Abstract
Heat shock proteins (Hsps) are a class of highly conserved proteins produced in virtually all living organisms from bacteria to humans. Hsp60 and Hsp10, the most important mitochondrial chaperones, participate in environmental stress responses. In this study, the full-length complementary DNAs (cDNAs) of Hsp60 (PmHsp60) and Hsp10 (PmHsp10) were cloned from Penaeus monodon. Sequence analysis showed that PmHsp60 and PmHsp10 encoded polypeptides of 578 and 102 amino acids, respectively. The expression profiles of PmHsp60 and PmHsp10 were detected in the gills and hepatopancreas of the shrimps under pH challenge, osmotic stress, and heavy metal exposure, and results suggested that PmHsp60 and PmHsp10 were involved in the responses to these stimuli. ATPase and chaperone activity assay indicated that PmHsp60 could slow down protein denaturation and that Hsp60/Hsp10 may be combined to produce a chaperone complex with effective chaperone and ATPase activities. Overall, this study provides useful information to help further understand the functional mechanisms of the environmental stress responses of Hsp60 and Hsp10 in shrimp.
Keywords: Hsp60, Hsp10, pH challenge, Osmotic stress, Heavy metal exposure, Penaeus monodon
Introduction
Heat shock proteins (Hsps), also known as stress proteins and extrinsic chaperones, are a suite of highly conserved proteins produced in all living organisms, especially when exposed to environmental stresses, such as heat shock, UV irradiation, and heavy metals or other toxic chemicals (Roberts et al. 2010). Hsps are involved in a variety of biological functions, such as protein folding, damaged protein repair, nascent protein transport, and immune presentation (Welch 1991; Gething and Sambrook 1992; Freeman et al. 2000; Quintana and Cohen 2011; Pons et al. 2013). Based on their molecular mass, Hsps are generally classified into six major families, i.e., Hsp110, Hsp90, Hsp70, Hsp60, Hsp40, and small Hsps (Tang et al. 2011; Music et al. 2014).
Hsp60 is a homolog of phage growth E large (GroEL) in Escherichia coli (Xu et al. 2014). Over the last several decades, multiple Hsp60 functions have been discovered. For example, Hsp60 is capable of activating the innate immune response to atherosclerosis and other inflammatory disorders (Kol et al. 2000; Grundtman et al. 2011). Ghosh et al. (2008) demonstrated that Hsp60 is involved in apoptosis and cell cycle regulation. Hsp60 is able to substantially accelerate procaspase-3 maturation by different upstream activator caspases (Xanthoudakis et al. 1999). The possible roles of Hsp60 in certain cellular processes, such as sperm cell differentiation, reproduction development, and environment stress response, have also been elucidated by other researches (Meinhardt et al. 1995; Paranko et al. 1996; Kozlova et al. 1997; Neuer et al. 2000; Qian et al. 2012). Much of the research on organism protection under environmental stress in the last decades has examined the involvement of Hsp60 in multiple pathophysiological processes. For instance, Hsp60 was observed to be activated to prevent protein denaturation under heat stress (Martin et al. 1992). Xu et al. (2014) also found that Hsp60 is involved in the heat shock response in the sea cucumber Apostichopus japonicas. In addition, Hsp60 plays important roles in salinity stress mediation in Portunus trituberculatus (Xu and Qin 2012). Hsp60 participates in immune and stress responses under different environmental stresses in Litopenaeus vannamei (Huang et al. 2011). Hsp10 (GroES in E. coli), as the co-chaperonin of Hsp60, exerts its biological functions through combining with Hsp60 in many situations (Cappello et al. 2006; Jha et al. 2011; Cappello et al. 2014; Boettinger et al. 2015). Hsp60 and Hsp10 form a folding cage through their rings and produce large and efficient protein-editing machinery that facilitates proper folding and assembly of mitochondrial-imported proteins and corrects misfolded polypeptides (Hansen et al. 2003; Magen et al. 2008). Although the Hsp60/Hsp10 complex system has been well studied in many species; however, the regulatory mechanism of this system, especially in crustaceans, remains unclear (Costa et al. 2011; Johnson et al. 2014; Nisemblat et al. 2014). The chaperon folding cage is composed of two parts: a dome-shaped heptameric ring of Hsp10 (GroES) subunits and Hsp60 (GroEL) cylinder with ATPase (Saibil and Ranson 2002). The cylinder contains three domains: an equatorial ATP-binding domain, an intermediate hinge domain, and an apical domain that binds nascent polypeptides and Hsp10 (GroES) (Saibil et al. 2013). Hsp10 and nascent polypeptides undergo cycles of binding and release, regulated allosterically by the Hsp60 ATPase (Hayer Hartl et al. 2015). The ATP hydrolysis drives conformational changes of cage, creating a protected environment favoring protein folding (Clare and Saibil 2013). The chaperone activity is generally determined by ability to prevent thermal aggregation of citrate synthase (CS), luciferase (Luc), alcohol dehydrogenase (ADH) or malate dehydrogenase (MDH) (Tsai et al. 2012). In the present study, mitochondrial citrate synthase was applied to identify the mitochondrial Hsp60 chaperon activity with a view to better understand the defense mechanism of Hsps when black tiger shrimps (Penaeus. monodon) are under stress.
The black tiger shrimp (P. monodon) is one of the most important aquatic commercial animals in Asia, especially in South China. Considering the enormous economic losses induced by environmental stressors in the shrimp industry, studies focusing on heat shock proteins have become increasingly popular because of the important roles these proteins play in resistance to stress (Cesar and Yang 2007; Cui et al. 2010; Li et al. 2012; Qian et al. 2012). Unfortunately, only three Hsps, namely, Hsp90, Hsp70, and Hsp21, have been reported in P. monodon (Lo et al. 2004; Huang et al. 2008; Jiang et al. 2009). In order to understand the mechanisms of stress tolerance and disease resistance of P. monodon better, we characterized the complete compelemantary DNA (cDNA) sequences of Hsp60 and its co-chaperonin, Hsp10, in this species and investigated the messenger RNA (mRNA) expression of the two genes when exposed to pH challenge, osmotic stress, and heavy metal.
Materials and methods
Experimental animals
Healthy black tiger shrimps (P. monodon) weighing an average of 21 ± 1 g were obtained from the experimental base of South China Sea Fisheries Research Institute in Shenzhen (Guangdong, China). The shrimps were reared in tanks containing aerated 3.3 % seawater for 3 days at 25 ± 1 °C before commencement of the experiments. About two thirds of the water in each tank was renewed daily. The shrimps were fed with commercial diets during acclimation until 24 h before treatment.
Tissue distribution analysis
Ten tissue types, including the gills, heart, ovary, stomach, lymph, intestines, hepatopancreas, thoracic ganglion (thorax), muscle, and brain, were collected from three shrimps to determine the tissue distribution of PmHsp60 and PmHsp10 transcripts. All of the samples were kept in RNAlater (Ambion, CA, USA) and stored at −80 °C until RNA extraction.
pH challenge
Two solutions of pH 7.0 ± 0.1 and pH 9.0 ± 0.1 were prepared using 1.0 mol · L−1 HCl and 1.0 mol · L−1 NaOH. Fifty shrimps were divided amongst the solutions after acclimation to normal seawater (pH 8.0 ± 0.1). pH was measured using a pH electrode (Shanghai Shuangxu Electronics Co., Ltd., Shanghai, China). The gills and hepatopancreas of six shrimps in normal seawater were collected as control samples. Three individuals in the experiment groups were collected 6, 12, 24, 48, and 96 h after the pH challenge. All of the samples were kept in RNAlater and stored at −80 °C until RNA extraction.
Osmotic stress challenge
Fifty shrimps were divided into two groups with different salinity conditions. Low salinity (2.3 %) was achieved through addition of freshwater from the normal level (3.3 %). Salinity was measured using a salinity meter (WZ211, Shanghai JL Optics Instrument Co., Ltd., Shanghai, China). The second group of shrimps was maintained in 4.3 % seawater, which was obtained through addition of sea salt to normal seawater. The gills and hepatopancreas of three individuals were collected 4, 8, 16, and 32 h post-osmotic stress exposure. Three shrimps reared in normal seawater were collected and served as controls. All of the samples were kept in RNAlater and stored at −80 °C until needed.
Heavy metal exposure
Three heavy metals, namely, copper (Cu), zinc (Zn), and cadmium (Cd ), were selected as stressors. The concentrations of Cu, Zn, and Cd were set to 0.18, 0.506, and 7.73 mg · L−1, respectively, in accordance with previous studies (Chen and Lin 2001; Wang et al. 2001; Qian et al. 2012). Stock solutions of Cu, Zn, and Cd at concentrations equivalent to 100 times the respective experimental concentrations were prepared by dissolving the water-soluble salts CuSO4 · 5H2O, ZnSO4 · 7H2O, and CdCl2 · 2.5H2O in deionized Milli-Q water. The gills and hepatopancreas of six shrimps in normal seawater were collected as control samples. Three individuals in the experiment groups were collected 6, 12, 24, 48, and 96 h after heavy metal exposure. All of the samples were kept in RNAlater and stored at −80 °C until RNA extraction.
RNA isolation and reverse transcription
Total RNA was isolated from dissected tissues (about 50 mg) using TRIzol reagent (Invitrogen, Shanghai, China), following the manufacturer’s protocols. The extracted RNA was re-suspended in DEPC-treated water and stored at −80 °C until needed. RNA quantity, purity, and integrity were verified by both native RNA electrophoresis on 1.2 % agarose gel and the ratio of UV absorbance at 260 and 280 nm (NanoDrop Technologies, DE, USA). cDNA was synthesized from total RNA by using a PrimeScript™ Reverse Transcriptase kit (TaKaRa, Dalian, China), following the manufacturer’s protocol.
Hsp60 and Hsp10 cloning
Partial sequences of PmHsp60 and PmHsp10 were obtained from the splicing transcriptome of P. monodon established in our laboratory (unpublished). Two pairs of primers (Table 1) were designed based on the partial cDNA sequences to identify the PmHsp60 and PmHsp10 partial sequences. The PCR reaction was performed using a Gradient Mastercycler (Eppendorf, Hamburg, Germany) system with a total volume of 25 μL of PCR mixture containing 2.5 μL 10 × reaction buffer with 15 mmol · L−1 MgCl2, 2 μL of 10 mmol · L−1 dNTP mix, 1.5 μL of 25 μmol · L−1 of each primer, 1 μL template cDNA, 16.5 μL MilliQ water, and 0.5 μL BioReady ExTaq DNA Polymerase (5 U · μL−1) (TaKaRa, Dalian, China). The PCR cycles were conducted as follows: an initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min, and a final extension at 72 °C for 10 min. The PCR products were analyzed through electrophoresis in 1.2 % agarose gel and subsequently purified using an agarose gel DNA purification kit (Sangon, Shanghai, China). Afterward, the products were ligated into the pMD18-T vector (TaKaRa, Dalian, China) and sequenced (Invitrogen, Guangzhou, China). The full-length cDNA of PmHsp60 and PmHsp10 were amplified through the rapid amplification of cDNA ends (RACE) approach. RACE-PCR was performed using a SMARTTM RACE cDNA amplification kit (Clontech, Takara, Dalian, China) with specific primers (Table 1).
