Abstract
Objective
Literature database search is a crucial step in the development of clinical practice guidelines and systematic reviews. In the age of information technology, the process of literature search is still conducted manually, therefore it is costly, slow and subject to human errors. In this research, we sought to improve the traditional search approach using innovative query expansion and citation ranking approaches.
Methods
We developed a citation retrieval system composed of query expansion and citation ranking methods. The methods are unsupervised and easily integrated over the PubMed search engine. To validate the system, we developed a gold standard consisting of citations that were systematically searched and screened to support the development of cardiovascular clinical practice guidelines. The expansion and ranking methods were evaluated separately and compared with baseline approaches.
Results
Compared with the baseline PubMed expansion, the query expansion algorithm improved recall (80.2% vs. 51.5%) with small loss on precision (0.4% vs. 0.6%). The algorithm could find all citations used to support a larger number of guideline recommendations than the baseline approach (64.5% vs. 37.2%, p<0.001). In addition, the citation ranking approach performed better than PubMed’s “most recent” ranking (average precision +6.5%, recall@k +21.1%, p<0.001), PubMed’s rank by “relevance” (average precision +6.1%, recall@k +14.8%, p<0.001), and the machine learning classifier that identifies scientifically sound studies from MEDLINE citations (average precision +4.9%, recall@k +4.2%, p<0.001).
Conclusions
Our unsupervised query expansion and ranking techniques are more flexible and effective than PubMed’s default search engine behavior and the machine learning classifier. Automated citation finding is promising to augment the traditional literature search.
Keywords: Information Retrieval, PubMed, Practice Guideline, Medical Subject Headings, Natural Language Processing
1. Introduction
The practice of evidence based medicine requires integrating individual clinical expertise and the best available evidence in making decisions about patient care. However, health care practitioners have little time to keep up with the rapid growth in the biomedical literature. In 2009, there were about 25,400 peer-reviewed journals and the number increases 3.5% a year (1). Citations indexed in PubMed have grown from 4 million (pre 1975) to 22 million today (2). Each year, about 3000 clinical trial studies have posted results in ClinicalTrial.gov (3). Fraser and Dunstan showed that it’s almost impossible to keep up with the medical literature even within a narrow specialty (4). In a review of information-seeking behavior, Davies showed that clinicians’ lack of time, issues with information technology, limited search skills are top barriers for information searching (5). As a result, most clinical questions raised by clinicians at the point of care remain unanswered. In a recent systematic review, Del Fiol et al. showed that clinicians raised roughly one question out of every two patients seen and over 60% of these questions were not answered (6). To cope with information overload, clinicians rely on existing expert-compiled resources such as clinical practice guidelines (CPG) to fulfill their information needs (7). However, the development and update of CPGs is costly, slow and unable to keep up with the rate of new evidence in the medical literature. In a 2003 survey of guideline developers, the average cost for CPGs development was $200,000 per guideline in the United States (8). High quality guidelines that meet strict quality criteria (9, 10) require more time and resources. Time required for finishing peer-review for a cardiology guideline published by The American College of Cardiology (ACC) and American Heart Association (AHA) was from 12 to 18 months (11). In summary, the rapid pace of new published literature can quickly make the CPGs outdated and suboptimal for clinical decision-making.
In guideline development, experts perform systematic reviews of the available evidence, which involves a series of scientifically rigorous steps (11). The two first and important steps are a systematic literature search followed by screening for relevant citations. Literature search involves identifying possibly relevant studies from electronic literature databases. Citation screening involves quickly scanning abstract and full-text manuscripts to assess the eligibility of studies. Informatics research has investigated automated and semi-automated methods to aid with citation screening (12–16). Fiszman et al. were among the first research groups introducing informatics solutions to support clinical guideline development (15, 16). They developed a semantic filter to automatically classify relevant citations. Similarly, Cohen et al. investigated a machine learning approach to solve a classification problem in drug effectiveness reviews (12, 17). To meet the needs of citation screening, those methods aimed for a balance between recall and precision. However, recall is more important than precision in systematic literature search. The 2011 ACCF/AHA’s manual for clinical guideline development described the need for literature search to be comprehensive, and key to the development of valid guidelines (11). The Cochrane handbook for systematic reviews highlights that “searches should seek high sensitivity, which may result in relatively low precision.” (18) In the present study, we investigated the literature search stage and aimed to maximize recall while controlling the impact on precision. We developed and assessed query expansion and ranking methods to enhance information retrieval performance in the context of clinical guideline development. The solution was based on an extension of PubMed’s search engine, optimized to retrieve and rank relevant studies for cardiovascular guidelines.
There have been previous works that we leveraged to inform our system (15–17, 19–21). Fiszman‘s gold standard included citations that were used to support 30 clinical questions (16). Our work sought for a larger gold standard, which includes citations to support more than 600 guideline recommendations. Research on query expansion showed that using MeSH concepts and MeSH hierarchy can improve performance of image retrieval and biological question retrieval (19, 20). Our query expansion method was also based on finding relevant MeSH concepts, but was optimized to retrieve guideline conditions.
Traditional information retrieval or question answering systems rank documents by relevance or similarity to the user query. Generic queries (e.g., “heart failure”) can generate thousands of documents that share the search keywords. PubMed by default sorts the results by recently added date, without considering relevancy and scientific quality. Informatics research has investigated machine learning approaches to prioritize citation screening in systematic reviews (14, 22, 23). Yet, machine-learning approaches are arguably not flexible since they require sufficient high-quality training data and often do not generalize well to new domains. Unsupervised ranking methods have been investigated in the citation retrieval studies by Jonnalagadda et al. (24, 25). Their method assigned weights based on journal impact measures; however, the method validation was limited to the “heart failure” topic. In the present research, we developed novel unsupervised query expansion and citation ranking methods with a larger gold standard that includes cardiovascular conditions. We then compared the performance of these methods with PubMed’s query expansion and ranking, and a machine learning classifier.
