Abstract
It is known that four common inbred mouse strains show defects of the forebrain commissures. The BALB/cJ strain has a low frequency of abnormally small corpus callosum, while the 129 strains have many animals with deficient corpus callosum. The I/LnJ and BTBR T+ tf/J strains never have a corpus callosum, while half of I/LnJ and almost all BTBR show severely reduced size of the hippocampal commissure. Certain of the F1 hybrid crosses among these strains are known to be less severely abnormal than the inbred parents, suggesting the parent strains have different genetic causes of commissure defects. In this study, all hybrid crosses among the four strains were investigated. The BTBR x I/Ln hybrid expressed almost no defects of the hippocampal commissure, unlike its inbred parent strains. Numerous 3-way crosses among the four strains yielded many mice with no corpus callosum and severely reduced hippocampal commissure, which shows that the phenotypic defect can result from several different combinations of genetic alleles. The F2 and F3 hybrid crosses of BTBR and I/LnJ had almost 100% absence of the corpus callosum but about 50% frequency of deficient hippocampal commissure. The 4-way hybrid cross among all four abnormal strains involved highly fertile parents and yielded a very wide phenotypic range of defects from almost no hippocampal commissure to totally normal forebrain commissures. The F2 and F3 crosses as well as the 4-way cross provide excellent material for studies of genetic linkage and behavioral consequences of commissure defects.
Keywords: inbred strains, reciprocal hybrid cross, complementation test, complex trait, 4-way hybrid cross
Introduction
Four major inbred mouse strains exhibit reduced size or total absence of two large forebrain commissures, the corpus callosum (CC) and the hippocampal commissure (HC). Average severity of the phenotypic defect differs among the strains (Table 1). Hybrid crosses between certain of the strains indicate that the F1 hybrids are less severely afflicted than either parent strain and therefore they differ genetically at two or more loci pertinent for commissure formation (Livy et al., 1997; Livy & Wahlsten, 1991; Wahlsten et al., 2003b). Data on certain F1 crosses appeared in the literature over several years and involved different 129 substrains and a BALB substrain that no longer exists (BALB/cWah1). Furthermore, different F1 hybrids from previous studies were collected in various years in different labs, and there is concern that the lab environment may influence the results. Here we examine all four strains and numerous hybrid crosses in a single study conducted in the same time period in one lab with larger sample sizes than previous reports and extend the investigation to include many 3-way crosses among the four strains as well as the 4-way cross.
Table 1.
Forebrain commissure defects in inbred mouse strains assessed with similar histological methods and criteria for small commissure
| Strain | n | Small CC | No CC | Small HCf |
|---|---|---|---|---|
| BALB/cByJc,d | 68 | 1 | 1 | 0 |
| 129P1/ReJe | 82 | 27 | 9 | 2 |
| 129P3/Ja,b | 126 | 54 | 31 | 1 |
| 129S1/SvImJd | 20 | 2 | 4 | 1 |
| I/LnJb,e | 58 | 0 | 58 | 38 |
| BTBR T+ tfJd | 18 | 0 | 18 | 17 |
Mice with small HC are a subset of those with no CC.
The principal goal of the study was to identify crosses useful for future studies of genetic linkage and phenotypic correlates of forebrain commissure defects. It was expected that, if F1 hybrids have a lower frequency of severe commissure defects than the inbred parents, F2 as well as 3-way and 4-way crosses involving those strains should yield a wide range of phenotypic defects, including brains that are even more severely defective than either inbred parent, perhaps even brains with no hippocampal commissure at all, as well as normal brains, and therefore should be most promising for genetic linkage analysis.
All of the data presented in this report were newly collected for this study, with the exception of earlier results shown in one figure for the strain I/LnJ that was in very short supply recently and strain BTBR where the current sample was not large. Data for the inbred strains and certain of the F1 hybrid crosses replicate previously published data. This is an important observation for future genetic linkage studies, because there is extreme phenotypic variation within several isogenic groups. Here we show that the entire distribution of commissure sizes, ranging from totally absent to normal, is itself a stable, replicable characteristic for several genotypes.
A much larger set of hybrid crosses is studied here than in previous experiments, but for practical limitations it was not feasible to test all 44 = 256 possible crosses of four strains across two generations when each mouse has four grandparents. Some kind of compromise must be made to reduce the number of crosses. In the present study, many of the reciprocal crosses that would be required to prove the importance of maternal environment effects (Carlier et al., 1992), transgenerational epigenetic effects (Champagne, 2008; Wahlsten, 2011), and X-linked genes were not included. Table 2 summarizes the genotypes of most of the 1,089 mice that were studied anatomically.
Table 2.
Crosses of inbred strains and F1 hybridsa
| F1 hybrid cross; two abnormal strains | Crosses involving BTBR and I/Ln | 3-way; all abnormal strains (no BALB) [84] |
|---|---|---|
| 129 x BALB* [52] | (BTBR x I/Ln) x BTBR* [24] | 50% 129 alleles [9] |
| 129 x BTBR* [35] | (BTBR x I/Ln) F2 [55] and F3 [17] | 50% BTBR alleles [58] |
| 129 x I/Ln [20] | 3-way cross; 25% C57 alleles | 50% I/Ln alleles [17] |
| BALB x BTBR* [33] | (C57 x 129) x BALB* [32] | 3-way; all abnormal strains (no BTBR) [66] |
| BALB x I/Ln [17] | (C57 x 129) x BTBR* [16] | 50% 129 alleles [10] |
| BTBR x I/Ln [19] | (C57 x 129) x I/Ln [32] | 50% BALB alleles [20] |
| F1 hybrid cross; C57 x abnormal strain | (BALB x C57) x BTBR [17] | 50% I/Ln alleles* [36] |
| C57 x 129 [12] | (BALB x C57) x I/Ln [22] | 3-way; all abnormal strains (no I/Ln) [69] |
| C57 x BALB* [12] | 3-way; all abnormal strains (no 129) [72] | 50% 129 alleles; (BALB x BTBR) x 129 [32] |
| C57 x BTBR [12] | 50% BALB alleles* [30] | 50% BALB alleles; (129 x BTBR) x BALB [17] |
| C57 x I/Ln [21] | 50% BTBR alleles* [20] | 50% BTBR alleles; (129 x BALB) x BTBR [20] |
| Backcross involving C57 | 50% I/Ln alleles* [22] | 4-way cross among all abnormal strains [46] |
| (C57 x 129) x 129 [15] | (BALB x BTBR) x (129 x I/Ln) [35] | |
| (C57 x BALB) x BALB [18] | (BALB x I/Ln) x (129 x BTBR) [11] | |
| (C57 x BTBR) x BTBR* [29] |
Sample sizes in brackets. Abbreviations: 129, 129S1/SvImJ; BALB, BALB/cJ; BTBR, BTBR T+ tfJ; C57, C57BL/6J; I/Ln, I/LnJ. Genotype of female parent is given first; e.g., BTBR x I/Ln is BTBR female mated with I/Ln male. (BALB x I/Ln) x (129 x BTBR) 4-way cross is (BALB x I/Ln) female by (129 x BTBR) male.
