Haplotyping of human chromosomes is a prerequisite for cataloguing the full repertoire of genetic variation. We present a microfluidics-based linked-read sequencing technology that can phase and haplotype germline and cancer genomes using nanograms of input DNA. This high-throughput platform prepares barcoded libraries for short read sequencing and computationally reconstructs long range haplotype and structural variant information. We generate haplotype blocks in a nuclear trio that are concordant with expected inheritance patterns and phased a set of structural variants. We also resolve the structure of the EML4/ALK fusion in the NCI-H2228 cancer cell line using phased exome sequencing. Finally, we assign genetic aberrations to specific megabase-scale haplotypes generated from whole genome sequencing of a primary colorectal adenocarcinoma. This approach resolves haplotype information using up to 100 times less genomic DNA than some existing methods and enables the accurate detection of structural variants.
The human genome is diploid, with each cell containing a copy of both the maternal and paternal chromosomes. A comprehensive understanding of human genetic variation requires identifying the order, structure, and origin of these sets of alleles and their variants across the genome1. Haplotypes, the contiguous phased blocks of genomic variants specific to one homologue or another, are essential to such an analysis. Genome-scale haplotype analysis has many advantages for improving genetic studies. Phasing of germline variants can be used to identify causative mutations in pedigrees, determine the structure of genomic rearrangement events and unravel cis-versus trans-relationships of ostensibly linked variants. In the case of cancer genomes, phasing studies provide insight into the haplotype context of somatic mutations, genomic rearrangements critical for oncogenesis and aneuploidy-state of chromosomes2.
Traditionally, haplotype analysis has relied on statistical inference using genomic sequences of related individuals3. More recently, experimental sequencing approaches have been used1, 2, 4–9. Next generation sequencing (NGS) methods for determining haplotype either require sample dilution into a limited number of wells in 96 or 384 format microtiter plates for sequencing library preparation6–9 or subcloning into fosmids1, 2. In dilution haplotyping, the low molarity of molecules per given partition reduces the likelihood that one DNA molecule has overlapping sequence with another and thus haplotypes can be derived. However, existing dilution methods require complicated experimental protocols, high amounts of starting DNA, extensive manipulation of DNA and microwell plates with restricted partitioning capacity6–9. Recently, experimentally phased genome assemblies have also been demonstrated with long-read technology10, but high input DNA requirements, lower sequence read accuracy, and cost burdens create substantial barriers for its widespread adoption.
Here we develop a microfluidic technology that circumvents these complex experimental protocols and generates haplotype-resolved genome sequences using small amounts of input DNA. Approximately 300 genomic equivalents, or 1 ng of high molecular weight (HMW) genomic DNA, is distributed across over100,000 droplet partitions, where it is barcoded and subjected to random priming and polymerase amplification. Barcode-tagged DNA molecules are released from each droplet, after which a modified library preparation process is performed. The resulting libraries then undergo standard Illumina short-read sequencing. A computational algorithm uses the barcodes to link sequencing reads to the originating HMW DNA molecule, enabling the construction of contiguous segments of phased variants. Loading nanogram amounts of DNA into large number of gel beads minimizes the chance of coincidental barcode overlap and improves overall phasing performance. Furthermore, the increased number of barcodes per amount of genomic DNA improves detection of structural variants. Using exome enrichment, we determine gene-level phasing in a nuclear family trio and define the complex structure of an oncogenic rearrangement in a cancer cell line. Finally, we generate a phased genomic analysis of a primary colorectal cancer originating from clinical tissue sample.
RESULTS
Microfluidics and gel bead partitioning
This high-throughput, droplet-based reagent delivery system uses hydrogel beads (gel beads) to deliver barcoded oligonucleotides. Within the confines of a microfluidic chip, monodispersed gel beads are deformable and can be delivered with a single gel bead fill rate of over 85% per droplet11. An individual gel bead is functionalized with millions of copies of the same barcoded oligonucleotide. Droplet partitions number more than 100,000 and this feature exceeds what can be achieved with microwell systems or virtual partitioning9 by orders of magnitude.
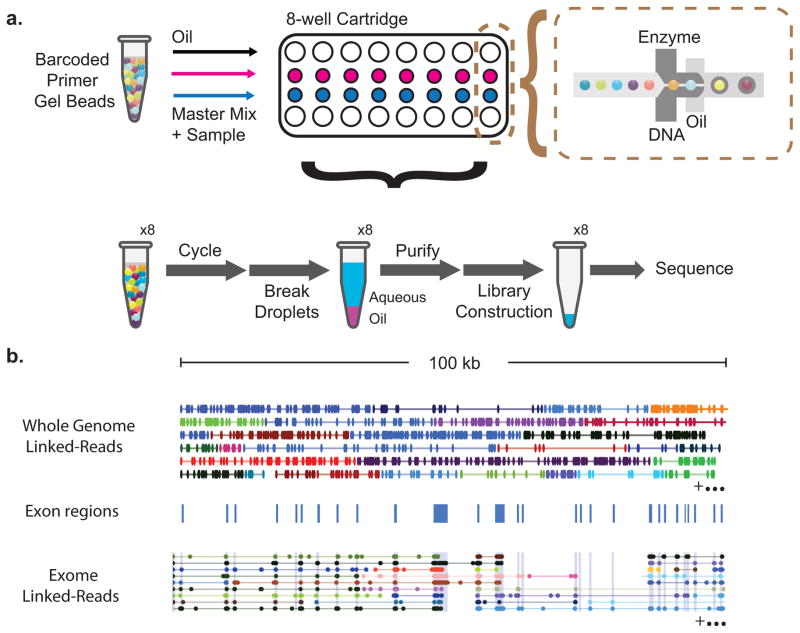
Reagent delivery and sample partitioning are performed within a plastic microfluidic consumable cartridge, which processes eight samples simultaneously. Cartridge reservoirs are loaded with gel beads, sample and reagent mixture and oil-surfactant solution. Reagents are delivered from the reservoirs via a network of microfluidic channels to a microfluidic “double-cross” junction (Fig. 1a). The first junction combines a close-packed aqueous slurry of gel beads with the sample and reagent mixture, and the second junction delivers the oil-surfactant solution. Droplet generation occurs at a rate of ~1 kHz at the second junction, resulting in more than 100,000 droplets loaded with greater than 85% single occupancy per droplet partition. The droplets flow to a collection reservoir where they are subsequently transferred to a conventional 96-well plate. A chemical reducing agent in the reaction mix dissolves the gel beads, triggering the release of the barcoded oligonucleotides from the gel matrix. As a result, independent solution-phase reactions are conducted without further addition of reagents.
Figure 1. Overview of the technology for generating linked-reads.
(a) Gel beads loaded with primers and barcoded oligonucleotides are first mixed with DNA and enzyme mixture, and subsequently mixed with oil-surfactant solution at a microfluidic “double-cross” junction. Gel bead-containing droplets flow to a reservoir where gel beads are dissolved, initiating whole genome primer extension. The products are pooled from each droplet. The final library preparation requires shearing the libraries and incorporation of Illumina adapters. (b) Top panel, linked-reads of the ALK gene from the NA12878 WGS sample. Each line represents linked-reads with the same barcode, with dots representing reads, and color depicting reads with different barcodes. Middle panel, blue blocks showing exon boundaries of the ALK gene. Bottom panel, linked-reads of ALK gene from the NA12878 exome data. Although there are only reads in exon regions, reads from neighboring exons are linked because of common barcodes. Only a very small fraction of linked-reads is presented here to conserve space.
In our study, 300 genomic equivalents (1 nanogram) are dispersed into >100,000 droplet partitions with different barcodes; thus, only a small number of genomic equivalents are loaded per partition. As we report later, it is highly unlikely that two distinct high-molecular weight molecules that cover the same loci but of opposing haplotype will share the same barcode (p<0.002). The use of fewer barcodes would require even smaller amounts of input DNA – equivalent to only a few genome equivalents – to maintain low rates of intermolecular genomic overlap within the same barcoded partition. Such low amounts of input DNA would lead to issues with significant allelic dropout and coverage gaps throughout the genome.
