Abstract
The yeast cell wall integrity MAPK Slt2 mediates the transcriptional response to cell wall alterations through phosphorylation of transcription factors Rlm1 and SBF. However, the variety of cellular functions regulated by Slt2 suggests the existence of a significant number of still unknown substrates for this kinase. To identify novel Slt2 targets, we generated and characterized an analog-sensitive mutant of Slt2 (Slt2-as) that can be specifically inhibited by bulky kinase inhibitor analogs. We demonstrated that Slt2-as is able to use adenosine 5′-[γ-thio]triphosphate analogs to thiophosphorylate its substrates in yeast cell extracts as well as when produced as recombinant proteins in Escherichia coli. Taking advantage of this chemical-genetic approach, we found that Slt2 phosphorylates the MAPK phosphatase Msg5 both in the N-terminal regulatory and C-terminal catalytic domains. Moreover, we identified the calcineurin regulator Rcn2, the 4E-BP (translation initiation factor eIF4E-binding protein) translation repressor protein Caf20, and the Golgi-associated adaptor Gga1 as novel targets for Slt2. The Slt2 phosphorylation sites on Rcn2 and Caf20 were determined. We also demonstrated that, in the absence of SLT2, the GGA1 paralog GGA2 is essential for cells to survive under cell wall stress and for proper protein sorting through the carboxypeptidase Y pathway. Therefore, Slt2-as provides a powerful tool that can expand our knowledge of the outputs of the cell wall integrity MAPK pathway.
Keywords: cell wall, mitogen-activated protein kinase (MAPK), phosphorylation, signaling, yeast, ATPγS analog, Slt2, cell wall integrity
Introduction
Protein phosphorylation is a key regulatory event in signal transduction in eukaryotic cells. As a result, protein kinases, a family of enzymes that catalyze phosphorylation of substrate proteins, play a major role in signaling pathways. Among them, MAPK pathways contain a three-tiered protein kinase cascade that consists of a mitogen-activated protein (MAP)3 kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAP kinase (MAPK) that are sequentially phosphorylated at their activation loop upon stimulation (1). Once activated, the MAPK in turn phosphorylates specific protein targets on serine and threonine residues within a consensus T/SP motif (2). These MAPK substrates are effectors of the cellular response triggered by the extracellular signal.
The model yeast Saccharomyces cerevisiae has five MAPK pathways that are involved in the regulation of mating, filamentous and invasive growth, osmoregulation, cell wall integrity (CWI), and spore wall assembly (3, 4). When the integrity of the cell wall is threatened, a compensatory salvage mechanism is activated to strengthen this vital structure. This cellular response is mainly mediated by the CWI pathway, which is essential for survival under cell wall stress conditions. The MAPK of this pathway, Slt2, directly phosphorylates the transcription factor Rlm1 (5), which is responsible for the major transcriptional response (6). The cell cycle transcriptional regulator SBF (7), the silencing protein Sir3 (8), the PKA regulatory subunit Bcy1 (9), and cyclin C (10) have also been reported to be phosphorylated by Slt2. Phosphorylation-based feedback loops that are exerted by Slt2 on upstream components of the CWI pathway, such as the Rho1-GDP-GTP exchange factor Rom2 and the MAPKKs Mkk1 and Mkk2 as well as on its negative regulator, the protein phosphatase Msg5, have also been described (11). However, although Slt2 regulates other cellular functions not mediated by its known targets, such as pexophagy, mitophagy, or the endoplasmic reticulum stress response (12, 13), no other bona fide Slt2 direct substrates have been identified to date.
Over the past few years, several novel methods for kinase substrate identification have been developed. On the one hand, large scale MS-based quantitative phosphoproteomic approaches allow the detection of proteins that become specifically phosphorylated upon activation of a signal transduction pathway (14). This is a very powerful technique that also brings about the identification of the phosphorylation sites within the protein, although the information regarding the kinase responsible for the phosphate transfer is uncoupled from the phosphorylation event. On the other hand, one technique that provides a direct coupling of kinase activity to its substrate relies on the use of analog-sensitive (as) kinases and orthogonal unnatural ATP analogs for the selective labeling of direct substrates by in vitro kinase assays (15). In this approach the active site of the kinase is engineered to allow the enzyme to accept a bulky ATP analog in which the γ-phosphate is replaced with a thiophosphate moiety. Such an ATP analog is not recognized by other wild-type kinases, and therefore, only the modified kinase is able to transfer the thiophosphate group to its target proteins. Then, thiophosphorylated proteins can be specifically immunodetected or affinity-purified (16). This strategy can be used to confirm direct phosphorylation of candidate substrates by a particular kinase or to identify direct substrates in complex samples, like cell extracts. In addition, as-kinases are valuable tools for functional studies because they allow their specific inhibition with bulky inhibitor analogs (17).
By using a quantitative stable isotope labeling by amino acids in cell culture (SILAC)-based phosphoproteomic approach, we previously identified a number of putative Slt2 targets that displayed enhanced phosphorylation at (S/T)P sites upon CWI pathway stimulation triggered by Pkc1 hyperactivation (18). Here, by using an analog-sensitive version of this MAPK (Slt2-as) and an ATPγS analog in in vitro kinase assays, we confirm that three of these candidates, Rcn2, Gga1, and Caf20, are bona fide substrates of Slt2.
Experimental Procedures
Microorganisms and Culture Conditions
For the cloning and amplification of plasmid DNA, the Escherichia coli strain DH5α (supE44, ΔlacU169, hsdR17, recA1, endA1, gyrA96, thi-1, relA1) was used. RosettaTM cells [F− ompT hsdSB (rB−mB−) gal dcm pRARE2 (CamR)] (Novagen) were selected for the expression of recombinant GST-fused proteins in E. coli. S. cerevisiae strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Euroscarf) and the isogenic BY4741 slt2Δ (19), Y00993 (BY4741 isogenic, slt2Δ::kanMX4) (Euroscarf), and YMM1 (BY4741 isogenic strains, slt2Δ::kanMX4, RLM1::6MYC::LEU2) (20) were used. Strains DD1–2D (MATa his2Δ1 leu2Δ0 trp1Δ0 ura3Δ0 ade1Δ0; msg5Δ::LEU2) (21), YMF3 (1783 isogenic, SLT2::6MYC::LEU2) (22), and those carrying the plasmids expressing the ORFs tagged at the N terminus with GST and expressed under the control of the GAL1/10 promoter (Open Biosystems), DBY746 (MATα trp1-289 leu2-3,112 ura3-52 his3Δ1), DBY746DK (DBY746 isogenic slt2::URA3) (23), 1783 (MATa leu2-3,112 ura3-52 trp1-1 his4 canlr), and DL454 (1783 isogenic mpklΔ::TRP1) (24) were used. Double mutant strains in the BY4741 background were generated by integrating the gga1Δ::NATr or gga2Δ::KanMX4 disruption cassettes, amplified with the corresponding oligonucleotides (Table 1) into BY4741 or BY4741 slt2Δ. Deletion transformants were selected with 60 μg/ml nourseothricin (Werner BioAgents) or 200 μg/ml G418 (Gibco Life Technologies). Yeast cultures were performed in either YPD (1% yeast extract, 2% peptone, 2% dextrose) or selective synthetic dextrose media (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose supplemented with the required amino acids), broth, or agar. Synthetic galactose and synthetic raffinose media were synthetic dextrose with 2% galactose and raffinose, respectively, instead of dextrose (25). When necessary, Congo red (CR) (Merck), the PP1 inhibitor analog 2,3-DMB-PP1 (4-amino-1-tert-butyl-3-(2,3-dimethylbenzyl)pyrazolo[3,4-d]pyrimidine) (Merck-Millipore Calbiochem), zymolyase 20T (ImmunOTM, MP Biomedicals, Inc.), caffeine (Sigma), sodium dodecyl sulfate (Duchefa Biochemie), or the alkaline phosphatase chromogenic substrate BCIP (5-bromo-4-chloro-3-indolyl-phosphate) (Roche Applied Science) were added at the indicated concentrations.
TABLE 1.
Oligonucleotides used in this work
Restriction sites are underlined, and mutated codons are in bold.