Asterisk (*) indicates data combined from more than one specific cross, reciprocal crosses in most cases.
Materials and methods
Mice
All inbred mice used as parents were obtained at six to eight weeks of age from the Jackson Laboratory (Bar Harbor, ME, USA) and then bred in our colony when at least 10 weeks old. Four inbred strains known to have defects of the corpus callosum served as the founders for subsequent hybrid crosses: 129S1/SvImJ (129), BALB/cJ (C or BALB), BTBR T+ tf/J (BTBR), and I/LnJ (I/Ln). There are many inbred strains derived from a 129 ancestor (Simpson et al. 1997; Threadgill et al. 1997), and all reports to date have indicated a moderate frequency of CC defects but very few HC defects (Table 1) (Wahlsten et al., 2001). The 129S1/SvImJ strain was used in this study because it is the only 129-derived strain in Tier 1 of the Mouse Phenome Database. Only male I/Ln mice were obtained for breeding because of short supply and that strain’s poor breeding and maternal performance. The C57BL/6J (B6 or C57) inbred strain served as a normal control and was used as a parent in several crosses. Two F1 hybrid mouse strains from the Jackson labs were used for certain crosses: (C)B6F1/J and (B6)129SF1/J . Brains were also studied for several other strains from the Jackson Labs: A/J, BALB/cByJ, C3H/HeJ, DBA/2J, FVB/NJ and SJL/J.
Husbandry and breeding
Animals were group housed with a maximum of four mice per cage in opaque shoebox cages lined with 7090 Aspen Sani-Chips®. Food (7002 Teklad 6% mouse/rat diet, Harlan, Madison, WI, USA) and Greensboro city tap water were provided ad libitum along with a fresh single nestlet with each cage change. Colony rooms had lights on for 12 h (07.00–19.00). Ambient temperature (21.5°±1°C) and humidity (48.5 ± 4%) were controlled within narrow ranges. A single male was mated with one to three females in a single cage when all mated mice were at least eight weeks old. Female weights were recorded at the time of mating, and a female was removed and singly housed when she had gained at least 4 g. Pregnant females were observed daily for births. Pups were weaned at 21 days after birth and group housed by sex. Some offspring were then mated at eight to 10 weeks of age.
Hybrid crosses
A wide variety of hybrid crosses was produced (Table 2). For any particular cross, the genotype of the female parent is always given first, followed by lower case x, followed by male genotype. When either the female or male parent was itself a hybrid, its origin is given in parentheses. For example, a mouse of genotype (129 x BTBR) x I/Ln represents offspring from a female hybrid cross of 129 and BTBR mated with an I/Ln male. The reciprocal crosses (129 x BTBR) and (BTBR x 129) would have the same complement of autosomal genes, but (129 x BTBR) would have mitochondria from strain 129 and Y chromsome from BTBR, whereas (BTBR x 129) would have BTBR mitochondria and a 129 Y chromosome. In order to make the list of crosses in Table 2 more compact, instances where reciprocal crosses were obtained are indicated by an asterisk (*).
Among the three-way crosses of abnormal strains, there was always one of the abnormal strains absent from any particular cross, and in almost all cases the inbred parent was the male while the female was the more fertile F1 hybrid. For example, the (BALB x I/Ln) x BTBR cross had no 129 alleles, and 50% of its autosomal alleles were from strain BTBR. The three-way crosses in Table 2 are grouped according to which strain was not a parent or grandparent. Within each of those groupings, crosses were formed so that some had 50% of autosomal alleles from each of the other three strains. For example, among crosses with no BALB ancestor, some had 50% 129 alleles, some had 50% BTBR, and others had 50% I/Ln. The mice with 50% BTBR alleles but no BALB included the (129 x I/Ln) x BTBR and (BTBR x I/Ln) x (129 x BTBR) crosses. Three-way crosses were then analysed according to which strain was missing from the cross and which contributed 50% of its alleles.
Sample sizes depended on the number of viable litters obtained. In almost every cross the data came from at least two litters, and in the large majority of instances they were from first litters produced by a female. When second litters were obtained, mice were re-mated only after the first litter had been weaned. Parental age of the breeders ranged widely, especially for the male studs from strains that were used to obtain F1 hybrids as well as three-way crosses or backcrosses. In a few instances a breeder female was used to obtain F1 hybrids and then was mated to a different genotype to obtain a three-way cross. All breeders were eventually euthanized and their brains stained and measured.
In almost all cases, sample sizes exceeded 20 mice per breeding combination when reciprocal crosses were pooled (Table 2). Fewer were obtained in the F1 hybrid crosses and backcrosses involving C57BL/6J because it has been well established in previous studies that commissure defects involving that strain are completely recessive and multigenic. Large sample sizes are most important in a cross where a very low frequency of commissure defects is expected. One would not want to conclude that the offspring of any particular kind of cross are completely normal unless a substantially large sample is studied. For the 3-way and 4-way crosses, a high frequency of commissure defects was expected and observed. Among the F1 hybrid crosses, at least one abnormal brain was seen in every cross.
Histology and measures
Brains were extracted when mice were a minimum of eight weeks old. Animals were euthanized by CO2 gas asphyxiation followed by cervical dislocation. Body weight was recorded and then whole brains were immediately removed and immersed in a 10% phosphate-buffered formalin fixative (Fisher Scientific, Hampton, NH, USA) for a minimum of 48 h. Staining and measurements were performed using methods described previously (Wahlsten et al., 2003a). Briefly, fixed whole brains had the spinal cord cut caudal to the medulla oblongata and olfactory bulbs removed; brains were then sagittally bisected at midline with the longitudinal cerebral fissure as the guide. After the two hemispheres were weighed, one hemisphere was immersed in a 0.2% gold chloride stain solution (Sigma, St. Louis, MO, USA) at 37°C. Staining was stopped when the commissures became obviously dark and nearby tissue started to darken (Fig. 1), whereupon the half brain was immediately immersed in 2.5% sodium thiosulfate fixative (Fisher Scientific, Hampton, NH, USA). Stained half brains were stored in 10% formalin until measures were taken. An image of the midsagittal plane was obtained using a Canon MP-E 65mm 1–5x Macro Lens with a EOS Rebel T1i camera body (Canon Inc., Tokyo, Japan). The unadjusted cross-sectional area in mm2 and maximum length in mm of the corpus callosum (CC), hippocampal commissure (HC), and anterior commissure (AC) at the mid-sagittal plane were measured using ImageJ version 1.4.3.67 software (http://rsbweb.nih.gov/ij/). The dorsal commissure of the fornix, when present, was not included in the size of either the CC or the HC. Measurements were made by an observer who knew the ID code of the animal written on the vial at the time of the measurement but not its genotype.