Barcode sequencing
To create barcoded DNA molecules for sequencing, we perform an optimized droplet-based assay that introduces a barcode-containing sequencing adapter into new fragments (Methods). High molecular weight DNA templates, ranging from ten to several hundred kb in size, are randomly distributed in picoliter reaction volumes across greater than 100,000 droplets. Within an individual droplet, gel bead dissolution releases the amplification primer into the partitioned solution. The primer contains the following components: i) an Illumina P5 sequence, ii) a designed 14bp barcode, iii) an Illumina R1 sequence and iv) a 10bp random primer sequence (Supplementary Fig. 1). Amplification is non-processive, and the length of molecules produced range from several hundred to several kilobases. After thermocycling the droplets, the emulsion is broken, the pooled aqueous fractions are recovered and the library preparation is completed with the addition of other adapter components. Additional details of library preparation and metrics of DNA loading are noted in the online Methods.
Whole genome sequencing with linked-reads
To assess the performance of linked-read sequencing for haplotyping, we relied on three HapMap samples that form a nuclear trio: NA12878 (mother), NA12877 (father) and NA12882 (child). These samples have been experimentally phased and have haplotypes statistically derived from inheritance patterns9, 12.
We generated barcode sequencing libraries from this trio. Approximately 1 ng of HMW DNA from each sample was processed on a 10× GemCode instrument and sequencing libraries were prepared. The barcode libraries underwent WGS to approximately 30× mean coverage. We used an Illumina Hiseq 2500 with 2×98 paired-end reads (Supplementary Table 1) for all sequencing analysis in this report. Reads were trimmed to remove the first 10 bp of each paired read to remove primer extension artifacts, aligned to the human reference genome (hg19) with BWA13, and PCR duplicates were marked utilizing the barcode information and alignment position (Methods, Supplementary Fig. 1). Overall, library quality was exceptionally high; greater than 95% of reads were mapped, more than 90% of the genome was covered at least once, with most gaps shorter than 50 kb (Table 1, Supplementary Table 1, Supplementary Fig. 2). PCR duplication-rates were minimal at less than 1.4% (Table 1). Library insert size averaged around 200 bp, and the number of binding events was estimated to be around 45,000 within each partition (Supplementary Table 2, Methods). Barcodes were uniformly distributed over 100,000 partitions based on an effective number of barcodes adjusted for unevenness of barcode counts (Methods, Table 1, Supplementary Fig. 2). The GC distribution is provided in Supplementary Figure 2d and shows the predictable effect of the non-processive amplification step.
Table 1.
Summary of phasing results.
| Phased whole genome sequencing | NA12878a | NA12877a | NA12882a | NA20847b | Patient 1532 Normalc | Patient 1532 Tumorc |
|---|---|---|---|---|---|---|
| Coverage | 37 | 34 | 36 | 32 | 31 | 32 |
| % aligned | 97 | 97 | 96 | 95 | 96 | 96 |
| % duplication | 1.38 | 1.38 | 1.19 | 6.45 | 0.5 | 0.7 |
| Relative genomic equivalents per partition | 0.0015 | 0.002 | 0.0019 | 0.0044 | 0.0015 | 0.0011 |
| % barcodes correctly assigned | 92 | 87 | 85 | 91 | 88 | 86 |
| Effective barcode diversity | 111808 | 151552 | 126326 | 144442 | 136888 | 125694 |
| Number of molecules (Million) | 15.0 | 34.5 | 23.4 | 43.7 | 30.0 | 20.6 |
| Length-weighted mean molecules length (kb) | 60.53 | 40.20 | 47.35 | 75.69 | 44.35 | 38.31 |
| % SNPs phased | 99 | 97 | 99 | 98 | 95 | 95 |
| % genes phased (<100 kb) | 97 | 92 | 97 | 95 | 93 | 91 |
| N50 phase block (kb) | 2834.44 | 890.08 | 1726.40 | 2577.04 | 1570.40 | 962.11 |
| Longest phase block (kb) | 14557.82 | 7016.43 | 11545.49 | 12729.94 | 8729.79 | 6706.66 |
| SNV short switch error rate (%) | 0.01% | 0.48% | 0.20% | 0.93% | N/A | N/A |
| SNV long switch error rate (%) | 0.01% | 0.03% | 0.02% | 0.09% | N/A | N/A |
| Phased exome sequencing | NA12878a | NA12877a | NA12882a | NA20847b | NCI-H2228 |
|---|---|---|---|---|---|
| Coverage | 239 | 497 | 443 | 185 | 244 |
| % bases on target | 57 | 62 | 59 | 62 | 52 |
| % aligned | 99 | 99 | 99 | 99 | 99 |
| % duplication | 8.09 | 18.03 | 13.79 | 4.76 | 8.26 |
| Relative genomic equivalents per partition | 0.0007 | 0.0010 | 0.0011 | 0.0008 | 0.0007 |
| % barcodes correctly assigned | 92 | 92 | 92 | 87 | 87 |
| Effective barcode diversity | 149170 | 138086 | 150115 | 154682 | 121906 |
| % genes phased (<100 kb) | 95 | 95 | 96 | 92 | N/A |
| N50 phase block (kb) | 136.54 | 103.42 | 113.65 | 83.94 | N/A |
| Longest phase block (kb) | 2086.24 | 1217.01 | 1199.03 | 957.57 | N/A |
| SNV short switch error rate (%) | 0.47% | 1.10% | 0.99% | 1.36% | N/A |
| SNV long switch error rate (%) | 0.05% | 0.11% | 0.07% | 0.06% | N/A |
Some droplets may coincidentally receive identical barcodes; performance for haplotyping is maintained by loading only a small amount of genomic DNA into each droplet partition. Conversely, significantly increasing DNA input increases the coincidental loading of overlapping DNA molecules to the same droplet and thus, barcode; the overlap among DNA templates obscures haplotype analysis. As a metric to gauge coincidental loading, we calculated the relative genome loading per a droplet partition. From nuclear trio WGS data, the relative genomic loading was less than 0.002 genome equivalents per partition (Table 1). Thus, these loading issues are minimized given the low molarity distribution of DNA per a droplet.
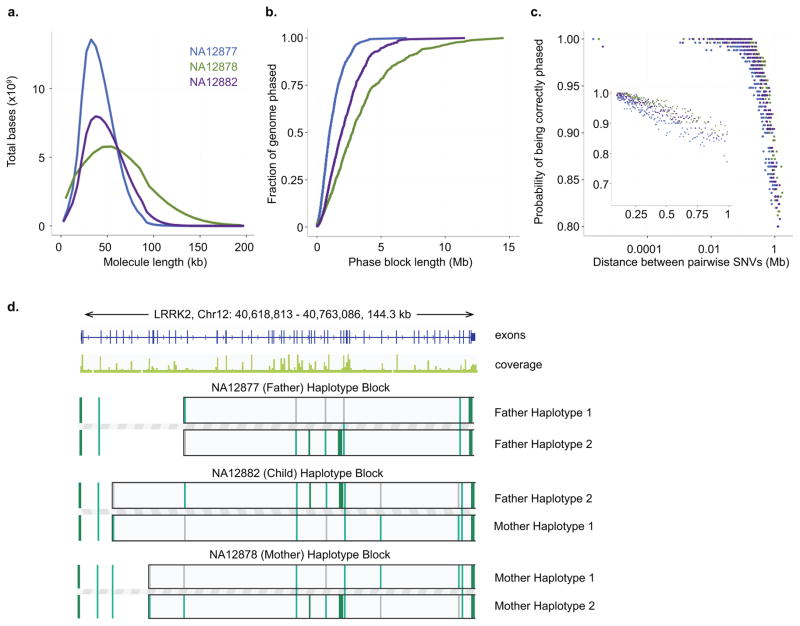
A feature of our analysis involves the concept of linked-reads; this term refers to sequences with the same barcode and determined to be in physical proximity based on alignment (Fig. 1b, Methods). This type of data enables the inference of input DNA length, phasing of haplotype blocks, and identification of DNA molecules with a new structural change (for example, genomic fusion) compared to the reference. The mean linked-reads per DNA molecule was about 15 for the trio libraries. With linked-reads, we determined that these libraries had an inferred length-weighted mean DNA molecule length of at least 40 kb, with maximum length reaching up to 200 kb (Fig. 2a). This is consistent with the distribution of input DNA detected by gel electrophoresis (Supplementary Fig. 2b).
Figure 2. Phasing performance of NA12878 trio analysis.
