Abstract
Many genetic liver diseases present in newborns with repeated, often lethal, metabolic crises. Gene therapy using non-integrating viruses such as AAV is not optimal in this setting because the non-integrating genome is lost as developing hepatocytes proliferate1,2. We reasoned that newborn liver may be an ideal setting for AAV-mediated gene correction using CRISPR/Cas9. Here we intravenously infuse two AAVs, one expressing Cas9 and the other expressing a guide RNA and the donor DNA, into newborn mice with a partial deficiency in the urea cycle disorder enzyme, ornithine transcarbamylase (OTC). This resulted in reversion of the mutation in 10% (6.7% – 20.1%) of hepatocytes and increased survival in mice challenged with a high-protein diet, which exacerbates disease. Gene correction in adult OTC-deficient mice was lower and accompanied by larger deletions that ablated residual expression from the endogenous OTC gene, leading to diminished protein tolerance and lethal hyperammonemia on a chow diet.
An X-linked deficiency of the OTC enzyme in humans causes recurrent and life-threatening episodes of hyperammonemia3,4. In males hemizygous for OTC deficiency, the first metabolic crisis usually occurs in the newborn period and is associated with up to 50% mortality, with survivors typically undergoing liver transplantation in the first year of life5. An animal model of OTC deficiency, the male sparse fur ash (spfash) mouse, has a G-to-A point mutation at the donor splice site at the end of exon 4 of the OTC gene, which leads to abnormal splicing and a 20-fold reduction in OTC mRNA and protein6. Affected animals have 5% residual OTC activity and can survive on a chow diet, but they develop hyperammonia that can be lethal when provided a high-protein diet.
In vivo genome editing of disease-causing mutations is a promising approach for the treatment of genetic disorders7–17. We developed a strategy using a dual-AAV system based on AAV8, which has high liver tropism to correct the point mutation in newborn spfash mice using Cas9 enzyme from Staphylococcus aureus (SaCas9)11–13. Prior to incorporating the individual components of the system into AAV8 vectors, we searched for protospacer-adjacent motif (PAM) sequences (NNGRRT) in proximity to the spfash mutation of the OTC gene and identified potential 20-nt protospacer sequences. Three sequences, sgRNA1–3 (Fig. 1a), were further evaluated following transfection of puromycin-containing plasmids into a mouse MC57G cell line. Evidence for double-strand breaks (DSBs) and the formation of indels at the desired site was demonstrated using the SURVEYOR assay (Supplementary Fig. 1a). One protospacer located within the adjacent intron (i.e., sgRNA3) failed to yield indels in this in vitro assay, while the others generated indels at the desired sites (Supplementary Fig. 1a). We selected the protospacer with a PAM within the adjacent intron (sgRNA1) because non-homologous end joining (NHEJ) without homology directed repair (HDR) within an exon could ablate residual OTC activity of the hypomorphic spfash mutation, thereby reducing residual ureagenesis. A plasmid cassette co-expressing the sgRNA1 guide RNA and SaCas9 was co-transfected with a plasmid containing a donor DNA template with approximately 0.9 kb of sequence flanking each side of the mutation. We mutated the corresponding PAM sequence in the donor template to reduce re-cleavage after HDR and included an AgeI site facilitate detection of HDR, which was achieved with high-efficiency (Supplementary Fig. 1b).
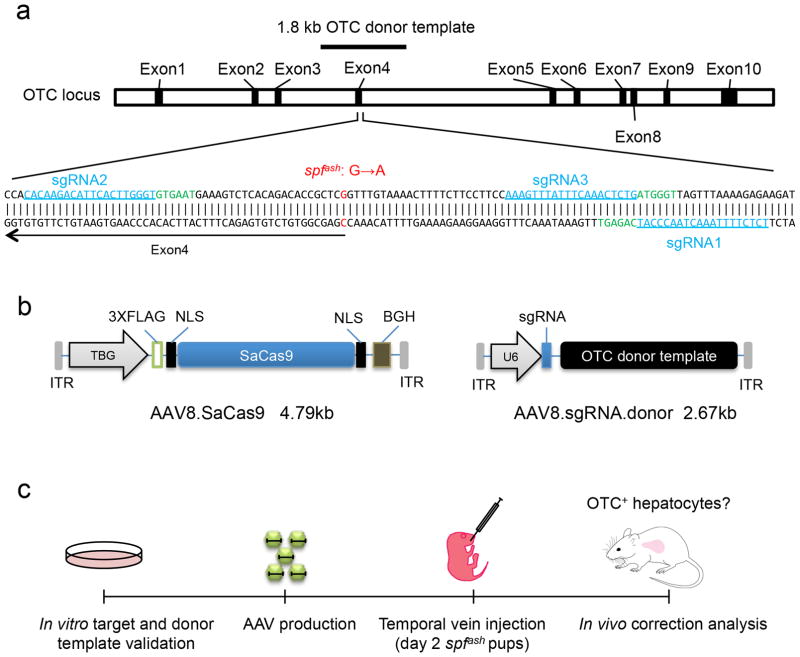
Figure 1. In vivo gene correction of the OTC locus in the spfash mouse liver by AAV.CRISPR-SaCas9.
(a) Schematic diagram of the mouse OTC locus showing the spfash mutation and three SaCas9 targets. spfash has a G-to-A mutation at the donor splice site at the end of exon 4 indicated in red on the top strand. The three selected SaCas9-targeted genomic sites (20 bp each) are in blue and underlined with the PAM sequences marked in green. The black line above exon 4 indicates the 1.8 kb OTC donor template. (b) Dual AAV vector system for liver-directed and SaCas9-mediated gene correction. The AAV8.sgRNA1.donor vector contains a 1.8-kb murine OTC donor template sequence as shown in (a) with the corresponding PAM sequence mutated and an AgeI site inserted. (c) Flowchart showing the key steps of AAV8.CRISPR-SaCas9-mediated gene correction in the neonatal OTC spfash model.