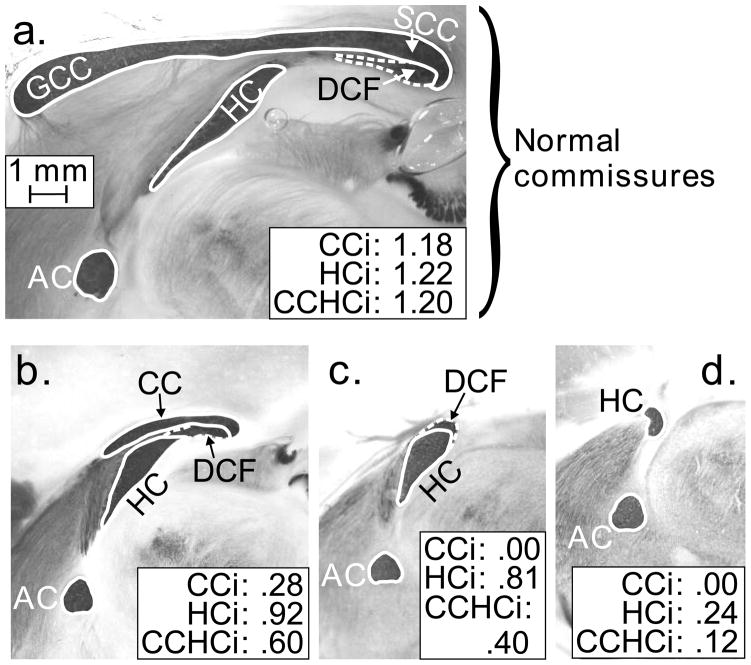
Fig. 1. Forebrain commissures at the mid-sagittal plane stained with gold chloride to reveal myelin.
Borders of each structure are shown with white lines. Commissure size is best indicated as cross-sectional area. An index of commissure size is the ratio of actual area to area expected from brain weight in strains of mice that never show absent or reduced CC. Further details of the indices are provided in the text. a. Normal brain with indices near 1.0. b. Brain with normal HC but abnormally small CC. c. Brain with HC size in the normal range but no CC at all. The DCF is located directly above the HC. d. Highly abnormal brain with no CC or DCF and a very small HC. Abbreviations: AC, anterior commissure; CC corpus callosum; CCHCi, combined index of CC and HC abnormality; CCi, index of CC abnormality; DCF, dorsal commissure of the fornix; GCC, genu of the CC; HC, hippocampal commissure; HCi, index of HC abnormality; SCC, splenium of the CC.
Measurements of each commissure were made by filling the measurement window in the ImageJ screen with the specific commissure, such that its maximum length was usually represented by at least 500 pixels for a normal structure. Maximum length of a structure was determined by placing two points on opposite sides of the stained commissure. At high magnification, the demarcation between stained and unstained tissue was usually clear and spanned a zone of only one to three pixels. When the zone of staining was less sharp, the observer placed the point midway between a dark area that was clearly stained and a nearby area that clearly was not stained. It was sometimes necessary to test different pairs of points to determine which entailed the greatest length. This was a particularly great challenge for the AC. Area of a structure was determined by specifying a large number of points along the periphery of the stained zone, using a higher density of points in portions of the commissure that had a smaller radius of curvature. A previous study where normal rat brains were measured twice with measures one week apart (Wahlsten et al., 2003a), found a standard error of the difference of about 0.1 mm for length of the CC and HC, and about 0.1 mm2 for their areas, and differences between the measures were usually less than 1% of length or area. Another study (Bishop & Wahlsten, 1999) reported test-retest reliability of 0.98 for commissure size in a sample of normal mice measured with less precise methods, and the value would be even higher if abnormally small commissures were included in the sample.
Criteria for abnormality
The range of ages when the brains were extracted varied from a minimum of eight weeks to over a year for some mice employed as breeders. Brain size differs substantially among mouse strains, is larger in hybrid crosses than their inbred parents, shows marked maternal environment effects (Wahlsten, 1983), increases substantially with age of the animal (Wahlsten, 1975; Wahlsten, 1984), and is also influenced by litter size (Bulman-Fleming & Wahlsten, 1988). It is well known from the principle of allometry that a larger brain tends to have larger parts (Gould, 1966). In normal mice, commissure size is markedly larger in larger brains. In order to remove the variance in commissure size arising from the wide range of brain sizes, an abnormality index was computed that had an average value of about 1.0 for normal brains and 0.0 for a brain lacking a commissure altogether. First a linear regression equation based on a large sample of mice from strains that never show commissure defects was used to find the expected commissure size for a given brain weight. Equations derived previously (Livy et al., 1997) were E(CC) = −0.1 + 2.2(brain weight) and E(HC) = 0.1 + 0.4(brain weight), where brain weight is grams. The index of abnormality for an animal was then computed as the ratio of the actual commissure cross-sectional area in mm2 (CC or HC) to the value expected from its brain weight: CCi = CC/E(CC) and HCi = HC/E(HC). An unusually small CC has a CCi value less than 0.7, and any brain with CCi < 0.5 is clearly abnormal. Likewise for the HC, the criterion for abnormally small commissure in relation to brain size is HCi < 0.7. If the HCi is 0.7 or less, it is almost certain that CCi = 0. This observation makes it meaningful to combine the CCi and HCi indices into a single index of commissure abnormality by taking their average: CCHCi = (CCi + HCi)/2. Any brain with CCHCi < 0.35 must have no CC and an abnormally small HC. When CCHCi is in the range 0.36 to 0.5, most brains will have a normal HC but absent or severely reduced CC. Brains with normal HC but abnormally small CC will almost always have 0.5 < CCHCi < 0.75.
Examples of different degrees of abnormality are shown in Fig. 1. The validity of the index relies on the fact that the HC is never reduced in size unless the CC is totally absent (Livy & Wahlsten, 1997; Wahlsten et al., 2006). Anatomically, it is supported by observations of an intimate relation between the early formation of the hippocampal commissure and associated glial cells at the cerebral midline and the later crossing of midline by axons of the corpus callosum in the embryo (Donahoo & Richards, 2009; Livy & Wahlsten, 1997; Ren et al., 2006; Wahlsten et al., 2006).
Statistical analysis
All statistical analyses were performed using Systat 12 (Systat Software Inc., PointRichmond, CA, USA) software. In cases where data were normally distributed or nearly so, parametric tests including analysis of variance (ANVOA) and multiple regression were used to evaluate significance. In a few cases where data in at least one group were markedly non-normal, the Kolmogorov-Smirnov two sample test was employed. Formal tests were not conducted when a large difference between groups was visually obvious in a graph. Criterion for significance was set at α= .01. Instances where .05 > P > .01 were considered marginally significant.
All procedures performed with live mice were done according to the Guidelines of the National Institutes of Health and protocols reviewed and approved by the Institutional Animal Care and Use Committee of UNCG.