Table 1.
PCR primers used in the experiment
| Primer name | Sequence (5′ → 3′) | Sequence information |
|---|---|---|
| Hsp60-F | GGCGAGTCGTCTGCTGCTCATT | Cloning of PmHsp60 partial cDNA sequence |
| Hsp60-R | CCTCGCTGAGACTGTCCAGGGTTA | Cloning of PmHsp60 partial cDNA sequence |
| Hsp10-F | TCTCACGGTTCTGCTGCGTTAATC | Cloning of PmHsp10 partial cDNA sequence |
| Hsp10-R | TTAGATGGACAAAACATTCATGCCC | Cloning of PmHsp10 partial cDNA sequence |
| Hsp60-5'GSP1 | TTGGGACTGCCCCAGCTCTGCTC | 5′RACE |
| Hsp60-5'GSP2 | CGAGCGACGGGAGTTCGCAATAAA | 5′RACE |
| Hsp60-3'GSP1 | AGCGTATGGCTCGTCTGGCCTCAG | 3′RACE |
| Hsp60-3'GSP2 | GAGTGGCTTCCCTCCTCACCACAGC | 3′RACE |
| Hsp10-5'GSP1 | CTTCTCCTCCAGGGTAACCTTTGTGCC | 5′RACE |
| Hsp10-5'GSP2 | CACGGTCGAACAGGGGAACAAACTTCT | 5′RACE |
| Hsp10-3'GSP1 | TGGTGGCACAAAGGTTACCCTGGAG | 3′RACE |
| Hsp10-3'GSP2 | AGTCTGCGGCCTGTTTCAGTTCACAA | 3′RACE |
| Hsp60-exF | GGAATTCCATATGCATCGCGCAGCCTCTTT | Recombinant expression |
| Hsp60-exR | CGCGGATCCCATCATGCCGCCCATGCCTC | Recombinant expression |
| Hsp10-exF | GGAATTCCATATGGCTGGTGCTTTGAAGAAGT | Recombinant expression |
| Hsp10-exR | CCCAAGCTTCTCGTTCTTCATCTTGGCCAG | Recombinant expression |
| qHsp60F | GCCAACAACACCAATGAA | Quantitative real-time PCR |
| qHsp60R | CATAACTCCACGCCTGAT | Quantitative real-time PCR |
| qHsp10F | CTGTTGGTGATGAAGTGA | Quantitative real-time PCR |
| qHsp10R | ATCTGCCTACGAATTGAC | Quantitative real-time PCR |
| qEF-1αF | AAGCCAGGTATGGTTGTCAACTTT | Quantitative real-time PCR |
| qEF-1αR | GCTTCGTGGTGCATCTCCACAGAC | Quantitative real-time PCR |
Sequence analysis, multiple sequence alignment, and phylogenetic analysis
The full cDNA sequences of PmHsp60 and PmHsp10 were analyzed for homology with the BLAST programs (Altschul et al. 1997) at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The deduced amino acid sequences were obtained using BioEdit software (https://www.bioedit.com/). Motifs were predicted using SMART (http://smart.embl-heidelberg.de/). Molecular mass and theoretical isoelectric point were predicted using the compute pI/Mw tool (http://web.expasy.org/compute_pi/), and subcellular localization predictions were performed on the PSORT II server (http://psort.hgc.jp/form2.html). Multiple sequence alignments were created using ClustalW software (Thompson et al. 1997) (http://www.clustal.org/). Bootstrapped neighbor-joining trees were constructed to illustrate PmHsp60 and PmHsp10 phylogenetic relationships using Mega software (version 5.03) (http://www.megasoftware.net/). The functional domain was predicted by InterPro (http://www.ebi.ac.uk/interpro/).
Spatial and temporal gene expression analysis
The PmHsp60 and PmHsp10 gene-specific primers F and R (Table 1), which produce 132 and 164 bp products, respectively, were used during QRT-PCR to detect the temporal expression of the genes in black tiger shrimp. The products were sequenced and confirmed to be the correct form of PmHsp60 and PmHsp10 genes. The housekeeping gene elongation factor-1 alpha (EF-1α) (GenBank: DQ021452.1) was selected as the internal control. Quantitative real-time PCR amplifications were performed in triplicate on a 384-well rotor with a total volume of 10 μL containing 5 μL of 2 × SYBR Premix Ex Taq (TaKaRa, Dalian, China), 2 μL of the cDNA template, 0.8 μL of 10 mmol · L−1 of each forward and reverse primer, and 1.4 μL of MilliQ water. The cycling protocol for real-time PCR was 94 °C for 30 s and 40 cycles of 94 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s. Melt curve analysis of the PCR products was performed from 65 to 95 °C at the end of each run to ensure that each single product was amplified. The relative expression levels of the target genes were calculated via the 2−ΔΔCt method and normalized to EF-1α.
Recombinant expression and purification of PmHsp60 and PmHsp10
The open reading frames (ORFs) of PmHsp60 and PmHsp10 were amplified with Hsp60-exF/R and Hsp10-exF/R primers, respectively (Table 1). After amplification, the PCR products were ligated into pMD18-T vector (TaKaRa, Dalian, China) and the sequences were confirmed. pMD18T-PmHsp60 was cleaved with NdeΙ and HindIII and then cloned into pET28a + (Invitrogen, Shanghai, China) at NdeΙ and HindIII sites, whereas pMD18T-PmHsp10 was cleaved with NdeΙ and BamHΙ. The resultant recombinant plasmids were transformed into E. coli BL21 (DE3) strain. E. coli BL21 (DE3) cells were cultured in LB medium with 50 mg · mL−1 kanamycin, and the protein was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when OD600 of bacteria concentration reached to 0.6. The recombinant PmHsp60 and PmHsp10 proteins were purified and refolded through Ni-NTA affinity chromatography on the resin with the chelated nickel ions according to the instruction for the His-bind Purification Kit (Novagen, Shanghai, China) as described by Cui et al. (2011). The refolding step was performed using the ion-exchange chromatography refolding method (Suttnar et al. 1994). The resultant protein was separated by reducing 12 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue R250. The concentrations of the purified PmHsps were quantified using a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, Shanghai, China), and the purified PmHsps were stored at −80 °C for use in subsequent experiments.
Western blot analysis
PmHsps were separated by 12 % SDS-PAGE, and the products were transferred onto nitrocellulose membranes. Membranes were blocked with 5 % skim milk and 0.3 % Tween 20 in PBS (PBST) at 4 °C for 12 h and incubated with His-Tag horseradish peroxidase (HRP) conjugated rabbit polyclonal antibody (Huaan Biotechnology, Hangzhou, China) diluted to 1:2000 at 37 °C for 1 h. The membrane was washed thrice with PBST, and immune reactive bands were identified through development of blots with HRP-diaminobenzidine (DAB) (Tiangen, Beijing, China).
PmHsp60 ATPase activity assay
ATPase activity was assayed by measuring phosphate release, and measurement was performed according to the reagent kit instructions (Nanjing Jiancheng Institute of Biological Engineering, Nanjing, China). PmHsp protein concentrations were adjusted to 100 μg · mL−1, and the reactions were performed for 10 min at 0, 25, 30, 37, 42, and 60 °C. ATPase activity was calculated as follows: PmHsp60 ATPase activity = (A1–A2) ÷ (A3–A4) ÷ X × Y × 6 ÷ C μmol Pi/mg protein/h, where A1, A2, A3, and A4 are the absorbances of the sample, control, standard, and blank, respectively, at 636 nm; X is the inorganic phosphate content of the standard, Y is the dilution number of the sample, and C is the protein concentration of the sample.
Chaperone activity assay
Citrate synthase (CS, from porcine heart, Sigma, Shanghai, China) aggregation was applied to identify chaperone activity; this procedure has been described in detail in a previous study (Huang et al. 2008). Briefly, the aggregation level of CS was monitored in HEPES buffer (50 mM HEPES, pH 8.0, 25 mM NaCl, 0.5 mM DTT) at 45 °C. Buffer containing different concentrations of recombinant PmHsp60 (or PmHsp60 and PmHsp10) was preincubated for 10 min at 45 °C before addition of CS to a final concentration of 20 mg · mL−1. Reactions were continuously monitored at 320 nm by an ELISA reader (Molecular Devices, MDS Analytical Technologies, CA, USA). The absorbance of CS alone at 90 min of heating was defined as 100 % aggregation.
Results
Identification and characterization of the PmHsp60 and PmHsp10 genes
The nucleotide sequence of PmHsp60 (Fig. 1) was deposited in the GenBank database under accession number KT001211. The cDNA sequence contained a 5′-untranslated region (UTR) of 129 bp, a 3′-UTR of 486 bp with a poly (A) tail, and an ORF of 1737 bp encoding 578 amino acids. The molecular weight of PmHsp60 was 60.99 kDa, and its theoretical isoelectric point (pI) was 5.48. Multiple sequence alignments indicated that PmHsp60 has a highly conserved Chaperonin 60 (cpn60)-domain (shown in gray) and a Gly-Gly-Met repeat at the C-terminal (shown as waved lines). Ring oligomerization interfaces (circles), polypeptide binding sites (e.g., Hsp10), and ATP/Mg binding sites (underline) were found in the PmHsp60 sequence. No signal peptide was found in the deduced amino acid sequence of PmHsp60.
Fig. 1.
Nucleotide and deduced amino acid sequence of PmHsp60. The deduced amino acid sequence is shown below the nucleotide sequence. The initiation code (ATG) and the termination code (TAA) are boxed. The conserved cpn60-domain is shown in gray and the classical mitochondrial Hsp60 (mtHsp60) signature motifs were bold. The ATP-binding motif was underlined and the ring oligomerisation interfaces were shown in circles. The typical GGM repeat motif at the C terminus were shown waved lines
The nucleotide sequence of PmHsp10 (Fig. 2) was deposited in the GenBank database under accession number KT001212. The PmHsp10 full-length cDNA was 744 bp, including a 309-bp ORF, a 102-bp 5′-UTR, and a 333-bp 3′-UTR. The ORF encoded a protein composed of 102 amino acids with a predicted molecular mass of 11.05 kDa and a theoretical pI of 7.8. A putative Chaperonin 10 (cpn10) conserved domain (shown in gray) was also detected. A mobile loop (underline) functioning as an interaction surface with the large subunit of the chaperonin complex (e.g., cpn60) was found in the PmHsp10 sequence.
Fig. 2.
