Abstract
Rho proteins are small GTP/GDP-binding proteins primarily involved in cytoskeleton regulation. Their GTP/GDP cycle is often tightly connected to a membrane/cytosol cycle regulated by the Rho guanine nucleotide dissociation inhibitor α (RhoGDIα). RhoGDIα has been regarded as a housekeeping regulator essential to control homeostasis of Rho proteins. Recent proteomic screens showed that RhoGDIα is extensively lysine-acetylated. Here, we present the first comprehensive structural and mechanistic study to show how RhoGDIα function is regulated by lysine acetylation. We discover that lysine acetylation impairs Rho protein binding and increases guanine nucleotide exchange factor-catalyzed nucleotide exchange on RhoA, these two functions being prerequisites to constitute a bona fide GDI displacement factor. RhoGDIα acetylation interferes with Rho signaling, resulting in alteration of cellular filamentous actin. Finally, we discover that RhoGDIα is endogenously acetylated in mammalian cells, and we identify CBP, p300, and pCAF as RhoGDIα-acetyltransferases and Sirt2 and HDAC6 as specific deacetylases, showing the biological significance of this post-translational modification.
Keywords: acetylation; acetyltransferase; histone deacetylase (HDAC); post-translational modification (PTM); Ras homolog gene family, member A (RhoA); Rho (Rho GTPase); sirtuin; guanine-nucleotide-dissociation inhibitor alpha; lysine acetylation; lysine acetyltransferase
Introduction
Rho proteins are guanine nucleotide-binding proteins (GNBPs)3 predominantly regulating the actin and microtubule cytoskeleton (1, 2). They are molecular switches and cycle between a GDP-bound inactive and a GTP-bound active conformation. In the GTP-bound state, Rho proteins bind to effector proteins regulating essential cellular processes: maintenance of cell architecture, intracellular transport, cell migration, cell movement, cytokinesis, and signal transduction. Rho protein dysfunction results in severe cellular disorders, such as neurodegenerative diseases, metastasis, and tumor invasion (3).
Rho proteins show a low intrinsic GTP hydrolysis and nucleotide exchange rate, which is strongly accelerated by RhoGTPase-activating proteins and Rho guanine nucleotide exchange factors, respectively (4). In the GTP-bound state, they are mostly bound to the plasma membrane via a polybasic region and a prenyl group (farnesyl or geranylgeranyl) forming a thioether with the C-terminal CaaX box cysteine side chain. About 80 different RhoGTPase-activating proteins and 80 Rho guanine nucleotide exchange factors have been described in humans to date (5, 6).
Another key regulator of Rho function is the Rho guanine nucleotide dissociation inhibitor (RhoGDI) that couples the GTP/GDP cycle to a membrane/cytosol cycle. Only three RhoGDIs have been found in mammals. RhoGDIα is ubiquitously expressed, RhoGDIβ is mainly expressed in hematopoietic cells, and RhoGDIγ is present in the brain, lung, kidney, and testis (7). This led to the hypothesis that RhoGDIs are housekeeping regulators of Rho proteins. However, recently, it has been found that RhoGDIs play more complex roles than originally expected. They are highly regulated by phosphorylation, can bind cytosolic GDP- and GTP-loaded Rho guanine nucleotide-binding protein (RhoGNBPs), are capable of transporting Rho proteins specifically to different cellular membranes, and regulate their turnover (8–10).
The interaction of Rho proteins and GDIs has been studied functionally and structurally. The crystal structures of full-length RhoGDIα alone and in complex with RhoA, Cdc42, and Rac1 have been solved by NMR and by x-ray crystallography (11–14). These studies revealed a modular structure of RhoGDIα, a C-terminal immunoglobulin (Ig) domain forming a hydrophobic pocket accommodating the prenyl group of the RhoGNBPs and an N-terminal intrinsically unfolded domain. This domain adopts a helix-turn-helix conformation upon binding the lipidated RhoGNBP contacting the switch I and II regions essential for effector binding.
For membrane extraction and membrane relocation of RhoGNBPs by RhoGDIα, a two-step reaction mechanism has been postulated, supported by its modular structure (7). In the first step of delivery, positively charged patches in the Ig domain of RhoGDIα are electrostatically attracted to the negatively charged membrane phospholipids. In the second step, the RhoGNBP inserts its lipid moiety into the membrane. An electrostatic network encompassing the negatively charged RhoGDIα N terminus competes with the membrane phospholipids for binding to the positively charged C terminus of the RhoGNBP (polybasic region) (15). It is still unclear how the tight Rho·GDP·RhoGDIα complexes are dissociated for RhoGNBPs to be reactivated by GEF-catalyzed GTP loading (14, 16).
It was shown that RhoGDIα is targeted by phosphorylation and lysine acetylation (10, 17–21). Some phosphorylation sites are in the direct vicinity of the identified lysine acetylation sites. Phosphorylation of RhoGDIα Ser-174 and Ser-101 by PAK1 upon stimulation by PDGF or EGF releases Rac1 but not RhoA and Cdc42 from its complex with RhoGDIα (17). RhoA phosphorylation at Ser-188 and Cdc42 at Ser-185 by PKA/PKG leads to stabilization of its complexes with RhoGDIα, translocation to the cytosol, and its protection from proteasomal degradation (22–24).
Recently, RhoGDIα has been found to be SUMOylated at Lys-138, leading to a stabilization of RhoA·RhoGDIα, resulting in decreased cancer cell motility (25). Several RhoGDIα lysine acetylation sites have been found in various quantitative proteomic screens performed in diverse cell and tissue types (20, 21, 26–28). For one site, RhoGDIα Lys-141, it has been shown by site-directed mutagenesis (K141Q as an acetylation mimic) that it leads to formation of thickened actin stress fibers and filopodia in HeLa cells (21).
Functionally, the acetylation sites in RhoGDIα identified by quantitative mass spectrometry have only marginally been characterized so far. Here we present the first comprehensive study using a combined synthetic biological, biophysical, and cell biological approach to unravel how lysine acetylation regulates RhoGDIα function. Our results reveal general mechanisms of how lysine acetylation regulates protein function and might open up new therapeutic strategies.
Experimental Procedures
Expression and Purification of Proteins
All proteins were expressed as GST fusions using the pGEX-4T5/Tev vector derived from pGEX-4T1 (GE Healthcare) or as His6-tagged fusion proteins (pRSF-Duet; Merck) in Escherichia coli BL21 (DE3) cells. For protein expressions, cells were grown to an A600 of 0.6 (37 °C; 160 rpm). The expression was induced by the addition of 100–300 μm isopropyl-β-d-thiogalactopyranoside and done overnight (18–20 °C; 160 rpm). Afterward, the cells were harvested (4000 × g, 20 min) and resuspended in buffer A (50 mm Tris/HCl, pH 7.4, 100 mm NaCl, 5 mm MgCl2, 2 mm β-mercaptoethanol) containing 200 μm Pefabloc protease inhibitor mixture. After cell lysis by sonication, the soluble fraction (20,000 × g, 45 min) was applied to the equilibrated affinity chromatography column. Washing was done with buffer B for the GSH column (50 mm Tris/HCl, pH 7.4, 300 mm NaCl, 5 mm MgCl2, 2 mm β-mercaptoethanol) and buffer C (buffer B plus 10 mm imidazole) for the Ni2+-NTA column. Tev protease cleavage was performed when suitable on the column or in batch overnight at 4 °C. After Tev cleavage, the protein was concentrated by ultrafiltration and applied to a size exclusion chromatography column (GE Healthcare). The concentrated fractions were shock-frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined measuring the absorption at 280 nm using the protein's extinction coefficient. For nucleotide-bound RhoGNBPs, the concentration was determined by a Bradford assay (Expedeon).