Results
Commissure sizes and criteria for abnormality of CC and HC
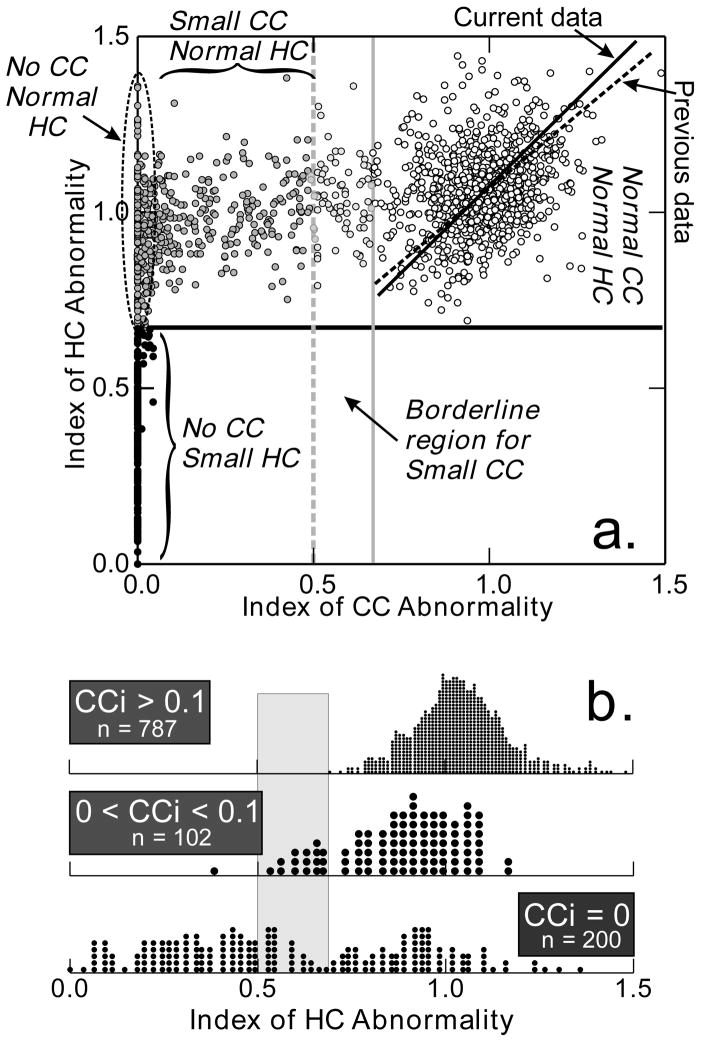
Examples of brains from the current study that have normal and abnormally small CCi and HCi values are shown in Fig. 1. In the present study, the relation between the HCi and CCi in the normal range (Fig. 2A) was closely comparable to that seen in an earlier study done with a wider range of genotypes (Wahlsten et al., 2003b). The HCi was always greater than 0.7 when CC was normal or somewhat small. For mice with very small CC (Fig. 2B), however, there were a few that had CCi > 0 but HCi < 0.7. Careful examination of the brains in the borderline region revealed that the problem was partly anatomical. In those near the borderline region with 0.6 < HCi < 0.7, it was sometimes difficult to discriminate clearly among myelinated axons of the CC, DCF, and HC.
Fig. 2. Index of CC abnormality versus index of HC abnormality.
a. Data for 1,089 brains included in this study. There are fewer dots because many values of the HC index overlap when the CC index is 0. Linear regression line for brains in the normal range is shown by a solid line for current data and a dashed line for a previous sample (Wahlsten et al., 2003b). Solid gray line shows the lower limit of CCi = 0.7 for a normal CC size, while the dashed gray line shows CCi = 0.5, below which all brains clearly have abnormally small CC. The solid black line shows the limit for normal HC with HCi = 0.7. No brain with any CC present at the midsagittal plane ever showed a severe reduction in the HC. One brain showed total absence of the HC and another had HCi = 0.04. Among the 12 brains with HCi < 0.1, eight were from the F2 or F3 hybrid cross of I/Ln and BTBR (distributions in Fig. 4). b. Overlapping points for mice at or near CCi = 0 are unpacked in separate dot distograms. For mice with CCi > 0.1, all have a normally distributred HC index > 0.7. The range for mice with no CC at all is very wide indeed. For those in the borderline region of 0 < CCi < 0.1, most have normal HC index but a few are in a grey zone.
Inbred strains
Several of the inbred strains examined in this study never showed abnormally small CCor HC: A/J, C3H/HeJ, C57BL/6J, DBA/2J, FVB/NJ, SJL/J. This finding confirms previous results from the same laboratory (Wahlsten et al., 2003b), data for which are available in the Mouse Phenome Database, and the new data are not presented here. The samples of BALB/cByJ and BALB/cJ brains in this study also were entirely normal (Fig. 3). It is highly likely that larger samples would reveal a few abnormalities (Table 1).
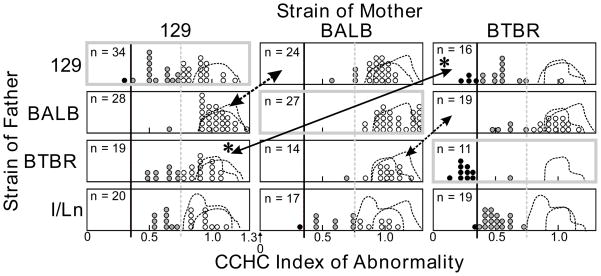
Fig. 3. Frequency distributions of the CCHC index of abnormality for several crosses.
Panels outlined in bold gray lines are inbred strains. Heavy black lines indicate the value of CCHCi = 0.35, below which an animal has no CC and an abnormally small HC; those animals are shown as black dots. Dashed gray line shows CCHCi = 0.75. Animals in the range 0.35 to 0.75 (gray dots) have a normal HC but small or absent CC. Animals above CCHCi = 0.75 (unfilled dots) have normal commissure sizes. F1 hybrid crosses with the normal strain C57BL/6J yielded entirely normal offspring and those data are not shown, but distributions of scores for each cross with C57BL/6J are represented by an outline with a fine dashed line. For crosses involving other strains, the outlines for the same strains crossed with C57BL/6J are superimposed to show the range of scores for anatomically normal mice. Arrows join reciprocal hybrid crosses. For the cross involving 129 and BTBR (shown as *), commissures were much more abnormal when BTBR was the mother.
Results for three inbred strains (Fig. 3) with more severe abnormalities replicated previous findings. All of the 11 BTBR T+ tf/J mice showed total absence of the CC and only one showed a HC in the normal range. The present sample of strain I/LnJ was reduced to one male with absent CC after death of another breeder, whereas a previous study of 50 I/LnJ brains found that all lacked a CC and about half had abnormally small HC (Livy et al., 1997). Data from that earlier study are reproduced in Fig. 4. The 129S1/SvImJ strain showed about 45% with abnormally small CC but only one of 33 with deficient HC, which is similar to values seen with other 129 strains (Table 1).
