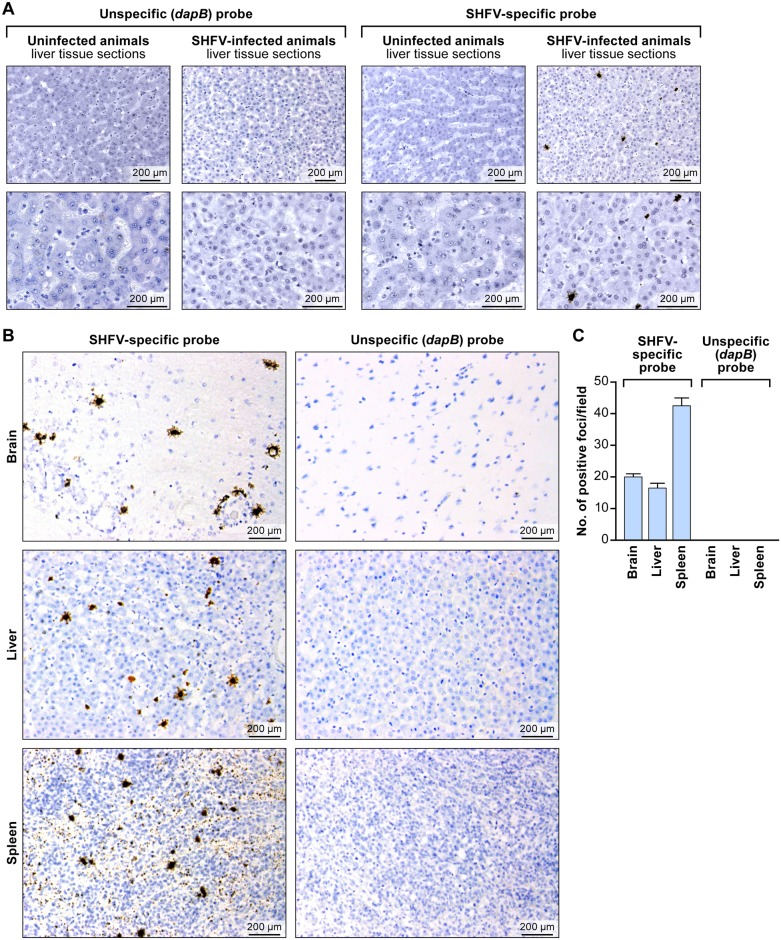
Next, we evaluated the usefulness of the RNAscope® assay for detection of SHFV in FFPE tissues. We hybridized rhesus monkey liver sections with the SHFV target probe or control target probes as described above. Again we only detected SHFV-positive (brown) foci in samples from SHFV-infected animals exposed to the SHFV target probe. Negative results were obtained for SHFV-infected samples exposed to control target probe and for uninfected samples exposed to either target probe (Fig 3A and 3B). These results indicate that RNAscope® assay is sufficiently specific to detect SHFV RNA in FFPE cells despite SHFV’s limited tissue tropism and spread.
Fig 3. In situ detection of SHFV RNA from tissue sections from an SHFV-infected rhesus monkey using RNAscope® in situ hybridization.
(A) Liver sections from an uninfected or SHFV-infected rhesus monkey labeled with unspecific (dapB) or SHFV-specific target probes. Top: all images were originally taken at 200X magnification. Bottom: all images were originally taken at 400X magnification. Positive results manifest as brown staining. (B) Detection of SHFV RNA in brain, spleen, and additional liver sections of the same animal (original magnification 400X). Positive results manifest as brown staining after amplification. (C) Quantification of SHFV RNA-positive foci in brain, liver, and spleen sections by counting; four fields were counted per tissue section of 200X-magnified images (p value calculated by multiple t-test analysis with GraphPad Prism 6 software).
Finally, we applied RNAscope® to brain and spleen tissue sections stemming from the same SHFV-infected animal [10]. Consistent with electron-microscopic and immunohistochemical results described previously for SHFV-infected rhesus monkeys [10], positive staining was obtained for all tested tissue types (liver, spleen, brain) with the SHFV target probe, but no signal was present when the control target probe was used (Fig 3B). We quantified the number of SHFV RNA-positive foci manually using LAS v3.8 in four fields per section (Fig 3C). Our data indicate that the number of SHFV-infected cells may be higher in spleen than in brain or liver (multiple t-test analysis: p <0.05 for spleen compared to brain or liver).