A two-vector approach was necessary to incorporate all components into AAV (Fig. 1b). Vector 1 expressed the SaCas9 gene from a liver-specific TBG promoter (subsequently referred to as AAV8.SaCas9), while vector 2 contained the sgRNA1 sequence expressed from the U6 promoter and the 1.8 kb donor OTC DNA sequence (referred to as AAV8.sgRNA1.donor). In all experiments, spfash pups were injected intravenously on postnatal day 2 with mixtures of vector 1 and vector 2 and subsequently evaluated for indel formation and functional correction of the spfash mutation (Fig. 1c).
We obtained liver samples from treated spfash animals, untreated spfash (spfash controls), wildtype littermates, and spfash mice administered AAV8.SaCas9 with a modified AAV8.control.donor without guide RNA (untargeted) at 1, 3, and 8 weeks following vector infusion. Pilot experiments elucidated optimal conditions of vector infusion with respect to doses and ratios of the two vectors (Supplementary Fig. 2). We administered 5×1011 genome copies (GC) of AAV8.sgRNA1.donor (or AAV8.control.donor) and 5×1010 GC of AAV8.SaCas9 in all newborn mouse experiments.
We analyzed the targeted region of the OTC gene by deep sequencing of PCR amplicons of liver tissue harvested 3 weeks (n=3) and 8 weeks (n=3) after vector treatment, and one untreated spfash mouse (Supplementary Table 1). More detailed descriptions of the actual indels from a subset of these animals is summarized in Supplementary Table 2. Following gene correction, indels were detected in 31% (26.5% – 35.5%) of OTC alleles from the 6 treated animals (Supplementary Table 1). More detailed studies in two treated mice indicated that over 90% of the deletions were less than 20 bp and only 1% extended into the adjacent exon (Supplementary Table 3). HDR-based correction of the G-to-A mutation was observed in 10% (6.7% – 20.1%) of OTC alleles from 6 treated animals (Supplementary Table 1). Analysis of amplified DNA between the G-to-A mutation and the donor-specific, altered PAM located 51 nt into the adjacent intron showed that approximately 83% of corrected alleles contained only donor derived-sequences between these two landmarks (reads with perfect HDR, Supplementary Table 1), while 3.5% of total OTC alleles had evidence of incomplete HDR events (reads with partial HDR, Supplementary Table 1). HDR-mediated targeted modifications were also estimated by the presence of a restriction-fragment length polymorphism (RFLP) introduced into the donor DNA in three animals harvested at each of the three time points. The average rate of HDR was 2.6% at 1 week, 18.5% at 3 weeks, and 14.3% at 8 weeks, confirming the high rate of HDR observed by deep sequencing (Supplementary Fig. 3a).
The algorithm described in www.benchling.com identified 49 potential off-target sites for sgRNA1. The top 16 sites most likely to create DSBs were amplified by PCR and deep sequenced (Supplementary Table 4). Samples from treated animals did not show indel rates above background (indel rates in untreated animals due to sequencing error, usually a fraction of a percent).
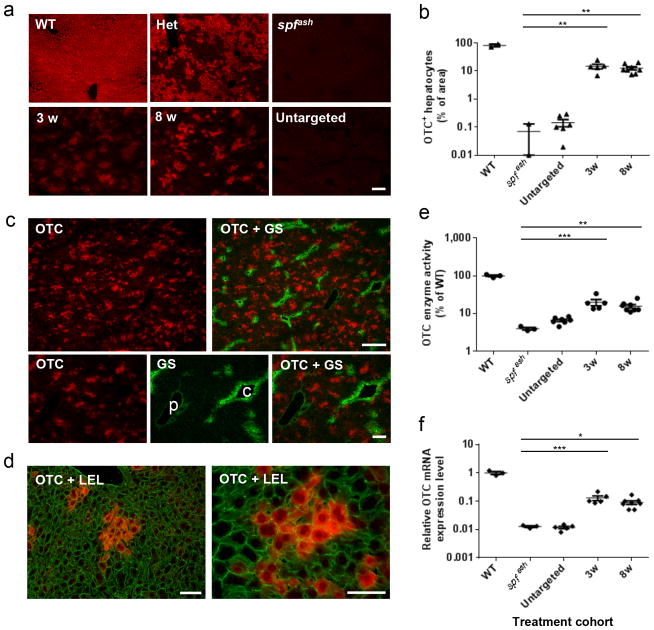
Tissue sections of liver were analyzed by immunohistochemistry for OTC expression. No signal (<1%) was observed in the spfash controls, while analysis of heterozygotes showed the predicted mosaicism (Fig. 2a). Morphometry indicated over 100-fold higher numbers of OTC-expressing cells in treated groups than found in the spfash control groups (Fig. 2b; 15% (6.8% – 24.4%) at 3 weeks and 13% (7.5% – 20.1%) at 8 weeks). Treated animals showed patches of OTC-expressing cells (Fig. 2c) that localized within all portions of the portal axis except around central veins, as predicted for endogenous OTC18 Higher magnification showed clusters of OTC-expressing hepatocytes consistent with correction followed by clonal expansion in the context of the growing liver (Fig. 2d). Direct measurements of OTC enzyme activity from liver homogenates and OTC mRNA from total cellular liver RNA revealed similarly high levels of correction in treated animals, resulting in 20% (13.4% – 33.7%) and 16% (11.0% – 25.4%) of normal OTC enzyme activity at 3 and 8 weeks, respectively (Fig. 2e), and 13% (8.6% – 21.8%) and 9% (5.0% – 16.8%) of normal OTC mRNA at 3 and 8 weeks, respectively (Fig. 2f). Despite the decrease in OTC+ hepatocytes, OTC enzyme activity, and OTC mRNA expression from 3 to 8 weeks, none of these differences were statistically significant (Fig. 2b; p=0.4828, Fig. 2e; p=0.2723, Fig. 2f; p=0.1475, respectively). OTC protein levels in liver of most treated animals were higher than in spfash controls but did not reach the levels found in wild-type mice (Supplementary Fig. 3b). Overall, there was good correlation of the estimates of correction based on histology, protein, and mRNA within individual animals.