(a) Length-weighted molecule size histogram of the trio WGS data. The Y-axis represents the DNA mass in the molecule length bin, which is calculated to be the product of fraction of molecules in the length bin and median of the length bin. (b) Cumulative distribution function of phase block length of the trio WGS samples. (c) Phasing accuracy. For all pairs of SNVs that are on the same phasing block, the probability of correct phasing of a pair is plotted as a function of its distance. The insert shows SNV pairs that are at least 0.1Mb away from each other. (d) Haplotype blocks of LRRK2 gene of the trio exome libraries, demonstrating Mendelian inheritance. While most of this gene is phased in all trio samples, the beginning of the gene is not phased (represented by SNVs not as part of the haplotype block). For this gene, NA12882 (child) inherited one allele from Haplotype 2 from NA12877 (father), and Haplotype 1 from NA12878 (mother). Grey bars in the phase blocks represent reference alleles, and green bars represent alternative alleles.
Using low genomic fractions per barcode achieves a high number of linked reads per an individual DNA molecule. We load approximately 3 Mb of genomic DNA per partition. In comparison, the CPT-Seq phasing method averages about 21 to 62 Mb genomic DNA per partition9. Phasing variants becomes substantially more robust; the high number of barcode partitions in combination with the low number of haploid genome equivalents reduces the probability of a partition having two high-molecular weight molecules overlapping the same genomic loci but of opposing haplotype. As a result, we achieve excellent phasing performance, require low amounts of genomic DNA and rely on a standard WGS coverage to achieve larger haplotypes as described later. Further decreasing the amount of genomic DNA per partition will accordingly increase the linked reads per molecule. However, the variance in the amount of genomic DNA loaded per partition substantially increases, resulting in allelic dropout and uneven coverage. We have empirically observed that our approach is robust within two-fold of the optimal DNA loading amount (data not shown).
Without using standard WGS and after SNV calling14, our method can achieve a 0.93 SNV calling sensitivity (which is a measure of true positive rate) and 0.99 PPV (positive predictive value, which is a measure of precision) at 30× sequencing of NA12878. In contrast, a TruSeq library at 0.99 PPV has a SNV calling sensitivity of 0.98. We compared the coverage distribution between barcode libraries and Illumina TruSeq libraries (Supplementary Fig. 3). Overall, 90% of all genomic bases are covered by both samples. We note that the genome coverage of GemCode libraries is broader and biased against GC-rich regions (Supplementary Figure 2d). However, the metrics observed with standard TruSeq libraries require substantially higher input DNA; Illumina TruSeq libraries made from 1ng of input generally suffers from high proportions of PCR duplicates.
Experimentally derived haplotypes from control genomes
We utilized linked-reads to phase single nucleotide variants (SNVs) that were previously annotated (Table 1, Supplementary Table 4) across all three samples in the nuclear trio. We used a maximum likelihood approach to find near-optimal local haplotype assignments based on read and barcode support (Methods, Supplementary Note 1). A phasing score (PQ) is assigned to each SNV by comparing the likelihood of the observed data in both phasing states of the variant (Methods).
As an indicator of haplotyping accuracy, we determined the N50 value from our linked-read analysis. This phasing metric reflects the haplotype block interval where larger blocks represent 50% of the overall phased genome sequence1, 9. We identified N50 phase block lengths ranging from 0.9 Mb to 2.8 Mb, with over 97% of SNVs being phased (Table 1, Fig. 2b).
Long switch errors occur when a variant position is incorrectly phased in the context of the adjacent flanking variants9, 12. We used this well-established metric as another indicator of the genome-wide accuracy of our haplotyping analysis (Methods). The overall long switch error rate was less than 0.03% (Table 1) for the nuclear trio when compared to phasing information generated via trio sequencing15. As an additional indicator of phasing accuracy, we examined the distance between all pairwise SNVs per a given haplotype, and the probability that these pairs were accurately assigned. Using this metric, phasing accuracy remained above 95% out to an interval distance greater than 0.5 Mb (Fig. 2c).
We investigated the impact of lower sequencing coverage on phasing performance. We determined the haplotypes of NA12878 using down-sampled proportions of the linked-read data. At approximately 30 Gb of sequence (~10× average coverage), 93% of SNVs were phased, the N50 phase block length was 1.1 Mb, and long switch error rates were less than 0.03% (Supplementary Table 3).
Phasing performance decreases in regions with small number of heterozygous variants. As shown in Figure 2c, we demonstrate that the probability of correctly phasing SNVs scales with the pairwise distance between variants. We examined the phasing scores at low-density (5 SNVs per 100kb) heterozygous SNV positions derived from pre-called gold-standard variants of NA1287815. We observed only 1,407 variants that fit this criterion, corresponding to less than 1% of all the heterozygous SNVs. From this set of sparsely distributed SNVs, 94% were confidently phased past the threshold score of PQ>23. In addition, over 60% of the low-density variants were phased with a maximal phasing score of 255 – this represents exceptionally high phasing quality even for genomic regions with a sparse distribution of SNVs. As a reference, we have also examined all heterozygous SNVs in NA12878; 99% of them were phased accurately and exceeded the threshold score of PQ>23 and 93% were phased with a maximal, highest quality phasing score of 255. We can potentially increase the phasing performance in low-density heterozygous regions if we increase input molecule length, increase sequencing depth, or decrease the amount of genomes loaded per partition.
As another comparison, we analyzed the genome of the Hapmap subject NA20847, an individual of Gujarati Indian descent. This genome had been sequenced and haplotyped using a fosmid approach1. Library quality and phasing performance were similar to results obtained on the nuclear trio (Table 1). More than 98% of SNVs were phased, with N50 phase block size of ~2.5 Mb. We calculated a long switch error rate of less than 0.09% when compared to fosmid-based haplotyping study.
Discovery of phased germline structural variants (SVs)
With traditional short read sequencing approaches, the discovery of SVs is computationally difficult, particularly in highly repetitive regions of the genome. We used linked-read data to call breakpoints in large-scale SVs and assign them to specific haplotypes. We developed an algorithm that efficiently searches all pairs of genomic loci for regions of large numbers of overlapping barcodes. We identify non-adjacent candidate positions of structural variation (Fig. 3a, b, Supplementary Fig. 4). Each breakpoint is assigned a Phred-like quality score, which describes the likelihood of the breakpoint being called by chance (Methods, Supplementary Note 2). Linked-read data provides sequencing information spanning the region around a breakpoint up to 10s of kb or higher. Thus, linked-read analysis can differentiate between a putative breakpoint created by a true SV and a false positive confounded by repetitive sequences (Fig. 3b).
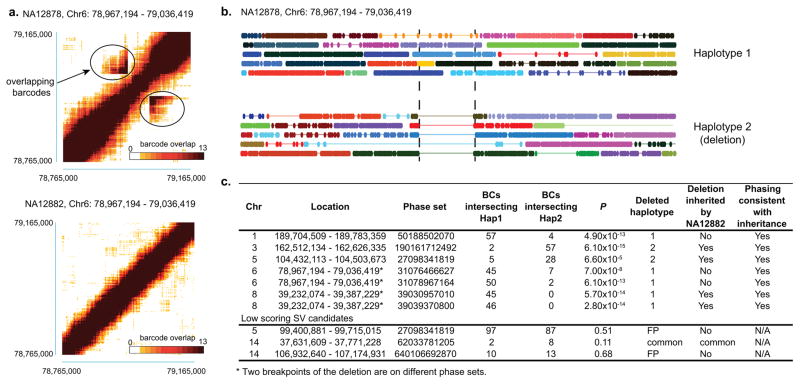
Figure 3. Detecting genomic deletions in NA12878.
(a) Heat map of overlapping barcodes is plotted for a deletion on Chr6: 78,967,194 – 79,036,419 in NA12878 (top). The areas circled in black represent overlapping barcodes near the breakpoints. The deletion is not observed in NA12882, and the heap map of barcodes in the same region is shown in the bottom as a negative control. (b) linked-read data of NA12878 WGS sample spanning Chr6: 78,967,194 – 79,036,419. Each line represents linked-reads with the same barcode, with dots representing reads, and color depicting reads with different barcodes. Dashed vertical black lines represent the breakpoints. The top panel represents the haplotype without a deletion. In this case, overlapping barcodes will only be observed in contiguous regions. Bottom panel represents the haplotype with a deletion, as shown by the gap in the linked-reads. In contrast to regions without a deletion, barcodes in the region before the gap will overlap with barcodes in the region after the gap. (c) Summary of 8 deletion candidates, including supporting evidence from overlapping barcode count, phasing of the deletion breakpoints, and inheritance support in NA12882. While all 5 high scoring SV candidates have support from each type of evidence, two of three lower scoring SV candidates lack support from any evidence included targeted sequencing. Haplotype assignment in one phase block is not necessarily the same as the haplotype assignment in a different phase block.