Conclusion
Together, our data indicate that RNAScope® is a suitable assay for simian arterivirus RNA detection both in vitro and in situ, and that the assay can differentiate between SHFV and KRCV-1, which are highly divergent members of the same viral clade. Future refinements will be necessary to pinpoint and eliminate target probe sequence components that lead to unspecific background staining, and to determine the assay’s limit of detection of simian arterivirus RNAs and its specificity in context of infection with simian arteriviruses that are more closely related to each other than SHFV and KRCV-1. A similar assay developed to cover additional simian arteriviruses would be especially useful because wild red colobus monkeys have been found to be co-infected with at least two simian arteriviruses: KRCV-1 and KRCV-2 [6]. Similarly, two other simian arteriviruses, Kibale red-tailed guenon viruses 1 and 2 (KRTGV-1/2) co-circulate in a wild red-tailed guenon (Cercopithecus ascanius) population [7]. However, the development of such an advanced RNAscope® assay will have to wait until tissue samples from infected wild primates become available or until the various simian arteriviruses other than SHFV and KRCV-1 have been isolated in cell culture and experimental animal studies have been performed. Likewise, it will be interesting to see for how long simian arterivirus RNA can be detected with RNAscope® in FFPE tissues. Our study suggests that at least in the case of SHFV, detection in such tissues is possible more than 5 years post-fixation.
In the meantime, the RNAscope® assay presented here could be expanded to include simultaneous detection of viral and cellular nucleic acids using a dual-color system as described in [27, 28]. Such a mixed detection assay could aid in the identification of the specific cell types infected by simian arteriviruses in vivo/in situ and thereby lead to a better definition of SHFV cell and tissue tropism.
Acknowledgments
We thank Jennifer Hufton and Larry J. Faucette of the IRF-Frederick for experimental support, and are grateful to Laura Bollinger and Jiro Wada of the IRF-Frederick for critically editing the manuscript and creating figures, respectively.
Data Availability
All relevant data are within the paper.
Funding Statement
The content of this publication does not necessarily reflect the views or policies of the US Department of Defense, the US Department of Health and Human Services, the US Department of the Army, or the institutions and companies affiliated with the authors. This work was funded in part through Battelle Memorial Institute’s prime contract the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I (S.Y., Y.C., C.L., E.P., S.M., J.C.J., P.B.J., J.H.K.). S.Y. and J.C.J. performed this work as employees of Battelle Memorial Institute. Subcontractors to Battelle Memorial Institute who performed this work are: Y.C., C.L., E.P., and J.H.K, employees of Tunnell Government Services, Inc.; and S.M., an employee of MRIGlobal. Battelle Memorial Institute, Tunnell Government Services, and MRIGlobal provided support in the form of salaries, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. All work under contract HHSN272200700016I is performed under the agreement/understanding that it will belong to the client and will be affiliated directly to the NIH/NIAID Integrated Research Facility at Fort Detrick. The specific roles of these authors are articulated in the ‘author contributions’ section. This work was also supported, in part, by the NIAID Division of Intramural Research (R.F.J., P.B.J.), by National Institutes of Health (NIH) grant TW009237 as part of the joint NIH-NSF Ecology of Infectious Disease program, grant R01 AI077376, and by the Office of Research Infrastructure Programs (ORIP) grant P51OD011106 (A.L.B., M.L., T.L.G., D.H.O’C., T.C.F.). These funders did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lapin BA, Shevtsova ZV. [To the 50th anniversary of the discovery of the simian hemorrhagic fever and SHF virus]. Voprosy virusologii. 2015;60(1):5–11 [in Russian]. . [PubMed] [Google Scholar]
- 2.Lauck M, Alkhovsky SV, Bào Y, Bailey AL, Shevtsova ZV, Shchetinin AM, et al. Historical outbreaks of simian hemorrhagic fever in captive macaques were caused by distinct arteriviruses. Journal of virology. 2015;89(15):8082–7. 10.1128/JVI.01046-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tauraso NM, Shelokov A, Palmer AE, Allen AM. Simian hemorrhagic fever. 3. Isolation and characterization of a viral agent. The American journal of tropical medicine and hygiene. 1968;17(3):422–31. . [PubMed] [Google Scholar]
- 4.Bailey AL, Lauck M, Sibley SD, Pecotte J, Rice K, Weny G, et al. Two novel simian arteriviruses in captive and wild baboons (Papio spp.). Journal of virology. 2014;88(22):13231–9. 10.1128/JVI.02203-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, et al. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PloS one. 2014;9(3):e90714 10.1371/journal.pone.0090714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, et al. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PloS one. 2011;6(4):e19056 10.1371/journal.pone.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, et al. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. Journal of virology. 2013;87(1):688–91. 10.1128/JVI.02433-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl-Jensen V, Johnson JC, Lauck M, Weinfurter JT, Moncla LH, Weiler AM, et al. Divergent simian arteriviruses cause simian hemorrhagic fever of different severity in macaques. mBIO. 2016;7(1):e02009–15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen AM, Palmer AE, Tauraso NM, Shelokov A. Simian hemorrhagic fever. II. Studies in pathology. The American journal of tropical medicine and hygiene. 1968;17(3):413–21. . [DOI] [PubMed] [Google Scholar]