Figure 2. Efficient restoration of OTC expression in the liver of spfash mice treated at neonatal stage by AAV8.CRISPR-SaCas9-mediated gene correction.
AAV8.SaCas9 (5×1010 GC/pup) and AAV8.sgRNA1.donor (5×1011 GC/pup) were administrated to postnatal day 2 (p2) spfash pups via the temporal vein. spfash mice were sacrificed at 3 (n=5) or 8 weeks (n=8) after treatment. Untargeted spfash mice received AAV8.SaCas9 (5×1010 GC/pup) and AAV8.control.donor (5×1011 GC/pup) at p2, and livers were harvested 8 weeks post treatment (n=6). Untreated WT (n=3) and spfash mice (n=3) were included as controls. (a) Immunofluorescence staining with antibodies against OTC on liver sections from spfash mice treated with the dual AAV vectors for CRISPR-SaCas9-mediated gene correction. Stained areas typically represent clusters of corrected hepatocytes. Untreated controls show livers from wild type, spfash heterozygous, and spfash hemizygous mice. Scale bar, 100μm. (b) Quantification of gene correction based on the percentage of area on liver sections expressing OTC by immunostaining as presented in panel a. (c) Random distribution of clusters of corrected hepatocytes along the portal-central axis shown by double immunostaining against OTC (red) and glutamine synthetase (GS, green), which is a marker of central veins (p, portal vein; c, central vein). Scale bars, 300 μm (upper panel) and 100 μm (lower panel). (d) Groups of corrected hepatocytes expressing OTC (red) shown by immunofluorescence on sections co-stained with fluorescein-labeled tomato lectin (Lycopersicon esculentum lectin, LEL; green) which outlines individual hepatocytes. Scale bar, 50μm. (e) OTC enzyme activity in the liver lysate of spfash mice at 3 and 8 weeks following dual vector treatment. (f) Quantification of OTC mRNA levels in the liver by RT-qPCR using primers spanning exons 4–5 to amplify wild-type OTC. Mean ± SEM are shown. * P < 0.05, ** P < 0.01, *** P < 0.001, Dunnett’s test.
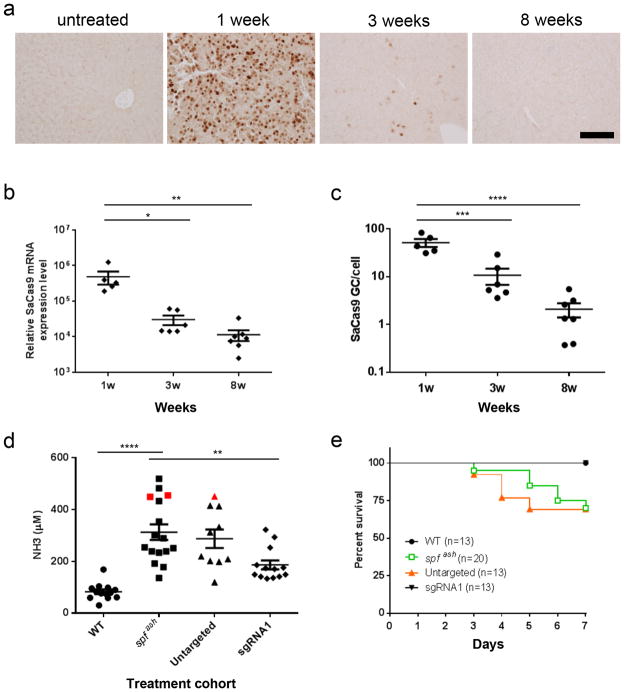
One concern about using AAV to deliver SaCas9 is the virus’s propensity to achieve stable transgene expression, which is not necessary to accomplish editing and may in fact contribute to immune and/or genome toxicity. Western blot analysis showed high level SaCas9 protein at 1 week that declined to undetectable levels by 8 weeks (Supplementary Fig. 3b). Furthermore, immunohistochemistry revealed nuclear-localized SaCas9 protein in 21% of hepatocytes at one week, which declined to undetectable levels (<0.1% hepatocytes) by 8 weeks (Fig. 3a). SaCas9 mRNA declined 43-fold during this 7-week period, to very low but still detectable levels (Fig. 3b). A 25-fold reduction in SaCas9 DNA during this same time interval indicates that elimination of vector genomes in the setting of the proliferating newborn liver is a primary contributor to the desired decline of SaCas9 expression (Fig. 3c). SaCas9 mRNA/vector copy did not change over time, indicating that the TBG promoter was still active.
Figure 3. Time course of SaCas9 expression following neonatal vector administration and functional improvement following high-protein diet challenge.
(a) Immunostaining with antibodies against FLAG on liver sections from an untreated mouse or treated spfash mice at 1, 3, or 8 weeks following neonatal injection of the dual AAV vectors for CRISPR-SaCas9-mediated gene correction. AAV8.SaCas9 (5×1010 GC/pup) and AAV8.sgRNA1.donor (5×1011 GC/pup) were administrated to p2 spfash pups via the temporal vein. Nuclear staining of FLAG-tagged SaCas9 were abundant at 1 week (n=5) but dramatically reduced at 3 weeks (n=6) and became scarce at 8 weeks (n=7) after vector injection. Scale bar, 100 μm. (b) Quantification of SaCas9 mRNA levels in liver by RT-qPCR. Mean ± SEM are shown. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, Dunnett’s test. (c) Quantification of SaCas9 vector genome in liver by qPCR. (d, e) Plasma ammonia levels and survival curves in control or dual AAV vector-treated spfash mice after a one-week course of high-protein diet. Seven weeks following neonatal treatment with the dual AAV vectors, mice were given high-protein diet for 7 days. (d) Plasma ammonia levels were measured 7 days after the high-protein diet. Plasma ammonia levels in WT mice (n=13) and AAV8.SaCas9 + AAV8.sgRNA1.donor-treated spfash mice (n=13) were significantly lower than untreated spfash mice (n=16) after a 7-day high-protein diet. Red squares indicate samples obtained from moribund untreated spfash mice 6 days after high-protein diet; red triangle indicates sample obtained from a moribund spfash mouse treated with untargeted vector (AAV8.control.donor with no sgRNA1, n=10) 5 days after high-protein diet. ** P< 0.01, **** P< 0.0001, Dunnett’s test. (e) Untreated spfash mice (n=20) or spfash mice treated with untargeted vectors (AAV8.control.donor, n=13) started to die 3 days after high-protein diet. All WT (n=13) and AAV8.SaCas9 + AAV8.sgRNA1.donor-treated mice (n=13) survived. * P< 0.05, Mantel-Cox test.