| Name | Primers |
|---|---|
| FwSLT2 | 5′-CAATTCTGGGAGATGGCTG-3 |
| RvSLT2 | 5′-CAAACTTCCGCGGAGTACG-3 |
| Fwslt2E108G | 5′-GGACTATATCTTTATGGTGAACTTATGGAATGTG-3′ |
| Rvslt2E108G | 5′-CACATTCCATAAGTTCACCATAAAGATATAGTCC-3′ |
| FwNco1-BamH1 SLT2 | 5′-CCATGGGATCCATGGCTGATAAGATAGAGAG-3′ |
| RvBamH1STOPNco1 SLT2 | 5′-GGATCCTAACCCATGGAAAAATATTTTCTATCTAATCCAAAC-3′ |
| FwBamH1-msg51–227 | 5′-CCCGGGATCCATGCAATTTCACTCAG-3′ |
| RvBamH1-msg51–227 | 5′-CCCCGGGTCAGGATCCCGTTCGTTGATAAACTCTTGC-3′ |
| FwBamH1 msg5228–489 | 5′-CCCCCCGGGGATCCAGGAAGAAACATCATCTG-3′ |
| RvBamH1-msg5228–489 | 5′-CCCCCGGGATCCGTTTATTTAGATGGACCATTG-3′ |
| FwBamH1 GGA1 | 5′-CCGGATCC ATGCCACAAAGAATTGAGCTTAC-3′ |
| RvXba1 GGA1 | 5′-CCTCTAGATTATATTGTGGGCAAACTGGTC-3′ |
| FwBamH1 SOD1 | 5′-CCGGATCCATGGTTCAAGCAGTCGCAG-3′ |
| RvEcoR1 SOD1 | 5′-CCGAATTCTTAGTTGGTTAGACCAATGAC-3′ |
| FwBamH1 RCN2 | 5′-CCGGATCCATGGCAAACCAAAAGCAAATG-3′ |
| RvEcoR1 RCN2 | 5′-CCGAATTCCTAATGGAAAAACTCGTTAAC-3′ |
| Fwrcn2S255A | 5′-GCCTCAAATCCTCCAAAAGCTCCAAGCATAACGGTTAAC-3′ |
| Rvrcn2S255A | 5′-GTTAACCGTTATGCTTGGAGCTTTTGGAGGATTTGAGGC-3′ |
| Fwrcn2S152A/S160A | 5′-CTCTTCATTAGCTCCGGATAAATCATCTCTAGAAGCGCCCACAATG-3′ |
| Rvrcn2S152A/S160A | 5′-CATTGTGGGCGCTTCTAGAGATGATTTATCCGGAGCTAATGAAGAG-3′ |
| Fwrcn2S152A | 5′-ATAAGGGAGGCTCTTCATTAGCTCCGGATAAATCATCTCTAGA-3′ |
| Rvrcn2S152A | 5′-TCTAGAGATGATTTATCCGGAGCTAATGAAGAGCCTCCCTTAT-3′ |
| Fwrcn2S160A | 5′-CGGATAAATCATCTCTAGAAGCGCCCACAATGTTGAAGCTTTC-3′ |
| Fwrcn2S160A | 5′-GAAAGCTTCAACATTGTGGGCGCTTCTAGAGATGATTTATCCG-3′ |
| FwBamH1 CAF20 | 5′-CCGGATCCATGATCAAGTATACTATCGATG-3′ |
| RvHindIII CAF20 | 5′-CCAAGCTTTTATGCTTCGTCGTCTTCGTC-3′ |
| Fwcaf20T102A | 5′-GAAGAAGAAACAGAAACCGCACCAACTTCTACTGTACCAG-3′ |
| Rvcaf20T102A | 5′-CTGGTACAGTAGAAGTTGGTGCGGTTTCTGTTTCTTCTTC-3′ |
| Fwgga1::NATr | 5′-GGACAAGTCACTACTTCAAGTATAACCCAGACAAGAGTCTTTTAAAACATGGAGGCCCAGAATACCC-3′ |
| Rvgga1::NATr | 5′-TCTCTGTAATATAATATGGCATCTACTTTTTTTTCAACTTCTCTACCGAATTTGACAGTATAGCGACCAGCATTCAC-3′ |
| K2 gga1::NATr | 5′-GGAGAAAACTTGTTAAGTCC-3′ |
| Fwgga2::KanMX4 | 5′-CCCTGTTTTTTTTCTGTAATCACG-3′ |
| Rvgga2::KanMX4 | 5′-CCTTTACACGATCTAGCATTGC-3′ |
| K2 gga2::KanMX4 | 5′-CCCTTTTCAAGACAGTCTGA-3′ |
DNA Manipulation and Plasmids
General DNA methods were performed using standard techniques. The oligonucleotides are described in Table 1.
To obtain plasmid pRS316-SLT2, the EcoRI-SalI 2.2-kb fragment bearing SLT2 from pHR0 (26) was cloned into EcoRI-SalI sites of pRS316 (27). FwSLT2 and RvSLT2 primers and mutagenic primers Fwslt2E108G and Rvslt2E108G were used to carry out an overlapping PCR using pRS316-SLT2 as the template. The HindIII-HindIII fragment from the amplified DNA was verified by sequencing and used to substitute the corresponding fragment of pRS316-SLT2 to yield plasmid pRS316-slt2-as.
The plasmids for expression of GST or GST-Slt2-as in yeast were generated via a two-step process. First, SLT2 or slt2-as was amplified by PCR with primers FwNco1-BamH1-SLT2 and RvBamH1STOPNco1-SLT2 using plasmids pRS316-SLT2 or pRS316-slt2-as as templates, respectively. These were then cloned into pGEMT (Promega) to generate the pGEMT-SLT2 and pGEMT-slt2-as plasmids. Second, the NcoI-BamHI fragments from these plasmids were subcloned into pEG(KG) (28) to yield pEG(KG)-SLT2 and pEG(KG)-slt2-as, respectively.
Non-phosphorylatable Myc-tagged versions of Msg5 were expressed in S. cerevisiae from Ycplac22-based plasmids (29). To this end, the custom-made plasmids pUC57-msg5N9A and pUC57-msg5C7A were obtained from GenScript bearing the MSG5 N-terminal and the C-terminal coding region, respectively, with mutations to change every serine or threonine followed by proline to alanine. Plasmid YCplac22-msg5N9Am was obtained by substituting a 1.4-kb ClaI fragment of YCplac22-MSG5m (22) by the corresponding fragment from pUC57-msg5N9A. The NsiI-NotI fragment from pUC57-msg5C7A was cloned into NsiI-NotI sites of YCplac22-MSG5m or YCplac22-msg5N9Am to yield YCplac22-msg5C7Am or YCplac22-msg516Am, respectively.
To express GST-fused proteins in E. coli, the DNA fragments encoding the corresponding proteins were amplified using PCR with the primers described in Table 1 using wild-type BY4741 genomic DNA as a template and cloned into pGEMT. Then, the restriction sites indicated in the primers were used to subclone these fragments into pGEX-KG (30). Plasmids pGEX-KG-rlm1 (31) or pGEX-KG-msg5C319A (22) were used to express the proteins GST-Rlm1 or GST-Msg5C319A, respectively, in E. coli. This plasmid and the primers FwBamH1-msg51–227, RvBamH1-msg51–227, FwBamH1-msg5228–489, and RvBamH1-msg5228–489 were used to amplify, via PCR, the N-terminal and C-terminal coding regions, respectively. The BamHI-BamHI fragment from the amplified DNA was verified by sequencing and used to substitute the corresponding fragment of pGEX-KG-msg5C319A to yield pGEX-KG-msg5NT or pGEX-KG-msg5CTC319A plasmids.
To express GST-Msg5 in S. cerevisiae, the plasmid pEG(KG)-MSG5 (22) was employed. pMLP1-LacZ (32) was used for reporting activity through the CWI pathway activity. Mutagenesis of specific residues was carried out by PCR site-directed mutagenesis (33).
Yeast Growth Assays
Growth assays on solid media were performed by culturing cells in selective synthetic dextrose medium to an A600 = 0.5 and spotting samples (5 μl) of 10-fold dilutions of the cell suspensions onto the surface of YPD plates followed by incubation at 30 °C for 2 days. The minimal inhibitory concentration of Congo red in the presence of 2,3-DMB-PP1 was estimated using the microdilution method in microplates of 96 wells with YPD medium, incubated at 30 °C for 48 h.
Preparation of Yeast Extracts and Immunoblotting Analysis
The procedures used to obtain yeast extracts, fractionation by SDS-PAGE, and transfer to nitrocellulose membranes have been described previously (25). Immunodetection of G6PDH (glucose-6-phosphate dehydrogenase) and Myc- and GST-tagged proteins and thiophosphate ester-labeled proteins was carried out using rabbit polyclonal anti-G6PDH (Sigma, catalog #A9521), mouse monoclonal anti-Myc (clone 9E10, Covance, catalog #MMS150P), rabbit polyclonal anti-GST (Santa Cruz Biotechnology, catalog #SC-459), and rabbit monoclonal anti-thiophosphate ester (Clone 51-8, Epitomics, catalog #2686-1) antibodies, respectively. Rabbit polyclonal anti-phospho-p44/42 (Thr-202/Tyr-204) MAPK antibody (Cell Signaling, catalog #9101) was used to specifically recognize dually phosphorylated ERK-type MAPKs as described previously (25). The primary antibodies were detected using fluorescently conjugated secondary antibodies IRDye 800CW goat anti-rabbit (Li-Cor Biosciences, P/N 926-32211), IRDye 680CW goat anti-rabbit (Li-Cor Biosciences, P/N 926-68021), or IRDye 680CW goat anti-mouse (Li-Cor Biosciences, P/N 926-68020) with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Carboxypeptidase Y (CPY) Colony Blotting
Yeast strains cultured in 96-well plates with liquid YPD were transferred to YPD plates with a pinning tool and incubated at 30 °C for 2 days. Then the round patches were replicated to a fresh YPD plate and overlaid with a nitrocellulose membrane (Hybond, Amersham Biosciences) before being incubated at 30 °C for a further 24 h. The nitrocellulose membranes were washed several times with PBS, blocked in PBS-milk, and immunoblotted with mouse monoclonal 10A5 anti-CPY antibody (Molecular Probes, catalog #A-6428).
β-Galactosidase Activity Assays
β-Galactosidase activity was determined as described (32).
Expression of Recombinant GST Fusion Proteins in E. coli
E. coli Rosetta cells transformed with the corresponding pGEX-KG-based plasmids were cultured in LB with ampicillin to an A600 = 0.5, and then isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm for 3 h. Bacteria pellets were collected and lysed by sonication in lysis buffer (1 mm DTT, 1 mm PMSF and protease inhibitors mixture (CompleteTM; Roche Applied Science) in PBS), and then the cell extracts were clarified by centrifugation.