2. Materials and Methods
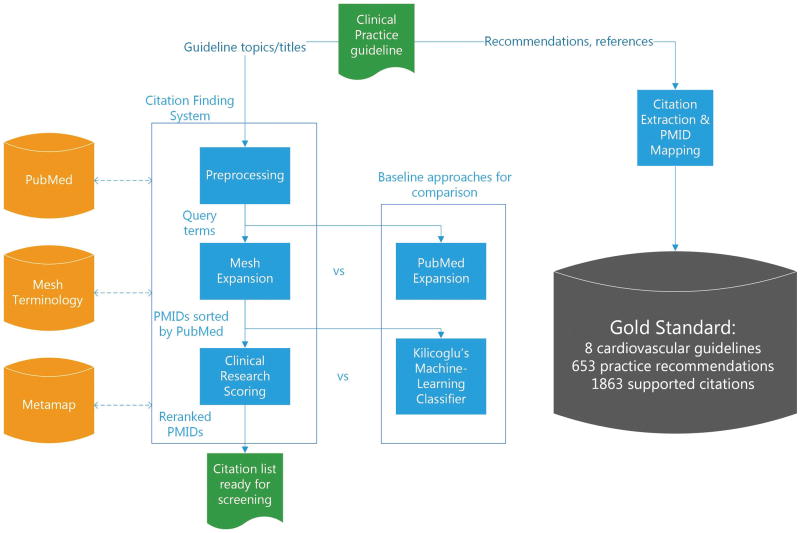
Our study design consisted of three main parts: (1) development of a gold standard composed of studies used in the development of cardiovascular guidelines; (2) iterative development of a citation finding system composed of two main components: query expansion and citation ranking; and (3) evaluation of each system component using standard information retrieval metrics and comparison with baseline approaches. Figure 1 depicts the summarization of our system architecture and study design.
Figure 1.
Overview of the citation finding system and the study design.
2.1 Gold standard
The gold standard consisted of citations that have been used to support guideline practice recommendations. We focused on the cardiovascular guidelines published by the American College of Cardiology (ACC) and the American Heart Association (AHA). The full revision cardiovascular guidelines developed by the ACC/AHA and published from 2010 to 2014 were retrieved using a PubMed search. Since the majority of guideline topics are about complete management of a condition, we focused on retrieving condition topics in this study. Topics about interventions or diagnostic procedures are reserved for future research. For those guidelines discussing the comprehensive management of cardiovascular conditions, we performed the following steps to build the gold standard: (1) Extracted all the citations listed in the “References” section of the guideline; (2) extracted the guideline recommendations whose evidence sources were provided in the guideline and the citations that were used as evidence sources to support each recommendation; and (3) automatically mapped those citations in free-text to PubMed IDs using the NCBI Batch Citation Matcher tool (26). Manual mapping was performed to supplement the citation IDs that could not be matched by the NCBI tool. Table 1 shows examples of guideline recommendations, supporting citations, and their corresponding PMIDs.
Table 1.
Examples of extracted guideline recommendations, supported citations, and PMID mappings for the “Guideline for the Management of Patients With Atrial Fibrillation” (2014)
| Guideline recommendations | Supported citations |
|---|---|
| Selection of antithrombotic therapy should be based on the risk of thromboembolism irrespective of whether the AF pattern is paroxysmal, persistent, or permanent (167–170). | 167. New oral anticoagulants for stroke prevention in atrial fibrillation: impact of gender, heart failure, diabetes mellitus and paroxysmal atrial fibrillation (27). PMID: 23253272 168.
Distribution and risk profile of paroxysmal, persistent, and permanent atrial fibrillation in routine clinical practice: insight from the real-life global survey evaluating patients with atrial fibrillation international registry (28). PMID: 22787011 169. Efficacy and safety of dabigatran compared to warfarin in patients with paroxysmal, persistent, and permanent atrial fibrillation: results from the RE-LY (Randomized Evaluation of Long-Term Anticoagulation Therapy) study (29). PMID: 22361407 170. Prevention of stroke in patients with atrial fibrillation: current strategies and future directions (30). PMID: 25534093 |
| Control of the ventricular rate using a beta blocker or nondihydropyridine calcium channel antagonist is recommended for patients with paroxysmal, persistent, or permanent AF (267–269). | 267. Ventricular rate control in chronic atrial fibrillation during daily activity and programmed exercise: a crossover open-label study of five drug regimens (31). PMID: 9973007 268. Efficacy of oral diltiazem to control ventricular response in chronic atrial fibrillation at rest and during exercise (32). PMID: 3805530 269. The Atrial Fibrillation Follow-up Investigation of Rhythm Management (AFFIRM) study: approaches to control rate in atrial fibrillation (33). PMID: 15063430 |
| Intravenous administration of a beta blocker or nondihydropyridine calcium channel blocker is recommended to slow the ventricular heart rate in the acute setting in patients without preexcitation. In hemodynamically unstable patients, electrical cardioversion is indicated (270–273). | 270. Efficacy and safety of esmolol vs propranolol in the treatment of supraventricular tachyarrhythmias: a multicenter double-blind clinical trial (34). PMID: 3904379 271. A placebo-controlled trial of continuous intravenous diltiazem infusion for 24-hour heart rate control during atrial fibrillation and atrial flutter: a multicenter study (35). PMID: 1894861 272. Intravenous diltiazem is superior to intravenous amiodarone or digoxin for achieving ventricular rate control in patients with acute uncomplicated atrial fibrillation (36). PMID: 19487941 273. Esmolol versus verapamil in the acute treatment of atrial fibrillation or atrial flutter (37). PMID: 2564725 |
2.2 System Overview
The system is an extension of PubMed’s search engine to enhance the ability to retrieve citations for clinical guideline development. The system has a preprocessing stage and two other main stages: query expansion and document ranking. The query expansion stage aims to improve recall while the document ranking aims to improve precision on top-ranked documents.