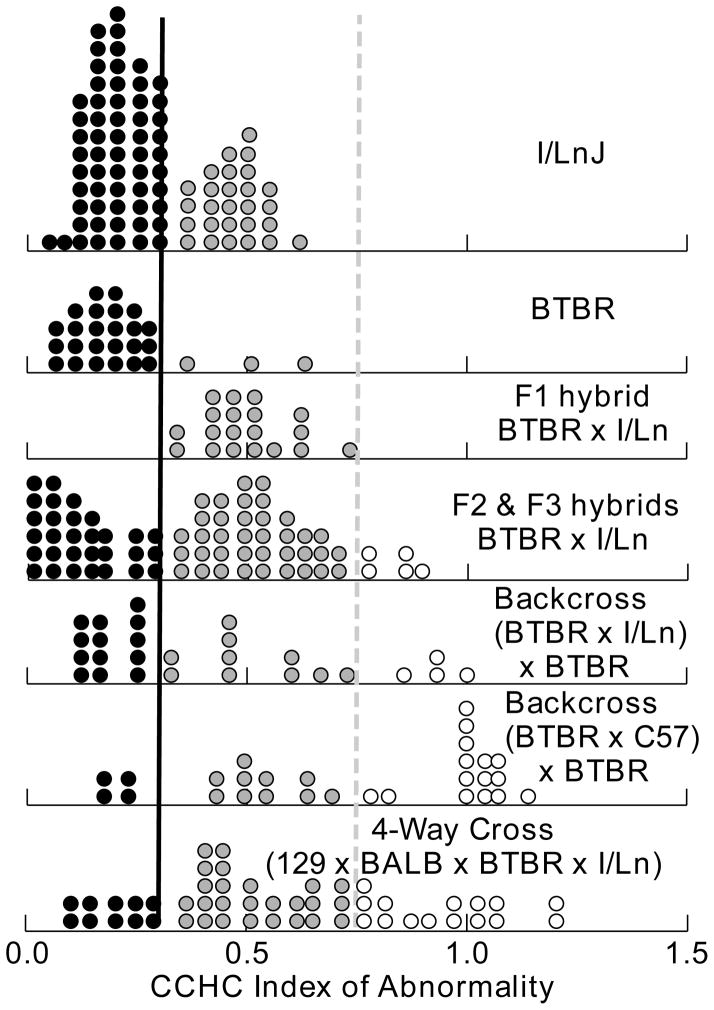
Fig. 4.
Distributions of the CCHC index of abnormality for crosses involving the inbred strains I/LnJ and BTBR T+ tfJ. Individuals shown as black dots (CCHCi < 0.35 below heavy black line) had no CC and abnormally small HC, where as those shown as gray dots (CCHCi < 0.75, dashed gray line) had normal size HC but abnormally small or absent CC. Animals shown as unfilled circles had normal CC and HC. Samples for the inbred parent strains were augmented with data from previous studies HC (Livy et al., 1997; Wahlsten et al., 2003). The distribution for the four-way cross among the four inbred stains prone to CC defects combines data for two specific kinds of crosses: (BALB x BTBR) x (129 x I/Ln) and (BALB x I/Ln) x (129 x BTBR).
F1 hybrid crosses
Crosses with C57BL/6J
Crossing any of the four abnormal strains with C57BL/6J indicated that commissure defects are completely recessive with respect to alleles possessed by that strain. Backcrosses of a hybrid derived from C57 to an abnormal parent strain revealed no abnormal offspring involving either BALB or 129 strains (data not shown), whereas there were many abnormalities in the backcross to BTBR (Fig. 4).
Crosses with BALB/cJ
Although BALB/cJ itself showed no commissure defects, the BALB x 129 and 129 xBALB crosses had a few brains with small CC, as did the crosses of BALB with the much more severely abnormal BTBR strain. The cross of BALB x I/Ln was far less affected than I/Ln but dramatically more affected than BALB. Thus, when BALB was crossed with any of the other three abnormal strains, the offspring clearly had more defective brains than when any of the same three strains were crossed with C57BL6J.
Crosses among 129, BTBR and I/Ln
For the pairing of the two most abnormal strains (Fig. 3), the BTBR x I/Ln F1 hybrid was far less affected than either abnormal parent; only one of 23 offspring had an abnormally small HC, compared with about 50% in I/Ln itself (Table 1, Fig. 4). This suggests that the two strains differ by at least two genes that contribute to commissure abnormalities. Crossing I/Ln with BALB yielded more defective brains than when BALB was crossed with the more anatomically severe BTBR strain, which also supports the existence of substantial genetic differences between BTBR and I/Ln. The 129 x I/Ln hybrid was less afflicted than strain 129. The cross of 129 and BTBR showed a maternal effect, where mice with a BTBR mother and 129 father had substantially more abnormalities than those with a 129 mother and BTBR father. This pattern supports an earlier finding of an X-linked gene in BTBR that contributes to corpus callosum defects (Kusek et al., 2007). The 129 x BTBR cross was less severely affected than strain 129, whereas the BTBR x 129 cross was more severely abnormal than strain 129.
F2 and F3 hybrid crosses of two abnormal strains
Because of the interesting result for the BTBR x ILn F1 hybrid cross, those animals were then mated to obtain F2 and F3 hybrids. The situations in the F2 and F3 were very similar and their data are therefore pooled in Fig. 4. The data point to genetic segregation at more than one locus. The range in the F2 and F3 mice was considerably greater than in either inbred parent or the F1 hybrid. Several animals showed total or almost total absence of the HC, something never found in either inbred parent. The bimodal distribution of the CCHCi index was not a purely genetic phenomenon because isogenic I/Ln mice also show bimodality. Almost all of the F2 and F3 mice could be divided into two phenotypic classes: those with no CC and a very small or absent HC, and those with no CC but a normal HC. A small number of mice showed normal HC and a small CC. The backcross of BTBR x I/Ln to BTBR had no animals with severely deficient HC.
Three-way crosses
Crosses among three abnormal strains
No matter which strain was omitted from a 3-way cross of abnormal strains (Fig. 5A, B), severe defects were seen, including animals with no CC and a very small HC. Likewise, a considerable number of anatomically normal brains resulted, regardless of which parent strain did not contribute to the cross. This indicates that severe abnormalities of the forebrain commissures as well as normal development can arise from a variety of combinations of genetic defects carried by the inbred parents.
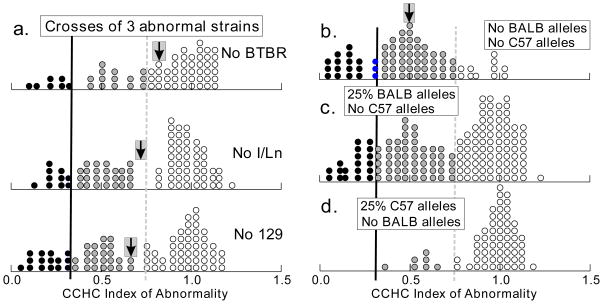
Fig. 5. Distributions of CCHC index for three-way crosses.