In assessing the impact of gene correction on the clinical manifestations of OTC deficiency, we evaluated the tolerance of spfash mice to a one-week course of a high-protein diet and found that blood ammonia was elevated from 83 ± 9 μM (n=13) in wild type controls to 312 ± 30 μM (n=16) in the spfash controls at the end of the diet course (Fig. 3d; p<0.0001). Substantial variation in blood ammonia levels was found in untreated spfash animals after the one-week diet course, which is consistent with findings in OTC-deficient patients, who show large fluctuations in ammonia over relatively short periods of time3. There was no significant difference between untreated spfash and untargeted spfash controls (Fig. 3d; p=0.83). In contrast, we observed a statistically significant 40% reduction in ammonia in treated as compared to untreated spfash animals (Fig. 3d; p=0.0014), and treated spfash mice showed a survival improvement (Fig. 3e; p=0.03). During the course of the high-protein diet, 30% of both untreated spfash (n=20) and untargeted spfash animals (n=13) developed clinical signs of hyperammonemia and died or had to be euthanized, while all of the wild type mice (n=13) and treated spfash mice (n=13) survived (Fig. 3e). Detailed histological analyses of liver and transaminase levels (both alanine and aspartate aminotransferase, ALT and AST, respectively) in SaCas9-treated spfash mice harvested at the end of the high-protein diet challenge failed to reveal any pathology or toxicity (Supplementary Fig. 4).
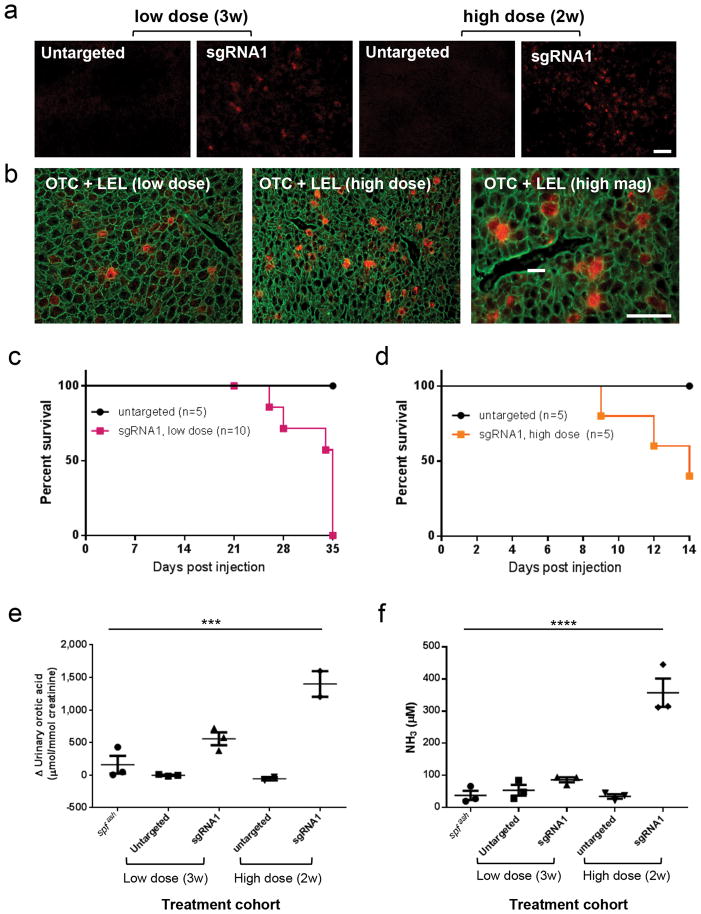
Based on encouraging results in newborn spfash mice, we then conducted similar studies in adult spfash mice, which were infused with two different doses of the AAV vectors and euthanized 2 to 3 weeks later. Sequence analysis of on-target DNA amplicons revealed remarkably high frequencies of indels at both low and high vector doses (Table 1; 44.6% (38.5% – 50.3%) and 42.0% (34.0% – 48.5%), respectively) but low levels of gene correction as measured by reversion of the G-to-A mutation (Table 1; 0.3% (0.2% – 0.3%) and 1.7% (1.3% – 2.1%), respectively). Additional evidence of correction was provided by immunofluorescence analysis of liver for OTC expression that showed 2.2% (0.71%, 2.51%, 3.45%) and 6.0% (3.11%, 5.05%, 9.86%) positive cells in low- and high-dose animals, respectively (Fig. 4a; n=3 per group). Adults showed a pattern of isolated OTC-positive cells (Fig. 4b), while clusters of OTC-positive cells were present in newborns (Fig. 3c). Between 3 to 4 weeks after treatment with low-dose vectors, the animals unexpectedly became sick and by week 5 all had to be euthanized (Fig. 4c). This toxicity was more severe in the high-dose animals, requiring termination of the study at 2 weeks (Fig. 4d). Further analysis of symptomatic animals demonstrated dose-dependent elevations of urine orotate (Fig. 4e) and plasma ammonia (Fig. 4f) on a chow diet. These findings suggested a compromise of residual ureagenesis that could not be explained by liver damage since liver histology appeared normal (Supplementary Fig. 5a) and serum transaminases were slightly, but not statistically, elevated over control groups (Supplementary Fig. 5b). Deep sequencing of the targeted region of the OTC gene revealed a surprising number of large deletions in adults as compared to what was observed in newborns, with 6.5% extending into the adjacent exon in adults as opposed to 1% in newborns (Supplementary Table 3). The more complex and extensive indels in adult spfash mice were unlikely to have been caused by higher and/or more persistent Cas9 expression since Cas9 mRNA was lower in 3-week adults (Fig. 4g) than in any liver tissues harvested up to 8 weeks after injection in newborns (Supplementary Fig. 6a and b). We speculate that different NHEJ mechanisms may exist in non-dividing adult hepatocytes versus dividing newborn hepatocytes that affects the quality of the DNA repair response.
