Abstract
A meta-analysis was performed with the aim of re-evaluating the role of the peroxisome proliferator activated receptor alpha (PPARA) gene intron 7 G/C polymorphism (rs4253778) in athletes’ high ability in endurance sports. Design: A meta-analysis of case control studies assessing the association between the G/C polymorphisms of the PPARA gene and endurance sports was conducted. The Cochrane Review Manager software was used to compare the genotype and allele frequencies between endurance athletes and controls to determine whether a genetic variant is more common in athletes than in the general population. Five studies, encompassing 760 endurance athletes and 1792 controls, fulfilled our inclusion criteria. The pooled odds ratio (and confidence intervals, CIs) for the G allele compared to the C allele was 1.65 (95% CI 1.39-1.96). The pooled OR for the GG genotype compared to the GC genotype was 1.79 (95% CI 1.44-2.22), and for the GG genotype compared to the CC genotype 2.37 (95% CI 1.40-3.99). There was no evidence of heterogeneity (I2 =0%) or of publication bias. Athletes with high ability in endurance sports had a higher frequency of the GG genotype and G allele.
Keywords: Peroxisome proliferator-activated receptors, Genetic polymorphism, Sports, Meta-analysis
INTRODUCTION
Genetics and its interaction with the environment determine an individual's athletic ability. Around 66% of the variance in athlete status is explained by genetic factors[1]. The remaining variance is due to other factors such as training, nutrition, equipment, motivation, sleeping and epigenetics. The search for genetic variants contributing to success in sports has been challenging because of the involvement of several genes that contribute in a minor way [2]. The small contributions of each gene are difficult to identify due to the small sample sizes of studies, which in consequence have insufficient statistical power to demonstrate statistically significant effects and therefore replication [3]. The impact of these limitations can be reduced by pooling single studies in meta-analyses [4, 5].
Several genetic studies have focused on endurance sports (e.g., marathon running, triathlons, rowing), which are highly aerobic sports. Of the polymorphisms associated with sports endurance, the angiotensin-converting enzyme (ACE) and the alpha actinin-3 (ACTN3) polymorphisms have been the most frequently studied,[6] and meta-analyses have confirmed the associations [7]. Many other genetic variants have been studied; however, most of the genes have been assessed only in one study, and therefore the results have not been replicated [6]. Several single studies assessing sports endurance have focused on the peroxisome proliferator activated receptor alpha gene (PPARA); however, the results have been inconsistent,[8–12] possibly due to small sample sizes.
The PPARA gene is located on chromosome 22 (22q12-q13.1 [12]. The most frequently analyzed genetic variant in this gene is a polymorphism located in intron 7 (G/C, rs4253778). The PPARA gene has been a good candidate gene to study athletic ability due to its role in lipid metabolism, glucose energy homeostasis and vascular inflammation [13]. It is activated under conditions of energy deprivation, promoting uptake, utilization, and catabolism of fatty acids [14]. There is evidence that this gene is involved in the immune responses of the human body to endurance training, as it enables activation of the FAO mitochondrial pathway [15]. The PPARA gene is expressed at high levels in tissues that catabolize fatty acids, such as the liver, skeletal muscle, heart and muscle [16].
The aim of this study was to re-evaluate whether there is an association of the PPARA G/C (rs4253778) polymorphism with success in endurance sports by conducting a systematic review and meta-analysis.
MATERIALS AND METHODS
PubMed was searched for all publications available up to April 2015 studying the association between the PPARA G/C polymorphism (rs4253778) and endurance sports, using the following search terms: Peroxisome Proliferator-Activated Receptors[Mesh] AND (athletes OR sport OR exercise OR endurance OR strength) AND (polymorphism OR gene OR genotype). References from retrieved publications were checked for any additional studies. The Cochrane Review manager (RevMan) version 5.2 was used to perform the meta-analysis (Nordic Cochrane Centre, Cochrane Collaboration, Copenhagen, Denmark). The method of moments was applied to calculate the odds ratio (OR) and 95% confidence intervals as outlined by DerSimonian and Laird in a random effects model for the pooled data [17]. The degree of heterogeneity between the study results was assessed with the I2 statistic [18]. Sources of heterogeneity were explored in sensitivity analyses by removing one study at a time and calculating pooled ORs on the remaining studies [19]. RevMan was also used to produce funnel plots to examine publication bias [20]. The method is based on the fact that precision in the estimation of the true effect increases as sample sizes of the studies increase. OR, representing the relative effects estimated from individual studies, is plotted against the standard error on a logarithmic scale as a measure of each study sample size. This takes into account that the statistical power of a trial is determined by both its total sample size and the number of outcomes [20]. Study selection and data extraction were performed in duplicate by two independent investigators. P values lower than 0.05 (two-tailed) were considered statistically significant. The Haploreg (version 4) database [21] was used for in silico prediction of functional annotations (transcription factor-binding motifs) for the intronic SNP rs4253778 in the PPARA gene.