As an initial test we examined eight large genomic deletion candidates with sizes greater than 70 kb as previously identified and validated in NA12878 16–18. Five were highly ranked by our prediction algorithm, while three had lower scores (Fig. 3c). Barcode count analysis showed that the five high-scoring breakpoints were also consistent with a loss-of-heterozygosity (LOH) as would be expected for a deletion (Supplementary Fig. 5).
We phased these deletions by overlapping the barcodes supporting a breakpoint with barcodes from neighboring haplotype blocks (Fig. 3b). Counting the haplotype-specific barcodes provides another type of score for additional vetting of the putative deletion. Five of these candidate deletions had flanking haplotype blocks that cover both sides of the deletion (Fig. 3c). In contrast, the breakpoints of two putative candidates with a low score could not be phased. As additional evidence of the accuracy of our haplotype deletion calls, we examined the consistency of haplotype blocks with Mendelian inheritance for NA12878, the mother and NA12882, the child in the nuclear trio. Among the five deletions with the highest ranked score, three were inherited in the child (Fig. 3c, Supplementary Table 5a,b). For the highest ranking cases, when the analysis of the child showed the deletion, it also inherited the related haplotype; when the child did not inherit the deletion, the other maternal haplotype was inherited (Fig. 3c, Supplementary Table 6).
We validated these breakpoints with targeted sequencing 19, 20. For this study, we used targeting probes that tile across the genomic intervals where the two breakpoints are created by a deletion (Methods, Supplementary Table 7, Supplementary Fig. 6). Targeted reads were examined for evidence of a breakpoint junction and deletion. In general, the chimeric junctions of the high scoring deletion breakpoints were validated and showed the appropriate inheritance pattern (Supplementary Fig. 6, Supplementary Table 6). In contrast, two of the lower scoring deletion candidates could not be confirmed, one of which is in a VDJ recombination region (Supplementary Table 6). One of the lower scoring candidates did have targeted sequencing results pointing to a deletion that was seen among all three individuals.
Linked-read data reveals other types of structural rearrangements besides deletions. Overall, our SV algorithm called 20 structural variants in NA12878, of which 11 were identified as deletions in a recent de novo assembly10, two were reported as inversions16, and one was identified as a retro-transposon insertion21 (Supplementary Fig. 4, Supplementary Table 8, Methods). To assess the sensitivity of our method, we compared our calls to a set of deletions identified from both conventional short-read sequencing18 and long-read-assisted assembly10. There were only 3 such deletions, all of which were detected and validated (Fig. 3c, Methods).
Exome-based phasing
Generating barcode exome libraries is straightforward and enables the haplotyping of genes with linked-reads (Methods, Fig. 1b). Using approximately 1 ng of DNA from NA12878, NA12877 and NA12882, barcode libraries underwent exome enrichment with Agilent SureSelect kits and were sequenced to a depth of greater than 185× (Table 1). After data pre-processing and alignment, we observed that over 99% of the linked-reads aligned to the human genome reference, greater than 57% of the bases were on-target and the data had a PCR duplication rate up to 18% at 450× (Table 1). Our algorithm phased more than 95% of genes under 100 kb and we observed N50 phase block lengths greater than 103 kb for the nuclear trio (Table 1, Supplementary Fig. 2e, f). In addition, haplotype blocks were consistent with Mendelian inheritance across the trio (Fig. 2d).
Detection of an EML4/ALK rearrangement via exome phasing
SVs such as cancer rearrangements frequently occur in intronic sequences rather than exons and can lead to chimeric gene products. Exome sequencing does not detect gene fusions for which the breakpoint is more than a few hundred base pairs from an exon without custom targeting assays and extremely high sequencing coverage22, 23. To overcome these issues, we used exome linked-reads to detect a clinically actionable cancer rearrangement. The lung cancer cell line NCI-H2228 contains an EML4/ALK fusion24, 25 in which exons 1–6 of EML4 are fused to exons 20–29 of ALK. This rearrangement leads to constitutive activation of ALK26, an oncogenic kinase driver. We prepared a barcode sequencing library from approximately 1ng of genomic DNA, conducted exome enrichment, then sequenced to an average sequencing coverage of 204× after duplication filtering (Table 1).
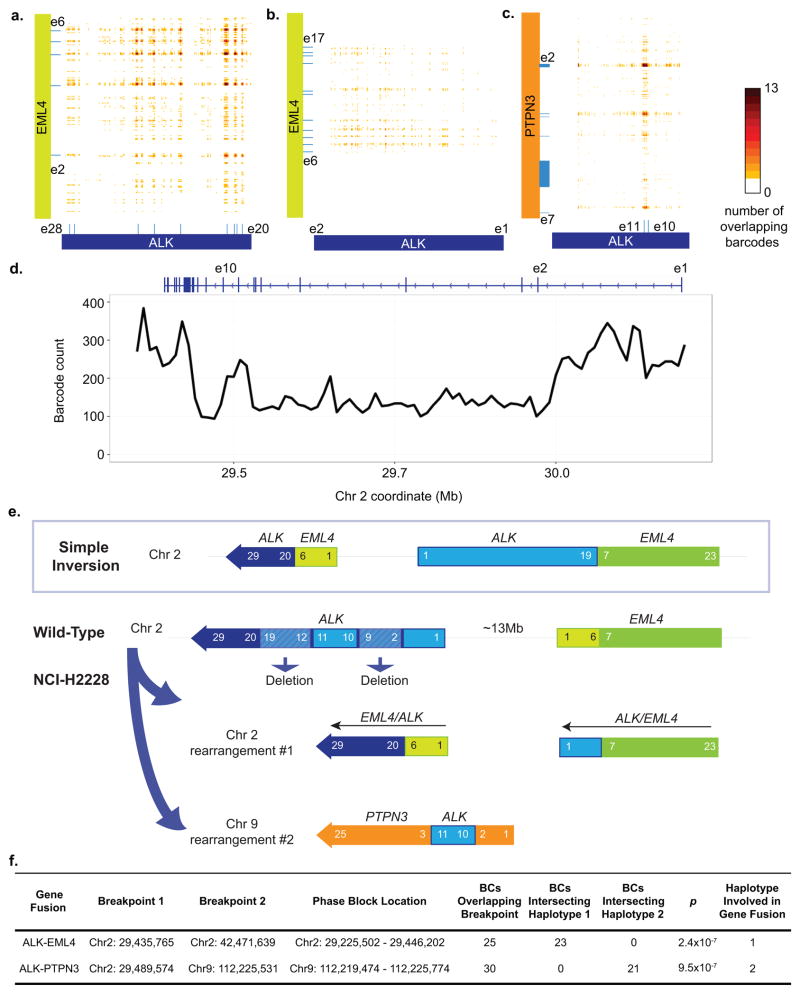
We correctly identified an EML4/ALK fusion (Fig. 4a–d, Supplementary Fig. 7a, b, Supplementary Table 9); our exome linked-read data showed that the rearrangement occurs between exons 20–26 of ALK and exons 2–6 of EML4 (Fig. 4a), consistent with previous reports and our own validation (Supplementary Fig. 7). A simple inversion would predict corresponding overlap between exon 19 of ALK with exon 7 of EML4 (Fig. 4e). Our results showed overlap of exon 1 of ALK and exon 7 of EML4 (Fig. 4b), suggesting a deletion of exons 2–19 of ALK and a more complex structure than a simple inversion. In addition, we identified an additional insertion of ALK exons 10–11 in the gene PTPN3 on chromosome 9 (Fig. 4c, Supplementary Fig. 7c, d, Supplementary Table 9) as has been previously reported27.
Figure 4. Rearrangement detection of an EML4-ALK gene fusion from exome sequencing of NCI-H2228.