Incorporation of N-(ϵ)-Acetyl-l-lysine
The site-specific incorporation of N-(ϵ)-acetyl-l-lysine was done by the addition of 10 mm N-(ϵ)-acetyl-l-lysine (Bachem) and 20 mm nicotinamide to inhibit the E. coli CobB deacetylase to the E. coli BL21 (DE3) culture at an A600 of 0.6 (37 °C). Cells were grown for an additional 30 min before protein expression was induced by the addition of 100–300 μm isopropyl-β-d-thiogalactopyranoside. Acetylated RhoGDIα was expressed from a pRSF-Duet-vector also encoding the synthetically evolved Methanosarcina barkeri tRNACUA and the acetyl-l-lysyl-tRNA-synthetase as described previously (29). The incorporation of acetyl-l-lysine in E. coli is done cotranslationally as a response to an amber stop codon.
In Vitro Farnesylation
Geranylgeranylated proteins are prone to aggregation and are poorly soluble at micromolar concentrations needed for biophysical studies. Therefore, we used an in vitro farnesylation approach. Purified RhoA L193A/Cdc42 L191A was enzymatically farnesylated by recombinantly expressed and purified human farnesyltransferase using farnesylpyrophosphate as substrate. The in vitro farnesylation was done in 1 ml of buffer containing 100 mm NaCl, 50 mm Tris/HCl, pH 7.4, 5 mm MgCl2, 2 mm tris(2-carboxyethyl)phosphine, and 10 μm ZnCl2 by incubating 200 μm protein with a 1.5-fold molar excess of farnesylpyrophosphate (Jena Bioscience) and 6 μm farnesyltransferase (1 h at 30 °C, 1 h on ice). Finally, farnesylated proteins were purified by size exclusion chromatography (Superdex 75 10/300, GE Healthcare).
Fluorescence Measurements of GEF-catalyzed Nucleotide Dissociation
For nucleotide exchange reactions, RhoA-F was loaded with mantGDP by incubating the protein with a 10-fold excess of fluorescently labeled nucleotide in the presence of 10 mm EDTA. Redundant nucleotide was removed by size exclusion chromatography, and loading of RhoA-F was checked by HPLC. Nucleotide exchange reactions were done at 25 °C using a PerkinElmer Life Sciences LS55 spectrofluorimeter. All measurements were performed in standard buffer A containing a 50-fold molecular excess (final concentration 50 μm) of unlabeled GDP. After 1:1 complexes of RhoGDIα and RhoA-F·mantGDP (final concentration 1 μm) had been formed, the reaction was started by adding 500 nm mouse Dbs-GEF (PH (pleckstrin homology) domain-DH (dibble homology) domain; aa 624–960). Nucleotide exchange reactions were followed by fluorescence quenching as a function of time.
Plasmids, Enzymes, and Antibodies
For expression in mammalian cells, the expression plasmids pcDNA4/TO/MRGS-His6, pcDNA3.1-HisA, and pEGFP-N3 were used. Mutations were introduced by site-directed mutagenesis according to the QuikChange protocol (Agilent Technologies). The expression vectors for Myc-tagged lysine acetyltransferases (KATs) and for Myc-tagged Sirt2 and HDAC6 were purchased from transOMIC technologies. The rabbit polyclonal anti-Rac antibody was obtained from Sigma. For SUMO1 detection, the supernatant of a hybridoma cell line (clone 21C7-f) producing IgG against human SUMO1 was used. The anti-CD71 antibody was purchased from Santa Cruz Biotechnologies, Inc. Anti-RhoGDIα, anti-RhoA, anti-tubulin, anti-acetyl-l-lysine, anti-His6, anti-Sirt2, anti-HDAC6, anti-GAPDH, and anti-Myc antibodies were purchased from Abcam. For immunofluorescence, the secondary antibodies labeled with DyLight®488 (Abcam) and CF568-phalloidin (Biotium) were used. Both recombinant KATs (CBP, p300, pCAF, Tip60, and Gcn5) and lysine deacetylases (KDACs) (SIRT2 and HDAC6) were purchased from Biomol.
In Vitro SUMOylation Assay
For the in vitro SUMOylation assay, recombinantly expressed and purified proteins/enzymes were used. The reactions were performed in a buffer containing 50 mm Tris/HCl, pH 7.4, 100 mm NaCl, 5 mm MgCl2, 2 mm DTT, 1 mm PMSF, and 5 mm ATP. 100 ng/μl RhoGDIα was mixed with 3 ng/μl E1 (human Aos1/Uba2; both full-length), 3 ng/μl E2 (full-length human Ubc9), and 300 ng/μl human SUMO1. The reactions were incubated overnight at 30 °C and terminated by adding SDS sample buffer. Proteins were analyzed by SDS-PAGE and IB.
Immunoprecipitation, Pull-down, and Immunoblotting
For immunoprecipitation of acetylated proteins, cells were sonicated in lysis buffer (10 mm Tris/HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% (v/v) Triton X-100, and protease inhibitor mixture from Sigma). Lysates were incubated with anti-acetyl-l-lysine-agarose beads (ImmuneChem) at 4 °C overnight. The beads were washed three times in lysis buffer, and acetylated proteins were eluted by incubating the beads in elution buffer (50 mm Tris/HCl, pH 7.4, 150 mm NaCl, 0.1% (w/v) SDS, 1% (v/v) Triton X-100, 6 m urea) for 20 min at room temperature. For analysis of His6-tagged proteins, 12 h after transfection, cells were harvested and lysed in binding buffer (10 mm Tris/HCl, pH 8.0, 100 mm NaH2PO4, 300 mm NaCl, 2 mm β-mercaptoethanol, 0.05% (v/v) Tween 20, 8 m urea, and 10 mm imidazole). Lysates were incubated with Ni2+-NTA magnetic beads (5 Prime) for 2 h at 4 °C with rotation. Subsequently, the beads were washed three times in binding buffer supplemented with 20 mm imidazole, and His6-tagged proteins were eluted with binding buffer containing 250 mm imidazole for 20 min at room temperature. Eluates were then resolved by SDS-PAGE and analyzed by immunoblotting performed using a standard protocol. Bound antibodies were visualized by using enhanced chemiluminescence (Roth). Immunoblotting analysis was done by measuring mean gray intensities using ImageJ software. For statistical analyses, a two-tailed Student's t test was applied.