Preprocessing
This step takes the title of the guideline as input and extracts the conditions of interest. Since there is little variation among guideline titles, we used simple regular expression rules such as words following “Patients With”, “diagnosis and treatment of”, and “management of” to extract main conditions from guideline titles (e.g “Guideline for the Management of Patients With Atrial Fibrillation”, “Guideline for the diagnosis and treatment of hypertrophic cardiomyopathy”, “Guidelines for the diagnosis and management of patients with Thoracic Aortic Disease”). This step also detects whether a particular guideline focuses on one or more conditions. For instance, the phrase “Extracranial Carotid and Vertebral Artery Disease” was broken into two conditions: “Extracranial Carotid Disease” and “Vertebral Artery Disease”.
Query Expansion
Based on the extracted condition terms, we conducted a search using PubMed’s default search behavior. When entering a query on the PubMed search interface, PubMed automatically expands the query to maximize recall. For instance, PubMed expands the query “atrial fibrillation” by injecting additional MeSH terms and keywords: "atrial fibrillation"[MeSH concepts] OR ("atrial"[All Fields] AND "fibrillation"[All Fields]) OR "atrial fibrillation"[All Fields]. We used the results of PubMed expansion as the baseline to compare with our expansion approach. Our approach aims to find relevant and meaningful MeSH terms of the condition topics. Additional MeSH terms were injected to original query using the Boolean OR operator.
Common Filter
We consistently applied a set of filters (i.e., publication date, human study and English language) for all queries generated. We considered other filters such as hasabstract and the Haynes clinical filters(38), but those filters led to missing important eligible studies.
MeSH expansion
We developed an algorithm (Figure 2) to expand the seed query using MeSH resources (MeSH descriptors, MeSH Tree), and a natural language processing application (Metamap (39)). The algorithm takes input as a single search query and outputs the expanded query. If there are multiple queries (multiple conditions), they were joined by the Boolean OR operator. Eventually, the query is adjusted by the common filter and applied the PubMed sorting mechanisms. To conduct a PubMed query, we formulated the PubMed query into the URL syntax and used the Entrez Programming Utilities (E-utilities) (40) to submit and retrieve results from the NCBI servers. The algorithm uses the following methods to find relevant MeSH concepts:
Figure 2.

Pseudo-code for the query expansion algorithm.
Disorder concept expansion: This step attempts to find MeSH concepts that best describe the condition of interest using a concept-mapping method. We used Metamap (41) to map narrative terms found in the Preprocessing stage into UMLS concepts. Metamap was restricted to the MeSH terminology. The UMLS concepts were translated to MeSH concepts by querying the MRCONSO table (42). We used the MeSH descriptors and MeSH Tree (43) to populate MeSH metadata and select concepts that have the semantic type “Disease or Syndrome”. Concepts whose ancestors have this semantic type were also extracted.
Statistical expansion: This method is based on the assumption that documents are likely relevant to a query if the extracted terms are mentioned in the document titles. The statistical expansion method first retrieved all articles that include the exact search term in the title. MeSH concepts of those articles were retrieved, aggregated and sorted by frequency. The highest frequency concept having the semantic type “Disease or Syndrome” was selected. The statistical expansion is triggered if the concept-mapping approach doesn’t recognize any concepts.
Body-part expansion: In some guidelines, the condition of interest is related to abnormalities in specific anatomical locations (e.g., heart valves, aortic valve). In exploratory work, we observed that using body-part concepts could improve recall in some queries. To find body-part concepts, we run Metamap on the disorder concept entry terms, filter out the generic concepts, and select concepts having the semantic type “Body Part, Organ, or Organ Component”.
Parent expansion: this step looks for direct parent concepts by iteratively traversing the MeSH Tree. Using parent concepts in some circumstances can improve recall, but may substantially impact precision. Hence, the algorithm only uses parent expansion when the expansion set has not reached a specific threshold, and disables expansion to other MeSH children (e.g. using tag [MESH: NOEXP]).
MeSH stop list: We maintain a stop list of MeSH concepts to be filtered out from expansion. The list contains three general concepts for cardiovascular topics: Disease, Heart Diseases, and Heart. We investigated the technique to generate the stop list automatically, but it was not quite successful as constructing manually. Our strategy is to test the algorithm in more diverse topics until we identify a pattern for a successful stop list.
Document Ranking
We presents three ways searchers can obtain a ranked list of citations: (1) Use PubMed’s sorting functionalities, (2) Use a general purpose machine learning classifier to identify clinical sound studies, and (3) Use our proposed scoring approach for clinical research studies.
PubMed sorting funtionalities
PubMed offers 7 ways to sort order for search results: Most Recent, Relevance Publication Date, First Author, Last Author, Journal, and Title. Most Recent is the PubMed’s default sorting that ranks citations by the time they were added to MEDLINE database. The Relevance sort uses PubMed’s internal algorithm to assign weight to citations depending on the frequency search terms are found and the fields they are found(44). We used and evaluated the Most Recent and Relevance sorts to compare with our proposed ranking approach. The other sorts based on publication time and alphabetical orders are less likely to identify relevant citations.
A machine learning approach
In 2009, Kilicoglu et al. implemented an ensemble approach combining several machine learning classifiers (Naïve Bayes, support vector machine (SVM), and boosting) to identify scientifically rigorous studies (45). The classifier was built on five basic features: words, MEDLINE metadata, sematic predications, relations, and UMLS concepts. In the original study, the classifier trained on 10,000 citations could achieve 82.5% precision and 84.3% recall on an unseen test set of 2000 citations. The classifier outputs the probability a citation is scientifically rigorous. We used this classifier as the baseline ranking approach.
Clinical research scoring approach
We propose an alternative method for ranking MEDLINE citations using three dimensions: MeSH majority, study design, and journal ranking. These dimensions attempt to capture three characteristics that are desirable for retrieved studies: relevancy, study quality, and study impact.
MeSH Majority: a PubMed document can be indexed with multiple MeSH concepts, but only a small subset are indexed as “major topic.” Using the expanded MeSH concepts from the query expansion stage, we assigned a MeSH score of 2.0 if one of the MeSH concepts or any of its children was tagged as a major topic. Otherwise, a MeSH score of 1.0 was applied.