(a) Overlap of barcodes between exons 2–16 of ALK and exons 2–6 of EML4. The heat map depicts the number of overlapping barcodes. Blue bars on ALK and EML4 represent exons. (b) Overlap of barcodes between exon 1 of ALK and exons 7–16 of EML4. (c) Overlap of barcodes between exons 10–11 of ALK and 5′ half of PTPN3. (d) Barcode counts in ALK region of NCI-H2228 WGS sample. The top blue bar represents the schematics of ALK gene structure, with e1, e2, and e10 denoting exon 1, exon 2, and exon 10, respectively. (e) Schematics illustrating complex chromosomal rearrangement involving ALK, EML4 and PTPN3. Instead of seeing the simple inversion reported in the literature, we observed a deletion, an inversion of ALK on Chr2 with EML4, and an insertion of ALK into PTPN3 on Chr9. (f) Phasing support around ALK and PTPN3 breakpoints in EML4-ALK, and ALK-PTPN3 gene fusion. Haplotype assignment in one phase block is not necessarily the same as the haplotype assignment in a different phase block.
Based on these results for this cell line, we inferred a refined structure of the overall structural rearrangement (Fig. 4e) covering the ALK deletion, EML4/ALK inversion, and insertion of exons 10–11 of ALK into PTPN3. Exons 20–29 of ALK are contained within a 220 kb phase block; only one haplotype overlaps with the EML4/ALK fusion. Similarly, exons 3–4 of PTPN3 are contained with a 40 kb phase block and there is a distinct segregation of the ALK insertion into only one haplotype of the PTPN3 gene (Fig. 4f). The rearrangement structure was separately verified with linked-reads whole genome sequencing (Supplementary Table 1, Supplementary Fig. 7c, d). Analysis of the barcode counts in the WGS data (Fig. 4d, f) revealed a coverage reduction consistent with a deletion in the region covering exons 2–19 of ALK.
Haplotype analysis of a primary colon adenocarcinoma
Whole genome haplotype analysis has been reported for the HeLa tumor cell line2, but the phasing analysis of primary tumor genome has remained difficult. Unlike cancer cell lines that are more homogeneous in their genetic composition, primary tumors derived from clinical samples pose a number of challenges. The amount of DNA from clinical samples is often limited and the cellular composition frequently includes normal stromal tissue. As a first time demonstration of a phasing analysis for a primary cancer genome, we analyzed a primary colon adenocarcinoma and identified three classes of genetic aberrations in the context of Mb-scale haplotypes: i) mutations; ii) copy number variants (CNVs); iii) rearrangements. First, we performed short read WGS on both the tumor and matched normal pair to an average sequencing coverage of 50× followed by variant calling (Supplementary Table 1). For both samples, we used ~2 million heterozygous SNVs determined from the short reads for subsequent phasing (Supplementary Table 4, Methods).
For the phased analysis of a primary cancer genome, we used ~1ng genomic DNA from the tumor and normal sample as direct input, without other preparative steps, for generating a barcode sequencing library. This tumor had 70% purity. We sequenced the barcode libraries to an average coverage of ~30×. After linked-read alignment, we determined that the genomic DNA had an inferred DNA length distribution with mean >50 kb (Fig. 5a). Over 94% of SNVs were phased with N50 phase block lengths of ~1.5 Mb for the normal sample and ~0.9 Mb for the tumor (Table 1, Fig. 5b). We generated haplotype blocks for both samples and examined the haplotype intersection intervals. From this intersection, 90% of the bases were shared between the two samples.
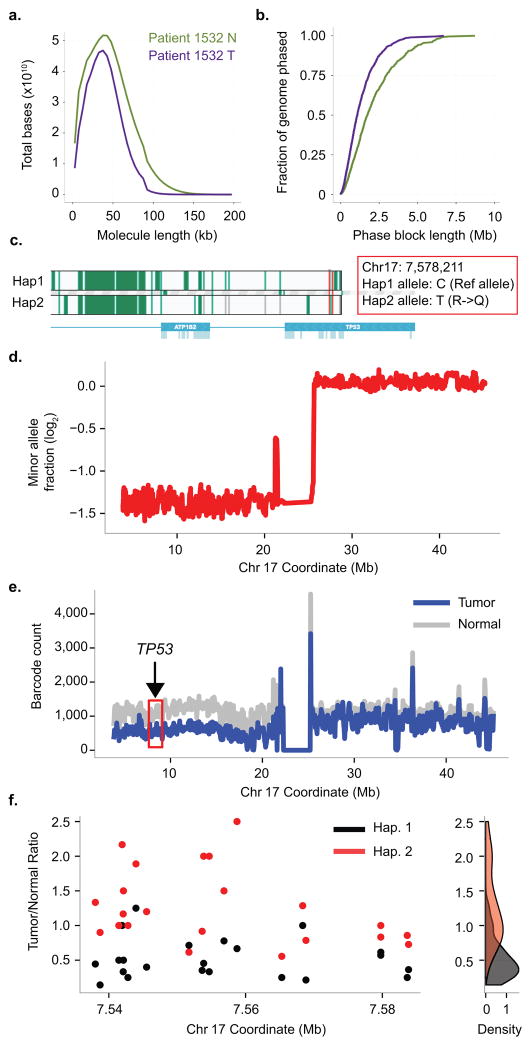
Figure 5. Phasing analysis of a primary colon cancer genome and structure of the TP53 driver event.
(a) Length-weighted molecule size histogram of Patient 1532 Normal (green) and Tumor (purple) samples. (b) Cumulative distribution function of phase block length of the Normal and Tumor pair. (c) Phased haplotype block showing TP53 C to T mutation on Haplotype 2. (d) Minor allele fraction of the tumor sample (relative to the matched normal) on Chr 17. There is a deletion in the p-arm of Chr 17 in the tumor sample with a copy number of 1. (e) Barcode count throughout Chr 17 between the tumor (blue) and matched normal (grey). The red box depicts the region where TP53 is located. (f) Phasing analysis of TP53 between tumor and matched normal. Left, Ratio of SNV counts between Tumor and Normal in TP53 region, Haplotype 1 in red and Haplotype 2 in black. Right, Density of SNV ratios of Haplotype 1 and Haplotype 2. Whereas the SNV density centers around 1 for Haplotype 2, most SNV ratios between Tumor and Normal is only 0.5 on Haplotype 2, indicating that LOH is on Haplotype 2.
Seventeen deleterious cancer mutations were identified per CADD scores 28 and assigned to specific haplotype blocks (Supplementary Table 10). A number of the mutations occurred in known colorectal cancer drivers such as TP53 and NRAS29. Using the linked-read analysis, we identified five rearrangements. Two are inter-chromosomal translocations (Supplementary Table 11). The short read WGS data provided validation of these events; the breakpoints of the five structural variants were confirmed by BreakDancer30 predictions and supported by reads covering the breakpoints (Supplementary Table 11). Analysis of the short read WGS for copy number alteration algorithm31, identified 26 copy number variant (CNV) intervals of which 24 were validated by counting the average number of barcodes and linked-reads (Supplementary Table 12). There were two CNV intervals (short-read analysis) not validated by barcode counting and occurred in centromeric and telomeric regions that had lower linked-read coverage.
We used linked-read analysis and haplotype blocks to explore the context and structural alterations affecting a critical driver mutation in TP53, namely a candidate deleterious non-synonymous R213Q mutation (Supplementary Table 10). Phasing analysis demonstrated that the R213Q mutation is within a 46 kb phase block on the haplotype 2 allele (Fig. 5c). Traditional short read WGS analysis indicated an LOH event represented by a hemizygous deletion in the p-arm of chromosome 17 (Fig. 5d). Barcode count analysis with linked-reads confirmed this observation and once considering the mutation allelic fraction, the corrected copy number at that region is 1. The LOH results from an extensive genomic deletion overlapping the TP53 mutation (Fig. 5e). The phased SNV frequencies in the haplotype 1 allele are reduced in the tumor compared to the normal, indicating that LOH in the tumor sample is associated with the loss of the haplotype 1 allele (Fig. 5f). Thus, the TP53 R213Q mutation is in trans with the deleted allele haplotype. As a result, the tumor contains only a single, inactivated copy of TP53. Taken together, this result shows the unambiguous biallelic inactivation of TP5332, 33.
DISCUSSION
We demonstrate the use of a high-throughput microfluidics technology to construct phased sequencing libraries from nanogram inputs of high molecular weight DNA. By using gel beads as the barcode delivery reagent, we demonstrate the robust loading of a single barcode to a microdroplet partition. This technology addresses issues that affect current experimental phasing approaches such as problems of Poisson-distributed barcode loading and limited partitioning with microwells. This is the first study that demonstrates a droplet-based system for whole genome phasing and structural variant analysis. In addition to phasing and structural variant calling, linked-reads can potentially also be applied to de novo genome assembly, remapping of difficult regions of the genome, detection of rare alleles, and elucidating complex structural rearrangements.