RESULTS
The search strategy retrieved 113 studies. The abstracts of all studies were reviewed and 14 complete studies were retrieved. Of the 14 studies, 10 were excluded (7 because another polymorphism was studied, and 3 because another type of sport was assessed). One study was identified by cross reference [12].
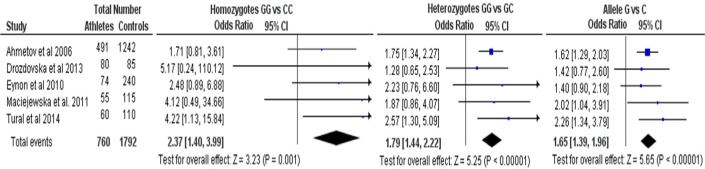
In total 5 studies were included, with a total of 760 endurance athletes and 1792 controls [9–12, 22]. Genotypic frequencies for both the cases and the controls in all studies were in Hardy-Weinberg equilibrium. Sports included were rowing, marathon, biathlon, triathlon, cross country skiing, swimming, skating (3,000-5,000) and road cycling. Athletes were chosen if they had participated in national and international championships, and both elite and non-elite athletes were included. Controls were sedentary individuals or individuals who had not participated in any competitive sport. Three studies found a statistically significant association between the G allele and sports endurance,[11, 12, 23] while 2 reported no statistically significant association.(9;10)
Figure 1 presents the OR and p values for the pooled analyses. Endurance athletes had a higher frequency of the GG genotype and G allele compared to controls. The pooled OR of the G allele compared to the C allele was 1.65 (95% CI 1.39-1.96). The pooled OR for the GG genotype compared to the GC genotype was 1.79 (95% CI 1.44-2.22), and for the GG genotype compared to the CC genotype it was 2.37 (95% CI 1.40-3.99). There was no evidence of heterogeneity (I2 =0%) or publication bias (not shown).
FIG. 1.
Meta-analysis for association studies for PPARA gene and endurance sports
The Haploreg database predicted that the intronic SNP rs4253778 in the PPARA gene leads to changes in transcription factor-binding motifs for the interferon regulatory factor (IRF) family of transcription factors.
DISCUSSION
In the present study, we pooled the data of 760 endurance athletes and 1792 control individuals to evaluate the association between the PPARA gene G/C polymorphism and endurance sports. The results demonstrated that athletes with high ability in endurance sports have a higher frequency of the GG genotype and G allele compared to controls.
The PPARA gene codes for the nuclear receptor protein peroxisome proliferator activated receptor alpha (PPAR-alpha), which is a transcriptional factor and a major regulator of lipid metabolism [24]. It is activated under conditions of energy deprivation and during metabolic and physiological stress, such as in fasting, [25] and hypothetically also during endurance sports. PPAR-alpha regulates the expression of genes involved in multiple steps of lipid metabolism, glucose metabolism, and energy homeostasis, and in addition it plays a role in the inflammatory process and vascular function [26]. The Haploreg database predicted that the rs4253778 SNP could lead to changes in binding sites for members of the IRF family of transcription factors (encoded by 9 genes in humans: IRF1-IRF9) [27]. This family of transcription factors has a growing role in the regulation of multiple cellular functions, including physiological processes in neural and muscular tissues [28, 29].
There are consistent findings that the PPARA gene G/C polymorphism is associated with sports endurance. In addition to our findings, there is evidence that the G allele is associated with increased fatty acid oxidation in skeletal muscles and an increased proportion of type I slow twitch fibres [30]; these fibres use oxygen in a more efficient manner during continuous muscle activity. Endurance athletes have relatively more type I slow twitch than fast twitch fibres in the trained musculature, which permits a sustained muscular contraction over a long period of time [12, 31]. Furthermore, the GG genotype was shown to be correlated with high values of oxygen pulse [32].