- 10.Johnson RF, Dodd LE, Yellayi S, Gu W, Cann JA, Jett C, et al. Simian hemorrhagic fever virus infection of rhesus macaques as a model of viral hemorrhagic fever: clinical characterization and risk factors for severe disease. Virology. 2011;421(2):129–40. 10.1016/j.virol.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vatter HA, Donaldson EF, Huynh J, Rawlings S, Manoharan M, Legasse A, et al. A simian hemorrhagic fever virus isolate from persistently infected baboons efficiently induces hemorrhagic fever disease in Japanese macaques. Virology. 2015;474:186–98. 10.1016/j.virol.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vatter HA, Brinton MA. Differential responses of disease-resistant and disease-susceptible primate macrophages and myeloid dendritic cells to simian hemorrhagic fever virus infection. Journal of virology. 2014;88(4):2095–106. 10.1128/JVI.02633-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caì Y, Postnikova EN, Bernbaum JG, Yú SQ, Mazur S, Deiuliis NM, et al. Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. Journal of virology. 2015;89(1):844–56. 10.1128/JVI.02697-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu SP, Zhang Z, Liu YG, Tian ZJ, Wu DL, Cai XH, et al. Pathogenicity and distribution of highly pathogenic porcine reproductive and respiratory syndrome virus in pigs. Transbound Emerg Dis. 2013;60(4):351–9. 10.1111/j.1865-1682.2012.01354.x . [DOI] [PubMed] [Google Scholar]
- 15.Trang NT, Hirai T, Ngan PH, Lan NT, Fuke N, Toyama K, et al. Enhanced detection of Porcine reproductive and respiratory syndrome virus in fixed tissues by in situ hybridization following tyramide signal amplification. J Vet Diagn Invest. 2015;27(3):326–31. 10.1177/1040638715579260 . [DOI] [PubMed] [Google Scholar]
- 16.Larochelle R, Magar R. Differentiation of North American and European porcine reproductive and respiratory syndrome virus genotypes by in situ hybridization. J Virol Methods. 1997;68(2):161–8. . [DOI] [PubMed] [Google Scholar]
- 17.Sirinarumitr T, Sorden SD, Morozov I, Paul PS. Double in situ hybridization for simultaneous detection of porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus (PCV). J Vet Diagn Invest. 2001;13(1):68–71. . [DOI] [PubMed] [Google Scholar]
- 18.Bexton S, Wiersma LC, Getu S, van Run PR, Verjans GM, Schipper D, et al. Detection of Circovirus in Foxes with Meningoencephalitis, United Kingdom, 2009–2013. Emerg Infect Dis. 2015;21(7):1205–8. 10.3201/eid2107.150228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamers SL, Rose R, Ndhlovu LC, Nolan DJ, Salemi M, Maidji E, et al. The meningeal lymphatic system: a route for HIV brain migration? Journal of neurovirology. 2015. Epub 2015/11/18. 10.1007/s13365-015-0399-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoddard DG Jr, Keeney MG, Gao G, Smith DI, Garcia JJ, O'Brien EK. Transcriptional activity of HPV in inverted papilloma demonstrated by in situ hybridization for E6/E7 mRNA. Otolaryngology-head and neck surgery: official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2015;152(4):752–8. Epub 2015/03/01. 10.1177/0194599815571285 . [DOI] [PubMed] [Google Scholar]
- 21.Saravia J, You D, Shrestha B, Jaligama S, Siefker D, Lee GI, et al. Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33. PLoS pathogens. 2015;11(10):e1005217 Epub 2015/10/17. 10.1371/journal.ppat.1005217 ; PubMed Central PMCID: PMCPmc4608776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brostoff T, Dela Cruz FN Jr, Church ME, Woolard KD, Pesavento PA. The raccoon polyomavirus genome and tumor antigen transcription are stable and abundant in neuroglial tumors. Journal of virology. 2014;88(21):12816–24. 10.1128/JVI.01912-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–9. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowling AJ, Pence HE, Church JB. Application of a novel and automated branched DNA in situ hybridization method for the rapid and sensitive localization of mRNA molecules in plant tissues. Appl Plant Sci. 2014;2(4):1400011 10.3732/apps.1400011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vatter HA, Di H, Donaldson EF, Baric RS, Brinton MA. Each of the eight simian hemorrhagic fever virus minor structural proteins is functionally important. Virology. 2014;462–463:351–62. 10.1016/j.virol.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauck M, Palacios G, Wiley MR, Lǐ Y, Fang Y, Lackemeyer MG, et al. Genome sequences of simian hemorrhagic fever virus variant NIH LVR42-0/M6941 isolates (Arteriviridae: Arterivirus). Genome Announc. 2014;2(5):e00978–14. 10.1128/genomeA.00978-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Su N, Wang LC, Wu X, Bui S, Nielsen A, et al. Dual-color ultrasensitive bright-field RNA in situ hybridization with RNAscope. Methods in molecular biology. 2014;1211:139–49. 10.1007/978-1-4939-1459-3_12 . [DOI] [PubMed] [Google Scholar]
- 28.O'Shannessy DJ, Somers EB, Wang LC, Wang H, Hsu R. Expression of folate receptors alpha and beta in normal and cancerous gynecologic tissues: correlation of expression of the beta isoform with macrophage markers. J Ovarian Res. 2015;8(1):29 10.1186/s13048-015-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.