Study Design: We assign a Study Design (SD) score to a study based on the publication type of the retrieved document (score 4.0: Practice Guideline, Guideline, Review with Meta-Analysis; score 3.0: Randomized Controlled Trial; score 2.0: Clinical Trial, Controlled Clinical Trial, Case-Control Studies, Cohort Studies, Longitudinal Studies, Cross-Sectional Studies, Cross-Over Studies, Observational Study, Evaluation Studies, Validation Studies, Comparative Study; and score 1.0: any other types). The rationale for the SD scoring was adapted from the GRADE system (18). If a study has multiple publication types, the maximum SD score found on the matrix is chosen. The SD score is increased with the presence of blinding methods (single-blinded method +0.1, double-blinded method +0.2) and setting (multicenter study +0.1).
Journal ranking: Journal ranking is an estimation of scientific quality and clinical impact of the study based on the popularity of the publishing source. We used the open-access SCImago Journal Rank (SJR), an impact factor metric, published by Scopus in 2012. The National Library of Medicine’s (NLM) journal records were mapped to Scopus’ records using the journal’s ISSN number, from which we retrieve the SJR metric.
Finally, the ranking score is calculated by multiplying all three metrics (ranking score= MeSHMajorScore * SD score * SJR). Since those metrics are independent, multiplication was considered to be the most appropriate method to aggregate the three metrics.
2.3 Evaluation
We used the gold standard described above to evaluate the query expansion and the ranking algorithms. We tested the following hypotheses: H1: the query expansion algorithm retrieves a perfect set of citations for a larger number of guideline recommendations than the PubMed expansion approach; and H2: the citation scoring approach has better recall at k than the machine learning classifier and the standard PubMed sort mechanisms.
In addition, we compared the algorithm performance in terms of standard information retrieval metrics. For the query expansion task, we measured recall and precision. The query expansion task was aimed to maximize recall while controlling impact on precision. We define the metric “Seeding Recall” to measure the ability of finding seed studies used to generate guideline recommendations. A practice recommendation can be synthesized from one or multiple studies. In the initial literature search, finding seed studies appeared in as many recommendations as necessary to understand the scope of the problem and guide future literature search.
To evaluate the ranking algorithms, we used the average precision metric. For a ranked list of documents, average precision is calculated by: where r is the number of relevant documents, and Rk is the position of the kth relevant document in the ranked list.
Precision at k (precision@k) and recall at k (recall@k) are defined as follows:
To test the H1 hypothesis, we convert the data to a binary outcome. We assigned TRUE if all citations for a recommendation were retrieved, and FALSE otherwise. The chi-square statistical test was used to assess the significance of the differences. To test the H2 hypothesis, we measured recall@k in all k positions and used the Wilcoxon signed rank test to assess the significance of the differences found.
3. Results
From 2010 to 2014, the American College of Cardiology (ACC) published 17 guidelines about cardiovascular topics. Four of them are Focus Update releases. We excluded those releases since the development process for the Focus Updates does not include a systematic search. Five guidelines were not on the comprehensive management of a condition and were also excluded. These guidelines covered narrower subtopics of diagnosis or treatment such as Secondary Prevention, Blood Cholesterol Treatment, and Coronary Artery Bypass Graft Surgery. Although it is possible to develop filters to target those subtopics, we decided not to cover them in this research. Eight guidelines met our inclusion criteria as summarized in Table 2. We were able to extract 653 practice recommendations, which cited 1863 citations. Of those, we were able to find PubMed IDs (PMIDs) in 1848 citations (99.2 %). A small portion of citations such as book chapters, online resources (e.g FDA site), and studies not indexed in MEDLINE did not have PMIDs.
Table 2.
Included cardiovascular guidelines along with their recommendations and the citations used to support recommendations.
| Authors | Published Year | Title | Recommendations | Citations w/ PMID | Citations w/o PMID |
|---|---|---|---|---|---|
| January et al(46) | 2014 | Guideline for the Management of Patients With Atrial Fibrillation | 62 | 132 | 1 |
| Brott et al(47) | 2010 | Guideline on the Management of Patients With Extracranial Carotid and Vertebral Artery Disease | 34 | 70 | 1 |
| Gersh et al(48) | 2011 | Guideline for the diagnosis and treatment of hypertrophic cardiomyopathy | 74 | 175 | 0 |
| Yancy et al(49) | 2013 | Guideline for the management of heart failure | 97 | 317 | 1 |
| O'Gara et at(50) | 2013 | Guideline for the management of ST- elevation myocardial infarction | 83 | 216 | 0 |
| Fihn et al(51) | 2012 | Guideline for the diagnosis and management of patients with stable ischemic heart disease | 123 | 407 | 4 |
| Hiratzka et al(52) | 2010 | Guidelines for the diagnosis and management of patients with Thoracic Aortic Disease | 63 | 156 | 4 |
| Nishimura et al(53) | 2014 | Guideline for the Management of Patients With Valvular Heart Disease | 117 | 375 | 4 |
The query expansion performance and comparison are summarized in Table 3. Overall, the query expansion algorithm achieved recall of 80.2% and seeding recall of 90.1%. In comparison with the default PubMed expansion, the algorithm improved recall by 28.7% and seeding recall by 26.5% with a 0.2% drop in precision. The ability to find seed studies (seeding recall) improved by 26.6%. Our query expansion algorithm could find all citations for more guideline recommendations than the default PubMed expansion (64.5% vs. 37.2%, p<0.0001).
Table 3.