Nucleotide and deduced amino acid sequence of PmHsp10. The deduced amino acid sequence is shown below the nucleotide sequence. The initiation codon (ATG) and termination codon (TAA) are marked by boxes. The conserved Cpn10-domain is shown in gray and the predicted mobile loop is underlined
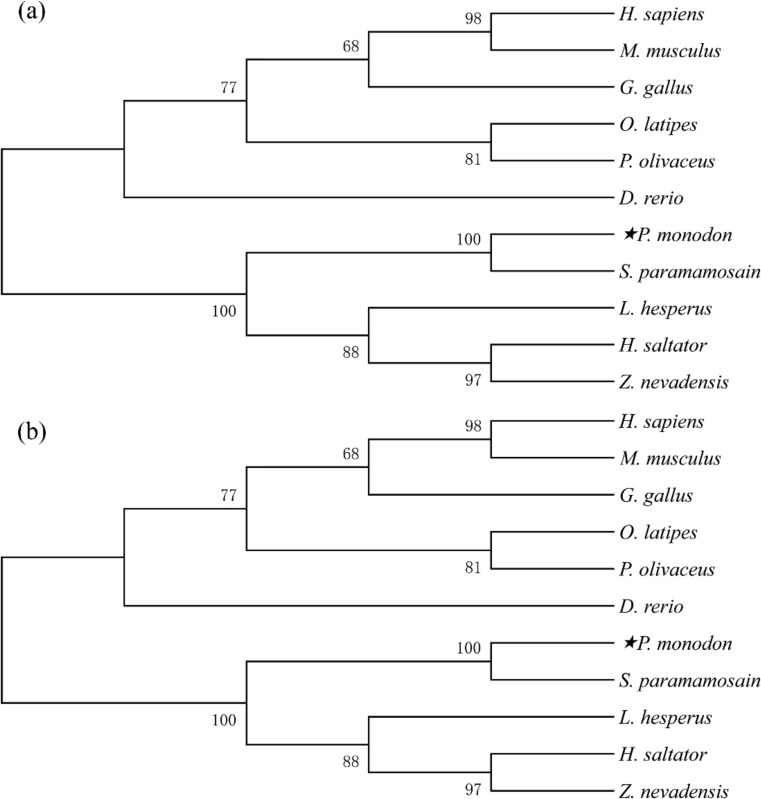
Phylogenetic analysis of PmHsp60 and PmHsp10
Eleven different Hsp60 sequences and nine Hsp10 sequences were downloaded from NCBI to determine their similarity with the deduced amino acid sequences of PmHsp60 and PmHsp10, respectively. The phylogenetic tree of Hsp60 showed that the proteins could be divided into two clusters (Fig. 3a). All invertebrates and arthropods were clustered together, and PmHsp60 was closely related to L. vannamei, which is in the branch of crustaceans. Similarly, the phylogenetic tree of Hsp10 could also be divided into two clusters of vertebrates and invertebrates (Fig. 3b). As expected, PmHsp10 was divided into the crustacean cluster and shared greater identity with the Scylla paramamosain than with other species under this cluster.
Fig. 3.
Phylogenetic analyses of PmHsp60 (a) and PmHsp10 (b). Alignment of amino acid sequences were produced by Clustal W, and the bootstrap neighbor-joining phylogeny tree was constructed by MEGA 5.03. GenBank accession numbers encoding Hsp60 are as follows: Litopenaeus vannamei (ACN30235), Scylla paramamosain (AGI74967.1), Drosophila melanogaster (NP_511115.2), Lissorhoptrus oryzophilus (AHE77384.1), Homo sapiens (ABB01006.1), Mus musculus (NP_034607.3), Misgurnus anguillicaudatus (AHB82279.1), Bos Taurus (NP_001160080.1), Paralichthys olivaceus (ABB76384.1), Danio rerio (NP_851847.1), Gallus gallus (NP_001012934.1); GenBank accession numbers encoding Hsp60 are as follows: Scylla paramamosain (AGI74966.1), Lygus Hesperus (AFX84557.1), Zootermopsis nevadensis (KDR14059.1), Harpegnathos saltator (XP_011146739.1), Danio rerio (NP_571601.1), Homo sapiens (NP_002148.1), Mus musculus (NP_032329.1), Gallus gallus (NP_990398.1), Oryzias latipes (NP_001098232.1), Paralichthys olivaceus (ABB76382.1)
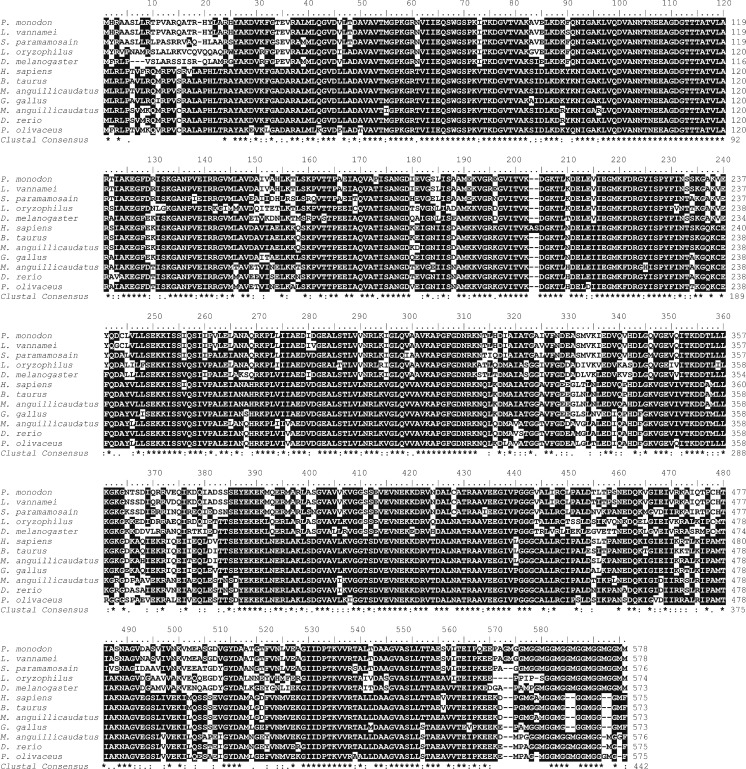
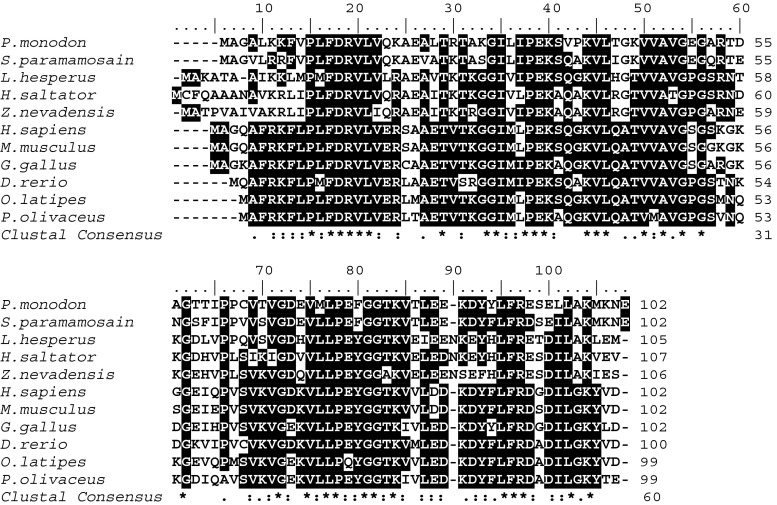
The deduced amino acid sequence of PmHsp60 showed the highest identity (98 %) with L. vannamei (ACN30235) as well as high similarity with Hsp60 from S. paramamosain (88 %, AGI74967.1), Drosophila melanogaster (78 %, NP_511115.2), and Lissorhoptrus oryzophilus (76 %, AHE77384.1). PmHsp60 also shared high homology with other vertebrates (Fig. 4). PmHsp10 revealed the highest identity with S. paramamosain (79 %, AGI74966.1) as well as high similarity with Lygus hesperus (61 %, AFX84557.1), Zootermopsis nevadensis (59 %, KDR14059.1), and Harpegnathos saltator (58 %, XP_011146739.1) (Fig. 5).
Fig. 4.
Multiple alignment of the deduced amino acid sequences of Hsp60 from Penaeus monodon and other species. Identical and similar amino acid residues in all the proteins are indicated by black and gray highlights respectively. The amino acid positions are shown on the right. The species names and GenBank accession numbers of Hsp60s are shown in Fig. 3
Fig. 5.
Multiple alignment of the deduced amino acid sequences of Hsp10 from Penaeus monodon and other species. Identical and similar amino acid residues in all the proteins are indicated by black and gray highlights respectively. The amino acid positions are shown on the right. The species names and GenBank accession numbers of Hsp60s are shown in Fig. 3
Recombinant expression and Western blot analysis of PmHsp60 and PmHsp10
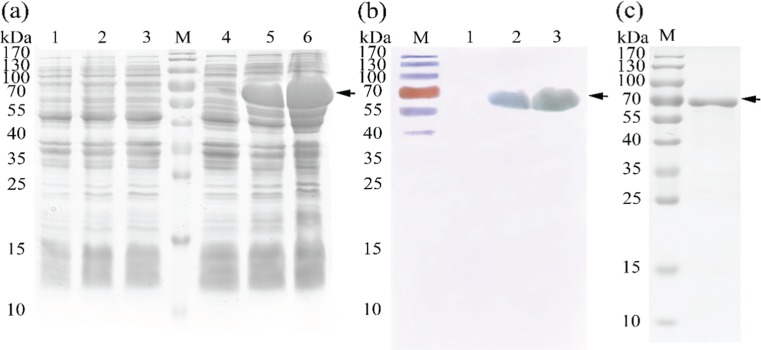
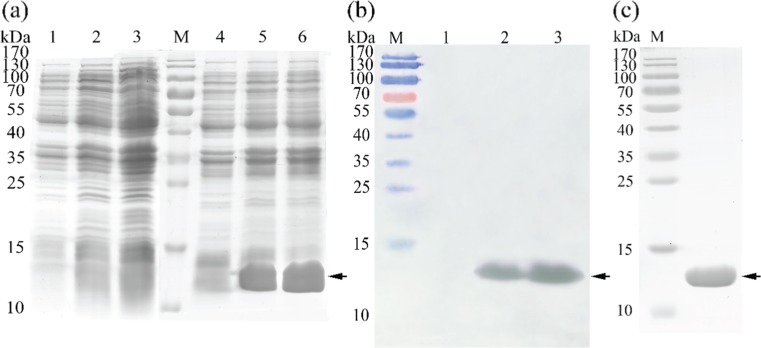
The recombinant plasmids of PET28a-PmHsp60 and PET28a-PmHsp10 were expressed in E .coli BL21 (DE3). A PmHsp60-6 × His tagged fusion protein about 64 kDa, consistent with the predicted molecular mass of PmHsp60 with the His tags added by the vector, was detected during SDS-PAGE (Fig. 6a). In the full-length PmHsp10 overexpression experiment, a protein with a molecular mass of 11 kDa was found during SDS-PAGE after isopropyl β-d-1-thiogalactopyranoside (IPTG) induction (Fig. 7a). No protein band was found at the same position in the non-induced controls of the two PmHsp genes. The fusion protein was detected through Western blot analysis. Figure 6b clearly shows a specific band of about 64 kDa (arrow) after Western blot analysis. The results of PmHsp10 Western blot analysis showed a specific protein band of about 11 kDa (arrow) (Fig. 7b). Purified and refolded PmHsps were obtained through affinity chromatography, and only one target band was detected by SDS-PAGE (Figs. 6c and 7c).