Figure 4. Gene targeting/correction in the liver of spfash mice treated as adults by AAV8.CRISPR-SaCas9 vectors.
Adult spfash mice (8–10 weeks old) received an intravenous injection of AAV8.SaCas9 (1×1011 GC) and AAV8.sgRNA1.donor (1×1012 GC), or higher dose of AAV8.SaCas9 (1×1012 GC) and AAV8.sgRNA1.donor (5×1012 GC), or untargeted vectors at the equivalent doses. (a) Immunofluorescence staining with antibodies against OTC on liver sections collected at 3 (low-dose, n=3) or 2 weeks (high-dose, n=3) after injection. Stained cells typically showed as single corrected hepatocytes. Scale bar, 100μm. (b) Isolated corrected hepatocytes expressing OTC (red) shown by immunofluorescence on sections co-stained with fluorescein-labeled tomato lectin (LEL; green) which outlines individual hepatocytes. Scale bar, 50μm. (c) Survival curve of the low-dose cohorts: sgRNA1 (n=10) or untargeted vector at the same dose (n=5). (d) Survival curve of the high-dose cohorts: sgRNA1 (n=5) or untargeted vector at the same doses (n=5). The experiment was terminated at 14 days post vector injection. (e) Change of urine orotic acid levels in adult spfash mice after treatment with high-dose gene targeting vectors (n=3 for untreated spfash and low-dose groups; n=2 for high-dose groups) (f) Elevation of plasma NH3 levels in adult spfash mice after treatment with high-dose gene targeting vectors (n=3 for each group). Mean ± SEM are shown. *** P < 0.001, **** P < 0.0001, Dunnett’s test.
A key challenge in using the CRISPR/Cas9 system to correct a mutation in vivo is delivering the three components of the system (sgRNA, Cas9, and donor DNA) into the same cell in a way that is safe and efficient19,20. One recent report in the literature approached a similar challenge by utilizing chemically modified mRNA to deliver therapeutic levels of another site-specific endonuclease into murine lung21. Here, we leveraged our experience with liver-directed gene replacement therapy with highly hepatotropic AAV vectors22 to move closer to realizing these goals in a model of liver metabolic disease. While the focus on newborn animals was driven by a compelling unmet need in patients with these lethal metabolic diseases, it also created some unique technical advantages. The surprisingly high level of correction in our newborn experiments is likely due to high expression of SaCas9 with abundant donor DNA in the context of dividing cells. Our previous studies of AAV8 gene transfer in newborn monkeys demonstrated the same high peak levels of transduction and gene transfer (i.e., 92% hepatocytes expressing GFP and 32 vector genomes per cell) as achieved in mice administered the same dose of GFP-expressing vector (i.e., 80% hepatocytes expressing GFP and 14 vector genomes per cell) with similar kinetics of decline, which is encouraging in terms of translation to larger species including humans2,23.
Issues of safety relate primarily to the expression of Cas9 in the context of an sgRNA that could create off-target DSBs with carcinogenic sequelae, although our findings in the adult mice suggest that large on-target deletions could potentially be a safety issue in some contexts. More extensive characterization of these potential toxicities is necessary before clinical translation can be considered24,25, although, in animals treated as newborns, we could not detect indels of likely off-target sites at the level of sensitivity achieved by deep sequencing. Cas9 could also elicit pathologic immune responses16, as has been observed in gene replacement therapies in which the transgene is a foreign protein. However, systemic delivery of AAV in a newborn helps mitigate potential immunologic adverse events for several reasons26–28. First, expression of the prokaryotic SaCas9 protein is transient because the non-integrated vector is lost during hepatocyte proliferation1. Furthermore, we have shown that exposure of newborn rhesus macaques to AAV-encoded proteins induces tolerance to these proteins, thereby circumventing toxicity caused by destructive adaptive immune responses29.
This study provides convincing evidence for efficacy in an authentic animal model of a lethal human metabolic disease following in vivo genome editing. Furthermore, our observation of dramatic differences in clinical outcome following HDR-mediated gene correction of newborn versus adult animals illustrates potential unintended consequences of NHEJ-mediated ablation of residual function in hypomorphic mutant genes that may complicate some applications of therapeutic genome editing.
Online Methods
Plasmid construction
The smaller-sized Cas9 from Staphylococcus aureus (SaCas9) is more suitable for packaging into an AAV vector. We codon-optimized FLAG-tagged SaCas9 according to human codon usage (hSaCas9) and constructed pX330.hSaCas9 by replacing the hSpCas9 and sgRNA scaffold in pX330 with hSaCas9 and SaCas9 sgRNA scaffold. Three 20-nt target sequences preceding a 5′NNGRRT PAM sequence were selected for OTC gene editing. A puromycin-resistance gene cassette was cloned into pX330.hSaCas9-derived plasmids for selection of transfected cells following in vitro transient transfection. To generate a dual AAV vector system for in vivo OTC gene correction by SaCas9, we constructed two AAV cis-plasmids: 1) the hSaCas9 was subcloned from pX330.hSaCas9 into an AAV backbone plasmid containing the full-length TBG promoter (two copies of enhancer elements of the α microglobulin/bikunin gene followed by a liver-specific TBG promoter) and the bovine growth hormone polyadenylation sequence, yielding AAV8.SaCas9; 2) the 1.8-kb OTC donor template was cloned into the pAAV backbone, and the U6-OTC sgRNA1 cassette was inserted into the AflII site, yielding AAV8.sgRNA1.donor. The PAM sequence in the donor template in AAV8.sgRNA1.donor was mutated to prevent re-cleavage by Cas9 after HDR, and an AgeI site was added to facilitate detection of HDR (Supplementary Table 5). The “untargeted” AAV8.control.donor differs from the “targeted” AAV8.sgRNA1.donor by eliminating the protospacer from the U6-OTC sgRNA1 cassette. All plasmid constructs were verified by sequencing.