Several studies have recently demonstrated high-throughput barcoding of droplet partitions34–36 for single-cell RNA-Seq and analysis of short bacterial 16S sequences. These other approaches use individual barcodes ranging up into the millions that are introduced into a specific partition. However, none of these droplet applications generate megabase-scale haplotypes from whole genome sequencing. As noted previously, there are a number of other genome sequencing approaches used for phasing1, 2, 5–9, 37, 38 and an overview is listed in Supplementary Table 13. Only one of these approaches uses droplets, and this method does not involve sequencing but rather relies on digital PCR counting methods to assess a single-plex candidate locus38.
To assess performance, we conducted a phased genome analysis on several well-defined genomes. With this technology, we phased over 95% of SNVs in all samples with N50 phase block sizes ranging from 0.8 Mb to 2.8 Mb, at a low switch error rate of less than 0.001. This phasing performance was achieved using existing variant datasets. We show that linked-read data can be used to phase de novo variants, although more coverage will be required to achieve parity with standard library preparation methods due to coverage biases against GC-rich regions (Supplementary Fig. 2d). Statistical inference of haplotypes from genomic intervals dominated by similar heterozygous variants among family members is an issue that experimental phasing overcomes. For example, in the NA12878 nuclear trio, about 10% of the total number of SNVs in the child are inherited from such regions with common genotypes39.
Our technology is compatible with standard downstream NGS assays, such as exome enrichment, as barcode information is introduced as the first step in the library preparation process. With the nuclear trio samples, over 95% of genes less than 100 kb were phased using this phased exome sequencing approach, which enables the economical use of phased analysis on large numbers of samples.
We used phasing and read barcode counts to identify structural variation such as large genomic deletions and rearrangements that were independently validated by multiple methods. Using exome linked-reads, we delineated the complex rearrangements such as the EML4/ALK inversion in the case of the NCI-H2228 cell line. In addition, we showed that linked-read phasing of structural variants distinguish true SVs from false predictions.
We also used this approach to phase a cancer genome derived from a primary tumor. The combination of somatic mutations, haplotype blocks and barcode counting identified the trans-relationship between a mutation in TP53 and a chromosome 17 p-arm loss in colon adenocarcinoma. We additionally generated haplotypes incorporating other critical genetic aberrations such as copy number alterations and rearrangements. We anticipate that phased cancer genomes will provide new insight into the underlying genomic structural alterations underlying tumor development and maintenance.
The identification of potentially pathogenic mutations and structural variants remains a challenge and linked-read sequencing provides a unique opportunity to improve our understanding of diseases such as cancer.
METHODS
Genomic DNA samples
The Institutional Review Board (IRB) at Stanford University School of Medicine approved the study. Informed consent was obtained and the samples were made available from the Stanford Cancer Center Tissue Bank. This study used a primary colorectal adenocarcinoma and matched normal tissue that were collected at time of surgical resection and flash frozen. Both samples had genomic DNA extracted with the E.Z.N.A. SQ DNA/RNA Protein Kit (Omega Bio-Tek). The genomic DNA did not require further size selection or processing. We quantified the DNA with Life Technologies Qubit.
For the commercially acquired genomic DNA, we size selected DNA molecules 20 kb or higher using the BluePippin (Sage Science) (NA12877 and NA12882 from Coriell, and NCI-H2228 from ATCC). In addition, we harvested immortalized human lymphocyte cells (GM12878 and GM20847 from Coriell) and genomic DNA was extracted using the Gentra Puregene Cell kit (Qiagen).
Sequencing library construction using the GemCode platform
A GemCode Instrument (10× Genomics) was used for sample preparation. The high-throughput nature of the platform allows construction of 8 sequencing libraries by a single person in a day. Sample indexing and partition barcoded libraries were prepared using a beta version of the GemCode Gel Bead and Library Kit (10× Genomics, Pleasanton, CA). One nanogram of sample DNA was used for GEM reactions where DNA molecules were partitioned into droplets to amplify the DNA and introduce 14-bp partition barcodes. With 1ng genomic DNA of 50 kb molecule length, there are ~100 molecules per droplet.
GEM reactions were thermal cycled (95°C for 5 min; cycled 18×: 4°C for 30 sec, 45°C for 1 sec, 70°C for 20 sec, and 98°C for 30 sec; held at 4°C). After amplification, the droplets were fractured and the library intermediate DNA purified using the 10× Genomics protocol. The DNA was subsequently sheared to either 250bp or 500bp using a Covaris M220 system (Supplementary Table 2) to construct sample-indexed libraries using 10× Genomics adaptors. The barcode sequencing libraries were quantified by qPCR (KAPA Biosystems Library Quantification Kit for Illumina platforms). Sequencing was conducted with an Illumina Hiseq2500 with 2×98 paired-end reads based on the manufacturer’s protocols.
To compare barcode libraries against standard short read libraries, we prepared a TruSeq library (Illumina) following manufacturer’s protocols, using 100ng of DNA. Both barcode and TruSeq libraries used GM12878 genomic DNA. Each library was sequenced to ~30× coverage. At 30× coverage, the coverage of molecule in each droplet is 0.1×, and the number of linked-reads per molecule is around 15.
Five micrograms of each barcode library was used for exome capture (Agilent SureSelect Human All Exon V5+UTRs) with the Agilent SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA) supplemented with modified blocking oligonucleotides for Illumina Dual Indexing (TS HT i5 and TS HT i7) from IDT. Captured libraries were quantified by qPCR (KAPA Biosystems). Again, sequencing was conducted with an Illumina Hiseq2500 with 2×98 paired-end reads based on the manufacturer’s protocols.
Alignment, barcode assignment and calculation of sequencing metrics
The GemCode analysis software was used for processing the sequenced data from barcode libraries. Fastq files from Illumina sequencing reads were trimmed (removing the first 10nt of all reads) and aligned to the human genome (hg19) using bwa (mem algorithm, version 0.7.10-r789). Barcodes were incorporated into the read information in the bam file and only reads associated with valid barcodes were considered for alignment and downstream analysis. For visualization and some analysis, the barcode counts were calculated using non-overlapping window size of 100 kb, over all positions. Only uniquely mapped, non-duplicated reads with mapping quality (MAPQ) of 60 are considered.
Reads were sorted by position using samtools (Version 0.1.19-96b5f2294a). PCR duplicates were marked if two sets of read-pairs shared both identical aligned genomic position and an identical associated barcode sequence. Linked-reads were inferred by clustering reads from the same barcode on the genome, and their boundaries were set by two nearest reads more than 50 kb apart. The phrase, “barcodes correctly assigned” is the fraction of barcodes matching a known barcode. “Relative genomic loading per partition” was calculated as the fraction of the amount of DNA in a partition relative to the size of the human genome. The number of binding events is estimated as the product of binding density and genome loaded per partition.
For a uniform distribution of barcode frequencies, the probability of drawing two identical barcodes is p = Σi frequency(BCi) * frequency(BCi) = N/N2 = 1/N where N is the number of unique barcodes. Thus, effective barcode diversity, which accounts for a non-uniform distribution of barcode frequencies, can be calculated as:
where BCi = i-th barcode.
To perform the variant calling analysis, we used Freebayes to call variants on 10× and Truseq libraries, down-sampling each library to 10×, 20× and 30× coverage. Then sensitivity and PPV of SNVs were evaluated against ground-truth variants published by Cleary et. al15.
Phasing linked-reads
See Supplementary Note 1.
Structural variant calling from linked-read data
See Supplementary Note 2.
Phasing of structural variants
Phasing of large-scale variants used the final probabilistic assignment of barcodes to haplotype blocks calculated as part of the phasing code. For each haplotype block within a 30 kb window of each of the two breakpoints defining a structural variant, barcodes supporting the structural variant call were assigned to one of the two haplotypes for that haplotype block. For each haplotype block, the counts of barcodes assigned to each of the two haplotypes were used to calculate a p-value under the two-tailed binomial test. Phase calls were made on a structural variant when the p-value was < 0.01.