Comparison between PubMed expansion and MeSH expansion algorithm.
| Default PubMed Expansion | MeSH Expansion | Mean Difference | |
|---|---|---|---|
| Recall % (SD) | 51.5 (35.5) | 80.2 (5.1) | 28.7 (31.7) |
| Seeding recall % (SD) | 63.5 (31.6) | 90.1 (6.1) | 26.5 (29.6) |
| Precision % (SD) | 0.6 (0.5) | 0.4 (0.5) | −0.2 (0.3) |
| Recommendations for which all citations were found % | 37.2 | 64.5 | 27.3 |
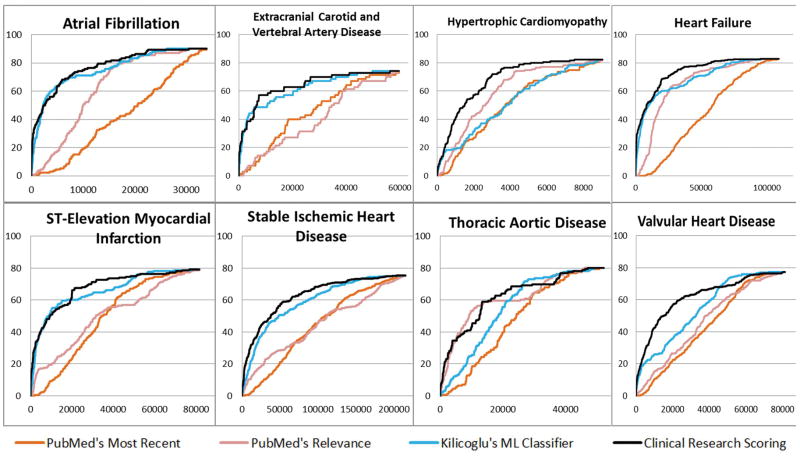
For citation ranking, the clinical research scoring approach had the best average precision of 7% compared to 2.1% machine-learning classifier, 0.9% PubMed’s sort by relevance, 0.5% PubMed’s sort by Most Recent. (Table 4). Similarly, the scoring approach had the highest average recall@k, improved 4.2% over the machine-learning classifier (66.2% vs 62%, p<0.001), 14.8% over PubMed’s sort by Relevance (66.2% vs. 51.4%, p<0.001), and 21.1% over PubMed’s sort by Most Recent (66.2% vs. 45.1%, p<0.001). In Figure 3, we illustrate the recall@k at various kth position in the ranked list. Overall, PubMed’s sorts essentially performed worse than machine learning classifier and the scoring approach. The curve of the scoring approach outperformed the machine-learning curve for most of the guidelines, especially at lower levels of k. However, the difference was significant in some guidelines (Hypertrophic Cardiomyopathy, Heart Failure , Thoracic Aortic Disease, Valvular Heart Disease), while only non-significantly improved in other guidelines.
Table 4.
Performance comparison among various ranking approaches.
| PubMed’s sorting by Most Recent | PubMed’s sorting by Relevance | Kilicoglu’s Machine learning classifier | Clinical research scoring approach | |
|---|---|---|---|---|
| Average precision % (SD) | 0.5 (0.7) | 0.9 (1.0) | 2.1(1.7) | 7.0 (4.8) |
| Recall@k % (SD) | 45.1 (26.2) | 51.4 (23.5) | 62 (18.6) | 66.2 (15.6) |
Figure 3.
Recall@k at various kth positions of 4 ranking methods in each of the cardiology guideline.
4. Discussion
Significance
We developed and evaluated an automated approach to retrieve relevant and high-quality citations from PubMed. The approach can be used to assist the development of clinical guidelines and systematic reviews. The results showed that our proposed method outperformed the default PubMed query expansion in terms of recall (80.2% vs. 51.5%) and seeding recall (90% vs. 63.5%), with a non-significant loss in precision (0.6% vs. 0.4%; p=0.09). In addition, the method could find all citations for a larger number of guideline recommendations than the PubMed expansion (64.5% vs. 37.2%, p<0.0001). The results reflect the goal of systematic search, that is to maximize recall to identify all relevant studies while controlling impact on precision to keep the results manageable.
We experienced a stable recall variance on all guideline topics (stddev = 5.1), however, the improving effect variance was high (stddev = 31.7). A subsequent analysis showed that three topics “Atrial Fibrillation”, “hypertrophic cardiomyopathy”, and “heart failure” had no improvement on recall, partially because the baseline PubMed expansion achieved good performance (avg recall 85.1%). All other topics had improvements in recall. The greatest improvement was seen in the topic “Extracranial Carotid and Vertebral Artery Disease” in which PubMed expansion did not perform well. The query expansion algorithm was able to find supporting MeSH terms such as “Carotid Artery Diseases”, “Vertebrobasilar Insufficiency”, ” Brain Ischemia”, and “Cerebrovascular Disorders”, and recall was improved by 70%.
The system achieved precision of 0.6% versus 0.8% with PubMed expansion. Therefore, we deem the system’s precision performance was acceptable and comparable with existing methods. Achieving good precision is difficult and secondary for systematic search. In fact, the manual search approach achieved precision below 1% (54–56). The poor precision can be attributed to the main goal of systematic search, which is to be exhaustive. Therefore, the queries were generally designed to be able to capture all potentially relevant candidates. In addition, some systematic reviews had specific inclusion/exclusion criteria which are not easily represented in the search queries without risking loss of recall. Further efforts to improve precision relates to previous works on document classification, in which training data to predict the inclusion/exclusion patterns are required (12, 17).
The citation ranking method proposed in this research used a simple light-weight approach that is independent of training data. Furthermore, the proposed approach improved ranking performance of the standard PubMed’s ranking by “most recent” (average precision +6.5%, recall@k +21.1%, p<0.001), PubMed’s ranking by “relevance” (average precision +6.1%, recall@k +14.8%, p<0.001), and the general purpose machine learning classifier (average precision +4.9%, recall@k +4.2%, p<0.001).
Implications
In the development of systematic reviews, manual search is considered as the state-of-the-art approach, but it does not guarantee perfect recall. The quality of the search is essentially impacted by skills, experience and domain knowledge of searchers on the review topics. A common approach to improve recall is to gather results from multiple sources either from different search strategies or from domain experts. The American College of Cardiology Foundation (ACCF) recommends clinicians to perform their own search along with systematic search by skilled librarians (11). Our method is not intended to completely replace the manual process. However, it can serve as starting point or as a reference list to augment the manual search approach. For example, taking our dataset, if reviewers screen the top 100 citations retrieved by our system, they would be able to find 16.2% of the citations included in the guidelines and seed citations for 24.4% of the guideline recommendations. Another potential approach is to use citation tracking by examining articles that cite or are cited by seed citations. The seeding recall metric used in our study provides a measure of algorithm performance in this respect. The system was able to find the seed studies for 90% of the guideline recommendations.