The PPARA gene G/C polymorphism is located in the non-coding region of the gene in intron 7, and therefore it is likely to be non-functional. However, there is a possibility that this polymorphism is in linkage disequilibrium with a functional variant in the promoter of the enhancer element of the PPAR-alpha gene that results in PPAR-alpha gene expression. In addition, there is evidence that this gene interacts with other polymorphisms in the peroxisome proliferator-activated receptors,[33–35] and with other genes such as the apoA-I and apoB genes [36]. Therefore further studies are needed to explore these interactions and their effects in endurance sports.
Case-control association studies have been widely used to identify susceptibility genes, and these remain the most common study design in sports genomics [23]. They determine whether one allele of a polymorphism is more common in a group of elite athletes than in the general population. However, one of the greatest limitations of these types of studies is their small sample size. As seen in the present study, this limitation can be overcome by performing a meta-analysis. Ioannidis and Lohmuller found that, when combining association studies with contradictory findings, approximately 20-30% of genetic association studies were statistically significant [3, 37]. The use of meta-analysis is an important step in the investigation of previously inconsistent results.
All the identified studies focused on studying the effect of single genes; therefore future studies are needed to understand the interaction of the PPARA gene with other genes and with the environment. To date, most of the studies assessing the genetic components of physical performance have focused on endurance and strength; future research should focus on identifying genetic markers associated with other sport phenotypes such as flexibility, coordination and temperament. A better understanding of sports phenotypes, their associated genes and their interaction with environmental factors will eventually help clinicians and coaches to identify individuals with genetic potential to be elite athletes, and identify individuals with a predisposition to potential health issues as well as physiological weakness and predisposition to injuries.
Acknowledgments
DAF was supported by research grants from VCTI-UAN and Colciencias.
Disclaimer
SLL works for Novartis Pharmaceuticals Corporation; the content of this article is solely the responsibility of the author.
REFERENCES
- 1.De Moor MH, Spector TD, Cherkas LF, Falchi M, Hottenga JJ, Boomsma DI, De Geus EJ. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res Hum Genet. 2007;10(6):812–20. doi: 10.1375/twin.10.6.812. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Padmanabhan S, Wolfarth B, Fuku N, Lucia A, Ahmetov II, Cieszczyk P, Collins M, Eynon N, Klissouras V, et al. Genomics of elite sporting performance: what little we know and necessary advances. Adv Genet. 2013;84:123–49. doi: 10.1016/B978-0-12-407703-4.00004-9. [DOI] [PubMed] [Google Scholar]
- 3.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33(2):177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 4.Forero DA, Arboleda GH, Vasquez R, Arboleda H. Candidate genes involved in neural plasticity and the risk for attention-deficit hyperactivity disorder: a meta-analysis of 8 common variants. J Psychiatry Neurosci. 2009;34(5):361–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA, van Duijn CM. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13(8):772–85. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- 6.Ahmetov II, Fedotovskaya ON. Current Progress in Sports Genomics. Adv Clin Chem. 2015;70:247–314. doi: 10.1016/bs.acc.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Ma F, Yang Y, Li X, Zhou F, Gao C, Li M, Gao L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS One. 2013;8(1):e54685. doi: 10.1371/journal.pone.0054685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmetov II, Mozhayskaya IA, Flavell DM, Astratenkova IV, Komkova AI, Lyubaeva EV, Tarakin PP, Shenkman BS, Vdovina AB, Netreba AI, et al. PPARalpha gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97(1):103–8. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 9.Drozdovska SB, Dosenko VE, Ahmetov II, Ilyin VN. The association of gene polymorphisms with athlete status in ukrainians. Biol Sport. 2013;30:163–7. doi: 10.5604/20831862.1059168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eynon N, Meckel Y, Sagiv M, Yamin C, Amir R, Sagiv M, Goldhammer E, Duarte JA, Oliveira J. Do PPARGC1A and PPARalpha polymorphisms influence sprint or endurance phenotypes? Scand J Med Sci Sports. 2010;20(1):e145–e150. doi: 10.1111/j.1600-0838.2009.00930.x. [DOI] [PubMed] [Google Scholar]