Membrane Extraction Assay
For separating membrane and cytoplasmic fractions, HEK293T cells were lysed in HEPES buffer (25 mm HEPES/NaOH, pH 8.0, 100 mm NaCl, 5 mm MgCl2, and protease inhibitor mixture). After preclearing (1000 × g, 10 min, 4 °C) lysates were centrifuged at 120,000 × g for 1 h at 4 °C. Subsequently, the membrane pellet was washed once and resupended in HEPES buffer. Membrane fractions were then incubated with 250 ng of recombinant RhoGDIα with rotation for 1 h at 4 °C and again centrifuged at 120,000 × g for 1 h at 4 °C. The supernatant was analyzed by immunoblotting.
In Vitro Acetylation/Deacetylation Assay
For in vitro acetylation, 85 pmol of purified RhoGDIα were incubated with 100 μm acetyl-CoA and 1 μl of recombinant acetyltransferase (α-TAT1, aa 1–194 was recombinantly expressed; full length: CBP, Gcn5, and TIP60; p300, aa 965–1810; pCAF, 165 aa from HAT domain; activities as purchased from Biomol) in transferase buffer (50 mm Tris/HCl, pH 7.3, 50 mm KCl, 5% (v/v) glycerol, 1 mm DTT, 0.1 mm EDTA) for 4 h at 25 °C in a total volume of 40 μl. Reactions were stopped by adding SDS buffer and boiling the samples for 5 min at 95 °C. In vitro deacetylase assays were performed in deacetylase buffer (25 mm Tris pH 8.0, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 0.1 mg/ml BSA, and 0.5 mm NAD+). 75 pmol of acetylated RhoGDIα were incubated with 0.5 μg of Sirt2 (aa 50–356) or HDAC6, respectively. Reactions were allowed to proceed for 4–6 h at 25 °C. Reaction products were analyzed by immunoblotting.
Cell Culture and Transfection
HEK293T and HeLaB cells were grown in DMEM or minimum essential medium (Life Technologies) supplemented with 10% fetal bovine serum (PAN-Biotech) in the presence of penicillin, streptomycin, and l-glutamine (Life Technologies). The stable cell line HeLa T-REx (pcDNA4/TO/MRGS-His6-SUMO1) for inducible His6-SUMO1 expression was grown in minimum essential medium containing the same supplements plus 150 μg/ml Zeocin and 5 μg/ml blasticidin (Life Technologies) (30). Transfections were performed with Lipofectamine®LTX with PLUSTM reagent (Life Technologies). For acetylation studies, HEK293T cells were grown in DMEM containing 2 μm SAHA, 1 μm trichostatin A, 10 mm nicotinamide, 10 μm sirtinol, and 1 mm sodium butyrate for 6 h.
Preparation of Liposomes and Cosedimentation Assay
The liposomes were prepared in glass vials rinsed with chloroform. Brain extract from bovine brain type I (Folch Fraction I, Sigma B1502) was mixed with chloroform to a final concentration of 1 mg/ml as described. The chloroform was evaporated using a slow argon flow to produce a thin lipid film on the vial. The vial was placed in a dessicator for at least 60 min to dry the film completely. 1 ml of buffer A was added to the vial and incubated for 5 min at room temperature for rehydration while gently agitating. The liposomes were formed during incubation at 40 °C for 5 min. The suspension was sonicated (10 min, 40 °C), vortexed, and dropped into liquid nitrogen for 2 min. The thaw and freeze steps were repeated five times. Uniformly sized liposomes were formed by extrusion through 0.2-μm polycarbonate filter membranes (Whatman catalog no. 800281) at least 21 times. The cosedimentation assay was done as described (31). In a total volume of 50 μl, 40 μl of freshly prepared liposomes (1 mg/ml) were mixed with the respective concentration of proteins and incubated on ice for 10 min in buffer A. The cosedimentation was done by ultracentrifugation at >100,000 × g (20 min, 4 °C). The obtained pellet and supernatant fractions were analyzed by SDS-PAGE and Coomassie staining or immunoblotting.
Immunofluorescence and Microscopy
HeLaB cells were grown on coverslips and fixed in 3% (w/v) paraformaldehyde for 20 min at room temperature. After permeabilization with 0.5% (v/v) Triton X-100, cells were blocked in PBS containing 5% (w/v) BSA and incubated with fluorescently labeled antibodies for 30 min at room temperature. Coverslips were embedded in ProLong® Gold Antifade (Life Technologies) and examined by using an UltraView Vox Spinning Disc confocal microscope (PerkinElmer Life Sciences) collecting Z-stack images. Actin quantifications were performed using ImageJ software. After Z-stack projection to maximum phalloidin intensity, each transfected cell was outlined, and mean intensities of phalloidin fluorescence were measured. Data were normalized by dividing the fluorescence intensity of every single cell by the average of those of non-transfected cells. Student's t test was employed for statistical analysis.
Isothermal Titration Calorimetry (ITC) Measurements
The interactions of purified proteins were thermodynamically characterized by isothermal titration calorimetry on an ITC200 instrument (GE Healthcare) based on Ref. 61. The measurements were done in buffer A at 25 °C at a concentration of 40–60 μm RhoGDIα (syringe) and 4–6 μm farnesylated RhoA/Cdc42 (cell), otherwise as indicated. The heating power per injection was plotted as a function of time until binding was saturated. A one-site binding model was fitted to the data using the MicroCal software. This resulted in the stoichiometry of binding (N), the enthalpy change (ΔH), and the equilibrium association constant (KA) as direct readout. ΔG, ΔS, and the equilibrium dissociation constant (KD) are derived. We used the standard EDTA-CaCl2 sample tests as described by MicroCal to assess the statistical significance of individual observations. These gave values within the manufacturer's tolerances of ±20% for KA values and ±10% in ΔH.
Crystallization
The RhoA-F·RhoGDIα Ac-Lys-178 crystals were obtained by the sitting drop vapor diffusion method in 0.2 m lithium sulfate, 0.1 m BisTris/HCl, pH 5.5, 20% (w/v) PEG3350 overnight at 20 °C. 30% (w/v) d-glucose was used as a cryoprotectant. The crystals belonged to the space group P61 with one heterodimer per asymmetric unit. The native data set was collected at the Swiss Light Source in Villingen at 100 K on beamline X06DA at a wavelength of 1 Å using a Pilatus 2M detector. The oscillation range was 0.1 °C, and 2700 frames were collected. The program XDS was used for indexing and integration (32). Scaling was done with Aimless (33). Initial phases were determined using the program Phaser within the program suite Phenix-dev-1893 and the Cdc42-G·RhoGDIα (PDB entry 1DOA) structure as a search model (34, 35). Refinement was done using the program phenix.refine (36). Restraints for the cysteine-farnesyl thioether were obtained by the program eLBOW in Phenix (37). The program Coot version 0.7.1 was used to build the model into the 2Fo − Fc and Fo − Fc electron density maps in iterative rounds of refinement with phenix.refine for RhoA-F·RhoGDIα Ac-Lys-178 (38, 39). In the final model, 100% of all residues are in the allowed regions of the Ramachandran plot as judged by the program Molprobity (40, 41). All structure figures presented here were made with PyMOL version 1.7.2.0 (42). Data collection and refinement statistics are given in Table 2. Rwork is calculated as follows: Rwork = Σ|Fo − Fc|/ΣFo. Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated as Rwork using the test set reflections only.
TABLE 2.