Expression of GST Fusion Proteins in S. cerevisiae
Yeast cells expressing GAL1-driven GST-fused proteins were cultured overnight at 30 °C in selective synthetic raffinose medium lacking uracil, refreshed in selective synthetic galactose medium lacking uracil at A600 = 0.3, and cultured at 30 °C for an additional 6 h. Cells were collected and broken as previously described (25).
Expression and Purification of GST-Slt2-as in S. cerevisiae and in Vitro Kinase Assays
Y00993 (slt2Δ) cells transformed with the plasmids pEG(KG) or pEG(KG)-slt2-as were cultured overnight at 30 °C in synthetic raffinose medium lacking uracil, refreshed in YPD at A600 = 0.3, and cultured at 30 °C for 3 h. Then, to activate Slt2, Congo red was added at a final concentration of 30 μg/ml, and cultures were maintained at the same temperature for an additional 4 h. Cells were glass bead-lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 10 mm EDTA, 300 mm NaCl, 10 mm NaF, 10 mm Na4P2O7, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors mixture (CompleteTM; Roche Applied Science)). Cell extracts were clarified, and GST or GST-fused proteins were captured with glutathione-Sepharose beads (GE Healthcare) for 2 h. Beads were washed three times with lysis buffer and two times with kinase buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 10 mm MgCl2,1 mm sodium orthovanadate, 5 mm sodium pyrophosphate, 50 mm NaF, and 80 mm β-glycerol phosphate). In the kinase assay, 25 μl of slurry containing glutathione-Sepharose beads bound to GST or GST-Slt2-as in kinase buffer were mixed with 4 μl of cell extract from E. coli containing the recombinant GST-fused protein or 15 μl of cell extract (∼70 mg protein/ml) from yeast expressing the GST-fused protein. The kinase reactions were initiated by adding 1 μl of 3 mm ATP and 1 μl of 10 mm N6-(phenylethyl)-ATPγS (Biolog Life Science Institute). After 30 min at 30 °C, reactions were stopped by adding EDTA at a final concentration of 20 mm. For Western blotting analysis of thiophosphorylated substrates, alkylation of these substrates was carried out by adding p-nitrobenzyl mesylate for a final concentration of 2.5 mm (Abcam Biochemicals) and leaving the samples at room temperature for 3 h. This thiol-specific alkylating agent generated a bio-orthogonal thiophosphate ester that is recognized by a specific anti-thiophosphate ester antibody (Epitomics). Anti-GST antibodies (Santa Cruz Biotechnology) were also used to monitor the amount of GST or GST-fused proteins.
Co-purification Assays
For in vitro binding assays, cells were collected and broken as described (25) in lysis buffer lacking SDS and Nonidet P-40. Yeast extracts were incubated with glutathione-Sepharose beads for 2 h. Beads were washed extensively with the same buffer and resuspended in SDS loading buffer, and the proteins were analyzed by SDS-PAGE and immunoblotting as previously described (22).
Results
Generation of a Functional Analog-sensitive Slt2 Mutant Version
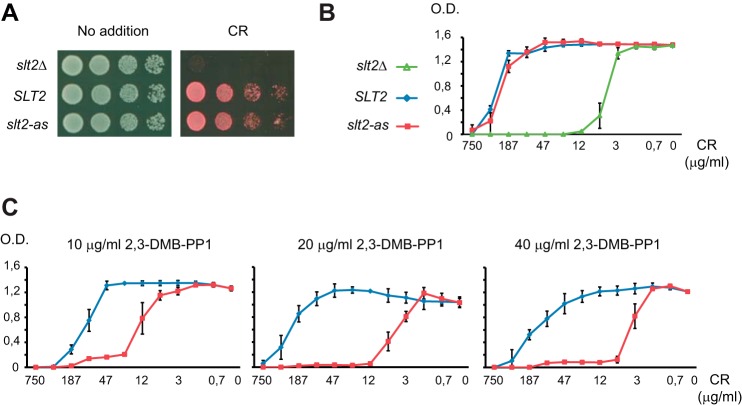
To generate an analog-sensitive version of Slt2, we mutated the gatekeeper Glu-108 residue within the ATP binding pocket of the MAPK to glycine according to the chemical-genetic approach developed by Shokat and co-workers (16). We then tested the ability of the slt2E108G mutant allele, expressed under the control of its own promoter from a centromeric plasmid, to complement the sensitivity of a slt2Δ strain to Congo red, a cell wall stressor compound known to stimulate the CWI pathway (34). As shown in Figs. 1, A and B, slt2Δ cells bearing Slt2E108G (hereinafter designated as Slt2-as) grew similarly to those expressing the wild-type Slt2 version under Congo red exposure. This result indicates that the Slt2-as mutant protein remained functional in the absence of kinase inhibitor analogs. We next tested the extent to which the slt2-as expressing cells were sensitive to Congo red in the presence of different concentrations of the pyrazolopyrimidine-based inhibitor 2,3-DMB-PP1 (17). As observed in Fig. 1C, treatment with 2,3-DMB-PP1 increased the Congo red sensitivity of the slt2-as expressing cells in a dose-dependent manner. The minimal inhibitory concentration of Congo red was lower on Slt2-as expressing cells than on cells expressing wild-type Slt2 at any of the tested concentrations of inhibitor, suggesting that this analog is able to greatly inhibit the catalytic activity of Slt2-as in vivo.
FIGURE 1.
Functionality of Slt2-as is inhibited by 2,3-DMB-PP1. A, 10-fold serial dilutions of the slt2Δ strain Y00993 transformed with the empty vector pRS316 (slt2Δ), pRS316-SLT2 (SLT2), or pRS316-slt2-as (slt2-as) were spotted onto YPD plates in the absence (No addition) or presence of Congo red (10 μg/ml) and incubated at 30 °C for 48 h. B, graphic showing the effect of distinct Congo red concentrations on growth in liquid medium after 48 h of the same transformants as in A. Results are the mean of three independent transformants. Error bars indicate S.D. C, graphic showing the effect of the indicated concentrations of 2,3-DMB-PP1 on growth of slt2Δ strain Y00993 transformed with plasmids pRS316-SLT2 (blue line) or pRS316-slt2-as (red line). Experiments were performed and data processed as in B.
We next assessed whether 2,3-DMB-PP1 effectively reduced the activation of the transcription factor Rlm1, which is a known Slt2 target. Phosphorylation of Rlm1 by Slt2 upon Congo red treatment triggers a wide transcriptional response that includes the induction of RLM1 gene expression (6). Therefore, Rlm1 activation can be monitored by a change in both its electrophoretic mobility and protein abundance (20). As shown in Fig. 2A, Rlm1 activation increased in the time in which slt2-as cells were exposed to Congo red in the absence of 2,3-DMB-PP1, whereas no significant activation was observed in the presence of the inhibitor. A progressive increase in the dual phosphorylation of Slt2 by its upstream MAPKKs occurred in such stimulating conditions, reaching a similar level in cells unexposed or exposed to this analog. According to these results, the lack of Rlm1 activation displayed by slt2-as cells is not due to a reduced ability of the Slt2-as mutant protein to become phosphorylated and thereby activated upon stimulation of the CWI pathway, but to the 2,3-DMB-PP1-dependent specific inhibition of the catalytic activity of Slt2-as. We also tested the induction of MLP1, a Rlm1-dependent gene that has been widely used as a typical transcriptional reporter of the CWI pathway (35). A comparative analysis of the MLP1-driven lacZ expression in slt2-as- and SLT2-bearing cells treated with Congo red confirmed that Rlm1 was fully activated in the absence of 2,3-DMB-PP1; however, the addition of this inhibitor significantly abrogated Rlm1 activation by Slt2-as upon CWI pathway stimulation (Fig. 2B).
FIGURE 2.
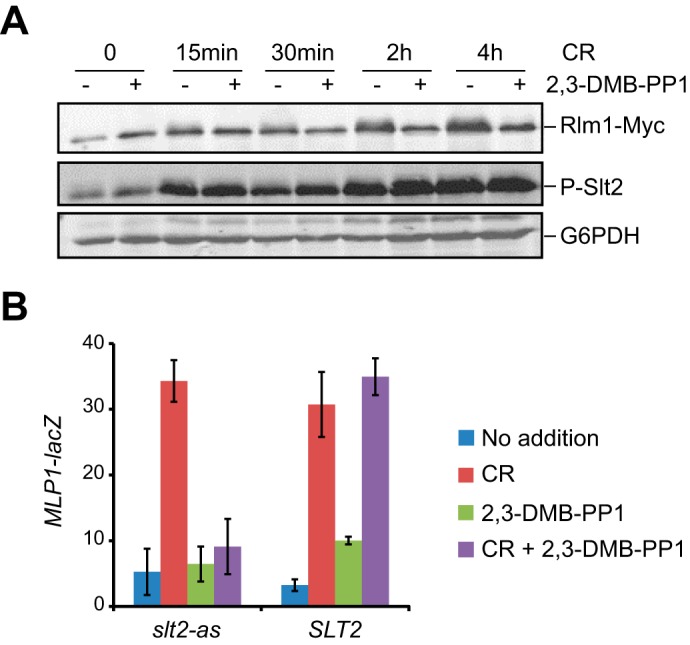
Rlm1 activation is inhibited by 2,3-DMB-PP1 in Slt2-as-expressing cells. A, Western blot analysis of extracts of YMM1 (slt2Δ RLM1::6MYC) cells transformed with pRS316-slt2-as treated with 30 μg/ml of Congo red for the indicated times in the presence (+) or absence (-) of 20 μm 2,3-DMB-PP1. Rlm1-Myc, phosphorylated Slt2, and G6PDH (as loading control) were detected with anti-Myc, anti-phospho-p44/42, and anti-G6PDH antibodies, respectively. B, β-galactosidase activity of cell extracts from Y00993 (slt2Δ) cells bearing pMLP1-LacZ and pRS316-SLT2 or pRS316-slt2-as. Cells were left untreated or treated for 4 h with 30 μg/ml Congo red and/or 20 μm 2,3-DMB-PP1 as indicated in the graphic. Data shown represent the average of β-galactosidase activity of three independent transformants. Error bars indicate S.D.