AAV vector production
All AAV8 vectors were produced by the Penn Vector Core at the University of Pennsylvania as previously described30. The genome titer (GC mL−1) of AAV vectors was determined by quantitative PCR (qPCR). All vectors used in this study passed the endotoxin assay using the QCL-1000 Chromogenic LAL test kit (Cambrex Bio Science).
Cell culture and transfection
MC57G cells (ATCC) were maintained in DMEM medium supplemented with 10% FBS and cultured at 37°C with 5% CO2. Cell lines were used directly upon receipt from ATCC and were not authenticated or tested for mycoplasma contamination. For in vitro target and/or donor template testing, plasmids were transfected into MC57G cells using Lipofectamine® LTX with Plus™ reagent (Life Technology) per manufacturer’s recommendations. Transfected cells were under puromycin (4 μg mL−1) selection for 4 days to enrich transfected cells.
Genomic DNA extraction and SURVEYOR assay
Genomic DNA from transfected MC57G cells was extracted using the QuickExtract DNA extraction solution (Epicentre Biotechnologies). The efficiency of each individual sgRNA was tested by the SURVEYOR nuclease assay (Transgenomics) as described previously31 using the PCR primers listed in Supplementary Table 5.
Animal studies
spfash mice were maintained in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care)-accredited and PHS (Public Health Service)-assured facility at the University of Pennsylvania, as described previously32. All animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. Mating cages were monitored daily for births. Newborn (postnatal day 2, p2) male pups received a temporal vein injection of the mixture of two vectors at the intended doses for each in a volume of 50 μl, as described33. Untreated wild-type (WT), spfash heterozygous (Het), and spfash hemizygous mice served as controls. Mice were sacrificed at 1, 3, or 8 weeks after vector treatment, and liver samples were harvested for analyses. Mice were genotyped at weaning or at the time of necropsy to confirm genotype.
For testing the efficacy of OTC correction, a high-protein diet (40% protein, Animal Specialties & Provisions) was given to 7-week-old mice for 7 days. After this time, plasma was collected for measurement of plasma NH3 using the Sigma Ammonia Assay Kit. The remaining samples were sent to Antech Diagnostics for measurements of ALT, AST, and total bilirubin.
Note that the entire litter of newborn male pups was injected with either the test or control vectors, and no specific randomization method was used. The following assays were performed in a blinded fashion in which the investigator was unaware of the nature of the vectors or vector dose: vector injection, OTC and Cas9 (FLAG) immunostaining and quantification, histopathology analysis on liver, OTC enzyme activity assay, and gene expression analysis and RT-qPCR.
The adult gene editing experiments were conducted in 8- to 10-week-old male spfash mice. Animals in low-dose groups received a tail vein injection of AAV8.SaCas9 (1×1011GC) and AAV8.sgRNA1.donor (1×1012 GC) or untargeted vectors at the same doses, and they were sacrificed at 3 weeks after injection for analyses. Animals in high-dose groups received a tail vein injection of AAV8.SaCas9 (1×1012 GC) and AAV8.sgRNA1.donor (5×1012 GC) or untargeted vectors at the same doses, and they were sacrificed at 2 weeks after injection for analyses.
OTC and Cas9 immunostaining
Immunofluorescence for OTC expression was performed on frozen liver sections. Cryosections (8 μm) were air dried and fixed in 4% paraformaldehyde (all solutions in phosphate-buffered saline) for 10 min. Sections were then permeabilized and blocked in 0.2% Triton containing 1% donkey serum for 30 min. A rabbit anti-OTC antibody34 (diluted 1:1000 in 1% donkey serum) was used to incubate the sections for 1 hr. After washing, the sections were stained with tetramethylrhodamine (TRITC)–conjugated donkey anti-rabbit antibodies (Jackson Immunoresearch Laboratories, Cat# 711–025–152) in 1% donkey serum for 30 min, washed, and mounted with Vectashield (Vector Laboratories).
Some sections were additionally stained with a monoclonal antibody against glutamine synthetase (BD Biosciences, clone 6, Cat# 610517) as a marker for pericentral hepatocytes followed by fluorescein isothiocyanate (FITC)-labeled donkey anti-mouse antibodies (Jackson Immunoresearch Laboratories, Cat# 715–095–150). Double staining was performed by mixing the two primary and secondary antibodies, respectively, and following the above protocol. Other sections were counterstained with fluorescein-labeled tomato lectin (Lycopersicon esculentum lectin, LEL; Vector Laboratories, Cat# FL-1171) by adding LEL to the secondary antibody solution at a dilution of 1:500.
Cas9 expression was detected on sections from paraffin-embedded livers via immunostaining for FLAG tag using monoclonal antibody M2 (Sigma, Cat# F1804). Paraffin sections were processed according to standard protocols with an antigen retrieval step (boiling for 6 min in 10 mM citrate buffer, pH 6.0). Staining was performed using a mouse-on-mouse (MOM) kit (Vector Laboratories) according to the manufacturer’s instructions.
To quantify percentages of OTC-expressing hepatocytes, 10 random images were taken with a 10x objective from each liver section stained for OTC expression. In some cases, where only a small liver section was available, only 5 pictures were taken. Using ImageJ software (Rasband W. S., National Institutes of Health, USA; http://rsb.info.nih.gov/ij/), images were thresholded for OTC-positive area (i.e. the OTC-positive area was selected) and the percentage of the OTC-positive area was determined for each image. In a second measurement the images were thresholded for “empty” area (e.g., veins and sinusoids) to determine the percentage of the area not occupied by liver tissue. This was possible as a result of the presence of weak background fluorescence of the liver tissue. The final percentage of OTC-positive liver tissue (i.e., OTC-positive hepatocytes) was then calculated per adjusted area (total area minus empty area), and the values were averaged for each liver.