Validation of genomic deletions with targeted sequencing
We validated a series of genomic deletions using targeted sequencing20. The methods are fully described by Hopmans et al.19. For this validation study, we relied on targeting assays that uses target-specific primer probes that hybridize to the target DNA molecule20. Afterwards, a polymerase extension captures the specific genomic target sequence. Previously, we demonstrated the utility of this method for confirming SVs, even in the context of genomic mixtures where a candidate rearrangement is present in only a fraction of the sample19. As a result of random fragmentation of genomic DNA in the library preparation, breakpoints of structural variants will be randomly distributed within a subset of the sequencing reads.
For this assay, we designed multiple primer probe sequences flanking each putative breakpoint associated with a structural variant candidate. This targeting method is generally successful at selecting sequences up to 1 kb if not further away from the primer probe. The primer probe sequences chosen were on both the forward and reverse strands surrounding both sides of a target putative breakpoint within a distance of 0.75 kb (Supplementary Fig. 6). Reads captured by primer probes upstream from the breakpoints should cross on the reverse strand; reads captured by primer probes downstream from the breakpoints should cross on the forward strand.
For the eight candidate deletions that were validated, we designed and synthesized 163 primer-probe oligonucleotides (Supplementary Table 7). Generally, all of these oligonucleotides were unique in terms of their representation in the genome. The only exception was for 15 probes intended to validate a deletion in chromosome 5 (position 99,400,335 – 99,713,992). This deletion occurs in an area of the genome that is highly repetitive, so only two of these 15 primer probes contain a 20mer that aligns uniquely to the human genome with no single-mismatch alignments.
Single-end alignment using bwa (mem algorithm, version 0.7.10) was performed on the individual reads from the mate-pairs. The targeting primer sequence is included in read 2 and used as an index for a given target segment. The captured sequence is in read 1 and were indexed based on the read 2 targeting primer. The read 1 sequences that completely aligned to the human genome were excluded. The remaining read 1 sequences were evaluated for evidence of breakpoint and counted. The reads that had breakpoints were concatenated to create a breakpoint sequence. Reads crossing breakpoints were generated by finding reads that included a soft-clipped section such that the aligning portion preceded or followed the breakpoint; soft-clipped reads that also included soft-clipping on the non-breakpoint side were excluded. Using this read set we counted 20mers that contained a chimeric junction containing sequence on both sides of the breakpoint candidate.
Evaluation of structural variant calls in NA12878
To assess the false discovery rate of our SV calling algorithm, we compared our structural variant calls in NA12878 against a recent de-novo assembly using genomic DNA from this individual10. We obtained a list of assembly-based deletion and insertion calls in NA12878 from the Genome in a Bottle website (ftp://ftp-trace.ncbi.nih.gov/giab/ftp/technical/NA12878_PacBio_MtSinai). We then constructed two deletion datasets: (a) a “confident” set containing deletions that were marked as “passing” by the study. These were deletions that were called by 3 or more out of the 7 methods used in that paper; (b) a “relaxed” set containing all deletions detected by at least one computational method in the de-novo assembly data.
We focused on deletion calls in the following comparison because 1) deletion calls are much easier to compare across datasets; 2) we omitted insertions from the above two sets because our algorithm is not designed to detect gaps in the reference genome. Out of the 20 calls that were made in NA12878 via linked-reads, 40% and 55% respectively matched those from the confident and relaxed Pendleton datasets to within 20 kb.
Besides deletion calls, our SV algorithm can detect other types of structural rearrangement. Indeed, two of our calls matched inversions reported in the literature16. One additional call is a retro-transposon insertion that has been found in Caucasian individuals21. Although the three calls were not explicitly called by Pendleton et. al., they were supported by long sequence reads (i.e. Pacific Biosciences sequencer) obtained from the same assembly work10. Altogether, this increases the percentage of the validated calls against the de-novo assembly to 70%. We have included a comparison of the calls in Supplementary Table 8.
RT-PCR validation of EML4-ALK fusion
RT-PCR was used to confirm the EML4-ALK and ALK-PTPN3 fusions in NCI-H2228 cancer cell line. We used the Cells-to-CT 1-Step Power SYBR Green Kit (Life Technologies) according to the manufacturer’s recommendations. The gene ACTB was assayed using SYBR Green Kit Control Kit (Life Technologies). As a negative control, NA12878 cells were assayed in parallel. Briefly, ~7500 cells were lysed and treated with DNase I in a total of 55 ul. Two ul of lysate was used for a 20 ul PCR reaction. The PCR products were visualized using the BioAnalyzer High Sensitivity DNA Kit (Agilent), with respectively the amplicons ACTB diluted 1:50, EML4/ALK 1:20, and ALK/PTPN3 diluted 1:3. The primers for the EML4/ALK amplicon are (F) 5′-GCATAAAGATGTCATCATCAACCAAG; (R) 5′-CGGAGCTTGCTCAGCTTGTA. The PCR primers for ALK/PTPN3 are: (F) 5′-TGGCTGCAGATGGTCGCATGG; (R) 5′-AGTCCACGGAGTCGTCATCAT.
Cancer whole genome sequencing with short reads and data processing
Whole genome libraries were made per the manufacturer’s protocol (Illumina). Sequencing libraries underwent cluster-generation on an Illumina cBot using paired end flowcells and Illumina TruSeq chemistry and sequenced at Illumina with the HiSeq 2500 for 2×100 cycle reads with indexing. Sequence reads were aligned to the human genome version hg19 using bwa13. The Genome Analysis Toolkit (GATK)14 was used to determine overall sequencing coverage and variant calls.
Cancer genome somatic mutation calling for coding mutations
The whole genome sequence data was aligned using bwa 0.7.513 aln and sampe with default parameters against NCBI human genome build 37. Data was sorted and duplicate marked using Picard’s AddOrReplaceReadGroups and MarkDuplicates functions respectively. Picard version 1.63 was used in all steps. The files were merged in the GATK14 RealignerTargetCreator step. This step and the IndelRealigner step were used to realign locally; IndelRealigner referred to dbSNP version 135. The BaseRecalibrator function used CycleCovariate and ContextCovariate as covariates and referred to dbSNP 135. At this point the realigned bam file of Patient 1532’s data was split up to allow for easier processing. GATK PrintReads was run on realigned bam files with the appropriate recalibration data table to produce recalibrated bam files. The GATK UnifiedGenotyper was then run with the parameters --dbsnp dbsnp_135.b37.vcf --max_alternate_alleles 11. These raw calls were then recombined. The GATK VariantRecalibrator was run on the raw VCF data, using the hapmap, omni, and dbsnp resources with standard priors and using HaplotypeScore, MQRankSum, ReadPosRankSum, FS, MQ and DP as filter elements. Finally, the ApplyRecalibration step was used to determine whether calls received a PASS value or not. Variants were called using GATK version 2.6–4. After variants were called, all SNV positions where the tumor and normal calls differed were submitted to CADD annotation28. SNVs were then filtered to require a somatic variant (positions where the normal tissue shows no variant and the tumor does or the normal tissue is heterozygous and the tumor has a homozygous variant) in a coding region with coverage depth >= 10 in both samples and a CADD phred score greater than or equal to 25 (Supplementary Table 10). Sequencing coverage was assessed with the GATK DepthOfCoverage tool at depths of 10, 20 and 30 (Supplementary Table 1).
SNVs were then extracted from the phased VCF files and their phasing status was assessed. The tumor haplotype is based on the haplotype of the first SNV in the local normal phase block: that haplotype is always arbitrarily assumed to be 1. The normal and tumor haplotypes are then set to be congruent to one another by comparing positions heterozygote in both samples. In case the normal region is not phased, the tumor haplotype is assumed to be 2. If the tumor SNV is a homozygote while the normal is a heterozygote, the haplotype is assigned to the wild-type haplotype of the normal.
Cancer genome allelic imbalance analysis
For assessment of loss-of-heterozygosity (LOH) events, our analysis relied on minor allelic frequency (MAF) data. The MAF is a ratio comparison of allelic read depths from heterozygous SNVs identified from the normal genome compared to the same position from the tumor. The input file is a VCF containing the normal and tumor reads. The calls are filtered to require a genotype quality (GQ) of 30 or greater in both normal and tumor at that position, an overall read depth of 10 or greater, and a minor allele depth of at least 3 in the normal genome. The allele depth ratio is calculated as the minor allele count divided by the major allele count. The MAF value is determined as follows: we divide the tumor allele depth ratio by the normal depth ratio and taking the log2 of the quotient. For graphic display, we used a smoothed MAF value based on a window average of 100 contiguous SNVs from each genome.