Ranking studies by relevancy and scientific rigor might be useful to help prioritize early stages in the development of systematic reviews. A good ranking mechanism increases the odds of finding relevant studies with less effort. Previous studies on work prioritization (22, 23) favor using machine learning methods, which use previous manual screening as labeled data to train classifiers. However, in systematic search, new questions are often raised that have insufficient historical data to train a competent machine learning model. As a result, searchers often rely on standard functionalities of search engines, or ML classifiers that were trained on broad topics. Our experiments showed that standard ranking methods of biomedical search engines and a general purpose ML classifier can be further improved using heuristics such as MeSH majority, research design, and journal ranking. These heuristics are independent of the training data and not specific to any particular guideline topic or domain.
This study focuses on cardiovascular guideline as our domain of interest; however, the proposed techniques are applicable to literature search in general. First, the system employed reusable expansion techniques to identify relevant MeSH concepts (concept-mapping, statistical, and MeSH Tree traversing) that are not specific to cardiology and should work well in any other domain related to treatment. For areas other than diseases (e.g., procedures), the algorithm could be adapted by using different semantic types. For example, a review topic focused on an intervention procedure could use semantic type “Therapeutic or Preventive Procedure”. Secondly, our ranking approach was based on three factors: MeSH Majority, Study Design, and Journal Ranking. MeSH Majority and Journal Ranking information can always be found in MEDLINE and Scopus. The assignment of the study design (SD) score is adapted from the GRADE approach(18), which is widely used in the assessment of evidence quality independent of clinical domain.
Limitations
This study has five main limitations. First, our gold standard consists of eight guidelines, which limits the generalizability of our findings. However, the guidelines we selected represent a broad coverage in the important field of cardiovascular diseases. In 2010, ACCF/AHA published a methodology manual that mandated all practice recommendations grade A and B to be accompanied with citations to the evidence sources. This practice will help expand the size and breadth of gold standards in future studies. Second, our research was limited to guidelines on the treatment of cardiovascular diseases, so it is unknown whether the results generalize to other domains. Yet, our approach did not use any methods that were specific to cardiovascular diseases, so it is expected that the methods will generalize to other domains and topics. Third, our query expansion algorithm uses an ad-hoc threshold (5000) for triggering parent concept expansion. The selection of this threshold was somewhat arbitrary and can be improved further based on heuristics such as the descriptive statistics of retrieved documents. Fourth, our system achieved low precision that is common and secondary in systematic search. Previous techniques based on automated and semi-automated document classification to support citation screening could be used to improve precision. Last, we didn’t re-train the Kilicoglu’s classifier with our dataset and use the classifier developed in their original research (45). In the early stage of literature search, the lack of labeled data made it difficult to train a competent machine learning classifier.
Future studies
Areas that warrant further investigation include improving overall precision using automated and semiautomated document classification techniques; expanding the gold standard beyond cardiovascular topics; improving the method to distinguish diagnosis and treatment topics; and applying the method to other types of systematic review, such as Cochrane systematic reviews, and drug effectiveness reviews.
5. Conclusions
We present informatics solutions to improve the retrieval performance of high quality studies to support the development of clinical guidelines in the cardiovascular domain. Overall, our methods are unsupervised and integrated over a widely used biomedical search engine (PubMed). The methods showed improved recall over standard PubMed’s query expansion and rankings, and a general-purpose machine learning classifier. The proposed approach could be used to aid the systematic search and screening process in the development of systematic reviews and clinical guidelines.
Hightlights.
Automated citation finding can augment manual literature search
We built a gold standard with citations from 653 guideline recommendations
The query expansion method vs. PubMed improved recall with non-significant loss on precision
The unsupervised citation ranking approach performed better than standard PubMed ranking and a machine learning classifier
Acknowledgments
This project was supported in part by grants 1R01LM011416-02 and 4R00LM011389-02 from the National Library of Medicine.
References
- 1.Ware M, Mabe M. An overview of scientific and scholarly journal publishing. The stm report. 2009 [Google Scholar]
- 2.Statistical Reports on MEDLINE®/PubMed® Baseline Data. Available from: https://www.nlm.nih.gov/bsd/licensee/baselinestats.html.
- 3.Trends, Charts, and Maps: ClinicalTrials.gov. Available from: http://clinicaltrial.gov/ct2/resources/trends.
- 4.Fraser AG, Dunstan FD. On the impossibility of being expert. BMJ (Clinical research ed ) 2010;341:c6815. doi: 10.1136/bmj.c6815. English. [DOI] [PubMed] [Google Scholar]
- 5.Davies K, Harrison J. The information-seeking behaviour of doctors: a review of the evidence. Health Info Libr J. 2007 Jun;24(2):78–94. doi: 10.1111/j.1471-1842.2007.00713.x. [DOI] [PubMed] [Google Scholar]
- 6.Del Fiol G, Workman TE, Gorman PN. Clinical Questions Raised by Clinicians at the Point of Care: A Systematic Review. JAMA internal medicine. 2014 doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 7.Smith R. Strategies for coping with information overload. Bmj. 2010;341:c7126. doi: 10.1136/bmj.c7126. [DOI] [PubMed] [Google Scholar]
- 8.Burgers JS, Grol R, Klazinga NS, Makela M, Zaat J, Collaboration A. Towards evidence-based clinical practice: an international survey of 18 clinical guideline programs. International journal for quality in health care : journal of the International Society for Quality in Health Care / ISQua. 2003 Feb;15(1):31–45. doi: 10.1093/intqhc/15.1.31. [DOI] [PubMed] [Google Scholar]
- 9.Schünemann HJ, Fretheim A, Oxman AD Research WACoH. Improving the use of research evidence in guideline development: 1. Guidelines for guidelines. Health Res Policy Syst. 2006;4(13):1–6. doi: 10.1186/1478-4505-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner T, Misso M, Harris C, Green S. Development of evidence-based clinical practice guidelines (CPGs): comparing approaches. Implementation science : IS. 2008;3(45):1–8. doi: 10.1186/1748-5908-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ACCF/AHA TaskForce on Practice Guidelines. Methodology Manual and Policies From the ACCF/AHA Task Force on Practice Guidelines. 2010 Available from: http://assets.cardiosource.com/Methodology_Manual_for_ACC_AHA_Writing_Committees.pdf.