Data collection and refinement statistics for RhoA-F·RhoGDIα AcK178 (PDB entry 5FR2)
| Parameter | Value |
|---|---|
| Data collection | |
| Space group | P62 |
| Cell dimensions | |
| a, b, c (Å) | 176.94, 176.94, 63.88 |
| α, β, γ (degrees) | 90.0, 90.0, 120.0 |
| Resolution (Å)a | 49.07-3.35 (3.62-3.35) |
| Rsym or Rmergeb | 0.365 (0.914) |
| I/σI | 4.9 (1.9) |
| Completeness (%) | 99.8 (99.3) |
| CC½c | 0.926 (0.631) |
| Redundancy | 6.0 (6.0) |
| No. of reflections | 99,152 (20,356) |
| Unique reflections | 16,656 (3367) |
| Refinement | |
| Resolution (Å) | 3.35 |
| No. of reflections | 16,650 |
| No. of “free” reflections | 835 |
| Rworkd/Rfreed | 23.97/26.63 |
| No. of atoms (non-hydogen) | |
| Protein | 2931 |
| GDP/ion (Mg2+) | 28/1 |
| Water | 17 |
| β-d-Glucose | 12 |
| Clashscoree | 3.57 |
| Ramachandran plot (%)e | |
| Favored | 98.04 |
| Allowed | 1.96 |
| Outliers | 0 |
| B-Factors (Å2) | |
| Protein | 55.08 |
| GDP/Mg2+ | 53.11/ (27.87) |
| β-d-Glucose | 77.52 |
| Water | 38.73 |
| Average B-factors (Å2) | |
| Main chain | 54.53 |
| Side chain | 55.48 |
| All atoms | 54.98 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (degrees) | 0.823 |
a Values for the highest resolution shell in parentheses.
b Rsym = ΣΣ I(hkl; j) − 〈I(hkl)〉|/ΣΣ〈I(hkl)〉 with I(hkl; j) being the jth measurement of the intensity of the unique reflection (hkl) and 〈I(hkl)〉 the mean overall symmetry-related measurements (62).
c CC½ correlation coefficient from Ref. 63.
d Rwork = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree is claculated similarly to Rwork using a random 5% of the working set of reflections (64).
e MolProbity (40).
Liquid Chromatography and Mass Spectrometry
Peptides derived from in-gel digestion were desalted using the stop-and-go extraction technique prior to mass spectrometry analysis (43). Liquid chromatography and tandem mass spectrometry instrumentation consisted of an nLC 1000 nano liquid chromatography coupled via a nano-electrospray ionization source to a quadrupole-based mass spectrometer, QExactive. Peptide separation was done by linear increase of buffer B within a binary buffer system: 1) 0.1% formic acid in water and 2) 0.1% formic acid in acetonitrile. Within 40 min, the content of buffer B was raised to 27% followed by a washing (95%) and re-equilibration (5%) step within 10 min. The mass spectrometer acquired spectra in a data-dependent mode. After MS1 acquisition at 70,000 resolution at 200 m/z (automatic gain control target, 3e6, maximum injection time, 20 ms), the 10 most intense peaks were selected for HCD fragmentation and measurement in the orbitrap mass analyzer (35,000 resolution at 200 m/z, AGC target: 5e5, maximum IT 120 ms). Dynamic exclusion was set to 25 s, limiting extensive refragmentation.
Proteomics Data Analysis
MaxQuant and the implemented Andromeda search engine were used to analyze raw data (44, 45). MS/MS spectra were correlated against the Uniprot E. coli database containing the target protein. Default mass tolerance settings, a peptide length of 7 amino acids, and a minimal score for modified and unmodified peptides of 0 were used. The false discovery rate was controlled at the peptide-spectrum match, and the protein level was controlled using the decoy approach by a revert algorithm to 1%. Acetylation at lysine residues, oxidation of methionine, and acetylation at the protein N terminus was set as a variable modification, whereas carbamidomethylation of cysteines was defined as a fixed modification. For data analysis, we used the raw intensities of specific acetylation sites for analysis and normalized these by label-free quantification of used protein. Ratios were calculated comparing these values with control experiments without any KAT. Hierarchical clustering was performed for visualization using the gplots package from R Bioconductor.
Results
RhoGDIα Is Endogenously Lysine Acetylated in Human Cells
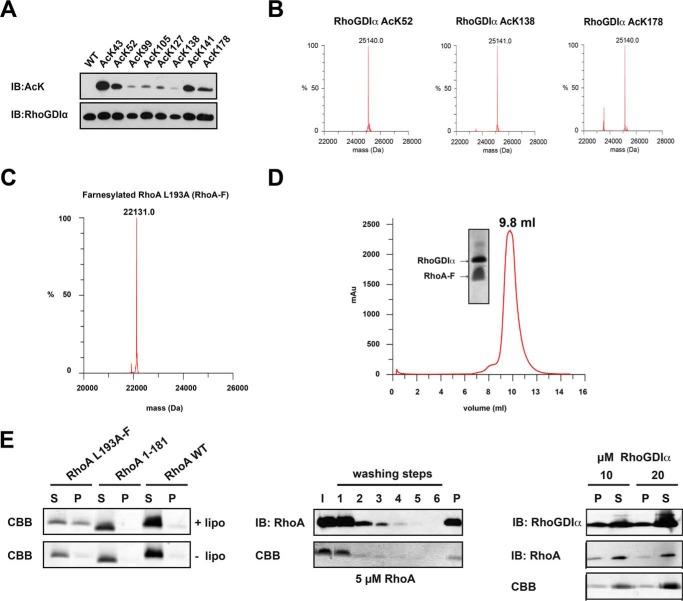
By quantitative mass spectrometry, RhoGDIα has been found to be lysine-acetylated at least eight distinct sites: Lys-43 and Lys-52 in the N-terminal domain as well as Lys-99, Lys-105, Lys-127, Lys-138, Lys-141, and Lys-178 in the Ig domain (Fig. 1A). We tested whether RhoGDIα is lysine-acetylated endogenously in human HeLaB and HEK293T cells. To this end, we performed an immunoprecipitation of acetylated proteins using an agarose bead-coupled anti-acetyl-l-lysine antibody followed by immunoblotting using an anti-RhoGDIα antibody. In HEK293T and HeLaB cells, we identified endogenously lysine-acetylated RhoGDIα. Moreover, the signal was increased upon treatment with several classical and sirtuin deacetylase inhibitors, indicating that endogenous RhoGDIα acetylation is regulated by KDACs (Fig. 1B; data not shown for HeLaB cells). As expected, the amount of acetylated tubulin, immunoprecipitated using the anti-acetyl-l-lysine antibody-coupled beads, increased upon KDAC inhibitor treatment (Fig. 1B, bottom, lane 4). Notably, the differences observed in the acetylation levels are not due to a variation in the expression levels because the amount of tubulin was similar in the samples treated with/without KDAC inhibitors (Fig. 1B, Input). Additionally, we transiently expressed His6-RhoGDIα in HEK293T cells. Again, we found acetylation for His6-RhoGDIα, and the acetylation level was increased upon the addition of a KDAC inhibitor mixture as shown by Ni2+-NTA pull-down (PD) and subsequent immunoblotting using an anti-acetyl-l-lysine antibody (Fig. 1C).
FIGURE 1.