Cancer genome copy number and structural variant analysis
To determine somatic copy number alterations and the affected genomic intervals from whole genome sequencing data, we used the SeqCBS method31. The software implementation is available as an open-source R package named SeqCBS (http://cran.r-project.org). The CNV analysis used an R script that reads a configuration file listing the sequence data sets to be compared, namely the case (tumor) versus the control (normal). The algorithm then performs the segmentation on these two files, compares them, and produces both local and whole-chromosome CNV plots. For any such region, there is a general test statistic and a relative gain or loss copy number value. Generally, we required a test statistic > 1,000 as a basic cutoff and a copy number value of greater than 2.5 or less than 1.6 as our thresholds for marking an event as a significant amplification or loss. We validated these calls with linked reads by counting the average number of barcodes-annotated reads over 50 kb window spanning across the length of each candidate.
To validate SV calls made by the GemCode software analysis of linked-reads we examined sequence data from the short read WGS dataset. We used BreakDancer30 with default setting to generate a set of SV candidates and then identified putative locations as predicted by the phased SV call set and associated quality score. In addition, we identified soft-clipped reads in the vicinity of the breakpoints, which are indicative of a structural variant breakpoint. Afterwards, we tabulated the number of reads directly supporting the breakpoint. Soft-clipped reads were manually curated in IGV to verify base quality, and were individually aligned in BLAT to verify the breakpoint locations.
Supplementary Material
Acknowledgments
This work was supported by the following grants from the NIH: NHGRI P01HG000205 to B.T.L., E.S.H, S.M.G., J.M.B. and H.P.J., NCI R33CA174575 to J.M.B., S.G. and H.P.J. and NHGRI R01HG006137 to H.P.J. The American Cancer Society provided additional support to S.G. and H.P.J. [Research Scholar Grant, RSG-13-297-01-TBG]. In addition, H.P.J. received support from the Doris Duke Clinical Foundation, the Clayville Foundation, the Seiler Foundation and the Howard Hughes Medical Institute.
Footnotes
Accession codes. Data have been deposited in the Short Read Archive (SRA) under accession number SRP051629 and dbGAP under the accession number phs000898.v1.p1.
AUTHOR CONTRIBUTIONS
B.T.L., M.S., M.J., J.M.B., C.M.H., S.K.P, L.M., R.B., A.J.M., Y.L., A.D.P., A.J.L., P.H., L.G., K.P.B., P.V. P., E.S.H., C.W., K.M.G, S.S., K.D.N., B.J.H. and H.P.J. designed the experiments. B.T.L., J.M.B., C.M.H., L.M., J.M.T., P.A.M., P.W.W., R.B., A.J.M., Y.L., P.B., A.D.P., A.J.L., P.J.M, G.M.V., L.M., M.L., L.G., D.E.B., K.P.B., P.V. P., E.S.H., C.W., J.P.D., I.W., H.S.O, J.Y.L., Z.K.B., K.M.G, G.P.D., Z.W.B., F.M., N.O.K., J.A.B., S.P.G., C.B., A.N.F., A.C. and B.J.H. conducted the experiments. D.A.M., R.B., A.J.M., S.W.S., S.K., J.A.B., A.P.K., K.D.N. and B.J.H. designed the instrument. M.S., M.J., C.M.H., P.W.W., R.B., A.J.M., Y.L., A.D.P., A.J.L., P.H., L.M., L.G., K.P.B., P.V. P., S.K., J.P.D., J.A.B., K.D.N. and B.J.H. designed reagents for phasing. B.T.L, J.M.B., E.S.H. and H.P.J. designed reagents for targeted sequencing analysis. G.X.Y.Z., M.S., S.K.P, P.J.M, G.K.L., D.L.S., W.H.H., R.T.W., S.S. and K.D.N. wrote the haplotype analysis algorithms. J.M.B. and S.G. wrote the analysis algorithms for short read sequencing analysis. M.S., P.J.M, A.W., G.K.L., D.L.S., W.H.H. and R.T.W. wrote the analysis software. G.X.Y.Z., B.T.L., M.S., M.J., J.M.B., C.M.H., S.K.P, J.M.T., R.B., A.J.M., Y.L., P.B., P.J.M, P.H., L.M., M.L., A.W., K.P.B., P.V. P., S.K., J.P.D., I.W., H.S.O, S.M.G., S.G., J.Y.L., Z.K.B., K.M.G, W.H.H., G.P.D., Z.W.B., F.M., J.A.B., S.P.G., C.B., A.N.F., H.H., A.C., S.S., K.D.N., B.J.H. and H.P.J. analyzed the data. G.X.Y.Z., B.T.L., M.S., M.J., S.G., B.J.H. and H.P.J. wrote the manuscript. H.P.J. oversaw the genetic analysis.
COMPETING FINANCIAL INTERESTS
The following authors, as listed by initials, are employees of 10× Genomics: G.X.Y.Z., M.S., M.J., C.M.H., S.K.P, D.A.M., L.M., J.M.T., P.A.M., P.W.W., R.B., A.J.M., Y.L., P.B., A.D.P., A.J.L., P.J.M, G.M.V., P.H., L.M., M.L., L.G., A.W., D.E.B., S.W.S., K.P.B., P.V. P., S.K., G.K.L., D.L.S., J.P.D., I.W., H.S.O, J.Y.L., Z.K.B., K.M.G, W.H.H., G.P.D., Z.W.B., F.M., N.O.K., R.T.W., J.A.B., S.P.G., A.P.K., C.B., A.N.F., A.C., S.S., K.D.N., B.J.H.
References
- 1.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adey A, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genomes Project C et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suk EK, et al. A comprehensively molecular haplotype-resolved genome of a European individual. Genome Res. 2011;21:1672–1685. doi: 10.1101/gr.125047.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duitama J, et al. Fosmid-based whole genome haplotyping of a HapMap trio child: evaluation of Single Individual Haplotyping techniques. Nucleic Acids Res. 2012;40:2041–2053. doi: 10.1093/nar/gkr1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaper F, et al. Whole-genome haplotyping by dilution, amplification, and sequencing. Proc Natl Acad Sci U S A. 2013;110:5552–5557. doi: 10.1073/pnas.1218696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvaraj S, JRD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pendleton M, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015 doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abate AR, Chen CH, Agresti JJ, Weitz DA. Beating Poisson encapsulation statistics using close-packed ordering. Lab Chip. 2009;9:2628–2631. doi: 10.1039/b909386a. [DOI] [PubMed] [Google Scholar]
- 12.Kuleshov V, et al. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleary JG, et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J Comput Biol. 2014;21:405–419. doi: 10.1089/cmb.2014.0029. [DOI] [PubMed] [Google Scholar]
- 16.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopmans ES, et al. A programmable method for massively parallel targeted sequencing. Nucleic Acids Res. 2014;42:e88. doi: 10.1093/nar/gku282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myllykangas S, Buenrostro JD, Natsoulis G, Bell JM, Ji HP. Efficient targeted resequencing of human germline and cancer genomes by oligonucleotide-selective sequencing. Nat Biotechnol. 2011;29:1024–1027. doi: 10.1038/nbt.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrider DR, et al. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 2013;9:e1003242. doi: 10.1371/journal.pgen.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipson D, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi YL, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 25.Koivunen JP, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 27.Jung Y, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–597. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- 28.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen JJ, Zhang NR. Change-point model on nonhomogeneous Poisson processes with application in copy number profiling by next-generation DNA sequencing. The Annals of Applied Statistics. 2012;6:476–496. [Google Scholar]
- 32.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 33.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. New England Journal of Medicine. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 34.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgstrom E, et al. Phasing of single DNA molecules by massively parallel barcoding. Nat Commun. 2015;6:7173. doi: 10.1038/ncomms8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vree PJ, et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol. 2014;32:1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 38.Regan JF, et al. A rapid molecular approach for chromosomal phasing. PLoS One. 2015;10:e0118270. doi: 10.1371/journal.pone.0118270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roach JC, et al. Chromosomal haplotypes by genetic phasing of human families. Am J Hum Genet. 2011;89:382–397. doi: 10.1016/j.ajhg.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.