In combination, these results indicate that mutation of the gatekeeper Glu-108 residue at the ATP binding pocket to glycine generates a fully functional mutant version of Slt2 that is able to accommodate bulky inhibitor analogs, such as 2,3-DMB-PP1, which are not recognized by the wild-type kinase but specifically inhibit the catalytic activity of the Slt2-as version.
Specific Labeling of Rlm1 by Slt2-as with N6-(Phenethyl)-ATPγS
Due to the modified active-site pocket, analog-sensitive kinases can also accept unnatural bulky N6-substituted ATPγS analogs and transfer the thiophospho group to its substrates in kinase reactions. Treatment of the resulting thiophosphorylated proteins with a thiol-specific alkylating agent generates the corresponding thiophosphate ester that can be detected by immunoblotting with a specific anti-thiophosphate ester antibody (16). To ascertain if the Slt2-as kinase was able to thiophosphorylate its target proteins, we performed an in vitro kinase assay using activated GST-Slt2-as purified from Congo red-treated yeast cells, N6-(phenethyl)-ATPγS as an ATP analog, and recombinant GST-Rlm1 produced in E. coli as a known Slt2 substrate (5). Previously, we confirmed that fusion to GST did not reduce either the kinase activity of Slt2-as or its inhibition by 2,3-DMB-PP1, as assessed by monitoring Congo red-induced MLP1-lacZ expression in cells overexpressing GST-Slt2-as from a galactose-inducible promoter in the absence or presence of the inhibitor, respectively (Fig. 3A). Remarkably, GST-Slt2-as promoted higher MLP1 induction than the wild-type version. As shown in Fig. 3B, GST-Slt2-as was capable of using N6-(phenethyl)-ATPγS to thiophosphorylate GST-Rlm1 with high specificity. In the absence of the as-kinase, very low background labeling was observed, indicating minimal usage of the ATP analog by other possible yeast or bacterial kinases remaining after GST-based purification.
FIGURE 3.
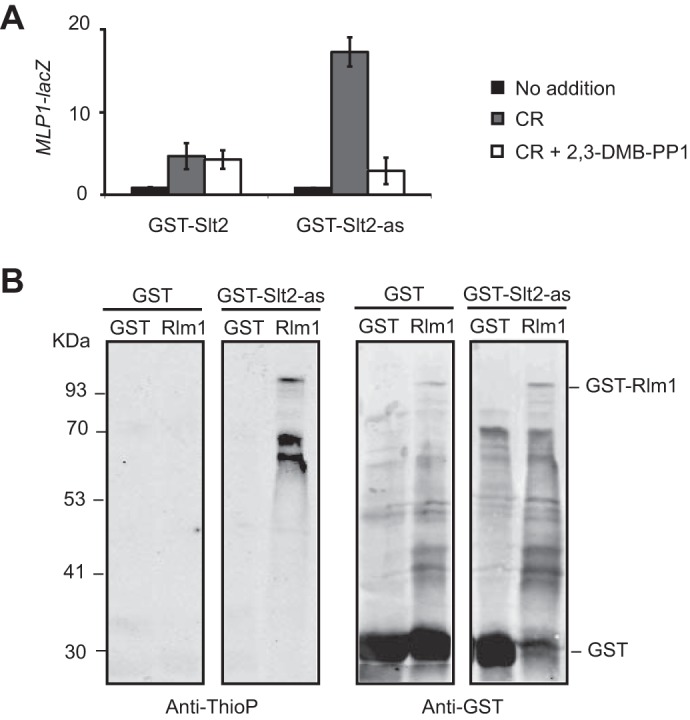
GST-Slt2-as thiophosphorylates recombinant GST-Rlm1. A, expression of MLP1-lacZ, determined as β-galactosidase activity of cell extracts from co-transformants carrying the plasmid pMLP1-LacZ and pEG(KG)-SLT2 or pEG(KG)-slt2-as. Cells were grown in synthetic galactose medium at 24 °C in the absence (No addition) or presence of Congo red (30 μg/ml) and 20 μm of 2,3-DMB-PP1 when indicated for 4 h. Data shown represent the average of β-galactosidase activity of three independent transformants. Error bars indicate S.D. B, in vitro kinase assay using GST or GST-Slt2-as purified from Congo red-treated yeast cells and E. coli extracts containing GST or GST-Rlm1. Anti-thiophosphate (Anti-ThioP) ester antibodies (left panel) and anti-GST (right panel) were used to detect thiophosphorylated proteins and GST or GST-Rlm1, respectively.
Slt2-as Thiophosphorylates the MAPK Phosphatase Msg5 Both at the Regulatory and the Catalytic Domains
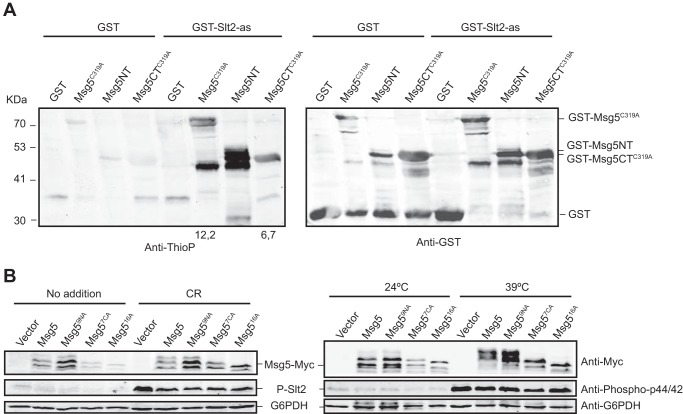
The dual specificity MAPK phosphatase Msg5 is another bona fide Slt2 substrate (22). Msg5 controls the CWI pathway by dephosphorylating Slt2, which in turn phosphorylates Msg5. As expected, a recombinant GST-tagged catalytically inactive version of Msg5 (Msg5C319A) was selectively thiophosphorylated by GST-Slt2-as in the kinase assay using N6-(phenethyl)-ATPγS, whereas only a very faint labeling was observed in the control reaction with GST (Fig. 4A). The use of an inactive phosphatase mutant version is needed to prevent both autodephosphorylation and dephosphorylation of the active kinase in vitro. This result confirms that Slt2 directly phosphorylates Msg5 and reinforces the value of this assay to detect Slt2 target proteins.
FIGURE 4.
GST-Slt2-as thiophosphorylates recombinant GST-Msg5 in both the N- and C-terminal domains. A, Western blot analysis of the kinase assay performed as in Fig. 3B but using as substrate E. coli extracts expressing the catalytically inactive version GST-Msg5C319A from plasmid pGEX-KG-msg5C319A or the two halves of this protein GST-Msg5NT (residues 1–227) and GST-Msg5CTC319A (residues 228–489) from plasmids pGEX-KG-msg5NT and pGEX-KG-msg5CTC319A, respectively. Anti-thiophosphate (Anti-ThioP) ester antibodies (left panel) and anti-GST (right panel) were used to detect thiophosphorylated proteins and GST or GST-Msg5, respectively. Numbers below the blot indicate the amount of thiophosphorylated GST-Msg5C319A and GST-Msg5TC319A in the kinase assay using GST-Slt2-as normalized with respect to their amount in the control kinase assay with GST. Due to signal saturation, the relative amount of thiophosphorylated GST-Msg5NT is not indicated. B, Western blot analysis of extracts from the mutant strain DD1–2D (msg5Δ) transformed with YCplac22 (vector), YCplac22-MSG5m, YCplac22-msg5N9Am, YCplac22-msg5C7Am, or YCplac22-msg516Am, growing at 24 °C under the absence (No addition) or presence of Congo red (30 μg/ml) for 3 h (left panels) or at 39 °C for 1 h (right panels) as indicated. Phosphorylated Slt2, Myc-tagged proteins, and G6PDH (as loading control) were detected with anti-phospho-p44/42, anti-Myc, and anti-G6PDH antibodies, respectively.
We also wanted to know whether this phosphorylation occurred in the N-terminal regulatory or the C-terminal catalytic regions of Msg5. To this end, we separately expressed the two halves of this protein in E. coli and used them as substrates in the kinase reaction. As shown in Fig. 4A, both Msg5 regions were thiophosphorylated by Slt2-as. In line with this result, analysis of the Msg5 amino acid sequence revealed the existence of 14 ((S/T)P) putative MAPK-phosphorylation sites (36), 9 in the N-terminal region (Ser-22, Ser-49, Ser-62, Thr-64, Ser-85, Thr-89, Ser-115, Ser-135, Thr-178), and five in the C-terminal domain (Ser-357, Ser-377, Ser-422, Thr-434, Thr-437).