RhoGDIα is lysine-acetylated. A, left, overview of reported lysine acetylation sites of RhoGDIα identified in recent quantitative proteomic screens in cells and tissues as indicated. Right, localization of the eight acetylated lysine residues in the RhoA·RhoGDIα structure (PDB entry 4F38). Lys-99, Lys-105, Lys-127, Lys-138, Lys-141, and Lys-178 are in the immunoglobulin domain, and Lys-43 and Lys-52 are in the N-terminal domain. Yellow, RhoA; gray, RhoGDIα. Acetylated lysines are highlighted in red. B, RhoGDIα is endogenously acetylated in HEK293T cells, as shown by immunoprecipitation of lysine acetylated proteins and probing of RhoGDIα. Acetylation is regulated by KDACs, as seen by the increase of acetylated RhoGDIα after incubating the cells with KDAC inhibitors for 6 h. IB for α-tubulin serves as control. +, with KDAC inhibitors; −, without KDAC inhibitors. C, transiently expressed His6-tagged RhoGDIα is acetylated in HEK293T cells, as shown by Ni2+-NTA pull-down (PD) and IB. The Ac-Lys signal increased upon KDAC inhibitor treatment (+). Anti-His IB serves as loading control.
RhoGDIα Acetylation Affects Actin Filament Formation in Mammalian Cells
To test whether RhoGDIα acetylation interferes with Rho signaling, we analyzed how RhoGDIα acetylation affects cellular actin filament (F-actin) formation. To this end, we transiently overexpressed Gln and Arg mutants of RhoGDIα-EGFP, mimicking the acetylated and non-acetylated state, in HeLaB cells and examined phalloidin-stained filamentous actin by immunofluorescence and subsequent fluorescence intensity quantification (Fig. 2, A and B). Notably, we observed by immunoblotting experiments that the expression levels for RhoGDIα and the mutants were nearly the same (data not shown). Overexpression of K178QG (superscript “G” for RhoGDIα) resulted in markedly thickened actin stress fibers and significantly increased the cellular F-actin content compared with cells expressing RhoGDIαWT or K178RG. In fact, the expression of K178RG reduced the actin filament level even below the value observed for the mock control, showing that neutralization of the charge and steric effects are phenotypically distinguishable. For K141QG, we also observed an increase in F-actin; however, in contrast to the reported data, it was not statistically significant compared with RhoGDIαWT (Fig. 2B, bottom) (21). The mutants K99Q/RG both led to a significant increase in total actin filament content. In contrast, from all of the acetylation mimetics analyzed, the RhoGDIα K52Q/R are the only mutants significantly reducing the cellular F-actin compared with RhoGDIαWT to levels observed for MOCK control (Fig. 2B, top). This suggests that K52Q/RG both switch off RhoGDIα function and that lysine acetylation at Lys-52G might exert similar effects, mechanistically by electrostatic as well as steric components.
FIGURE 2.
RhoA acetylation interferes with F-actin formation. A, RhoGDIα acetylation induces actin polymerization in HeLaB cells. HeLa cells transiently transfected with RhoGDIα (bottom panels) or RhoGDIα-EGFP (top panels) were stained for filamentous actin (F-actin; red). B, quantitative analysis of F-actin. Shown is the F-actin fluorescence intensity of cells from A normalized to non-transfected cells. The results are depicted as mean ± S.D. (error bars) from three independent experiments. At least 10 cells were examined per condition per experiment. *, p < 0.01; **, p < 0.001 for the indicated comparison. #, p < 0.05 compared with RhoGDIαWT-expressing cells. For statistical analyses, a two-sided Student's t test was performed. Scale bars, 20 μm.
Preparation of Site-specifically Lysine-acetylated RhoGDIα and Farnesylated RhoA
To explain the phenotypes observed in cells, we mechanistically analyzed the impact of lysine acetylation on RhoGDIα function. To this end, we used a synthetically evolved, orthogonal acetyl-l-lysyl-tRNA-synthetase/tRNACUA pair from M. barkeri in E. coli to site-specifically incorporate acetyl-l-lysine recombinantly into RhoGDIα (genetic code expansion concept) (29, 46, 47). We successfully produced homogeneously acetylated RhoGDIα in yields suitable for biophysical studies as judged from immunoblotting and electrospray ionization (ESI)-mass spectrometry (Fig. 3, A and B). Notably, the anti-acetyl-l-lysine antibody recognizes the acetylated proteins differentially, suggesting that residues in vicinity are important for antigen recognition (Fig. 3A).
FIGURE 3.
Farnesylated RhoA is functional in binding to RhoGDIα, liposome binding, and extraction by RhoGDIα. A, RhoGDIα is lysine-acetylated using the genetic code expansion concept. The acetyl-l-lysine incorporation was verified by anti-acetyl-l-lysine IB. B, quantitative incorporation of N-(ϵ)-acetyl-l-lysine into RhoGDIα, as judged by ESI-mass spectrometry. The molecular masses correspond exactly (±1 Da) to the protein mass calculated (His6-RhoGDIα, 25,099.1 Da; single-acetylated RhoGDIα, 25,099.1 ± 42 Da; double-acetylated RhoGDIα, 25,099.1 ± 84 Da). C, RhoA is homogeneously and quantitatively farnesylated, as judged by ESI-mass spectrometry (expected mass, 22,131.2 Da). D, analytical size exclusion chromatography of a RhoA-F·RhoGDIα complex (Superdex 75 10/300). RhoA-F binds tightly to RhoGDIα coeluting from the gel filtration column at an elution volume of 9.8 ml. Notably, RhoA-F alone elutes at 11.9 ml (data not shown). E, RhoA-F liposome binding and extraction by RhoGDIα. Left, RhoA binds to liposomes, dependent on the presence of the farnesyl moiety, as shown by a liposome cosedimentation assay. To this end, liposomes were prepared and incubated with RhoA-F, C-terminally deleted RhoA 1–181, and full-length but non-prenylated RhoA. After ultracentrifugation, only the farnesylated full-length RhoA-F was detectable in the pellet fraction and cosedimented with the liposomes, whereas the other RhoA proteins can only be detected in the soluble fraction, as shown by SDS-PAGE and staining with Coomassie Brilliant Blue (CBB). Middle, liposomes can be loaded with farnesylated RhoA-F. After loading of liposomes with farnesylated RhoA-F, non-bound RhoA-F can be removed from the supernatant by consecutive washing. After six washing steps, no RhoA-F can be detected in the supernatant, neither by immunoblotting using a specific anti-RhoA antibody nor by Coomassie Brilliant Blue staining. RhoA-F can be detected only in the pellet fraction after cosedimentation of the liposomes, showing that the liposomes are loaded with RhoA-F. Right, RhoGDIα can solubilize RhoA-F from RhoA-F-loaded liposomes in a concentration-dependent manner. RhoA-F-charged liposomes were treated with increasing concentrations of RhoGDIα, as indicated. With 20 μm RhoGDIα, there is more RhoA-F solubilized from the liposomes compared with the sample with 10 μm RhoGDIα. RhoGDIα is stained by immunoblotting using an anti-RhoGDIα antibody, and RhoA is stained by an anti-RhoA antibody. I, input; S, supernatant; P, pellet.