To determine the percentage of Cas9-positive hepatocytes, two sections from each liver were analyzed, one stained for Cas9 (via FLAG tag), the other section stained with hematoxylin to label all nuclei. Three images from every section were taken with a 10x objective, and the number of either Cas9-positive or hematoxylin-stained hepatocyte nuclei was determined using ImageJ’s “Analyze Particles” tool that allows one to select and count stained hepatocyte nuclei. Hematoxylin-stained nuclei from other cell types could be excluded based on size and circularity parameters. The percentage of Cas9-positive nuclei was then calculated based on the total number of hepatocyte nuclei visible in the hematoxylin-stained sections.
Histopathology
Hematoxylin and eosin (H&E) staining was performed on sections from paraffin-embedded liver samples processed and stained according to standard protocols. Sections were analyzed for any abnormalities compared to livers from untreated animals.
OTC enzyme activity assay
OTC enzyme activity was assayed in liver lysates as described previously with modifications35. Whole-liver fragments were frozen in liquid nitrogen and stored at −80°C until OTC measurements were performed. A homogenate of 50 mg liver tissue per mL was prepared in 50 mM Tris acetate buffer, pH 7.5, with a Polytron homogenizer (Kinematica AG). A total of 250 μg of liver tissue was used per assay tube, and assays were performed in duplicate. The protein concentration was determined on the remaining liver homogenate using the Bio-Rad Protein assay kit (Bio-Rad) according to the manufacturer’s instructions.
Western blot analysis
Western blot analyses were performed on liver lysates as described previously36. OTC protein was detected by a custom rabbit polyclonal antibody (1:10,000 dilution)34. Mouse anti-FLAG M2 antibody (1:2000 dilution, Sigma, Cat# F1804) and mouse anti-actin antibody (1:1,000 dilution, Cell Signaling Technology, Cat# 8457L) were used to detect Cas9 and actin. Blots were imaged with ChemiDoc MP system and analyzed using ImageLab 4.1 software (Bio-Rad).
Gene expression analysis and RT-qPCR
RNA was isolated using Trizol (Life Technology) and reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR to measure murine OTC, SaCas9, and GAPDH were performed using gene-specific primers (Life Technologies). Data were normalized to GAPDH levels.
On-target and off-target mutagenesis analyses
HDR-mediated targeted modifications were confirmed by restriction-fragment length polymorphism (RFLP) analysis, as described previously31. The HDR-Fwd and HDR-Rev primers were designed to anneal outside of the region of homology between the donor template and targeted genomic region. The PCR products were purified and digested with AgeI restriction enzyme. To further analyze the OTC intron 4 on-target site, the genomic region was amplified by nested PCR. Briefly, the genomic DNA was first amplified by the HDR-Fwd and HDR-Rev primers (Supplementary Table 5) using Q5® High-Fidelity DNA Polymerase (New England Biolabs) and gel purified to remove the residual AAV8.sgRNA1.donor in the genomic DNA. Then nested PCR was performed by using the purified 1st round PCR amplicon. Libraries were made from 250 ng of the 2nd PCR products using NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB) and sequenced on Illumina MiSeq (2×250 base pair (bp) paired end or 2×300 bp paired end, Genewiz). Data were processed according to standard Illumina sequencing analysis procedures. Processed reads were mapped to the expected PCR amplicons as reference sequences using custom scripts. Reads that did not map to reference were discarded. Insertions and/or deletions were determined by comparison of reads against reference using custom scripts. The indel sequences as summarized for the OTC intron 4 on-target site are presented in Supplementary Table 2.
The most likely off-target sites were determined using the algorithm described in www.benchling.com, referred to as OT1 through OT16 (Supplementary Table 4). Primers spanning these sites (Supplementary Table 6) were used to amplify relevant sequences by nested PCR. Purified PCR fragments were then subjected to deep sequencing as described above.
Frequencies of on-target and off-target indels and on-target correction of the spfash mutation were determined as follows. MiSeq reads were analyzed using custom scripts to identify indels by matching reads against reference, with indels involving any portion of the sequence within 15 nt upstream or downstream of the predicted CRISPR-Cas9 cleavage site (3 nt downstream of the PAM, within the protospacer) considered to be possible off-target effects. Reads for which there was any 18-nt sequence with more than 2 mismatches with the corresponding 18-nt portion of the reference sequence, either upstream or downstream of a candidate indel, were discarded as errors. All candidate indels for the OT1 through OT16 sites were manually curated for confirmation.
For the OTC intron 4 on-target site, a read was counted as having “Perfect HDR” if on the antisense strand there was a perfect match with a 51-nt sequence from the donor, starting with the donor-specific ‘CACCAA’ at the location of the PAM, through the donor-specific AgeI insert ‘ACCGGT’, and ending with the SNV ‘C’ at the spfash OTC mutation site. A read was counted as being a “Read with a ‘G’” if it either (1) met the criterion for “Perfect HDR” or (2) had the SNV ‘G’ on the sense strand in the expected spfash OTC mutation site 54 nt upstream of the predicted CRISPR-Cas9 cleavage site (accounting for the size of the donor-specific AgeI insert ‘ACCGGT’), with up to two mismatches with the 18-nt intronic portion of the reference sequence adjacent to the spfash OTC mutation site. A read was counted as having “Partial HDR” if it did not meet the criteria for “Perfect HDR” and “Read with a ‘G’” and if there was a perfect match with an 18-nt sequence from the donor, starting with the donor-specific ‘CACCAA’ at the 3′ end of the target site and ending with the donor-specific AgeI insert ‘ACCGGT’.
Statistical analyses
Test and control vectors were evaluated in at least 3 mice per group at each time point to ensure reproducibility. Sample sizes are noted in figure legends. All animals with successful temporal vein injection were included in the study analysis. Those animals with unsuccessful injection were excluded. Injection success was determined according to vector genome copies in liver via qPCR, where animals with vector genome copies <10% of the mean value of the dosing group at the same time point were considered to be unsuccessful.