We previously reported how the phosphorylation of Msg5 by Slt2 in vivo under cell wall stress conditions resulted in an SDS-PAGE mobility shift (22). We wanted to know which Msg5 domain is responsible for this mobility shift. Therefore, we investigated the effect of mutating all the putative MAPK phosphorylation sites in each half of the protein on the electrophoretic behavior of Msg5 upon treatment with Congo red (Fig. 4B, left panels) or at a temperature of 39 °C (Fig. 4B, right panels), two well known stimuli of the CWI pathway (20, 22). To this end, we obtained Mgs59NA (with the nine proline-directed S/T sites at the N-terminal part changed to alanine), Msg57CA (bearing the five (S/T)P sites at the C-terminal region as well as the Ser-420 and Ser-421 previous to the Ser-422 site mutated to alanine), and the Mgs516A mutant version containing all these substitutions. Changing the nine S/T residues at the N-terminal region did not alter the mobility shift after treatment either with Congo red or when treated at a temperature of 39 °C. In contrast, substitution of the seven S/T residues at the C-terminal domain strongly reduced the appearance of the lower mobility phosphorylated bands in both conditions, as also occurred in the case of the Mgs516A mutant version (Fig. 4B).
Therefore, although both Msg5 regions can be phosphorylated by Slt2, only phosphorylation at the C-terminal domain is responsible for the observed electrophoretic mobility shift. Strikingly, the loss of all the putative MAPK phosphorylation sites in Msg5 did not seem to alter the ability of this phosphatase to dephosphorylate Slt2. As shown in Fig. 4B, the high phospho-Slt2 levels displayed by an msg5Δ mutant, both in the absence or presence of the different stimuli, were reduced to the same extent by expression of Msg5 or any of the mutant Mgs59NA, Mgs57CA, and Mgs516A proteins.
Identification of Rcn2, Caf20, and Gga1 as Slt2 Substrates
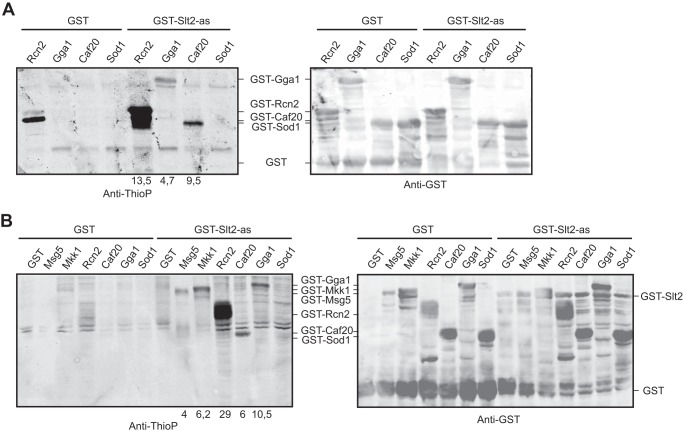
By using a quantitative SILAC (stable isotope labeling by amino acids in cell culture)-based phosphoproteomic approach, we previously identified 43 proteins containing phosphopeptides that display enhanced phosphorylation upon CWI pathway stimulation triggered by Pkc1 hyperactivation (18). Among them, 25 proteins displayed differentially phosphorylated (S/T)P residues, constituting putative MAPK targets. Therefore, we decided to test whether some of these proteins were direct Slt2 substrates by using the above validated Slt2-as-based thiophosphorylation in vitro kinase assay on 13 candidate target proteins produced in E. coli. Three of them, Rcn2, Gga1, and Caf20, were thiophosphorylated by Slt2-as (Fig. 5A), whereas the other 10, namely Sod1, Cdc33, Fba1, Ipp1, Pfk2, Pil1, Pst2, Pup2, Rnr4, and Tup1, did not display specific labeling in the Slt2-as kinase assay over the control assay using GST. Curiously, a protein likely corresponding to a degradation product of Rcn2 appeared thiophosphorylated in the absence of Gst-Slt2as; this was probably due to the activity of a kinase from E. coli (Fig. 5A).
FIGURE 5.
Identification of novel Slt2 phosphorylation targets. A, Western blot analysis of the in vitro kinase assay carried out as in Fig. 3B but using E. coli cell extracts containing recombinant GST-Rcn2, GST-Gga1, GST-Caf20, or GST-Sod1 as substrate. B, Western blot analysis of the in vitro kinase assay similar to that described in Fig. 3B but using yeast extracts containing GST, GST-Msg5, GST-Mkk1, GST-Rcn2, GST-Gga1, GST-Caf20, or GST-Sod1 as substrate. Anti-thiophosphate (Anti-ThioP) ester antibodies (right panels) and anti-GST (left panels) were used to detect thiophosphorylated proteins and GST-tagged proteins, respectively. Numbers below the blot indicate the amount of thiophosphorylated GST-Msg5, GST-Mkk1, GST-Rcn2, GST-Caf20, and GST-Gga1 in the kinase assay using GST-Slt2-as normalized with respect to their amount in the control kinase assay with GST.
We next explored the ability of Slt2-as to thiophosphorylate these proteins when expressed in yeast. To this end, we made use of a collection of plasmids bearing yeast genes encoding proteins N-terminally fused to GST under the control of the GAL1 promoter. Whole cell extracts from yeast expressing the proteins tested above, except Fba1, Cdc33, and Tup1, which were not present in the plasmid collection, were used in the in vitro kinase assay. GST-Slt2-as was able to selectively thiophosphorylate Rcn2, Gga1, and Caf20 (Fig. 5B), but it did not have the same effect on any of the other proteins tested. As expected, Gst-Slt2-as also thiophosphorylated the known Slt2 targets Msg5 and Mkk1, supporting the utility of the assay using yeast cell extracts for finding Slt2 substrates.
Co-purification assays of GST-fused Rcn2, Gga1, and Caf20 indicated that these proteins physically interact with Slt2-Myc in yeast cells (Fig. 6), reinforcing the idea that they are bona fide Slt2 substrates. Treatment of cells with Congo red did not modify the binding of any of these proteins with Slt2, suggesting that phosphorylation is not regulating their interaction with this MAPK.
FIGURE 6.
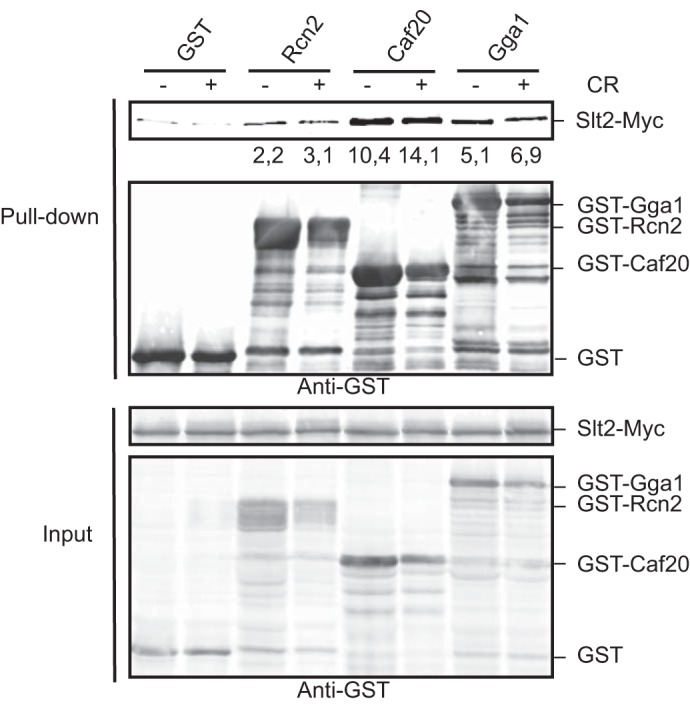
Rcn2, Caf20, and Gga1 physically interact with Slt2. Western blot analysis of the in vitro co-purification assays. The strain YMF3 (SLT2::6MYC) was transformed with plasmids expressing Rcn2, Caf20, or Gga1 fused to GST under control of the GAL1 promoter. Transformants were grown for 2 h in the presence of galactose and then Congo red for a final 30 μg/ml was added (+) or not (−), and cultures were incubated for a four additional hours. Cell extracts (input) were incubated with glutathione-Sepharose beads to purify GST-complexes (pull-down), and immunodetection was performed with anti-Myc and anti-GST antibodies that recognize Slt2-Myc and GST-fused proteins as indicated. Similar results were obtained in three different experiments, and selected images correspond to representative blots. Numbers below the anti-Myc blot indicate the amount of Slt2-Myc pulled down by GST-Rcn2, GST-Caf20, and GST-Gga1 normalized with respect to the amount pulled down by GST, both in the absence (−) or presence (+) of Congo red.
Identification of Slt2 Phosphorylation Sites in Caf20 and Rcn2
We analyzed the amino acid sequence of the proteins identified as Slt2 substrates to find ((S/T)P) putative MAPK-phosphorylation sites. Caf20 contains only one site, corresponding to Thr-102, which was found up-phosphorylated in our previous phosphoproteomic analysis (18). Rcn2 includes four potential sites at Thr-49, Ser-152, Ser-160 and Ser-255, whereas Gga1 comprises seven of these sites along the protein. We decided to change the putative phosphorylatable residue Thr-102 of Caf20 to alanine and perform a kinase assay on the mutated substrate produced in E. coli. As shown in Fig. 7A, the Caf20T102A mutant protein was not thiophosphorylated by Slt2-as, indicating that Thr-102 is the Caf20 phosphorylation site for the MAPK of the CWI pathway. In the case of Rcn2, each individual mutation at Ser-152, Ser-160, or Ser-255 reduced the thiophosphorylation of this protein by Slt2-as (Fig. 7B), but the most intense effect was observed when Ser-255, the only of these Rcn2 phosphosites previously detected as up-phosphorylated (18), was mutated. However, labeling was totally lost only in the mutant protein with all three residues substituted by alanine (Fig. 7B). This experiment confirmed that Rcn2 is a direct Slt2 substrate and revealed these three serines as phosphorylation sites in this protein, validating the Slt2-as kinase assay as a useful strategy for the identification of residues phosphorylated by this MAPK on its substrates.