It was shown earlier that prenylation of RhoGNBPs is essential for their biological function; however, it is independent of a specific isoprenoid modification (48). Importantly, in vivo or in vitro geranylgeranylated RhoA (RhoA-G) was prone to aggregation at micromolar concentrations, and the reported picomolar affinities of RhoA-G toward RhoGDIα are above the detection limit of ITC (data not shown) (14). To study the impact of RhoGDIα acetylation on RhoA function, we therefore used in vitro farnesylated RhoA as a model system. To this end, we used recombinantly expressed, purified human farnesyltransferase and farnesylpyrophosphate as a substrate to quantitatively farnesylate RhoA (RhoA-F) and Cdc42 (Cdc42-F) in vitro, as shown by ESI-mass spectrometry (Fig. 3C). RhoA-F was able to form a stable, low nanomolar 1:1 complex with RhoGDIα as shown by analytical size exclusion chromatography and ITC (Figs. 3D and 4A). Furthermore, it was able to bind to liposomes dependent on the presence of the farnesyl moiety, and it could be extracted by RhoGDIα in a concentration-dependent manner (Fig. 3E).
FIGURE 4.
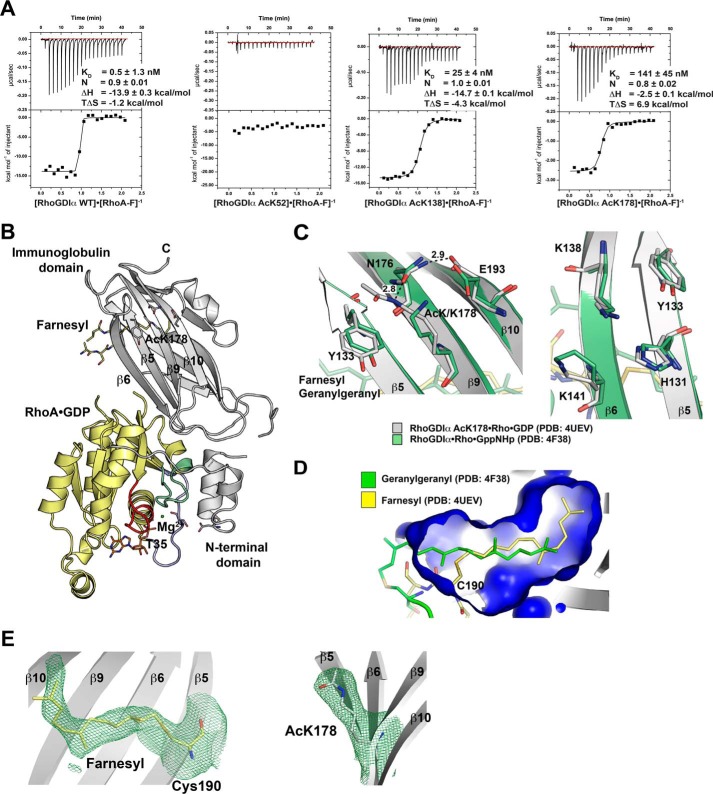
Lysine acetylation in RhoGDIα impairs binding toward farnesylated RhoA. A, thermodynamic characterization of selected RhoA-F-RhoGDIα interactions, as determined by ITC. Acetylation at Lys-52G abolishes binding toward RhoA-F, and Ac-Lys-138G and Ac-Lys-178G both lead to a reduction in affinity (KD = 141 nm (Ac-Lys-178) and 25 nm (Ac-Lys-138). B, ribbon representation of the RhoA-F·RhoGDIα Ac-Lys-178 complex. The conformation is almost unaltered compared with the RhoA-G·RhoGDIα structure (PDB entry 4F38). Yellow, RhoA; gray, RhoGDIα Ac-Lys-178. The GDP, Mg2+, and T35 from switch I (blue), switch II (green), and the P-loop (red) are highlighted. C, left, Structural coupling of RhoGDIα Ac-Lys-178. Ac-Lys-178G on β9 interacts directly with Tyr-133G on β5 by hydrophobic stacking. Ac-Lys-178G interacts indirectly with Glu-193G on β10, forming a hydrogen bond with Asn-176G on β9. Shown is a close-up from a superposition of the structure solved here (PDB entry 5FR2; gray) with the non-acetylated RhoA-G·RhoGDIα structure (PDB entry 4F38; green). The non-acetylated Lys-178G could not form these interactions. Green, geranylgeranyl from PDB 4F38; yellow, farnesyl from PDB 5FR2. Right, upon acetylation, Lys-138G on β6 might form interactions with His-131G and Tyr-133G on β5. Superposition is as in the left panel. D, the hydrophobic pocket of RhoGDIα Ac-Lys-178 (blue surface; PDB entry 5FR2) accommodates the farnesyl (yellow) of RhoA slightly differently than the geranylgeranyl (green) of RhoA-G·RhoGDIα (PDB entry 4F38). Shown is a superposition of RhoGDIα from the complexes indicated. E, FO − FC omit maps of the RhoA-F·GDP·RhoGDIα structure presented here (PDB entry 5FR2). Shown is the electron density of the farnesyl moiety (left) and the Ac-Lys-178 on β9 of RhoGDIα countered at 1.5 σ.
Functional Consequences of RhoGDIα Acetylation for the Interaction toward Prenylated RhoGNBPs
We examined whether acetylation of RhoGDIα impacts binding toward RhoA-F by ITC. RhoGDIα Ac-Lys-52 and the N-terminally truncated Δ66G mutant both strongly interfered with RhoA-F binding (Fig. 4A and Table 1). However, whereas Δ66G completely abolishes RhoA-F binding, Ac-Lys-52G showed at least residual, albeit 4 orders of magnitude reduced, binding toward RhoA-F as shown by ITC (KD = 4.1 μm; Table 1). Notably, Ac-Lys-52G does not interfere with the structural integrity of RhoGDIα because the protein behaved as RhoGDIαWT in size exclusion chromatography analyses (data not shown). These results strongly suggest a pivotal role of the RhoGDIα N terminus for the proper insertion of the prenyl moiety into the hydrophobic cavity of the Ig domain. The acetylation at Lys-138G leads to a 50-fold reduction in the affinity toward RhoA-F (Fig. 4A and Table 1). The importance of this residue is in agreement with recent findings showing that SUMOylation increases the affinity toward RhoA (25). Acetylation at Lys-138G would block the SUMOylation, strongly indicating a PTM cross-talk. Structurally, Ac-Lys-138G on β6 might favor interactions with His-131G and Tyr-133G on β5 (Fig. 4C, right). However, to finally judge how Ac-Lys-138G mechanistically exerts this effect, we would need further structural data. RhoGDIα Ac-Lys-178 showed a strongly reduced affinity toward both RhoA-F (141 nm) and Cdc42-F (98 nm) (Fig. 4A and Table 1). This supports previous findings showing that this region is of importance for Rho-binding because mutation of I177NG, directly adjacent to Lys-178G, leads to a 20-fold reduction in Cdc42 affinity (49, 50). Ile-177G is oriented toward the hydrophobic pocket of the Ig domain, and Lys-178G is surface-exposed. Acetylation at Lys-178G, by affecting the polarity and hydrophobicity of the Ig domain, might affect the Ig domain's conformation, thereby affecting binding toward prenylated RhoA. As a support, whereas the RhoGDIα Ac-Lys-178 RhoA-F interaction is enthalpically and entropically favored, all of the other interactions are entropically unfavored, indicating that acetylation of RhoGDIα at Lys-178 alters the binding mechanism toward RhoA-F (Table 1).