Limits for CCHCi and dot shading have the same meanings as in Fig. 3 and Fig. 4. Each distribution represents data pooled for two or more specific crosses listed in Table 2. Mean CCHC index is shown by an arrow for several groups, and one standard error above and below that mean is indicated by width of the grey box around the arrow. a. Crosses involving three of the four abnormal strains, each lacking alleles from one of the strains BTBR, I/Ln or 129. The distribution termed “No 129,” for example, involved six different three-way crosses among three of the four inbred strains that are prone to CC defects, but none of those crosses had a strain 129 ancestor. b. Three-way crosses among 129, BTBR and I/Ln with no BALB alleles. c. Data for three-way crosses having 25% BALB alleles but no C57 alleles. d. Three-way crosses with 25% C57 alleles had the other 75% of alleles from two of the three abnormal strains, not including BALB.
The average index of abnormality was strongly and significantly (F = 16.1, df = 3/284, P < .00001) influenced by which of the four abnormal strains was omitted from the 3-way cross (Fig. 5A, B). The proportion of commissure defects was clearly lower when BTBR was omitted (Fig. 5A), while many more brains were abnormal when BALB was omitted (Fig. 5B). Within a set of 3-way crosses having the same strain omitted as a parent, there was no consistent pattern resulting from which strain contributed 50% of the alleles. For the sets where either 129, BALB or BTBR was omitted, there were no significant effects of which of the other three strains contributed 50% of the alleles (all P > .01), whereas the set with I/Ln omitted showed more severe abnormalities when 129 contributed 50% of the alleles than when either BALB or BTBR contributed 50% (P < .01). This might indicate some kind of epistatic interaction, but in view of the relatively small sample sizes for any one cross and the imperfect balancing of the breeding design, no such conclusion is warranted.
3-way crosses where BALB/cJ was replaced by C57BL6J
Other comparisons confirmed that BALB alleles make a mouse vulnerable to commissure defects, even though that strain rarely shows anatomical defects. When one of the strains 129, BTBR or I/Ln was replaced with BALB (Fig. 5C), there were still many defective brains but severity was reduced compared with the 3-way cross of 129, BTBR, and I/Ln (Fig. 5B). If, on the other hand, either of 129, BTBR, or I/Ln was replaced by a C57 ancestor (Fig. 5D) and there was no BALB ancestor, almost all mice were normal. Among those 3-way crosses with one C57 grandparent and either 25% or 50% BALB alleles, all animals were normal (data not shown).
Four-way cross
The wide phenotypic range in the 4-way cross was remarkable (Fig. 4). Unlike the F2 cross of BTBR and I/Ln, there were many mice with anatomically normal brains. The 4-way cross could be divided into four phenotypic classes: mice with no CC and severely reduced HC, mice with normal HC but no CC, mice with normal HC and small CC, and mice with normal commissures. The wide range and genetic diversity make the 4-way cross especially promising for future genetic linkage studies as well as studies of phenotypic correlates of commissure abnormalities.
Despite the high frequency of commissure abnormalities in the F2 and F3 crosses of BTBR and I/Ln as well as the 4-way cross of abnormal strains, the reproductive performance of the hybrid parents was outstanding compared with inbred parents.
In view of the interesting results for the segregating crosses in this study, especially the wide phenotypic range of commissure sizes in many crosses, the material should be useful for genetic linkage analysis. Accordingly, tissue samples from all mice in this study have been carefully preserved at −70°C until PCR can be done.
Discussion
Defects of the corpus callosum in BALB strains and F1 hybrids with BALB
The frequency of abnormally small CC in one previous report of BALB/cByJ was one in 20 (Wahlsten et al., 2003b), and it was one in 48 in another (Wahlsten et al., 2001). Earlier data on BALB/cJ indicated a substantially higher frequency of about 39% absent and small CC but all normal HC (Wahlsten, 1977), and the discrepancy with current results is noteworthy. It is not known whether the change in BALB/cJ reflects modifications in the breeding regimen (Wahlsten, 1982a; Wahlsten, 1982b) or possibly new mutations that modulate CC formation. Whatever the case, it is clear from the F1 hybrids and 3-way crosses of BALB/cJ with other abnormal strains in the present study that the BALB/cJ genotype contains alleles that make mice prone to commissure defects, even though overt defects in BALB/cJ are now rare. This fact is evident only when the BALB alleles are combined with more severely abnormal alleles from another strain.
One strategy for future research would be to increase the overall frequency of defective animals in crosses involving BALB/cJ by mating each hybrid animal with BTBR, which would generate a 3-way cross similar to what is shown in Fig. 5A when there is no I/LnJ ancestor. Every animal would be heterozygous for BTBR alleles that contribute to commissure defects. This would greatly increase the frequency of commissure abnormalities and should make it much easier to map chromosomal regions important for the defects seen in BALB and 129.
F1 hybrids among abnormal strains
Several crosses point to meaningful genetic differences between the inbred parent strains that invite linkage analyses. The crosses of BTBR x 129 show a striking reciprocal difference that may arise from an X-linked gene. The same pattern of results was apparent in crosses of BTBR and BALB/cByJ in a previous study (Kusek et al., 2007). The present data for reciprocals of BALB x BTBR (Fig. 3) are mildly suggestive of an X-linked gene effects.
Two of the crosses with I/LnJ are not informative at this stage of the research. Strain 129 x I/Ln showed a distribution very similar to that of the 129S1/SvImJ parent, while BALB x I/Ln was midway between the two parent strains. The I/LnJ x BTBR cross, on the other hand, revealed some major genetic differences between the two strains. The two strains are not closely related in the genealogy of inbred strains (Petkov et al., 2004), even though they show very similar anatomical defects. Nevertheless, the HC of their F1 hybrid is almost invariably normal, while their F2 and F3 hybrid crosses have a bimodal distribution of HC size and a wider phenotypic range then either inbred parent. The F2 hybrid cross of these two strains should be valuable for mapping relevant genetic regions.
Development of the HC and CC
The developmental origins of the mouse HC are known at a gross anatomical level. Axons of the HC arise from the hippocampal formation and proceed in an anterior direction via the fimbria towards the medial septal region, where they form the columns of the fornix in a normal hybrid embryo weighing about 0.25 g. The first axons from the fornix cross midline to form the early HC in 0.3 g mouse embryos (Livy & Wahlsten, 1997), more than 24 h before the first CC axons arising from frontal cortex traverse the interhemispheric fissure in embryos weighing about 0.6 g (Ozaki & Wahlsten, 1992). In the strains BALB/cWah1 and 129P1/ReJ, formation of the HC can be markedly retarded but the CC will eventually form normally, provided the HC is present at midline when the first CC axons arrive (Livy & Wahlsten, 1997; Wahlsten & Smith, 1989). If HC formation is too severely retarded, however, the putative CC axons never cross midline (Wahlsten et al., 2006). Thus, it appears that absent CC is to a large extent a secondary consequence of faulty formation of the HC in these strains as well as I/Ln and BTBR.