FIGURE 7.
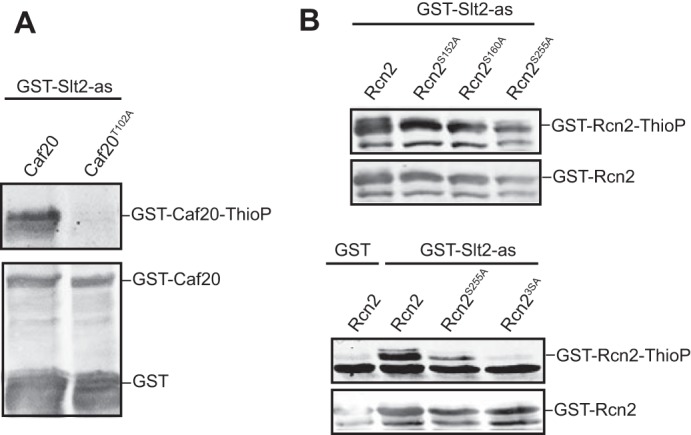
Determination of Slt2 phosphorylation sites on Caf20 and Rcn2. Western blot analysis of the in vitro kinase assay similar to Fig. 3B but with E. coli extracts containing recombinant GST-Caf20 and GST-Caf20T102A (A) or GST-Rcn2, GST-Rcn2S152A, GST-Rcn2S160A, GST-Rcn2S255A, and GST-Rcn23S-A (B) as substrates. Anti-thiophosphate ester antibodies (upper panels) and anti-GST (lower panels) were used to detect thiophosphorylated proteins and GST-tagged proteins respectively.
Gga1 and Gga2 Adaptors Contribute to Cell Wall Integrity
We wanted to know if any of these three Slt2 substrates participate in the CWI response. CAF20 codes for a 4E-BP (translation initiation factor eIF4E-binding protein) that acts as a cap-dependent translational repressor (37). Deletion of this gene did not increase the sensitivity of either wild-type or slt2Δ cells to cell wall altering agents (Congo red, zymolyase, caffeine, SDS). Rcn2 as well as Rcn1 belongs to the RCAN (regulator of the Ca2+ and calmodulin-dependent serine-threonine phosphatase calcineurin) conserved family of calcineurin regulators that seems to perform a complex regulatory role (38). Neither the single deletion of RCN1 or RCN2 nor the combination of both mutations led to a cell wall stress-sensitive phenotype. Furthermore, removal of any of these genes in an slt2Δ strain did not increase the sensitivity of this mutant to cell wall perturbing agents.
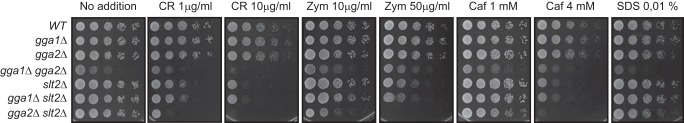
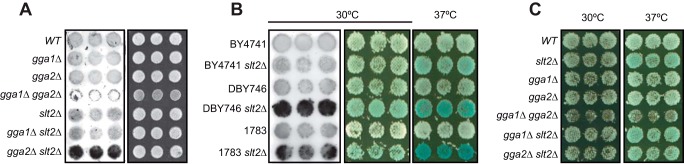
Gga1 is a Golgi-associated, γ-adaptin ear-containing, ARF-binding protein (GGA). GGA proteins load transmembrane protein cargos into transport vesicles and recruit clathrin and accessory proteins to multiprotein complexes on the nascent vesicles, regulating membrane traffic (39). GGA2 is a GGA1 paralog that also codes for a member of this family of adaptors. As observed in Fig. 8, single gga1Δ and gga2Δ mutants displayed no increased sensitivity to cell wall-altering agents compared with the wild-type strain. However, double gga1Δ gga2Δ mutants grew weakly and showed sensitivity to these compounds. Furthermore, in contrast to double gga1Δ slt2Δ mutants, gga2Δ slt2Δ cells presented a strong conditional negative genetic interaction under cell wall stress conditions (Fig. 8).
FIGURE 8.
Genetic interaction of gga1Δ, gga2Δ, and slt2Δ mutations in the absence and presence of cell wall stress. A, 10-fold serial dilutions of the BY4741 cell and the isogenic mutants strains gga1Δ, gga2Δ, gga1Δ gga2Δ, slt2Δ, gga1Δ slt2Δ, and gga2Δ slt2Δ. The cells were spotted onto YPD plates in the absence (No addition) or presence of Congo red (1 and 10 μg/ml), zymolyase (10 and 50 μg/ml), caffeine (1 and 4 mm), and SDS (0.01%) and incubated at 30 °C for 48 h.
Cells lacking both GGAs are defective for transport of yeast CPY to the vacuole and missort CPY to the periplasmic space (40). Therefore, we explored the involvement of Slt2 in trafficking through the CPY pathway by analyzing the secretion of CPY in single gga1Δ, gga2Δ, and slt2Δ mutants and the different double mutants resulting from the combination of these gene deletions. Colony blotting assays revealed a high secretion of CPY in double gga2Δ slt2Δ cells growing at 30 °C (Fig. 9A). The possibility of an increased level of extracellular CPY in this mutant due to cell lysis was ruled out through the analysis of the release of intracellular alkaline phosphatase (AP) to the medium by using the chromogenic AP substrate BCIP. Different levels of BCIP hydrolysis were observed in slt2Δ mutants from different genetic backgrounds, mainly when cells were incubated at 37 °C (Fig. 9B). Interestingly, the lowest AP release was observed in the BY4741 genetic background, which is the same background used to obtain the ggaΔ mutants. As shown in Fig. 9C, the double gga2Δ slt2Δ mutant did not show a significant release of AP to the extracellular medium in the conditions used for the CPY assay.
FIGURE 9.
Carboxypeptidase (CPY) secretion and alkaline phosphatase (AP) release of gga1Δ, gga2Δ, and slt2Δ mutants. A, patches of the BY4741 wild-type strain (WT) and the isogenic mutants gga1Δ, gga2Δ, gga1Δ gga2Δ, slt2Δ, gga1Δ slt2Δ, and gga2Δ slt2Δ were grown on YPD medium (right panel) and blotted onto a nitrocellulose membrane followed by immunodetection of the membrane with anti-CPY antibodies (left panel). B, dot blotting analysis of CPY secretion (black and white photograph) performed as in A and AP release detection in the indicated wild-type and isogenic slt2Δ strains at 30 °C and 37 °C (color photographs). For AP analysis, the patches were replicated to YPD supplemented with BCIP and incubated at 30 °C or 37 °C for 24 h. C, AP release analysis, performed as in B, in the same strains as in A.
Discussion
One of most powerful tools for functional studies of protein kinases is the use of analog-sensitive (as) versions, generated by mutating a natural bulky amino acid to smaller glycine or alanine at structurally conserved position in the active site. This modification allows the as-kinase to be potently and specifically targeted by inhibitor analogs with a bulky substituent complementing the enlarged ATP binding pocket. To be amenable to this chemical-genetic approach, the kinase of interest must fulfill two criteria. First, the kinase must tolerate the mutation of the gatekeeper residue without severe loss of the catalytic activity. Second, a potent inhibitor of the as-kinase must be identified (17). To obtain Slt2-as, we substituted the glutamic acid in position 108 with glycine. Slt2-as is fully active, as determined by its ability to phosphorylate and activate Rlm1, to drive Rlm1-dependent transcriptional activation, and to complement the slt2Δ mutant phenotype to the same extent as the wild-type kinase. In addition, we found that low concentrations of the PP1 analog 2,3-DMB-PP1 robustly inhibits Slt2-as. Therefore, Slt2-as satisfies the criteria to be a bona fide analog-sensitive kinase. Because PP1-analogs are stable, reversible, and cell-permeable inhibitors, Slt2-as can be considered as a valuable new research tool for the functional analysis of the CWI pathway.
As-kinases have also emerged as an efficient tool for identifying kinase substrates (41). Because a major goal of our laboratory was to identify novel targets of the MAPK Slt2, we focused on the use of Slt2-as for this purpose. Here, we first proved that Slt2-as was capable of using bulky ATPγS analogs to thiophosphorylate known substrates like the transcription factor Rlm1 and the MAPKK Mkk1. Using this methodology, we also gained insight into the phosphorylation of the MAPK phosphatase Msg5 by Slt2. Although Slt2-as thiophosphorylates in vitro both the N-terminal regulatory and the C-terminal phosphatase domains of MAPK phosphatase Msg5, only phosphorylation of the C-terminal region of the protein is responsible for the electrophoretic mobility shift we previously reported to occur in Msg5 upon CWI pathway activation (22). Mammalian MAPK phosphatase MKP1 has been shown to be phosphorylated by ERK1/2 in different positions. Strikingly, whereas ERK-mediated phosphorylation of the C-terminal domain of this phosphatase resulted in increased protein stability, phosphorylation of other positions, like Ser-296, led to the recruitment of a specific ubiquitin ligase, increasing the rate at which MKP1 is degraded (42, 43). However, we did not find significant differences in protein amount and functionality between the wild-type and any of the Msg5 versions lacking the putative MAPK phosphorylation sites either at the N terminus, the C terminus, or both parts of the phosphatase. This suggests a subtle and likely complex regulation of this phosphatase by Slt2.