TABLE 1.
Thermodynamic characterization of the RhoGDIα interactions with RhoA and Cdc42 as determined by ITC
KD is the equilibrium dissociation constant, ΔH is the enthalpy, ΔS is the entropy change, and N is the stoichiometry of the interaction. The farnesylated proteins were measured at 25 °C. NB, no binding.
| Interaction with RhoGDIα (40–60 μm) | KD | ΔH (kcal mol−1) | TΔS (kcal mol−1) | N |
|---|---|---|---|---|
| RhoA-F (4–6 μm) | ||||
| WT | 0.5 ± 1.3 | −13.9 ± 0.3 | −1.2 | 0.9 |
| Δ22 (20 °C) | 16 ± 5.1 | −7.5 ± 0.1 | 2.9 | 1.2 |
| Δ66 | NB | NB | NB | NB |
| AcK43 | 5.7 ± 4 | −23.2 ± 0.7 | −12.0 | 0.9 |
| AcK52 (20 °C) | NB | NB | NB | NB |
| AcK99 | 3.2 ± 2.3 | −10.1 ± 0.2 | 1.5 | 0.9 |
| AcK105 | 6.1 ± 2.2 | −15.5 ± 0.3 | −4.3 | 0.9 |
| AcK127 | 13.8 ± 3.7 | −13.2 ± 0.2 | −2.5 | 1.1 |
| AcK138 | 25 ± 3.8 | −14.7 ± 0.2 | −4.3 | 1.1 |
| AcK141 | 3.6 ± 1.3 | −19.7 ± 0.3 | −8.3 | 0.8 |
| AcK127,141 | 8.5 ± 2.7 | −17.3 ± 0.3 | −6.3 | 0.9 |
| AcK178 | 141 ± 45 | −2.5 ± 0.1 | 6.9 | 0.8 |
| RhoA-F (30 μm) | ||||
| AcK52 (20 °C) (300 μm) | 4100 ± 700 | −7.4 ± 0.4 | −0.2 | 0.7 |
| Δ66 (300 μm) | NB | NB | NB | NB |
| Cdc42-F (4–6 μm) | ||||
| WT | 4.6 ± 1.3 | −27.5 ± 0.3 | −16.1 | 1.1 |
| AcK52 | NB | NB | NB | NB |
| AcK105 | 3.2 ± 0.9 | −28.9 ± 0.3 | −17.3 | 1.1 |
| AcK127 | 8.0 ± 1.5 | −24.3 ± 0.2 | −13.3 | 1.1 |
| AcK141 | 2.9 ± 0.5 | −22.6 ± 0.1 | −10.9 | 1.3 |
| AcK127,141 | 10.8 ± 3.6 | −18.9 ± 0.3 | −8.0 | 1.1 |
| AcK178 | 98 ± 27 | −3.1 ± 0.04 | 6.5 | 1.2 |
Crystal Structure of RhoGDIα Ac-Lys-178 in Complex with Farnesylated RhoA·GDP
To show how Lys-178 acetylation of RhoGDIα alters the binding mechanism toward RhoA-F, we solved the structure of a RhoA-F·RhoGDIα Ac-Lys-178 complex at 3.35 Å resolution by x-ray crystallography (Fig. 4B and Table 2). The complex crystallized in the space group P62 with one heterodimer per asymmetric unit. We observed reliable electron density for the Ac-Lys-178 on β9 and also for the farnesyl moiety as judged by omit maps showing that Ac-Lys-178 and the farnesyl are correctly positioned (Fig. 4E). The overall conformation of the complex was almost unaltered compared with a previously solved structure of RhoA-G·RhoGDIα (PDB entry 4F38, overall root mean square deviation 0.501 Å; Table 3). However, the hydrophobic pocket of the RhoGDIα Ac-Lys-178 Ig domain has a slightly bigger volume, and the farnesyl group is positioned differently, penetrating deeper into the pocket compared with the geranylgeranyl group in the RhoA-G·RhoGDIα structure (PDB entry 4F38; Fig. 4D). Compared with the non-prenylated RhoA·RhoGDIα complex (PDB entry 1CC0), it showed conformational differences mostly to RhoGDIα (overall root mean square deviation 0.804 Å; Table 3). Ac-Lys-178 on RhoGDIα β9 structurally couples three β-strands of the Ig domain β5, β9, and β10) by forming a stacking interaction with Tyr-133G on β5 and a hydrogen bond with Asn-176G on β9. Asn-176G in turn forms a hydrogen bond to Glu-193G on β10 (Fig. 4C, left). These interactions are lacking in the non-acetylated structure, as shown by superposition of the structure presented here with the non-acetylated structure (Protein Data Bank entry 4F38; Fig. 4C, left). The Ig domain is able to switch between an open, prenyl-bound, and closed, prenyl-free, state by undergoing a rigid body reorientation on two invariant glycine residues (Gly-125G and Gly-147G). Glycine-based hinges are reported to be important for conformational flexibility also in other systems (51). By structurally coupling three β strands in the Ig domain, Ac-Lys-178G might interfere with the inherent Ig domain's conformational flexibility essential for the function of the Ig domain, resulting in the observed decrease in RhoA-F affinity.
TABLE 3.
Structural similarity of the RhoA-F·RhoGDIα AcK178 (PDB entry 5FR2) complex presented here and the complexes of RhoA-G·RhoGDIα (PDB entry 4F38) and RhoA·RhoGDIα (PDB entry 1CC0)
For comparison, the structural similarity between the complexes of RhoA·G·RhoGDIα (PDB entry 4F38) and RhoA·GDP·RhoGDIα (PDB entry 1CC0) is shown in the right column.
| RhoA-F·GDP·RhoGDIα AcK178 (PDB entry 5FR2) |
RhoA-G·RhoGDIα (PDB entry 4F38) |
||
|---|---|---|---|
| RhoA-G·RhoGDIα (PDB entry 4F38) | RhoA·GDP·RhoGDIα (PDB entry 1CC0) | RhoA·GDP·RhoGDIα (PDB entry 1CC0) | |
| RMSDa/Å | RMSDa/Å | RMSDa/Å | |
| RhoGDIα | 0.447 | 0.957 | 1.122 |
| RhoA | 0.380 | 0.350 | 0.582 |
| RhoA·RhoGDIα | 0.501 | 0.804 | 0.983 |
a Root mean square deviation; square root of the mean of the square of the distances between the matched atoms.