Molecular biology of axon guidance in the mouse forebrain
Many of the molecular components of the brain that are involved in axon guidance are well known. As pointed out by (Donahoo & Richards, 2009): “The environment along the extent of the callosal trajectory is complex, comprising multiple overlapping combinations of guidance factors.” More than 60 genes pertinent to axon guidance in the mouse forebrain have been identified through the use of targeted mutations (Demyanenko et al., 1999; Donahoo & Richards, 2009; Shen et al., 2008; Shu et al., 2003). Some of those mutations impair formation of all forebrain commissures, including the anterior commissure that is never abnormal in the four inbred strains studied here (Bilasy et al., 2011; Kassai et al., 2008; Piper et al., 2009a; Shen et al., 2002).
Other mutations reduce the HC and eliminate the CC but have lesser or no apparent effect on the AC (Donahoo & Richards, 2009; Magara et al., 1999). Mutations of the doublecortin (DCX) gene alter hippocampal development, reduces the HC, and gives rise to total absence of the CC in both mice and humans (Kappeler et al., 2007), and the anatomical defects appear to be very similar to what is seen in I/Ln and BTBR mice, although the AC is somewhat reduced in size and disorganized. A deletion of the DISC1 gene in humans is associated with absence of the CC (Osbun et al., 2011). The mouse Disc1 gene is highly expressed in the embyonic CC (Osbun et al., 2011), and mice with a targeted mutation of Disc1 also show reduced CC (Shen et al., 2008). A deletion in the Disc1 gene on mouse chromosome 8 is intriguing because it is found in certain 129 substrains (Koike et al., 2006) as well as strain BTBR (Clapcote & Roder, Mouse Gene Informatics MGI 3699429), but the deletion is also found in strain LP/J that shows a normal corpus callosum (unpublished data from Munn et al., 2011). Whether the genes involved in the four abnormal inbred strains examined here entail unusual alleles of any of the targeted mutations known to affect CC development remains to be investigated.
The corpus callosum and autism
Several human developmental disorders of the nervous system involve moderate to high frequency of absent corpus callosum (Paul, 2011). Among them, autism interests mouse researchers who are devising animal models of deficient social behavior that is typical of autistic children (Crawley, 2004). The inbred strain BTBR T+ tfJ has drawn attention because of reduced social interactions (Bolivar et al., 2007; McFarlane et al., 2008; Moy et al., 2007), and its lack of a corpus callosum has inspired thoughts about a possible contribution of CC deficits to autistic behavior. The 129S1/SvImJ strain also shows social deficits and has a moderately high frequency of absent CC (Moy et al., 2007). The correlation of absent CC with impaired social behavior may not represent a causal link, however. Lesion of the CC in neonates does not impair social behaviors, and the LP/J strain that is closely related to BTBR shares its social deficits but has a normal corpus callosum (Yang et al., 2009). The I/LnJ strain also expresses some unusual social interactions (Lipp & Waanders, 1990; Lipp & Wahlsten, 1992). Further study of the CC-behavior correlation may be informative using some of the hybrid crosses reported here. Deficiency of the CC may be a contributing cause but not a sufficient cause of deficient social behaviors.
Principal findings and directions for future research
The greatly reduced severity of hippocampal commissure defects in the F1 hybrids of I/Ln and BTBR mice and the bimodal distribution of HC size in their F2 and F3 hybrids point to a major genetic difference between the strains that should be amenable to linkage analysis.
The 4-way cross of the four abnormal inbred strains (Fig. 4) shows a very wide phenotypic range of commissure sizes that makes it an ideal genetic population for investigation of behavioral correlates of CC and HC sizes.
The 3-way crosses reveal that commissure size is a complex trait and may be amenable to QTL mapping. As more genotypes are involved in a cross, defects at any one locus may behave more like a quantitative trait where many different gene combinations can yield the same phenotypic value.
The data suggest a two-stage strategy is most promising for future research: first identify the major genes responsible for defects in the severely afflicted I/Ln and BTBR strains, and then to use this genetic information to evaluate linkage in additional crosses that combine alleles from the BALB and 129 strains with those known to be important in I/Ln and BTBR.
Acknowledgments
This research was supported by grant AA12714 (Wahlsten, PI) from the National Institute of Alcoholism and Alcohol Abuse. Technical assistance of Erika Hayes is greatly appreciated.
Footnotes
None of the authors has any conflict of interest with regard to the research reported here.
References
- Bilasy SE, Satoh T, Terashima T, Kataoka T. RA-GEF-1 (Rapgef2) is essential for proper development of the midline commissures. Neurosci Res. 2011;71:200–209. doi: 10.1016/j.neures.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Wahlsten D. Sex and species differences in mouse and rat forebrain commissures depend on the method of adjusting for brain size. Brain Res. 1999;815:358–366. doi: 10.1016/s0006-8993(98)01088-9. [DOI] [PubMed] [Google Scholar]
- Bolivar VJ, Walters SR, Phoenix JL. Assessing autism-like behavior in mice: variations in social interactions among inbred strains. Behav Brain Res. 2007;176:21–26. doi: 10.1016/j.bbr.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulman-Fleming B, Wahlsten D. Effects of hybrid maternal environment on brain growth and corpus callosum defects of inbred BALB/c mice: A study using ovarian grafting. Exp Neurol. 1988;99:636–646. doi: 10.1016/0014-4886(88)90180-x. [DOI] [PubMed] [Google Scholar]
- Carlier M, Nosten-Bertrand M, Michard-Vanhee C. Separating genetic effects from maternal environmental effects. In: Goldowitz D, Wahlsten D, Wimer RE, editors. Techniques for the Genetic Analysis of Brain and Behavior: Focus on the Mouse. Elsevier; Amsterdam: 1992. pp. 111–126. [Google Scholar]
- Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol. 2008;29:386–397. doi: 10.1016/j.yfrne.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment Retard Dev Disabil Res Rev. 2004;10:248–258. doi: 10.1002/mrdd.20039. [DOI] [PubMed] [Google Scholar]
- Demyanenko GP, Tsai AY, Maness PF. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J Neurosci. 1999;19:4907–4920. doi: 10.1523/JNEUROSCI.19-12-04907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahoo AL, Richards LJ. Understanding the mechanisms of callosal development through the use of transgenic mouse models. Semin Pediatr Neurol. 2009;16:127–142. doi: 10.1016/j.spen.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Allometry and size in ontogeny and phylogeny. Biol Rev. 1966;41:587–640. doi: 10.1111/j.1469-185x.1966.tb01624.x. [DOI] [PubMed] [Google Scholar]
- Kappeler C, Dhenain M, Phan Dinh TF, Saillour Y, Marty S, Fallet-Bianco C, Souville I, Souil E, Pinard JM, Meyer G, Encha-Razavi F, Volk A, Beldjord C, Chelly J, Francis F. Magnetic resonance imaging and histological studies of corpus callosal and hippocampal abnormalities linked to doublecortin deficiency. J Comp Neurol. 2007;500:239–254. doi: 10.1002/cne.21170. [DOI] [PubMed] [Google Scholar]