- 11.Maciejewska A, Sawczuk M, Cieszczyk P. Variation in the PPARalpha gene in Polish rowers. J Sci Med Sport. 2011;14(1):58–64. doi: 10.1016/j.jsams.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Tural E, Kara N, Agaoglu SA, Elbistan M, Tasmektepligil MY, Imamoglu O. PPAR-alpha and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Mol Biol Rep. 2014;41(9):5799–804. doi: 10.1007/s11033-014-3453-6. [DOI] [PubMed] [Google Scholar]
- 13.van Raalte DH, Li M, Pritchard PH, Wasan KM. Peroxisome proliferator-activated receptor (PPAR)-alpha: a pharmacological target with a promising future. Pharm Res. 2004;21(9):1531–8. doi: 10.1023/b:pham.0000041444.06122.8d. [DOI] [PubMed] [Google Scholar]
- 14.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20(5):649–88. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt B, Fluck M, Decombaz J, Kreis R, Boesch C, Wittwer M, Graber F, Vogt M, Howald H, Hoppeler H. Transcriptional adaptations of lipid metabolism in tibialis anterior muscle of endurance-trained athletes. Physiol Genomics. 2003;15(2):148–57. doi: 10.1152/physiolgenomics.00089.2003. [DOI] [PubMed] [Google Scholar]
- 16.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–66. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 17.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmetov II, Mozhayskaya IA, Flavell DM, Astratenkova IV, Komkova AI, Lyubaeva EV, Tarakin PP, Shenkman BS, Vdovina AB, Netreba AI, et al. PPARalpha gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97(1):103–8. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 23.Ahmetov II, Fedotovshaya ON. Sports genomics: Current state of knowledge and future directions. Cel and Mol Exercise Physiology. 2012;1:1–22. [Google Scholar]
- 24.Sher T, Yi HF, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32(21):5598–604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- 25.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–98. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fruchart JC, Duriez P, Staels B. Peroxisome proliferator-activated receptor-alpha activators regulate genes governing lipoprotein metabolism, vascular inflammation and atherosclerosis. Curr Opin Lipidol. 1999;10(3):245–57. doi: 10.1097/00041433-199906000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89(6-7):744–53. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. 2005;19(11):1498–500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- 29.Zhao GN, Jiang DS, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2015;1852(2):365–78. doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Ahmetov II, Mozhayskaya IA, Flavell DM, Astratenkova IV, Komkova AI, Lyubaeva EV, Tarakin PP, Shenkman BS, Vdovina AB, Netreba AI, et al. PPARalpha gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97(1):103–8. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 31.Ahmetov II, Williams AG, Popov DV, Lyubaeva EV, Hakimullina AM, Fedotovskaya ON, Mozhayskaya IA, Vinogradova OL, Astratenkova IV, Montgomery HE, et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum Genet. 2009;126(6):751–61. doi: 10.1007/s00439-009-0728-4. [DOI] [PubMed] [Google Scholar]
- 32.Ahmetov II, Egorova E, Mustafina LJ. The PPARA gene polymorphism in team sports athletes. Central European Journal of Sports Sciences and Medicine. 2013;1(1):19–24. [Google Scholar]
- 33.Liu M, Zhang J, Guo Z, Wu M, Chen Q, Zhou Z, Ding Y, Luo W. [Association and interaction between 10 SNP of peroxisome proliferator-activated receptor and non-HDL-C] Zhonghua Yu Fang Yi Xue Za Zhi. 2015;49(3):259–64. [PubMed] [Google Scholar]
- 34.Hai B, Xie HJ, Guo ZR, Wu M, Chen Q, Zhou ZY, Yu H, Ding Y. [Association of both peroxisome proliferator-activated receptor, gene-gene interactions and the lipid accumulation product] Zhonghua Liu Xing Bing Xue Za Zhi. 2013;34(11):1071–6. [PubMed] [Google Scholar]
- 35.Luo W, Guo Z, Wu M, Hao C, Hu X, Zhou Z, Zhou Z, Yao X, Zhang L, Liu J. Association of peroxisome proliferator-activated receptor alpha/delta/gamma with obesity, and gene-gene interaction, in the Chinese Han population. J Epidemiol. 2013;23(3):187–94. doi: 10.2188/jea.JE20120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hai B, Xie H, Guo Z, Dong C, Wu M, Chen Q, Zhou Z, Zhu Q, Liu M, Fan W, et al. Gene-Gene Interactions among Pparalpha/delta/gamma Polymorphisms for Apolipoprotein (Apo) A-I/Apob Ratio in Chinese Han Population. Iran J Public Health. 2014;43(6):749–59. [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]