Fig. 6.
Expression of recombinant PmHsp60 in E. coli and western blot analysis. a. SDS-PAGE of expressed protein. Lanes M, pre-stained molecular weight marker; 1, non-induced cells transformed empty plasmid; 2, 3-h-induced cells transformed empty plasmid; 3, 6-h-induced cells transformed empty plasmid; 4, non-induced cells transfected recombinant; 5, 3-h-induced cells transformed recombinant; 6, 6-h-induced cells transformed recombinant. b. Western blotting of PmHsp60. Lanes 1, non-induced; 2, 3-h-induced PmHsp60-6 × His fusion protein; 3, 6-h-induced PmHsp60-6 × His fusion protein. c. purified PmHsp60-6 × His fusion protein
Fig. 7.
Expression of recombinant PmHsp10 in E. coli and western blot analysis. a. SDS-PAGE of expressed protein. Lanes M, pre-stained molecular weight marker; 1, non-induced cells transformed empty plasmid; 2, 3-h-induced cells transformed empty plasmid; 3, 6-h-induced cells transformed empty plasmid; 4, non-induced cells transfected recombinant; 5, 3-h-induced cells transformed recombinant; 6, 6-h-induced cells transformed recombinant. b. Western blotting of PmHsp10. Lanes 1, non-induced; 2, 3-h-induced PmHsp10-6 × His fusion protein; 3, 6-h-induced PmHsp10-6 × His fusion protein. c. purified PmHsp10-6 × His fusion protein
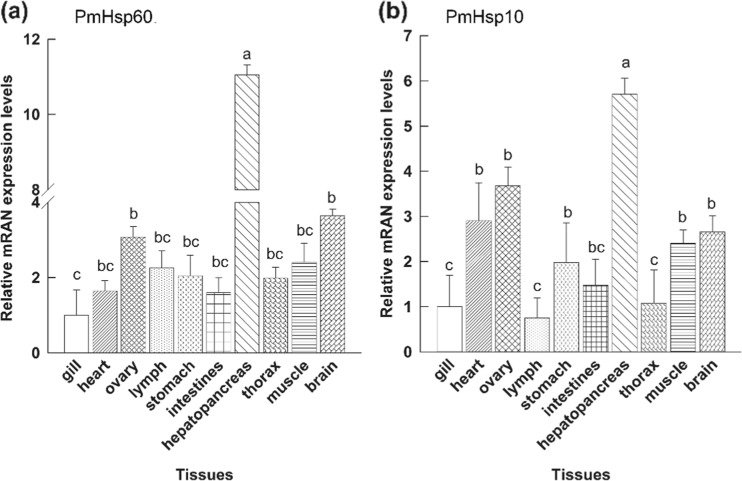
Tissue distribution of PmHsp60 and PmHsp10 analysis
Different tissues were collected to determine the distributions of PmHsp60 and PmHsp10 transcripts, and the results are shown in the Fig. 8a. The highest expression of the PmHsp60 transcript was detected in the hepatopancreas and the lowest expression was noted in the gills, heart, lymph, stomach, intestines, thorax, and muscle. Similarly, the highest expression level of the PmHsp10 transcript was found in the hepatopancreas, whereas the lowest expression was found in the lymph, gills and thorax (Fig. 8b).
Fig. 8.
Relative expressions of PmHsp60 (a) and PmHsp10 (b) transcripts in different tissues. Relative expressions of PmHsp60 and PmHsp10 in different tissues, gill, heart, ovary, lymph, stomach, intestines, hepatopancreas, thoracic ganglion (thorax), muscle and brain, by quantitative RT-PCR analysis. Relative expressions of PmHsp60 and PmHsp10 were calculated with the 2−ΔΔCt method. Each bar represent the mean ± SD (n = 3). Different lowercase letters indicate statistically significant differences (P < 0.05)
Transcription analysis of PmHsp60 and PmHsp10 post-environmental stress
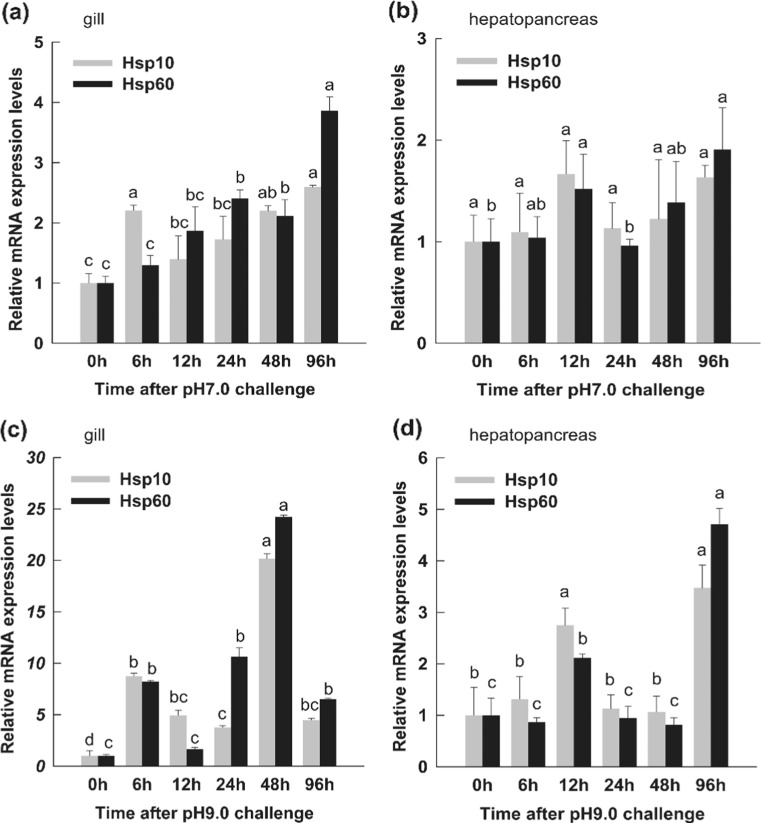
In the pH challenge experiments, the expression level of PmHsp60 mRNA was upregulated and reached peak values (3.8-fold higher compared with that of the control) in gills 96 h after pH-7.0 exposure. PmHsp10 was induced at 6 h, and maximum mRNA expression levels were found at 6, 48 and 96 h after pH 7.0 challenge (Fig. 9a). In the hepatopancreas, the expression level of PmHsp60 mRNA was upregulated at 12 and 48 h. No obvious variation in the expressions of PmHsp10 was observed after pH 7.0 challenge (Fig. 9b). Compared with the results of the pH 7.0 challenge, the PmHsp genes were induced immediately by pH 9.0 stress. The relative expressions of PmHsp60 and PmHsp10 transcripts were rapidly upregulated within 6 h after pH 9.0 exposure and declined at 12 h. Moreover, maximum expression levels were detected at 48 h in gills. At their peak, PmHsp60 and PmHsp10 expression levels were, respectively, 24- and 20-fold higher relative to the corresponding levels of the controls (Fig. 9c). In the hepatopancreas, PmHsp60 and PmHsp10 transcription levels were upregulated compared with that of the control within 12 h under pH 9.0 stress and subsequently declined to the same levels at 24 (PmHsp60) and 48 h (PmHsp10); peak values were observed at 96 h (Fig. 9d).
Fig. 9.
Relative expression levels of PmHsp60 and PmHsp10 in gills and hepatopancreas after pH challenge. a. In gill after pH7.0 challenge. b In hepatopancreas after pH7.0 challenge. c In gill after pH9.0 challenge. d In hepatopancreas after pH9.0 challenge. The untreated (0 h) group was considered as control. The EF-1α RNA was used as an internal control and the relative expression levels of PmHsp60 and PmHsp10 were obtained relative to EF-1α expression. Each bar represent the mean ± SD (n = 3). Different lowercase letters indicate statistically significant differences (P < 0.05)
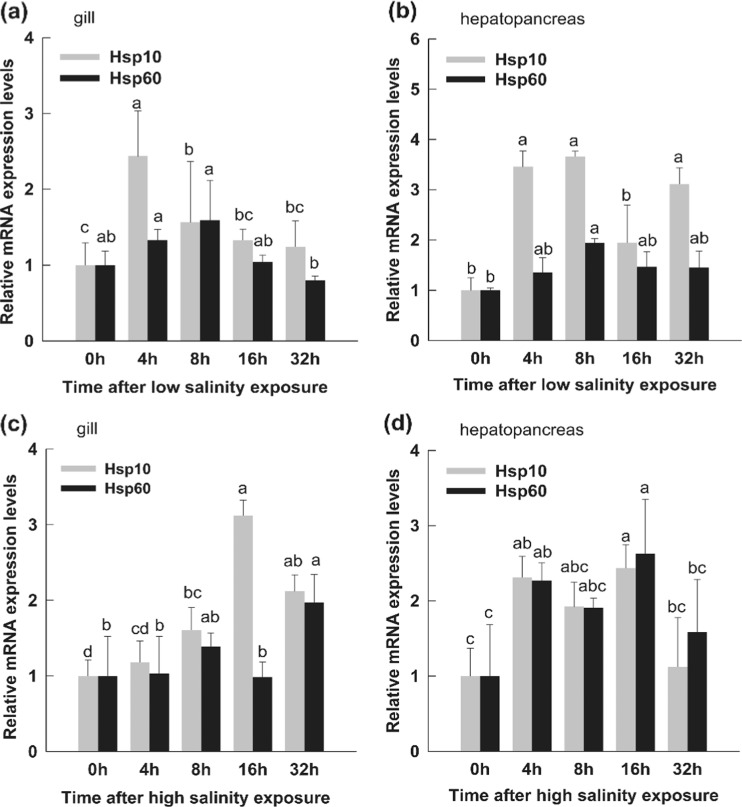
In the salinity stress experiment, PmHsp10 mRNA expression reached maximum levels after 4 h of low salinity exposure and declined after 8 h in the gills (Fig. 10a). In the hepatopancreas, PmHsp10 was induced at 4 h and reached maximum expression levels at 4, 8 and 32 h of low salinity stress (Fig. 10b). The PmHsp60 transcript was only slightly upregulated under low salinity stress in both tissues. The PmHsp60 transcript level increased to maximum levels (twofold higher compared with that of the control) at 32 h in the gills under high salinity; moreover, maximum PmHsp10 expression level was 3.2-fold higher than that of the control at 16 h (Fig. 10c). Both PmHsp60 and PmHsp10 transcripts were elevated at 4 h, and this level was maintained for 16 h in the hepatopancreas when the shrimps were stressed in pH 9.0 solution (Fig. 10d).
Fig. 10.