- Kassai H, Terashima T, Fukaya M, Nakao K, Sakahara M, Watanabe M, Aiba A. Rac1 in cortical projection neurons is selectively required for midline crossing of commissural axonal formation. Eur J Neurosci. 2008;28:257–267. doi: 10.1111/j.1460-9568.2008.06343.x. [DOI] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci USA. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusek GK, Wahlsten D, Herron BJ, Bolivar VJ, Flaherty L. Localization of two new X-linked quantitative trait loci controlling corpus callosum size in the mouse. Genes Brain Behav. 2007;6:359–363. doi: 10.1111/j.1601-183X.2006.00264.x. [DOI] [PubMed] [Google Scholar]
- Lipp HP, Waanders R. The acallosal mouse strain I/Ln: behvaioral comparisons and effects of cross-breeding. Behav Genet. 1990;20:728–729. [Google Scholar]
- Lipp HP, Wahlsten D. Absence of the corpus callosum. In: Driscoll P, editor. Genetically-Defined Animal Models of Neurobehavioral Dysfunction. Birkhauser; Boston: 1992. pp. 217–252. [Google Scholar]
- Livy DJ, Schalomon PM, Roy M, Zacharias MC, Pimenta J, Lent R, Wahlsten D. Increased axon number in the anterior commissure of mice lacking a corpus callosum. Exp Neurol. 1997;146:491–501. doi: 10.1006/exnr.1997.6564. [DOI] [PubMed] [Google Scholar]
- Livy DJ, Wahlsten D. Tests of genetic allelism between four inbred mouse strains with absent corpus callosum. J Hered. 1991;82:459–464. doi: 10.1093/oxfordjournals.jhered.a111128. [DOI] [PubMed] [Google Scholar]
- Livy DJ, Wahlsten D. Formation of the hippocampal commissure in normal and acallosal mouse embryos. Hippocampus. 1997;7:2–14. doi: 10.1002/(SICI)1098-1063(1997)7:1<2::AID-HIPO2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Magara F, Muller U, Li ZW, Lipp HP, Weissmann C, Stagljar M, Wolfer DP. Genetic background changes the pattern of forebrain commissure defects in transgenic mice underexpressing the beta-amyloid-precursor protein. Proc Natl Acad Sci USA. 1999;96:4656–4661. doi: 10.1073/pnas.96.8.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Perez A, Holloway LP, Barbaro RP, Barbaro JR, Wilson LM, Threadgill DW, Lauder JM, Magnuson TR, Crawley JN. Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav Brain Res. 2007;176:4–20. doi: 10.1016/j.bbr.2006.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn E, Bunning M, Prada S, Bohlen M, Crabbe JC, Wahlsten D. Reversed light-dark cycle and cage enrichment effects on ethanol-induced deficits in motor coordination assessed in inbred mouse strains with a compact battery of refined tests. Behav Brain Res. 2011;224:259–271. doi: 10.1016/j.bbr.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbun N, Li J, O’Driscoll MC. Genetic and functional analyses indentify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet. 2011;155:1865–1876. doi: 10.1002/ajmg.a.34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki HS, Wahlsten D. Prenatal formation of the normal mouse corpus callosum: A quantitative study with carbocyanine dyes. J Comp Neurol. 1992;323:81–90. doi: 10.1002/cne.903230107. [DOI] [PubMed] [Google Scholar]
- Paul LK. Developmental malformation of the corpus callosum: a review of typical callosal development and examples of developmental disorders with callosal involvement. J Neurodev Disord. 2011;3:3–27. doi: 10.1007/s11689-010-9059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Ding YM, Cassell MA, Zhang WD, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Moldrich RX, Lindwall C, Little E, Barry G, Mason S, Sunn N, Kurniawan ND, Gronostajski RM, Richards LJ. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 2009a;4:43. doi: 10.1186/1749-8104-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Anderson A, Shen WB, Huang H, Plachez C, Zhang J, Mori S, Kinsman SL, Richards LJ. Imaging, anatomical, and molecular analysis of callosal formation in the developing human fetal brain. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:191–204. doi: 10.1002/ar.a.20282. [DOI] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, McCaig CD, Riedel G, St CD. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Mani S, Donovan SL, Schwob JE, Meiri KF. Growth-associated protein-43 is required for commissural axon guidance in the developing vertebrate nervous system. J Neurosci. 2002;22:239–247. doi: 10.1523/JNEUROSCI.22-01-00239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Yee D, Matin A, Nadeau JH, Magnuson T. Genealogy of the 129 inbred strains: 129/SvJ is a contaminated inbred strain. Mamm Genome. 1997;8:441–442. doi: 10.1007/s003359900453. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Genetic variation in the development of mouse brain and behavior: Evidence from the middle postnatal period. Dev Psychobiol. 1975;8:371–380. doi: 10.1002/dev.420080411. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Heredity and brain structure. In: Oliverio A, editor. Genetics, Environment and Intelligence. Elsevier; Amsterdam: 1977. pp. 93–115. [Google Scholar]
- Wahlsten D. Deficiency of corpus callosum varies with strains and supplier of the mice. Brain Res. 1982a;239:329–347. doi: 10.1016/0006-8993(82)90513-3. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Mice in utero while their mother is lactating suffer higher frequency of deficient corpus callosum. Dev Brain Res. 1982b;5:354–357. doi: 10.1016/0165-3806(82)90135-3. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Maternal effects on mouse brain weight. Dev Brain Res. 1983;9:215–221. doi: 10.1016/0165-3806(83)90054-8. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Growth of the mouse corpus callosum. Dev Brain Res. 1984;15:59–67. doi: 10.1016/0165-3806(84)90140-8. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Mouse Behavioral Testing. Elsevier; London, UK: 2011. [Google Scholar]
- Wahlsten D, Bishop KM, Ozaki HS. Recombinant inbreeding in mice reveals thresholds in embryonic corpus callosum development. Genes Brain Behav. 2006;5:170–188. doi: 10.1111/j.1601-183X.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Colbourne F, Pleus R. A robust, efficient and flexible method for staining myelinated axons in blocks of brain tissue. J Neurosci Methods. 2003;123:207–214. doi: 10.1016/s0165-0270(02)00359-x. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Crabbe JC, Dudek BC. Behavioral testing of standard inbred and 5HT1B knockout mice: implications of absent corpus callosum. Behav Brain Res. 2001;125:23–32. doi: 10.1016/s0166-4328(01)00283-2. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Crabbe JC. Survey of 21 inbred mouse strains in two laboratories reveals that BTBR T/+ tf/tf has severely reduced hippocampal commissure and absent corpus callosum. Brain Res. 2003;971:47–54. doi: 10.1016/s0006-8993(03)02354-0. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Smith G. Inheritance of retarded forebrain commissure development in fetal mice: Results from classical crosses and recombinant inbred strains. J Hered. 1989;80:11–16. doi: 10.1093/oxfordjournals.jhered.a110781. [DOI] [PubMed] [Google Scholar]
- Yang M, Clarke AM, Crawley JN. Postnatal lesion evidence against a primary role for the corpus callosum in mouse sociability. Eur J Neurosci. 2009;29:1663–1677. doi: 10.1111/j.1460-9568.2009.06714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]