- 12.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc. 2006 Mar-Apr;13(2):206–19. doi: 10.1197/jamia.M1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace BC, Trikalinos TA, Lau J, Brodley C, Schmid CH. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics. 2010;11:55. doi: 10.1186/1471-2105-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonnalagadda S, Petitti D. A new iterative method to reduce workload in systematic review process. International journal of computational biology and drug design. 2013;6(1–2):5–17. doi: 10.1504/IJCBDD.2013.052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiszman M, Bray BE, Shin D, Kilicoglu H, Bennett GC, Bodenreider O, et al. Combining relevance assignment with quality of the evidence to support guideline development. Studies in health technology and informatics. 2010;160(Pt 1):709–13. [PMC free article] [PubMed] [Google Scholar]
- 16.Fiszman M, Ortiz E, Bray BE, Rindflesch TC. Semantic processing to support clinical guideline development. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2008:187–91. [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen AM, Ambert K, McDonagh M. A Prospective Evaluation of an Automated Classification System to Support Evidence-based Medicine and Systematic Review. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2010;2010:121–5. [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins JP, Green S. Cochrane handbook for systematic reviews of interventions. Wiley Online Library; 2008. [Google Scholar]
- 19.Crespo Azcarate M, Mata Vazquez J, Mana Lopez M. Improving image retrieval effectiveness via query expansion using MeSH hierarchical structure. Journal of the American Medical Informatics Association : JAMIA. 2013;20(6):1014–20. doi: 10.1136/amiajnl-2012-000943. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z, Kim W, Wilbur WJ. Evaluation of Query Expansion Using MeSH in PubMed. Inf Retr Boston. 2009;12(1):69–80. doi: 10.1007/s10791-008-9074-8. Epub 2009/09/24. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bui D, Redd D, Rindflesch T, Zeng-Treitler Q. An Ensemble Approach for Expanding Queries. DTIC Document. 2012 [Google Scholar]
- 22.Cohen AM, editor. AMIA annual symposium proceedings. American Medical Informatics Association; 2008. Optimizing feature representation for automated systematic review work prioritization. [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen AM, Ambert K, McDonagh M. Cross-topic learning for work prioritization in systematic review creation and update. Journal of the American Medical Informatics Association. 2009;16(5):690–704. doi: 10.1197/jamia.M3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moosavinasab S, Rastegar-Mojarad M, Liu H, Jonnalagadda SR. Towards Transforming Expert-based Content to Evidence-based Content. AMIA Summits on Translational Science Proceedings. 2014;2014:83. [PMC free article] [PubMed] [Google Scholar]
- 25.Jonnalagadda SR, PhD12 SM. Prioritizing journals relevant to a topic for addressing clinicians’ information needs. Medicine. 4:132. [Google Scholar]
- 26.NCBI. NCBI Batch Citation Matcher. Available from: http://www.ncbi.nlm.nih.gov/pubmed/batchcitmatch.
- 27.Ahmad Y, Lip GY, Apostolakis S. New oral anticoagulants for stroke prevention in atrial fibrillation: impact of gender, heart failure, diabetes mellitus and paroxysmal atrial fibrillation. Expert review of cardiovascular therapy. 2012 Dec;10(12):1471–80. doi: 10.1586/erc.12.148. [DOI] [PubMed] [Google Scholar]
- 28.Chiang CE, Naditch-Brule L, Murin J, Goethals M, Inoue H, O'Neill J, et al. Distribution and risk profile of paroxysmal, persistent, and permanent atrial fibrillation in routine clinical practice: insight from the real-life global survey evaluating patients with atrial fibrillation international registry. Circulation Arrhythmia and electrophysiology. 2012 Aug 1;5(4):632–9. doi: 10.1161/CIRCEP.112.970749. [DOI] [PubMed] [Google Scholar]
- 29.Flaker G, Ezekowitz M, Yusuf S, Wallentin L, Noack H, Brueckmann M, et al. Efficacy and safety of dabigatran compared to warfarin in patients with paroxysmal, persistent, and permanent atrial fibrillation: results from the RE-LY (Randomized Evaluation of Long-Term Anticoagulation Therapy) study. J Am Coll Cardiol. 2012 Feb 28;59(9):854–5. doi: 10.1016/j.jacc.2011.10.896. [DOI] [PubMed] [Google Scholar]
- 30.Hohnloser SH, Duray GZ, Baber U, Halperin JL. Prevention of stroke in patients with atrial fibrillation: current strategies and future directions. European Heart Journal Supplements. 2008;10(suppl H):H4–H10. [Google Scholar]
- 31.Farshi R, Kistner D, Sarma JS, Longmate JA, Singh BN. Ventricular rate control in chronic atrial fibrillation during daily activity and programmed exercise: a crossover open-label study of five drug regimens. J Am Coll Cardiol. 1999 Feb;33(2):304–10. doi: 10.1016/s0735-1097(98)00561-0. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg JS, Katz RJ, Bren GB, Buff LA, Varghese PJ. Efficacy of oral diltiazem to control ventricular response in chronic atrial fibrillation at rest and during exercise. J Am Coll Cardiol. 1987;9(2):405–11. doi: 10.1016/s0735-1097(87)80396-0. [DOI] [PubMed] [Google Scholar]
- 33.Olshansky B, Rosenfeld LE, Warner AL, Solomon AJ, O'Neill G, Sharma A, et al. The Atrial Fibrillation Follow-up Investigation of Rhythm Management (AFFIRM) study: approaches to control rate in atrial fibrillation. J Am Coll Cardiol. 2004 Apr 7;43(7):1201–8. doi: 10.1016/j.jacc.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 34.Abrams J, Allen J, Allin D, Anderson J, Anderson S, Blanski L, et al. Efficacy and safety of esmolol vs propranolol in the treatment of supraventricular tachyarrhythmias: a multicenter double-blind clinical trial. American heart journal. 1985 Nov;110(5):913–22. doi: 10.1016/0002-8703(85)90185-1. [DOI] [PubMed] [Google Scholar]