Statistical analyses were performed with GraphPad Prism 6.03 for Windows. The Dunnett’s multiple comparisons test was used to compare a number of variables with a single control. Due to the relatively small sample size, normality testing was not feasible. The Mantel-Cox test was used to test the survival distributions for differences. Group averages are presented as mean ± S.E.M.
Supplementary Material
Acknowledgments
We thank Penn Vector Core for supplying vectors, Penn Bioinformatics Core for assistance on deep sequencing data analysis, Y. Zhu and M. Nayalat for help on immunohistochemistry analysis, and L. Mays for assistance on manuscript preparation. This work was supported by NICHD P01-HD057247 (J.M.W.) and the Kettering Family Foundation (M.B.).
Footnotes
Author Contributions
L.W. and J.M.W. conceived this study. L.W., Y.Y., and J.M.W. designed the experiments. Y.Y., P.B., D.M., Z.H., J.W., H.Y., and C.X. performed the experiments. K.M. conducted the bioinformatics analysis of the deep sequencing data. J.M.W., L.W., Y.Y., P.B., H.M., K.M., and M.L.B. wrote and edited the manuscript.
Competing Financial Interests
J.M. Wilson is an advisor to REGENXBIO, Dimension Therapeutics, Solid Gene Therapy, and Alexion, and is a founder of, holds equity in, and has a sponsored research agreement with REGENXBIO and Dimension Therapeutics; in addition, he is a consultant to several biopharmaceutical companies and is an inventor on patents licensed to various biopharmaceutical companies.
References
- 1.Cunningham SC, Dane AP, Spinoulas A, Logan GJ, Alexander IE. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol Ther. 2008;16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Wang H, Bell P, McMenamin D, Wilson JM. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther. 2012;23:533–539. doi: 10.1089/hum.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batshaw ML, Tuchman M, Summar M, Seminara J. A longitudinal study of urea cycle disorders. Mol Genet Metab. 2014;113:127–130. doi: 10.1016/j.ymgme.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichter-Konecki U, Caldovic L, Morizono H, Simpson K. Ornithine Transcarbamylase Deficiency (1993–2013) Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews®. ( http://www.ncbi.nlm.nih.gov/books/NBK154378/)
- 5.Ah Mew N, Krivitzky L, McCarter R, Batshaw M, Tuchman M. Clinical outcomes of neonatal onset proximal versus distal urea cycle disorders do not differ. J Pediatr. 2013;162:324–329. e321. doi: 10.1016/j.jpeds.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodges PE, Rosenberg LE. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc Natl Acad Sci U S A. 1989;86:4142–4146. doi: 10.1073/pnas.86.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma R, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. doi: 10.1182/blood-2014-12-615492. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anguela XM, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122:3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barzel A, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedland AE, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther. 2015;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng R, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Dingemanse MA, et al. Development of the ornithine cycle in rat liver: zonation of a metabolic pathway. Hepatology. 1996;24:407–411. doi: 10.1002/hep.510240219. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt F, Grimm D. CRISPR genome engineering and viral gene delivery: a case of mutual attraction. Biotechnol J. 2015;10:258–272. doi: 10.1002/biot.201400529. [DOI] [PubMed] [Google Scholar]
- 20.Vasileva A, Jessberger R. Precise hit: adeno-associated virus in gene targeting. Nat Rev Microbiol. 2005;3:837–847. doi: 10.1038/nrmicro1266. [DOI] [PubMed] [Google Scholar]
- 21.Mahiny AJ, et al. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 22.Gao GP, et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, et al. AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta) Mol Ther. 2011;19:2012–2020. doi: 10.1038/mt.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;26:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L, Gaensler KM. Role of antigen-specific regulatory CD4+CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX. Gene Ther. 2013;20:987–996. doi: 10.1038/gt.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nivsarkar MS, et al. Evidence for contribution of CD4+ CD25+ regulatory T cells in maintaining immune tolerance to human factor IX following perinatal adenovirus vector delivery. J Immunol Res. 2015;2015 doi: 10.1155/2015/397879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LoDuca PA, Hoffman BE, Herzog RW. Hepatic gene transfer as a means of tolerance induction to transgene products. Curr Gene Ther. 2009;9:104–114. doi: 10.2174/156652309787909490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinderer C, et al. Neonatal Systemic AAV Induces Tolerance to CNS Gene Therapy in MPS I Dogs and Nonhuman Primates. Mol Ther. 2015;23:1298–1307. doi: 10.1038/mt.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lock M, et al. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moscioni D, et al. Long-term correction of ammonia metabolism and prolonged survival in ornithine transcarbamylase-deficient mice following liver-directed treatment with adeno-associated viral vectors. Mol Ther. 2006;14:25–33. doi: 10.1016/j.ymthe.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Daly TM. AAV-mediated gene transfer to the liver. Methods Mol Biol. 2004;246:195–199. doi: 10.1385/1-59259-650-9:195. [DOI] [PubMed] [Google Scholar]
- 34.Augustin L, Mavinakere M, Morizono H, Tuchman M. Expression of wild-type and mutant human ornithine transcarbamylase genes in Chinese hamster ovary cells and lack of dominant negative effect of R141Q and R40H mutants. Pediatr Res. 2000;48:842–846. doi: 10.1203/00006450-200012000-00023. [DOI] [PubMed] [Google Scholar]
- 35.Morizono H, et al. Expression, purification and kinetic characterization of wild-type human ornithine transcarbamylase and a recurrent mutant that produces ‘late onset’ hyperammonaemia. Biochem J. 1997;322(Pt 2):625–631. doi: 10.1042/bj3220625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, et al. Sustained correction of OTC deficiency in spf(ash) mice using optimized self-complementary AAV2/8 vectors. Gene Ther. 2012;19:404–410. doi: 10.1038/gt.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.