Relative expression levels of PmHsp60 and PmHsp10 in gills and hepatopancreas of shrimps under osmotic stress. a In gill after low salinity (2.3 %) exposure. b In hepatopancreas after low salinity (2.3 %) exposure. c In gill after high salinity (4.3 %) exposure. d In hepatopancreas after high salinity (4.3 %) exposure. The untreated (0 h) group was considered as control. The EF-1α RNA was used as an internal control and the relative expression levels of PmHsp60 and PmHsp10 were obtained relative to EF-1α expression. Each bar represent the mean ± SD (n = 3). Different lowercase letters indicate statistically significant differences (P < 0.05)
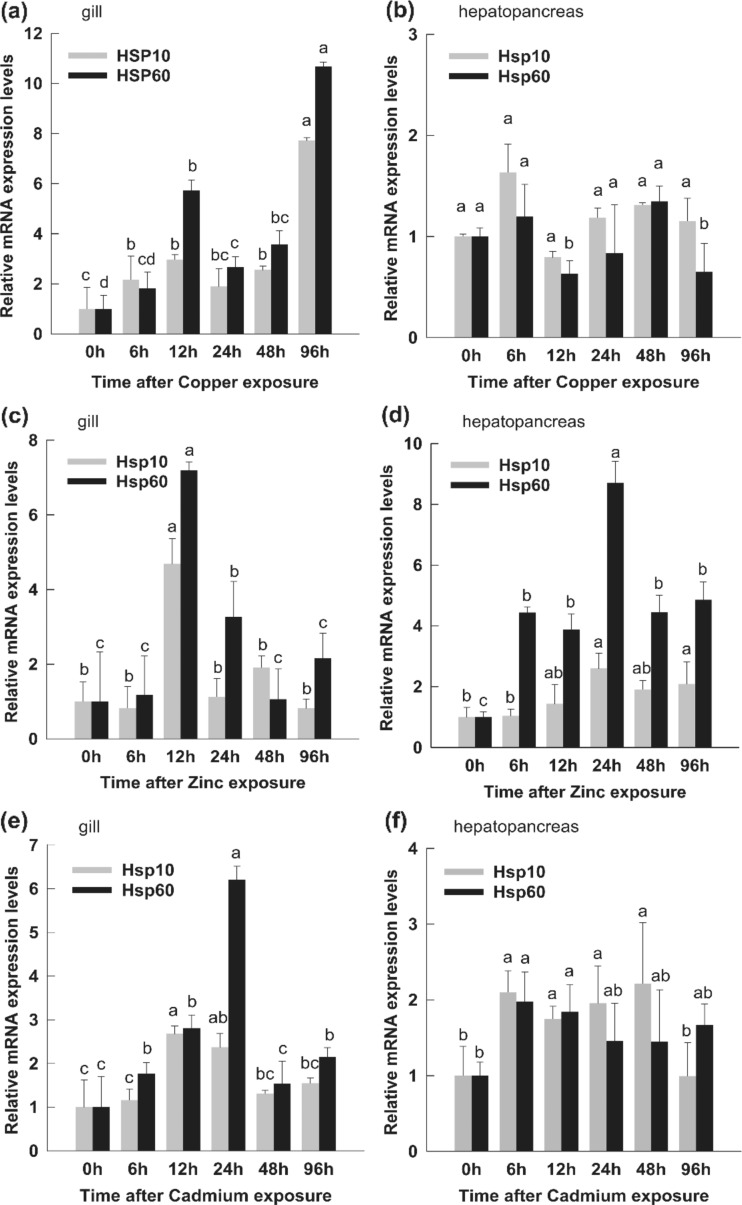
The relative mRNA expression levels of PmHsp genes in response to copper exposure are shown in Fig 11. In the gills, the PmHsp60 transcript level significantly increased at 12 h and decreased 24 and 48 h after the shrimps were exposed to copper stress. This parameter reached maximum levels at 96 h (10.7-fold higher compared with that of the control). PmHsp10 expression level was slightly induced before 48 h and reached maximum levels (7.8-fold higher compared with that of the control) at 96 h in the gills of shrimps under copper stress (Fig. 11a). In the hepatopancreas, PmHsps expression were maintained at levels similar to that of the control (Fig. 11b).
Fig. 11.
Relative expression levels of PmHsp60 and PmHsp10 in gills and hepatopancreas of shrimps under heavy metal exposure. a In gill after copper exposure. b In hepatopancreas after copper exposure. c In gill after zinc exposure. d In hepatopancreas after zinc exposure. e In gill after cadmium exposure. f In hepatopancreas after cadmium exposure. The untreated (0 h) group was considered as control; the untreated (0 h) group was considered as control. The EF-1α RNA was used as an internal control and the relative expression levels of PmHsp60 and PmHsp10 were obtained relative to EF-1α expression. Each Bar represent the mean ± SD (n = 3). Different lowercase letters indicate statistically significant differences (P < 0.05)
Similar tendencies of the PmHsp transcripts in the gills and hepatopancreas were observed during zinc stress exposure. In the gills, the maximal enhancement of PmHsp60 and PmHsp10 expression levels was reached at 12 h, which are about 7.2- and 4.8-fold higher compared with that of the control (Fig. 11c). In the hepatopancreas, upregulation of PmHsp60 and PmHsp10 transcripts was detected at the beginning of zinc exposure. Maximum PmHsp60 and PmHsp10 mRNA expression levels were observed at 24 h, which are 8.8- and 2.6-fold higher compared with that of the control, under zinc treatment (Fig. 11d). In the cadmium exposure experiment, the PmHsp60 transcript significantly increased at 24 h in the gills and reached maximum expression (6.3-fold higher compared with that of the control) thereafter. Furthermore, the maximum mRNA expression level of PmHsp10 was 2.7-fold higher compared with that of the control at 12 h in the gills (Fig. 11e). In the hepatopancreas, the two PmHsp genes were induced slightly, and the highest expression levels of PmHsp60 (2.0-fold higher compared with that of the control) and PmHsp10 (2.3-fold higher compared with that of the control) were detected at 6 and 48 h, respectively (Fig. 11f).
ATPase activity of PmHsp60
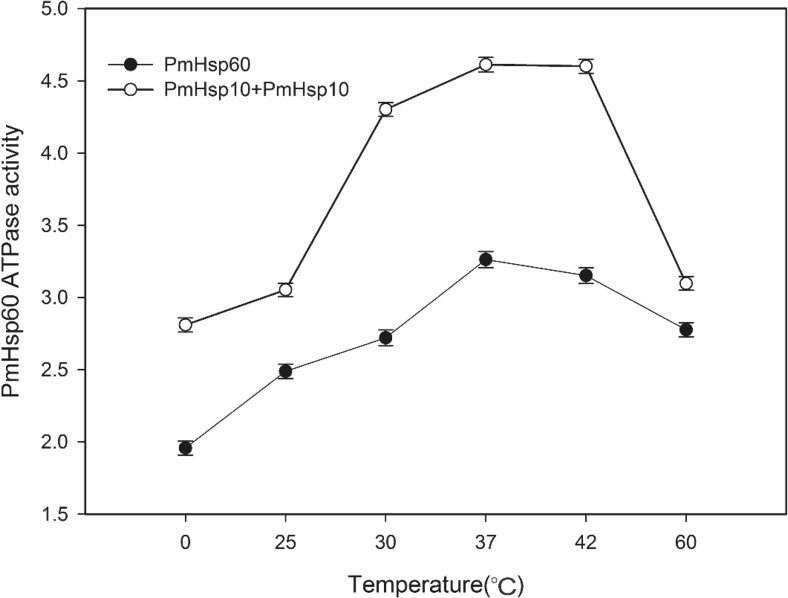
The ATPase of PmHsp60 is shown in Fig. 12. PmHsp60 revealed ATPase activity from 0 to 60 °C, and maximum activity was found at 37 °C. PmHsp60 ATPase activity increased at all temperature conditions when PmHsp10 was added to the reaction system. The maximum increase in activity was found over the temperature range of 37–42 °C, and the lowest increase in activity was detected at 60 °C.
Fig. 12.
ATPase activity of PmHSP60. PmHSP60 (black circle) and PmHSP60 + PmHSP10 (white circle) activity determination were performed at 0, 25, 30, 37, 42 and 60 °C. The light scattering of the solution was monitored at 636 nm
Chaperone activity of PmHsp60
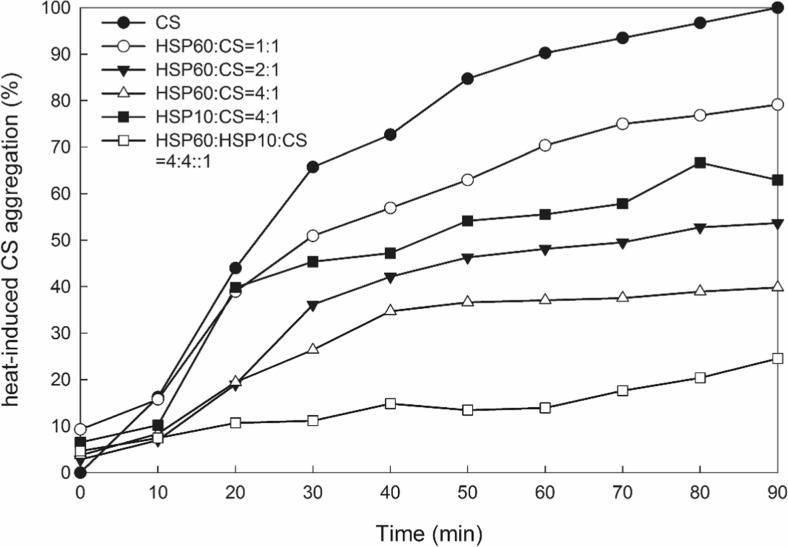
Heat-induced citrate synthase aggregation assay was performed to determine the chaperone activity of PmHsp60 (Fig. 13). At a 1:1 Hsp60/CS ratio (w/w), Hsp60 offered about 21 % protection. The extent of protection increased as the Hsp60 concentration increased. Approximately 57 % protection was observed at a 2:1 Hsp60/CS ratio. At a 4:1 ratio, over 60 % protection was observed. Only approximately 38 % protection was observed when the Hsp10/CS ratio was 4:1. However, over 76 % protection was observed at a 4:4:1 Hsp60/Hsp10/CS ratio.
Fig. 13.
Chaperone activity of PmHSP60 and PmHSP10. Thermally induced aggregation of citrate synthase (20 mg/ml) alone (black circle); PmHSP60/citrate synthase (w/w) at 1:1 (white circle); 2:1 (inverted black triangle) and 4:1 (upright triangle). PmHSP10/citrate synthase (w/w) at 4:1 (black box); PmHSP60/PmHSP10/citrate synthase (w/w/w) at 4:4:1 (white box). The light scattering of the solution was monitored at 320 nm
Discussion
In the present study, the full-length cDNAs of PmHsp60 and PmHsp10 were cloned from P. monodon. A Cpn60 conserved domain and seven Gly-Gly-Met repeats at the C terminus were found in the PmHsp60 sequence, in accordance with previous studies (Xu et al. 1997; Sanchez et al. 1999). The functional domains of the motif, such as ring oligomerization interfaces and ATP/Mg binding sites, were detected and similar characteristics were found in other species (Xu and Qin 2012; Xu et al. 2014). This finding suggested that PmHsp60 belongs to the mitochondrial Hsp60 (mtHsp60) chaperone family. As the PmHsp60 co-chaperone, PmHsp10 was cloned in the current study. A mobile loop was also found in the PmHsp10 sequence. The mobile loop mediates the interaction with the Hsp60 apical domains form a protein folding mechanism (Hayer Hartl et al. 2015). These motifs and structures could be used to research the functions of the Hsp60/Hsp10 complex in future studies.