- 35.Ellenbogen KA, Dias VC, Plumb VJ, Heywood JT, Mirvis DM. A placebo-controlled trial of continuous intravenous diltiazem infusion for 24-hour heart rate control during atrial fibrillation and atrial flutter: a multicenter study. J Am Coll Cardiol. 1991 Oct;18(4):891–7. doi: 10.1016/0735-1097(91)90743-s. [DOI] [PubMed] [Google Scholar]
- 36.Siu C-W, Lau C-P, Lee W-L, Lam K-F, Tse H-F. Intravenous diltiazem is superior to intravenous amiodarone or digoxin for achieving ventricular rate control in patients with acute uncomplicated atrial fibrillation*. Critical care medicine. 2009;37(7):2174–9. doi: 10.1097/CCM.0b013e3181a02f56. [DOI] [PubMed] [Google Scholar]
- 37.Platia EV, Michelson EL, Porterfield JK, Das G. Esmolol versus verapamil in the acute treatment of atrial fibrillation or atrial flutter. The American journal of cardiology. 1989 Apr 15;63(13):925–9. doi: 10.1016/0002-9149(89)90141-0. [DOI] [PubMed] [Google Scholar]
- 38.Haynes RB, McKibbon KA, Wilczynski NL, Walter SD, Werre SR. Optimal search strategies for retrieving scientifically strong studies of treatment from Medline: analytical survey. Bmj. 2005;330(7501):1179. doi: 10.1136/bmj.38446.498542.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronson AR. Metamap: Mapping text to the umls metathesaurus. Bethesda, MD: NLM, NIH, DHHS; 2006. pp. 1–26. [Google Scholar]
- 40.Sayers E. Entrez programming utilities help. 2009 http://www.ncbi.nlm.nih.gov/books/NBK25499.
- 41.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings / AMIA Annual Symposium AMIA Symposium. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 42.Medicine NLo. UMLS® Reference Manual. 2009 Available from: http://www.ncbi.nlm.nih.gov/books/NBK9685/
- 43.NLM. Medical Subject Headings. Files Available to Download. 2014 Available from: http://www.nlm.nih.gov/mesh/filelist.html.
- 44.NCBI. PubMed Help. 2015 Available from: http://www.ncbi.nlm.nih.gov/books/NBK3827/#pubmedhelp.Sorting_your_search_results.
- 45.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB. Towards automatic recognition of scientifically rigorous clinical research evidence. J Am Med Inform Assoc. 2009 Jan-Feb;16(1):25–31. doi: 10.1197/jamia.M2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.January CT, Wann LS, Alpert JS, Calkins H, Cleveland JC, Jr, Cigarroa JE, et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2014 Mar 28; doi: 10.1016/j.jacc.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 47.Brott TG, Halperin JL, Abbara S, Bacharach JM, Barr JD, Bush RL, et al. 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease. A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American Stroke Association, American Association of Neuroscience Nurses, American Association of Neurological Surgeons, American College of Radiology, American Society of Neuroradiology, Congress of Neurological Surgeons, Society of Atherosclerosis Imaging and Prevention, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of NeuroInterventional Surgery, Society for Vascular Medicine, and Society for Vascular Surgery. Circulation. 2011 Jul 26;124(4):e54–130. doi: 10.1161/CIR.0b013e31820d8c98. [DOI] [PubMed] [Google Scholar]
- 48.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011 Dec 13;124(24):e783–831. doi: 10.1161/CIR.0b013e318223e2bd. [DOI] [PubMed] [Google Scholar]
- 49.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr, Drazner MH, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013 Oct 15;62(16):e147–239. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 50.O'Gara PT, Kushner FG, Ascheim DD, Casey DE, Jr, Chung MK, de Lemos JA, et al. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013 Jan 29;127(4):e362–425. doi: 10.1161/CIR.0b013e3182742cf6. [DOI] [PubMed] [Google Scholar]
- 51.Fihn SD, Gardin JM, Abrams J, Berra K, Blankenship JC, Dallas AP, et al. 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and management of patients with stable ischemic heart disease: executive summary: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines, and the American College of Physicians, American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation. 2012 Dec 18;126(25):3097–137. doi: 10.1161/CIR.0b013e3182776f83. [DOI] [PubMed] [Google Scholar]
- 52.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE, Jr, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010 Apr 6;121(13):e266–369. doi: 10.1161/CIR.0b013e3181d4739e. [DOI] [PubMed] [Google Scholar]
- 53.Nishimura RA, Otto CM, Bonow RO, Carabello BA, Erwin JP, 3rd, Guyton RA, et al. 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014 Jun 10;63(22):e57–e185. doi: 10.1016/j.jacc.2014.02.536. [DOI] [PubMed] [Google Scholar]
- 54.Wiesbauer F, Domanovits H, Schlager O, Wildner B, Schillinger M, Blessberger H. The Cochrane Library. 2003. Perioperative beta-blockers for preventing surgery related mortality and morbidity. [Google Scholar]
- 55.De Lima LG, Saconato H, Atallah ÁN, da Silva EM. The Cochrane Library. 2014. Beta-blockers for preventing stroke recurrence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez-Briz E, Ruiz Garcia V, Cabello JB, Bort-Marti S, Carbonell Sanchis R, Burls A. Heparin versus 0.9% sodium chloride intermittent flushing for prevention of occlusion in central venous catheters in adults. The Cochrane database of systematic reviews. 2014;10 doi: 10.1002/14651858.CD008462.pub2. [DOI] [PubMed] [Google Scholar]