In addition, our labeling experiments using Slt2-as allowed us to confirm Gga1, Caf20, and Rcn2 to be direct substrates of Slt2 among a number of putative candidates that were identified in our previous phosphoproteomic analysis (18). Although thiophosphorylation-based approaches have been used in other cell types, to our knowledge this is the first evidence to indicate that this methodology can be successfully applied to identify yeast MAPK substrates. In fact, whereas not all as-kinases efficiently use ATPγS (44), Slt2-as readily in vitro thiophosphorylates its protein targets, both those produced as recombinant proteins in E. coli and those expressed in yeast and assayed in whole cell extracts. It is interesting to note that mutation of the gatekeeper residue of the homologue MAPK Erk2 leads to autoactivation due to enhanced autophosphorylation (45). This effect may also be operating in Slt2. In fact, especially when fused to GST, Slt2-as displays higher activity than wild-type Slt2, as determined by MLP1-LacZ reporter assays. This provides an additional advantage to kinase assays in which GST-Slt2-as is used.
Caf20 is a yeast 4E-BP translation repressor protein that interacts with the translation initiation factor eIF4E to hinder ribosome recruitment to the 5′ end of mRNAs. In mammalian cells, phosphorylation of specific serine and threonine residues modulates the affinity of 4E-BPs for eIF4E. For example, it has been clearly established that phosphorylation of 4E-BP1 by mTOR abrogates this interaction and promotes translation (46, 47). Interestingly, phosphorylation of 4E-BP1 by ERKs has been also reported (48). In yeast, translational regulation after mild hyperosmotic shock has been found to be extensively dependent on the MAPK Hog1 (49). However, the molecular mechanisms that regulate this process remain largely unknown. Our results, which demonstrate a physical interaction between Slt2 and Caf20 together with the identification of Thr-102 as a Slt2 direct phosphorylation site in this 4E-BP protein, provide new clues that can facilitate further investigation into how MAPKs modulate translation in response to distinct stresses in yeast.
Our findings also suggest that regulators of the Ca2+ and calmodulin-dependent serine-threonine phosphatase calcineurin (RCANs) could serve as a nexus for coupling the CWI pathway to calcium signaling. Slt2 has been reported to activate the calcineurin-mediated pathway in response to endoplasmic reticulum stress independently on known Slt2 targets (13). In addition, the calcineurin-regulated transcription factor Crz1 is known to induce the expression of several cell wall-related genes involved in the cell wall compensatory response (6, 50, 51). In mammals and fungi, the binding of RCANs to calcineurin can either stimulate or inhibit calcineurin signaling (52). In yeast, Rcn1 inhibits calcineurin-dependent responses when overexpressed (53) but stimulates calcineurin signaling after its phosphorylation by Mck1, a member of the GSK-3 family of protein kinases (54). Rcn2 has been identified as another yeast RCAN whose expression was strongly induced by calcineurin signaling (38); however, much less is known about its regulatory role. Here, we show that Slt2 binds Rcn2, confirming previous data obtained in a large scale approach (56), and identify the Rcn2 residues phosphorylated by this MAPK. These results are likely to help further efforts to unravel the complex interplay between calcium signaling and cell wall integrity.
Our genetic studies with GGA genes connect the CWI pathway with trafficking from the Golgi complex to the vacuole, in particular with the CPY pathway. The transport through this pathway is mediated by clathrin-coated vesicles (CCVs). GGA proteins and the AP-1 (adaptor protein-1) complex are two major clathrin adaptors that act at this level (57). Here, we show that Gga2 becomes essential to cope with cell wall stress when Slt2 is absent. This fits with the idea that Slt2 modulates the functionality of Gga1 by phosphorylation, making Gga2 essential when Gga1 is not properly regulated. Phosphorylation of GGA proteins have been shown to occur in mammalian cells (58). However, although it was widely accepted that interaction with cargos is regulated by GGA phosphorylation (59, 60), this regulatory mechanism was later put into question (61). We previously found the two putative MAPK phosphorylation sites Ser-375 and Ser-378 to be phosphorylated under the CWI pathway activating conditions (18). These two sites have been also found phosphorylated in another large scale analysis (62). Interestingly, these authors also found Gga1 to be ubiquitinated, suggesting a complex regulation of the functionality of this adaptor.
Our results suggest that a proper regulation of trafficking to the vacuole is important for cell wall integrity. The CPY pathway sorts vacuolar proteins including CPY, proteinase A (PrA) and subunits of the vacuolar ATPase. Of interest, mutations in genes affecting components of this ATPase complex render cells hypersensitive to cell wall stress (63). Furthermore, the intracellular pool of chitin synthase 3 (Chs3) is maintained by the cycling of this protein between the TGN and endosomes, dependent on the action of the clathrin adaptors AP-1 (adaptor protein-1), Gga1, and Gga2 and the epsin-related proteins Ent3 and Ent5 (64).
Interestingly, we have recently reported that mutants affected in another cargo adaptor, AP-2 (65), or in the conserved oligomeric Golgi (COG) complex (55), display negative genetic interactions with CWI mutants (19). It is very tempting to speculate that the CWI pathway could modulate vesicular trafficking at different levels. This regulation might be important in the adaptation to cell wall stress.
To summarize, we have generated a new powerful tool useful to identify substrates of the MAPK Slt2 and selectively inhibit its kinase activity. This will be of significant benefit to future functional studies of the CWI pathway. Further characterization of the identified target proteins will undoubtedly broaden our understanding about the connections of this signaling route to additional cellular processes.
Author Contributions
E. A.-R. conducted most of the experiments and analyzed the results, P. F.-P. initially characterized the Slt2-as version, developed the kinase assay, and conducted the experiments with recombinant E. coli proteins, A. S.-R. conducted the Msg5 phosphorylation analysis experiments, M. M. analyzed the results and wrote the paper, H. M. conceived the idea for the project, made the Slt2-as mutant, analyzed the results, and wrote the paper with M. M.
Acknowledgments
We thank Chao Zhang, Dorothea Fiedler, and Kevan M. Shokat for help in designing the analog-sensitive version of Slt2 and providing inhibitors and Javier Arroyo for materials used in this study. We also thank the people from Unit 3 of the Departamento de Microbiología II at UCM for useful comments and discussion throughout the work. We acknowledge the Servicio de Genómica y Proteómica (UCM, Madrid, Spain) for DNA sequencing.
This work was supported by Grants BIO2010-22369-C02-01 and BIO2013-44112-P from Ministerio de Ciencia e Innovación and Ministerio de Economía y Competitividad (Spain), respectively, and S2011/BMD-2414 from Comunidad Autónoma de Madrid (Spain). The authors declare that they have no conflicts of interest with the contents of this article.
- MAP
- mitogen-activated protein
- CWI
- cell wall integrity
- ATPγS
- adenosine 5′-[γ-thio]triphosphate
- as
- analog-sensitive
- CR
- Congo red
- 2,3-DMB-PP1
- 4-Amino-1-tert-butyl-3-(2,3-dimethylbenzyl)pyrazolo[3,4-d]pyrimidine
- BCIP
- 5-bromo-4-chloro-3-indolyl-phosphate
- G6PDH
- glucose-6-phosphate dehydrogenase
- CPY
- carboxypeptidase Y
- 4E-BP
- translation initiation factor eIF4E-binding protein
- GGA
- Golgi-associated, γ-adaptin ear-containing, ARF-binding protein
- RCAN
- regulator of the Ca2+ and calmodulin-dependent serine-threonine phosphatase calcineurin
- AP
- alkaline phosphatase.