Phylogenetic analyses of PmHsp60 and PmHsp10 were conducted by the neighbor joining method. The phylogenetic trees obtained revealed that PmHsp60 shows the highest identity with L. vannamei and the PmHsp10 is similar to S. paramamosain. Sequence alignment and phylogenetic analysis further suggested that PmHsp60 and PmHsp10 respectively belong to the Hsp60 and Hsp10 families. This finding provides some molecular evidence of the evolutionary and phylogenetic development of the black tiger shrimp.
In the present study, recombinant PmHsp60 and PmHsp10 were expressed in E. coli BL21 (DE3). A PmHsp60-6 × His tagged fusion protein of about 64 kDa, was detected via SDS-PAGE. A protein with a molecular mass of 11 kDa was found during SDS-PAGE after induction with 1 mM IPTG at 37 °C. There is no any nonspecific band that was detected in the PmHsps western results. PmHsp60 and PmHsp10 were rapidly obtained in large amounts in vitro. The results of the present study thus far provide a molecular foundation for proteomics and immunohistochemistry analyses to better understand the biological functions of PmHsps.
Aquatic animal health is well-known to be usually influenced by the external environment. Numerous economic losses in the aquaculture industry are caused by environmental stressors, such as heavy metal stress, heat stress, hypoxic stress, osmotic stress, and pH challenge. Thus, Hsps, as a series of molecular chaperones induced by environmental stress, play important roles in the innate immune responses of aquatic species. The highest transcripts of PmHsps were found in the hepatopancreas whereas the lowest transcripts were found in the gills of shrimp during distribution analysis (Fig. 8). The two tissues have similarly been reported to be environmentally sensitive tissues in other species (Zhou et al. 2009; Li et al. 2012; Shekhar et al. 2013). Therefore, PmHsp60 and PmHsp10 transcript levels in the hepatopancreas and gills were investigated upon exposure of P. monodon to different environmental stresses, including pH challenge, osmotic stress, and heavy metal exposure.
In the pH challenge experiment, the PmHsp60 and PmHsp10 transcripts were induced under pH 7.0 in the gills, and no obvious variation in their expression was observed in the hepatopancreas under the same conditions. However, the relative expressions of PmHsp60 and PmHsp10 mRNA in gills increased significantly 6 h after pH 9.0 exposure and reached maximum levels (24-fold higher compared with that of the control) at 48 h. Compared with the stable expressions of PmHsp60 and PmHsp10 at pH 7.0 in the hepatopancreas, transcript expressions peaked at 96 h after pH 9.0 exposure. We further found that the change tendencies of the PmHsp60 and PmHsp10 transcripts were similar in most cases. These results suggest that PmHsp60 and PmHsp10 may be involved in the defense against pH stress, especially alkalization and that gills are sensitive tissues to pH stress. A distinct change in the expressions of these genes has also been observed in other organisms post-acid shock. Helicobacter pylori Hsp70 is upregulated preferentially by pH rather than heat shock (Huesca et al. 1998). Phosphatidylinositol 3-kinase/Akt and MAPK regulate Hsp70 and Hsp27 induction in primary cultures of human esophageal microvascular endothelial cells in response to acid exposure (pH 4.5) (Rafiee et al. 2006). Four Hsps expression patterns were compared in response to pH challenge in L. vannamei (Qian et al. 2012). Acid or alkaline exposure may induce reaction oxygen species (ROS) and cooperatively upregulate manganese superoxide dismutase (MnSOD), catalase, glutathione peroxidase (GPx), and thioredoxin (Wang et al. 2009). Glutathione S-transferase (GST) is induced when ROS is produced (Dorts et al. 2009). Moreover, the Hsp60 folding machinery is crucial for the folding and functions of MnSOD (Magnoni et al. 2014). In the mitochondria, the combination of mitochondrial phospholipid hydroperoxide glutathione peroxidase and Hsp60/10 provides protection against damage resulting from simulated ischemia/reoxygenation injury (Hollander et al. 2003). These results suggest that Hsp60 and Hsp10 combine with antioxidant enzymes, such as MnSOD, GPx, and GST, to achieve protection against pH stress. However, the detailed mechanisms of such a combination require more investigation.
Osmotic stress is the most common environmental stress factor for aquatic organisms. The shrimps were exposed to low (2.3 %) and high salinity (4.3 %) to investigate the functions of PmHsp60 and PmHsp10. PmHsp10 expression in gills was found to reach maximum levels below 4 h under low salinity exposure; however, this expression was induced continuously in the hepatopancreas. The PmHsp60 transcript was upregulated slightly in these two tissues under low salinity stress. Under high osmotic stress, PmHsp60 and PmHsp10 mRNA expression reached maximum levels in the gills at 32 and 16 h, respectively. In the hepatopancreas, the transcripts of these two PmHsps were induced at 4 h and maintained until 16 h. The PmHsp60 and PmHsp10 transcript variations observed suggest that Hsp60 and Hsp10 may be involved in the refolding of misfolded proteins and maintenance of ionic homeostasis under osmotic stress. Hsps induced by salinity stress have been determined in numerous previous studies. For example, DnaK, together with GroEL, GroES, and a number of other Hsps, are induced during salt stress in Lactococcus lactis (Kilstrup et al. 1997). DnaK is required for K+ transport maintenance at high osmolality (Meury and Kohiyama 1991). T cells in experimentally induced salt-driven hypertension present a CD4 clonal response to Hsp70, which is overexpressed in the kidneys (Pons et al. 2013). Hsp21 was identified as a differentially expressed gene from a forward SSH cDNA library constructed from gut tissues of P. monodon showing significant increases in gene expression under osmotic stress (Shekhar et al. 2013). Ion uptake in crustaceans is mostly attributed to Na+/K+-ATPase and Ca2+-ATPase activity (Meyran and Graf 1986; Lima et al. 1997; Furriel et al. 2004; Masui et al. 2005). Several previous studied have found that Hsps could be involved in Na+/K+-ATPase stabilization (Burdon and Cutmore 1982; Deane and Woo 2005; Chatterjee et al. 2009). Hsp60 is essential to the functions of Ca2+-ATPase KlPmr1p (Uccelletti et al. 2005). Results of previous research and the expression levels of Hsp60 and Hsp10 determined in the present study suggest that these proteins play important roles in ion balance under osmotic stress. However, details of the regulation and stabilization mechanisms of these proteins under salinity stress are limited in crustaceans and require further research.
Increasing evidence has demonstrated that heavy metal pollution impairs the growth and development of aquatic organisms (Calabrese et al. 1977; Ahsanullah et al. 1981; Wong et al. 1995). Previous studies have also shown that P. monodon is influenced by heavy metal exposure (Chen HC Water quality criteria for farming the grass shrimp and Penaeus monodon. In 1985; Guhathakurta and Kaviraj 2000; Chen and Lin 2001; Mokhtar et al. 2009). Hsps, as a series of proteins that respond to environmental stress, play important roles in the defense against heavy metals. In the present study, the PmHsp60 transcript increased significantly at 12 h, and both PmHsp60 and PmHsp10 expression levels in gills reached peak values (10.7- and 7.8-fold compared with the control, respectively) at 96 h after copper exposure. Compared to the gills, the variation of PmHsps transcript was imperceptible in hepatopancreas. These results suggest that PmHsp60 may be involved in the protection against copper exposure and that the gills are tissues that sensitively responded to copper stress.
Zinc is physiologically essential for the normal growth and development of aquatic organisms. However, zinc may harm marine organisms when concentrations exceed actual physiological requirements (Wong et al. 1995). In the present study, the expression profiles of PmHsp60 and PmHsp10 were detected under zinc stress. The expression patterns observed showed that both PmHsp60 and PmHsp10 genes were induced significantly in the gills and reached maximum levels (7.2- and 4.8-fold higher compared with that of the control, respectively) at 12 h. In the hepatopancreas, the PmHsp60 and PmHsp10 transcripts were clearly upregulated at the beginning of zinc exposure. Maximum PmHsp60 (8.8-fold higher compared with that of the control) and PmHsp10 (2.6-fold higher compared with that of the control) mRNA expression levels were found 24 h after zinc treatment. These results indicate that more Hsps are required to defend zinc-induced stress and maintain normal physiological processes.
Cadmium is a widespread environmental pollutant which has received an increased worldwide concern. Cadmium toxicology has been studied in various aquaculture organisms (Wu et al. 2006; Seebaugh et al. 2012; Ma et al. 2013). and previous studies have demonstrated that Hsps are involved in the defense against cadmium toxicity (Jing et al. 2013; Jung and Lee 2013; Mazzei et al. 2015). However, very few studies have been conducted in P. monodon. In the current study, we investigated the expression patterns of PmHsp60 and PmHsp10 in shrimps exposed to cadmium stress. The PmHsp60 transcript increased significantly in gills at 24 h and reached peak levels (6.3-fold higher compared with that of the control) at 24 h. The maximum mRNA expression level of PmHsp10 in gills was 2.7-fold higher compared with that of the control at 12 h. In the hepatopancreas, both PmHsp genes were induced slightly under cadmium stress. The highest PmHsp60 expression was significantly induced by cadmium exposure, in accordance with previous studies (Qian et al. 2012). For example, the relative mRNA expression of LvHsp60 shows sensitive responses to cadmium exposure and reaches 21.9-fold that of the control group within 24 h. Similar phenomena have also been observed in mouse and chicken (Knoflach et al. 2011; Chen et al. 2012). These results suggest that Hsp60 may play an important role in cadmium tolerance and can be used as biomarker for cadmium pollution. PmHsp60 and PmHsp10 expressions were up- and downregulated with similar tendency. These results suggest that PmHsp60 and PmHsp10 probably perform their biological functions as a complex in response to environmental stress.
Previous studies have demonstrated that Hsp60 is one of the main Hsps to environment stress responses (Tsan and Gao 2004; Li et al. 2011). The ATPase and chaperone activities of Hsp60 play important roles in the folding of denatured proteins and responses to stress (Gupta et al. 2008). Figure 12 shows that the ATPase activity of PmHsp60 increased from 0 to 37 °C and decreased from 42 to 60 °C. Moreover, ATPase activity increased at every tested temperature after addition of PmHsp10 to the reaction system. Hsp10 interacts with a single ring of Hsp60 and forms a folding cage, and ATP binds to an open GroEL (Hsp60) ring before GroES (Hsp10) encapsulation (Magen et al. 2008; Tyagi et al. 2009). These results suggest that the Hsp60/Hsp10 complex produces an efficient ATPase machinery. The chaperone activity assay showed that Hsp60 facilitated proper protein folding activity (Fig. 13), and the extent of protection increased with variations of the mixing ratio (i.e., weight of chaperone/weight of target). Hsp60 was a more effective chaperone than Hsp10, and the Hsp60/Hsp10 complex demonstrated greater activity. Huang et al. (2011) also found that Hsp60 presents chaperone activity in L.vannamei, similar to our experimental results. The data suggest that Hsp60/Hsp10 functions as a major chaperone complex in response to environmental stresses.
In summary, the full-length cDNAs of PmHsp60 and PmHsp10 were cloned from P. monodon, and their expression profiles were detected when the shrimps were exposed to pH challenge, osmotic stress, and heavy metal stress. PmHsp60 and PmHsp10 were found to be involved in responses to pH, osmotic, and heavy metal stress. The results obtained from this study provide useful evidence of the ability of Hsp60/Hsp10 to defend shrimp against cellular stress and demonstrate how the complex participates in environmental stress responses.
Acknowledgments
This work was supported by the Special Fund for Fisheries-Scientific Research (A201300B03), the Special Scientific Research Funds for Central Non-profit Institutes (2015YD05, 2014TS12), the Guangdong Provincial Science and Technology Program (2013B040402016, 2014A020208039) and the Guangdong Province Marine Fishery Science and Technology and Industrial Development of Special Scientific and Technological Research and Development Projects (A201501A11).
References
- Ahsanullah M, Negilski D, Mobley M. Toxicity of zinc, cadmium and copper to the shrimp Callianassa australiensis III accumulation metals. Mar Biol. 1981;64:311–316. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettinger L, Oeljeklaus S, Guiard B, Rospert S, Warscheid B, Becker T. The mitochondrial heat shock protein 70 (Hsp70) and Hsp10 cooperate in the formation of Hsp60 complexes. J Biol Chem. 2015;290:11611–11622. doi: 10.1074/jbc.M115.642017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon RH, Cutmore CM. Human heat shock gene expression and the modulation of plasma membrane Na+, K+-ATPase activity. FEBS Lett. 1982;140:45–48. doi: 10.1016/0014-5793(82)80517-6. [DOI] [PubMed] [Google Scholar]
- Calabrese A, MacInnes J, Nelson D, Miller J. Survival and growth of bivalve larvae under heavy-metal stress. Mar Biol. 1977;41:179–184. doi: 10.1007/BF00394024. [DOI] [Google Scholar]
- Cappello F, et al. Hsp60 and Hsp10 down-regulation predicts bronchial epithelial carcinogenesis in smokers with chronic obstructive pulmonary disease. Cancer. 2006;107:2417–2424. doi: 10.1002/cncr.22265. [DOI] [PubMed] [Google Scholar]
- Cappello F, Marino Gammazza A, Palumbo Piccionello A, Campanella C, Pace A, Conway de Macario E, Macario AJ. Hsp60 chaperonopathies and chaperonotherapy: targets and agents. Expert Opin Ther Targets. 2014;18:185–208. doi: 10.1517/14728222.2014.856417. [DOI] [PubMed] [Google Scholar]
- Cesar JR, Yang J. Expression patterns of ubiquitin, heat shock protein 70, alpha-actin and beta-actin over the molt cycle in the abdominal muscle of marine shrimp Litopenaeus vannamei. Mol Reprod Dev. 2007;74:554–559. doi: 10.1002/mrd.20605. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Das P, Mazumder A, Nagdas SK, Sen PC. Localization and expression of a 70 kDa protein in goat spermatozoa having Na+, K + -ATPase inhibitory and arylsulphatase A activities. Mol Cell Biochem. 2009;321:85–94. doi: 10.1007/s11010-008-9922-2. [DOI] [PubMed] [Google Scholar]
- Chen HC (1985) Water quality criteria for farming the grass shrimp, Penaeus monodon. In 1985. Aquaculture Department, Southeast Asian Fisheries Development Center, p 165
- Chen JC, Lin CH. Toxicity of copper sulfate for survival, growth, molting and feeding of juveniles of the tiger shrimp. Penaeus monodon Aquac. 2001;192:55–65. doi: 10.1016/S0044-8486(00)00442-7. [DOI] [Google Scholar]
- Chen X, Zhu YH, Cheng XY, Zhang Z, Xu S. The Protection of Selenium against Cadmium-induced cytotoxicity via the heat shock protein pathway in chicken splenic lymphocytes. Molecules. 2012;17:14565–14572. doi: 10.3390/molecules171214565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Saibil HR. ATP-driven molecular chaperone machines. Biopolymers. 2013;99:846–859. doi: 10.1002/bip.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MP, et al. Molecular characterization of the Corynebacterium pseudotuberculosis hsp60-hsp10 operon, and evaluation of the immune response and protective efficacy induced by hsp60 DNA vaccination in mice. BMC Res Notes. 2011;4:243. doi: 10.1186/1756-0500-4-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, et al. A macrophage migration inhibitory factor like oxidoreductase from pearl oyster Pinctada fucata involved in innate immune responses. Fish Shellfish Immunol. 2011;31:173–181. doi: 10.1016/j.fsi.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Cui Z, Liu Y, Luan W, Li Q, Wu D, Wang S. Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunus trituberculatus) Fish Shellfish Immunol. 2010;28:56–64. doi: 10.1016/j.fsi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Deane E, Woo N. Evidence for disruption of Na+‐K+‐ATPase and hsp70 during vibriosis of sea bream, Sparus (=Rhabdosargus) sarba Forsskål. J Fish Dis. 2005;28:239–251. doi: 10.1111/j.1365-2761.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- Dorts J, Silvestre F, Tu HT, Tyberghein AE, Phuong NT, Kestemont P. Oxidative stress, protein carbonylation and heat shock proteins in the black tiger shrimp. Penaeus monodon, following exposure to endosulfan and deltamethrin. Environ Toxicol Pharmacol. 2009;28:302–310. doi: 10.1016/j.etap.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Michels A, Song J, Kampinga HH, Morimoto RI (2000) Analysis of molecular chaperone activities using in vitro and in vivo approaches. In: Stress Response, vol 2000. Springer, pp 393–419 [DOI] [PubMed]
- Furriel RP, Masui DC, McNamara JC, Leone FA. Modulation of gill Na+, K + -ATPase activity by ammonium ions: Putative coupling of nitrogen excretion and ion uptake in the freshwater shrimp Macrobrachium olfersii J Exp Zool A Comp. Exp Biol. 2004;301:63–74. doi: 10.1002/jez.a.20008. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- Grundtman C, Kreutmayer SB, Almanzar G, Wick MC, Wick G. Heat shock protein 60 and immune inflammatory responses in atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31:960–968. doi: 10.1161/ATVBAHA.110.217877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhathakurta H, Kaviraj A. Heavy metal concentration in water, sediment, shrimp (Penaeus monodon) and mullet (Liza parsia) in some brackish water ponds of Sunderban. India Mar Pollut Bull. 2000;40:914–920. doi: 10.1016/S0025-326X(00)00028-X. [DOI] [Google Scholar]
- Gupta RS, Ramachandra NB, Bowes T, Singh B (2008) Unusual cellular disposition of the mitochondrial molecular chaperones Hsp60, Hsp70 and Hsp10. In: Novartis Foundation symposium. Chichester; New York; John Wiley; 1999, p 59 [DOI] [PubMed]
- Hansen JJ, et al. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- Hayer Hartl M, Bracher A, Hartl FU. The GroEL-GroES chaperonin machine: A nano-cage for protein folding. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Hollander JM, Lin KM, Scott BT, Dillmann WH. Overexpression of PHGPx and HSP60/10 protects against ischemia/reoxygenation injury free radical. Biol Med. 2003;35:742–751. doi: 10.1016/s0891-5849(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Huang PY, Kang ST, Chen WY, Hsu TC, Lo CF, Liu KF, Chen LL. Identification of the small heat shock protein, HSP21, of shrimp Penaeus monodon and the gene expression of HSP21 is inactivated after white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. 2008;25:250–257. doi: 10.1016/j.fsi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Huang WJ, Leu JH, Tsau MT, Chen JC, Chen LL. Differential expression of LvHSP60 in shrimp in response to environmental stress. Fish Shellfish Immunol. 2011;30:576–582. doi: 10.1016/j.fsi.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Huesca M, Goodwin A, Bhagwansingh A, Hoffman P, Lingwood CA. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect Immun. 1998;66:4061–4067. doi: 10.1128/iai.66.9.4061-4067.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R, Vardhan H, Bas S, Salhan S, Mittal A. Chlamydia trachomatis heat shock proteins 60 and 10 induce apoptosis in endocervical epithelial cells. Inflamm Res. 2011;60:69–78. doi: 10.1007/s00011-010-0237-x. [DOI] [PubMed] [Google Scholar]
- Jiang S, Qiu L, Zhou F, Huang J, Guo Y, Yang K. Molecular cloning and expression analysis of a heat shock protein (Hsp90) gene from black tiger shrimp (Penaeus monodon) Mol Biol Rep. 2009;36:127–134. doi: 10.1007/s11033-007-9160-9. [DOI] [PubMed] [Google Scholar]
- Jing J, Liu H, Chen H, Hu S, Xiao K, Ma X. Acute effect of copper and cadmium exposure on the expression of heat shock protein 70 in the Cyprinidae fish Tanichthys albonubes. Chemosphere. 2013;91:1113–1122. doi: 10.1016/j.chemosphere.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson SM, et al. A biochemical screen for GroEL/GroES inhibitors. Bioorg Med Chem Lett. 2014;24:786–789. doi: 10.1016/j.bmcl.2013.12.100. [DOI] [PubMed] [Google Scholar]
- Jung MY, Lee YM. Expression profiles of heat shock protein gene families in the monogonont rotifer Brachionus koreanus-exposed to copper and cadmium. Toxicol Environ Health Sci. 2013;4:235–242. doi: 10.1007/s13530-012-0141-6. [DOI] [Google Scholar]
- Kilstrup M, Jacobsen S, Hammer K, Vogensen FK. Induction of heat shock proteins DnaK. GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoflach M, et al. Non-toxic cadmium concentrations induce vascular inflammation and promote atherosclerosis. Circ J. 2011;75:2491–2495. doi: 10.1253/circj.CJ-11-0196. [DOI] [PubMed] [Google Scholar]
- Kol A, Lichtman AH, Finberg RW, Libby P, Kurt-Jones EA. Cutting edge: heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. J Immunol. 2000;164:13–17. doi: 10.4049/jimmunol.164.1.13. [DOI] [PubMed] [Google Scholar]
- Kozlova T, Perezgasga L, Reynaud E, Zurita M. The Drosophila melanogaster homologue of the hsp60 gene is encoded by the essential locus l (1) 10Ac and is differentially expressed during fly development. Dev Genes Evol. 1997;207:253–263. doi: 10.1007/s004270050113. [DOI] [PubMed] [Google Scholar]