References
- 1. Qi M., and Elion E. A. (2005) MAP kinase pathways. J. Cell Sci. 118, 3569–3572 [DOI] [PubMed] [Google Scholar]
- 2. Ubersax J. A., and Ferrell J. E. Jr. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 3. Chen R. E., and Thorner J. (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773, 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Engelberg D., Perlman R., and Levitzki A. (2014) Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: state of the art after 25 years. Cell Signal. 26, 2865–2878 [DOI] [PubMed] [Google Scholar]
- 5. Jung U. S., Sobering A. K., Romeo M. J., and Levin D. E. (2002) Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46, 781–789 [DOI] [PubMed] [Google Scholar]
- 6. García R., Bermejo C., Grau C., Pérez R., Rodríguez-Peña J. M., Francois J., Nombela C., and Arroyo J. (2004) The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279, 15183–15195 [DOI] [PubMed] [Google Scholar]
- 7. Madden K., Sheu Y. J., Baetz K., Andrews B., and Snyder M. (1997) SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275, 1781–1784 [DOI] [PubMed] [Google Scholar]
- 8. Ray A., Hector R. E., Roy N., Song J. H., Berkner K. L., and Runge K. W. (2003) Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33, 522–526 [DOI] [PubMed] [Google Scholar]
- 9. Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., and Hall M. N. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin C., Strich R., and Cooper K. F. (2014) Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress. Mol. Biol. Cell 25, 1396–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Molina M., Cid V. J., and Martín H. (2010) Fine regulation of Saccharomyces cerevisiae MAPK pathways by post-translational modifications. Yeast 27, 503–511 [DOI] [PubMed] [Google Scholar]
- 12. Mao K., and Klionsky D. J. (2011) MAPKs regulate mitophagy in Saccharomyces cerevisiae. Autophagy 7, 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonilla M., and Cunningham K. W. (2003) Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell. 14, 4296–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thingholm T. E., Jensen O. N., and Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 15. Bishop A., Buzko O., Heyeck-Dumas S., Jung I., Kraybill B., Liu Y., Shah K., Ulrich S., Witucki L., Yang F., Zhang C., and Shokat K. M. (2000) Unnatural ligands for engineered proteins: new tools for chemical genetics. Annu. Rev. Biophys. Biomol. Struct. 29, 577–606 [DOI] [PubMed] [Google Scholar]
- 16. Hertz N. T., Wang B. T., Allen J. J., Zhang C., Dar A. C., Burlingame A. L., and Shokat K. M. (2010) Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr. Protoc. Chem. Biol. 2, 15–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang C., Lopez M. S., Dar A. C., Ladow E., Finkbeiner S., Yun C. H., Eck M. J., and Shokat K. M. (2013) Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology. ACS Chem. Biol. 8, 1931–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mascaraque V., Hernáez M. L., Jiménez-Sánchez M., Hansen R., Gil C., Martín H., Cid V. J., and Molina M. (2013) Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol. Cell Proteomics 12, 557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin H., Shales M., Fernandez-Piñar P., Wei P., Molina M., Fiedler D., Shokat K. M., Beltrao P., Lim W., and Krogan N. J. (2015) Differential genetic interactions of yeast stress response MAPK pathways. Mol. Syst. Biol. 11, 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marín M. J., Flández M., Bermejo C., Arroyo J., Martín H., and Molina M. (2009) Different modulation of the outputs of yeast MAPK-mediated pathways by distinct stimuli and isoforms of the dual-specificity phosphatase Msg5. Mol. Genet. Genomics 281, 345–359 [DOI] [PubMed] [Google Scholar]
- 21. Doi K., Gartner A., Ammerer G., Errede B., Shinkawa H., Sugimoto K., and Matsumoto K. (1994) MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flández M., Cosano I. C., Nombela C., Martín H., and Molina M. (2004) Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 279, 11027–11034 [DOI] [PubMed] [Google Scholar]
- 23. Torres L., Martín H., García-Saez M. I., Arroyo J., Molina M., Sánchez M., and Nombela C. (1991) A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol. Microbiol. 5, 2845–2854 [DOI] [PubMed] [Google Scholar]
- 24. Lee K. S., Irie K., Gotoh Y., Watanabe Y., Araki H., Nishida E., Matsumoto K., and Levin D. E (1993) A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell Biol. 13, 3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martín H., Rodríguez-Pachón J. M., Ruiz C., Nombela C., and Molina M. (2000) Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 1511–1519 [DOI] [PubMed] [Google Scholar]
- 26. Martín H., Arroyo J., Sánchez M., Molina M., and Nombela C. (1993) Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees. Mol. Gen. Genet. 241, 177–184 [DOI] [PubMed] [Google Scholar]
- 27. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitchell D. A., Marshall T. K., and Deschenes R. J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9, 715–722 [DOI] [PubMed] [Google Scholar]
- 29. Gietz R. D., and Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 30. Guan K. L., and Dixon J. E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267 [DOI] [PubMed] [Google Scholar]
- 31. Sanz A. B., García R., Rodríguez-Peña J. M., Díez-Muñiz S., Nombela C., Peterson C. L., and Arroyo J. (2012) Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity MAPK pathway. Mol. Biol. Cell 23, 2805–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. García R., Rodríguez-Peña J. M., Bermejo C., Nombela C., and Arroyo J. (2009) The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 284, 10901–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang W., and Malcolm B. A. (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions, and insertions using QuikChange site-directed mutagenesis. BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 34. de Nobel H., Ruiz C., Martin H., Morris W., Brul S., Molina M., and Klis F. M. (2000) Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase, resistance, and thermotolerance. Microbiology 146, 2121–2132 [DOI] [PubMed] [Google Scholar]
- 35. Arias P., Díez-Muñiz S., García R., Nombela C., Rodríguez-Peña J. M., and Arroyo J. (2011) Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics 12, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mok J., Kim P. M., Lam H. Y., Piccirillo S., Zhou X., Jeschke G. R., Sheridan D. L., Parker S. A., Desai V., Jwa M., Cameroni E., Niu H., Good M., Remenyi A., Ma J. L., Sheu Y. J., Sassi H. E., Sopko R., Chan C. S., De Virgilio C., Hollingsworth N. M., Lim W. A., Stern D. F., Stillman B., Andrews B. J., Gerstein M. B., Snyder M., and Turk B. E. (2010) Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3, ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altmann M., Schmitz N., Berset C., and Trachsel H. (1997) A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 16, 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mehta S., Li H., Hogan P. G., and Cunningham K. W. (2009) Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol. Cell. Biol. 29, 2777–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller G. J., Mattera R., Bonifacino J. S., and Hurley J. H. (2003) Recognition of accessory protein motifs by the γ-adaptin ear domain of GGA3. Nat. Struct. Biol. 10, 599–606 [DOI] [PubMed] [Google Scholar]
- 40. Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., and Bonifacino J. S. (2000) GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koch A., and Hauf S. (2010) Strategies for the identification of kinase substrates using analog-sensitive kinases. Eur. J. Cell Biol. 89, 184–193 [DOI] [PubMed] [Google Scholar]
- 42. Lin Y. W., and Yang J. L. (2006) Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. J. Biol. Chem. 281, 915–926 [DOI] [PubMed] [Google Scholar]
- 43. Caunt C. J., and Keyse S. M. (2013) Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS J. 280, 489–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Allen J. J., Li M., Brinkworth C. S., Paulson J. L., Wang D., Hübner A., Chou W. H., Davis R. J., Burlingame A. L., Messing R. O., Katayama C. D., Hedrick S. M., and Shokat K. M. (2007) A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emrick M. A., Lee T., Starkey P. J., Mumby M. C., Resing K. A., and Ahn N. G. (2006) The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. U.S.A. 103, 18101–18106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gingras A. C., Raught B., and Sonenberg N. (2001) Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15, 807–826 [DOI] [PubMed] [Google Scholar]
- 47. Richter J. D., and Sonenberg N. (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 [DOI] [PubMed] [Google Scholar]
- 48. Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., Sonenberg N., and Lawrence J. C. Jr. (1994) PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266, 653–656 [DOI] [PubMed] [Google Scholar]
- 49. Warringer J., Hult M., Regot S., Posas F., and Sunnerhagen P. (2010) The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol. Biol. Cell 21, 3080–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stathopoulos A. M., and Cyert M. S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11, 3432–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., and Levin D. E. (1998) Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davies K. J., Ermak G., Rothermel B. A., Pritchard M., Heitman J., Ahnn J., Henrique-Silva F., Crawford D., Canaider S., Strippoli P., Carinci P., Min K. T., Fox D. S., Cunningham K. W., Bassel-Duby R., Olson E. N., Zhang Z., Williams R. S., Gerber H. P., Pérez-Riba M., Seo H., Cao X., Klee C. B., Redondo J. M., Maltais L. J., Bruford E. A., Povey S., Molkentin J. D., McKeon F. D., Duh E. J., Crabtree G. R., Cyert M. S., de la Luna S., and Estivill X. (2007) Renaming the DSCR1/Adapt78 gene family as RCAN: regulators of calcineurin. FASEB J. 21, 3023–3028 [DOI] [PubMed] [Google Scholar]
- 53. Kingsbury T. J., and Cunningham K. W. (2000) A conserved family of calcineurin regulators. Genes Dev. 14, 1595–1604 [PMC free article] [PubMed] [Google Scholar]
- 54. Hilioti Z., Gallagher D. A., Low-Nam S. T., Ramaswamy P., Gajer P., Kingsbury T. J., Birchwood C. J., Levchenko A., and Cunningham K. W. (2004) GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev. 18, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Willett R., Kudlyk T., Pokrovskaya I., Schönherr R., Ungar D., Duden R., and Lupashin V. (2013) COG complexes form spatial landmarks for distinct SNARE complexes. Nat. Commun. 4, 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., and Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 57. Bonifacino J. S. (2004) The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5, 23–32 [DOI] [PubMed] [Google Scholar]
- 58. McKay M. M., and Kahn R. A. (2004) Multiple phosphorylation events regulate the subcellular localization of GGA1. Traffic 5, 102–116 [DOI] [PubMed] [Google Scholar]
- 59. Ghosh P., and Kornfeld S. (2003) Phosphorylation-induced conformational changes regulate GGAs 1 and 3 function at the trans-Golgi network. J. Biol. Chem. 278, 14543–14549 [DOI] [PubMed] [Google Scholar]
- 60. Doray B., Bruns K., Ghosh P., and Kornfeld S. A. (2002) Autoinhibition of the ligand-binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc. Natl. Acad. Sci. U.S.A. 99, 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cramer J. F., Gustafsen C., Behrens M. A., Oliveira C. L., Pedersen J. S., Madsen P., Petersen C. M., and Thirup S. S. (2010) GGA autoinhibition revisited. Traffic 11, 259–273 [DOI] [PubMed] [Google Scholar]
- 62. Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N. J., and Villén J. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. García R., Botet J., Rodríguez-Peña J. M., Bermejo C., Ribas J. C., Revuelta J. L., Nombela C., and Arroyo J. (2015) Genomic profiling of fungal cell wall-interfering compounds: identification of a common gene signature. BMC. Genomics 16, 683–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Copic A., Starr T. L., and Schekman R. (2007) Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol. Biol. Cell 18, 1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paczkowski J. E., Richardson B. C., and Fromme J. C. (2015) Cargo adaptors: structures illuminate mechanisms regulating vesicle biogenesis. Trends Cell Biol. 25, 408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]