- Li J, et al. Cloning of a heat shock protein 90 (HSP90) gene and expression analysis in the ridgetail white prawn Exopalaemon carinicauda. Fish Shellfish Immunol. 2012;32:1191–1197. doi: 10.1016/j.fsi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Myocardial ischemia activates an injurious innate immune signaling via cardiac heat shock protein 60 and Toll-like receptor 4. J Biol Chem. 2011;286:31308–31319. doi: 10.1074/jbc.M111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima AG, McNamara JC, Terra WR. Regulation of hemolymph osmolytes and gill Na+/K+-ATPase activities during acclimation to saline media in the freshwater shrimp Macrobrachium olfersii (Wiegmann, 1836)(Decapoda, Palaemonidae) J Exp Mar Biol Ecol. 1997;215:81–91. doi: 10.1016/S0022-0981(97)00016-6. [DOI] [Google Scholar]
- Lo WY, Liu KF, Liao IC, Song YL. Cloning and molecular characterization of heat shock cognate 70 from tiger shrimp (Penaeus monodon) Cell Stress Chaperones. 2004;9:332. doi: 10.1379/CSC-47R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, et al. Oxidative damages and ultrastructural changes in the sperm of freshwater crab Sinopotamon henanense exposed to cadmium. Ecotoxicol Environ Saf. 2013;98:244–249. doi: 10.1016/j.ecoenv.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Magen D, et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnoni R, Palmfeldt J, Hansen J, Christensen JH, Corydon TJ, Bross P. The Hsp60 folding machinery is crucial for manganese superoxide dismutase folding and function. Free Radic Res. 2014;48:168–179. doi: 10.3109/10715762.2013.858147. [DOI] [PubMed] [Google Scholar]
- Martin J, Horwich AL, Hartl FU. Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science. 1992;258:995–998. doi: 10.1126/science.1359644. [DOI] [PubMed] [Google Scholar]
- Masui DC, et al. Gill microsomal (Na+, K+)-ATPase from the blue crab Callinectes danae: interactions at cationic sites. Int J Biochem Cell Biol. 2005;37:2521–2535. doi: 10.1016/j.biocel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Mazzei V, et al. Metallothioneins and heat shock proteins 70 in Armadillidium vulgare (Isopoda, Oniscidea) exposed to cadmium and lead. Ecotoxicol Environ Saf. 2015;116:99–106. doi: 10.1016/j.ecoenv.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Meinhardt A, Parvinen M, Bacher M, Aumüller G, Hakovirta H, Yagi A, Seitz J. Expression of mitochondrial heat shock protein 60 in distinct cell types and defined stages of rat seminiferous epithelium. Biol Reprod. 1995;52:798–807. doi: 10.1095/biolreprod52.4.798. [DOI] [PubMed] [Google Scholar]
- Meury J, Kohiyama M. Role of heat shock protein DnaK in osmotic adaptation of Escherichia coli. J Bacteriol. 1991;173:4404–4410. doi: 10.1128/jb.173.14.4404-4410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyran J, Graf F. Ultrahistochemical localization of Na+-K+ ATPase, Ca2+-ATPase and alkaline phosphatase activity in a calcium-transporting epithelium of a crustacean during moulting. Histochemistry. 1986;85:313–320. doi: 10.1007/BF00493483. [DOI] [PubMed] [Google Scholar]
- Mokhtar MB, Aris AZ, Munusamy V, Praveena SM. Assessment level of heavy metals in Penaeus monodon and Oreochromis spp. in selected aquaculture ponds of high densities development area. Eur J Sci Res. 2009;30:348–360. [Google Scholar]
- Music E, Khan S, Khamis I, Heikkila JJ. Accumulation of heme oxygenase-1 (HSP32) in Xenopus laevis A6 kidney epithelial cells treated with sodium arsenite, cadmium chloride or proteasomal inhibitors. Comp Biochem Physiol C Toxicol Pharmacol. 2014;166:75–87. doi: 10.1016/j.cbpc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Neuer A, Spandorfer S, Giraldo P, Dieterle S, Rosenwaks Z, Witkin S. The role of heat shock proteins in reproduction. Hum Reprod Update. 2000;6:149–159. doi: 10.1093/humupd/6.2.149. [DOI] [PubMed] [Google Scholar]
- Nisemblat S, Parnas A, Yaniv O, Azem A, Frolow F. Crystallization and structure determination of a symmetrical 'football' complex of the mammalian mitochondrial Hsp60-Hsp10 chaperonins. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2014;70:116–119. doi: 10.1107/S2053230X1303389X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranko J, Seitz J, Meinhardt A. Developmental expression of heat shock protein 60 (HSP60) in the rat testis and ovary. Differentiation. 1996;60:159–167. doi: 10.1046/j.1432-0436.1996.6030159.x. [DOI] [PubMed] [Google Scholar]
- Pons H, Ferrebuz A, Quiroz Y, Romero-Vasquez F, Parra G, Johnson RJ, Rodriguez-Iturbe B. Immune reactivity to heat shock protein 70 expressed in the kidney is cause of salt-sensitive hypertension. Am J Physiol Renal Physiol. 2013;304:F289–F299. doi: 10.1152/ajprenal.00517.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Liu X, Wang L, Wang X, Li Y, Xiang J, Wang P. Gene expression profiles of four heat shock proteins in response to different acute stresses in shrimp. Litopenaeus vannamei. Comp Biochem Physiol C. 2012;156:211–220. doi: 10.1016/j.cbpc.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Cohen IR. The HSP60 immune system network. Trends Immunol. 2011;32:89–95. doi: 10.1016/j.it.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Rafiee P, et al. Human esophageal microvascular endothelial cells respond to acidic pH stress by PI3K/AKT and p38 MAPK-regulated induction of Hsp70 and Hsp27. Am J Physiol Cell Physiol. 2006;291:C931–C945. doi: 10.1152/ajpcell.00474.2005. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis. 2010;33:789–801. doi: 10.1111/j.1365-2761.2010.01183.x. [DOI] [PubMed] [Google Scholar]
- Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. J Mol Biol. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Saibil HR, Ranson NA. The chaperonin folding machine. Trends Biochem Sci. 2002;27:627–632. doi: 10.1016/S0968-0004(02)02211-9. [DOI] [PubMed] [Google Scholar]
- Sanchez GI, Carucci DJ, Sacci J, Resau JH, Rogers WO, Kumar N, Hoffman SL. Plasmodium yoelii: cloning and characterization of the gene encoding for the mitochondrial heat shock protein 60. Exp Parasitol. 1999;93:181–190. doi: 10.1006/expr.1999.4455. [DOI] [PubMed] [Google Scholar]
- Seebaugh DR, Wallace WG, L’Amoreaux WJ, Stewart GM. Carbon assimilation and digestive toxicity in naive grass shrimp (Palaemonetes pugio) exposed to dietary cadmium. Bull Environ Contam Toxicol. 2012;88:449–455. doi: 10.1007/s00128-011-0493-7. [DOI] [PubMed] [Google Scholar]
- Shekhar MS, Kiruthika J, Ponniah AG. Identification and expression analysis of differentially expressed genes from shrimp (Penaeus monodon) in response to low salinity stress. Fish Shellfish Immunol. 2013;35:1957–1968. doi: 10.1016/j.fsi.2013.09.038. [DOI] [PubMed] [Google Scholar]
- Suttnar J, Dyr J, Hamšíková E, Novak J, Vonka V. Procedure for refolding and purification of recombinant proteins from Escherichia coli inclusion bodies using a strong anion exchanger Journal of Chromatography B. Biomed Sci Appl. 1994;656:123–126. doi: 10.1016/0378-4347(94)00078-6. [DOI] [PubMed] [Google Scholar]
- Tang D, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YL, Chiang YR, Wu CF, Narberhaus F, Lai EM. One out of four: HspL but no other small heat shock protein of Agrobacterium tumefaciens acts as efficient virulence-promoting VirB8 chaperone. PLoS One. 2012;7:e49685. doi: 10.1371/journal.pone.0049685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsan M-F, Gao B. Heat shock protein and innate immunity. Cell Mol Immunol. 2004;1:274–279. [PubMed] [Google Scholar]
- Tyagi NK, Fenton WA, Horwich AL. GroEL/GroES cycling: ATP binds to an open ring before substrate protein favoring protein binding and production of the native state. Proc Natl Acad Sci U S A. 2009;106:20264–20269. doi: 10.1073/pnas.0911556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelletti D, et al. The Golgi Ca2+-ATPase KlPmr1p function is required for oxidative stress response by controlling the expression of the heat-shock element HSP60 in Kluyveromyces lactis. Mol Biol Cell. 2005;16:4636–4647. doi: 10.1091/mbc.E05-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang Y, Zhang Z, Hong H, Zheng W. Effects of Cu 2+, Zn 2+, Cd 2+ on the Phototaxic Response of Artemia Salina and Two Shrimp (Penaeus penicillatus and Penaeus monodon) Nauplius. J Xiamen Univ (Nat Sci)/Xiamen Daxue Xuebao. 2001;40:1336–1339. [Google Scholar]
- Wang WN, et al. Oxidative stress. Comp Biochem Physiol C. 2009;150:428–435. doi: 10.1016/j.cbpc.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Welch WJ. The role of heat-shock proteins as molecular chaperones. Curr Opin Cell Biol. 1991;3:1033–1038. doi: 10.1016/0955-0674(91)90125-I. [DOI] [PubMed] [Google Scholar]
- Wong C, Cheung J, Chu K. Effects of copper on survival, development and growth of Metapenaeus ensis larvae and postlarvae (Decapoda: Penaeidae) Mar Pollut Bull. 1995;31:416–419. doi: 10.1016/0025-326X(95)00142-A. [DOI] [Google Scholar]
- Wu SM, Jong KJ, Lee YJ. Relationships among metallothionein, cadmium accumulation, and cadmium tolerance in three species of fish. Bull Environ Contam Toxicol. 2006;76:595–600. doi: 10.1007/s00128-006-0961-7. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, et al. Hsp60 accelerates the maturation of pro‐caspase‐3 by upstream activator proteases during apoptosis. EMBO J. 1999;18:2049–2056. doi: 10.1093/emboj/18.8.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Sun L, Liu S, Zhang L, Ru X, Zhao Y, Yang H. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs and their expression analysis under thermal stress in the sea cucumber Apostichopus japonicus. Comp Biochem Physiol B. 2014;171:49–57. doi: 10.1016/j.cbpb.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Xu Q, Qin Y. Molecular cloning of heat shock protein 60 (PtHSP60) from Portunus trituberculatus and its expression response to salinity stress. Cell Stress Chaperones. 2012;17:589–601. doi: 10.1007/s12192-012-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP) 7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang WN, Wang AL, He WY, Zhou QT, Liu Y, Xu J. Glutathione S-transferase in the white shrimp Litopenaeus vannamei: characterization and regulation under pH stress. Comp Biochem Physiol C. 2009;150:224–230. doi: 10.1016/j.cbpc.2009.04.012. [DOI] [PubMed] [Google Scholar]