RhoGDIα Acetylation Directly and Indirectly Cross-talks with SUMOylation
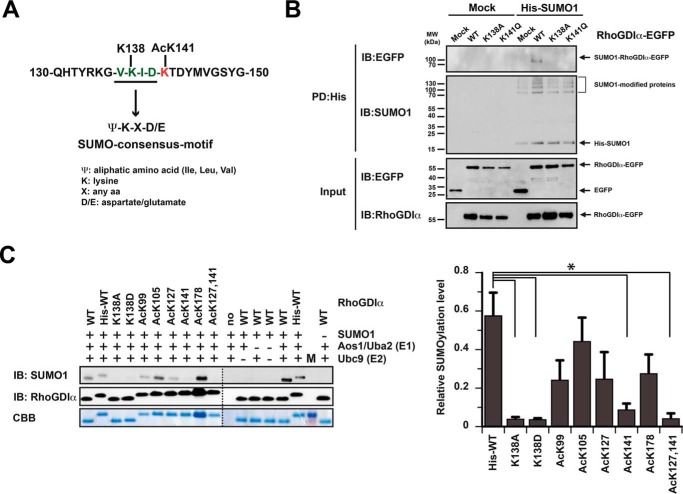
RhoGDIα is SUMOylated at Lys-138, which is also found to be lysine-acetylated (25). Because Lys-141G is located directly adjacent to the SUMOylation consensus motif (ΨKX(D/E); RhoGDIα: 137VKIDK141) (Fig. 5A), we analyzed whether the acetylation at Lys-141G influences SUMOylation of RhoGDIα at Lys-138.
FIGURE 5.
RhoGDIα acetylation directly and indirectly interferes with Lys-138 SUMOylation. A, SUMO consensus sequence of RhoGDIα (aa 130–150). B, RhoGDIα K141Q blocks K138-SUMOylation in vivo. HeLa T-REx cells stably expressing His6-SUMO1 or empty vector (mock) were transfected with indicated RhoGDIα-EGFP constructs. Shown is a pull-down (PD) of His6-SUMO1-modified proteins by Ni2+-NTA beads. Eluates and input were probed with the indicated antibodies. C, RhoGDIα Ac-Lys-141 blocks Lys-138 SUMOylation in vitro. Left, K138AG/K138DG were not SUMOylated (direct cross-talk), and Ac-Lys-141G abolishes SUMOylation (indirect cross-talk). Coomassie Brilliant Blue (CBB) staining and RhoGDIα IB served as loading control. Right, quantification of RhoGDIα SUMOylation. K138AG, K138DG, Ac-Lys-141G, and Ac-Lys-127,141G were significantly less SUMOylated as compared with RhoGDIαWT. Results are shown as mean ± S.E. (error bars) The experiment was performed independently three times. *, p < 0.05; two-sided Student's t test.
To this end, HeLa T-REx cells stably expressing His6-SUMO1 were transiently transfected with RhoGDIα-EGFP constructs, and His6-SUMO1-modified proteins were pulled down by Ni2+-NTA beads. Using an anti-GFP antibody, we showed that RhoGDIαWT is conjugated to SUMO1 in cells in contrast to RhoGDIα K141Q and K138A, which were not found to be SUMOylated (Fig. 5B).
To confirm these results and to quantify the SUMOylation efficiency, we additionally applied an in vitro SUMOylation assay. We used non-acetylated/lysine-acetylated RhoGDIα as substrate proteins and purified SUMO1, the E1 SUMO-activating enzyme Aos1/Uba2, and the E2 SUMO-conjugating enzyme Ubc9 to catalyze the reaction. Using an anti-SUMO1 antibody, non-acetylated RhoGDIαWT protein showed a clear SUMOylation signal (Fig. 5C). The SUMOylation of the K138A/DG mutants was completely abolished, confirming that Lys-138 is the major SUMOylation site in RhoGDIα directly competing with acetylation (25). RhoGDIα Ac-Lys-141 and RhoGDIα Ac-Lys-127,141 showed almost no SUMOylation, reflecting that acetylation at Lys-141G blocks RhoGDIα SUMOylation at Lys-138 (Fig. 5C). Thus, these results provide evidence for both a direct and an indirect cross-talk of RhoGDIα acetylation with its SUMOylation in vitro and in vivo.
Acetylation of RhoGDIα at Lys-52 Impairs RhoA Extraction from Cellular Membranes
To test whether acetylation of RhoGDIα interferes with its capability to extract endogenous RhoA from cellular membranes, we prepared membrane fractions from HEK293T cells and incubated those with site-specifically acetylated RhoGDIα proteins. Solubilized endogenous geranylgeranylated RhoA was separated from residual membrane-attached RhoA via ultracentrifugation and detected by immunoblotting of the supernatant (Fig. 6A, left). Most strikingly, RhoGDIα Ac-Lys-52 was not able to significantly extract RhoA compared with RhoGDIαWT (Fig. 6A, right). This supports our previous results showing that acetylation at Lys-52G abolishes RhoA binding. All of the other acetylated RhoGDIα proteins were capable of extracting modified RhoA to an extent comparable with RhoGDIαWT at least under the assay conditions used.
FIGURE 6.
Influence of RhoGDIα acetylation on RhoA membrane extraction and GEF-catalyzed nucleotide dissociation. A, extraction of endogenous RhoA from HEK293T membrane fractions by lysine-acetylated RhoGDIα proteins. The RhoA content extracted was analyzed by IB (left). RhoA of the supernatant was quantified using ImageJ software and normalized to the RhoGDIα amount. Results are shown as mean ± S.E. (error bars) from six independent experiments. Only Ac-Lys-52G shows a statistical significance compared with RhoGDIαWT. *, p < 0.05 for the indicated comparison (right). S, supernatant; M1, membrane fraction before solubilization; M2, membrane fraction after solubilization. B, influence of RhoGDIα acetylation on Dbs-GEF-catalyzed nucleotide exchange on RhoA. 1 μm RhoGDIα·RhoA·mantGDP-complex was incubated with 0.5 μm Dbs-GEF and a 50-fold molar excess (50 μm) of unlabeled GDP. The acetylation of RhoGDIα at Lys-52 decreases the inhibitory effect, whereas the other acetylation sites do not interfere with nucleotide exchange on RhoA. Data represent the mean from three independent experiments. C, position of Lys-52 and Lys-43 in the N-terminal RhoGDIα domain, as judged from the RhoA·RhoGDIα Ac-Lys-127,141 structure presented here (color code as in Fig. 5B). Lys-52G is an integral part of the N-terminal domain within hydrogen bond distance to Tyr-63R in switch II and the main chain carbonyl oxygen of Leu-41G. The side chain of Lys-43G is surface-exposed.
RhoGDIα Acetylation Interferes with GEF-catalyzed Guanine Nucleotide Exchange
To investigate how RhoGDIα acetylation interferes with GEF-catalyzed nucleotide exchange on RhoA, we performed a fluorescence-based nucleotide exchange assay. We used farnesylated RhoA loaded with fluorescently labeled mantGDP and preformed complexes with acetylated/non-acetylated RhoGDIα (concentration 1 μm). We followed the fluorescence quenching of the nucleotide exchange reactions catalyzed by 0.5 μm Dbs-GEF in presence of a 50-fold molar excess of unlabeled GDP (concentration 50 μm) (Fig. 6B). Surprisingly, we observed two phases in the exchange assays for all reactions with acetylated RhoGDIα but not for Ac-Lys-52G and also not for the reactions without RhoGDIα. This might reflect that the Dbs-GEF itself is able to act as a GDI displacement factor. As expected, the presence of RhoGDIαWT substantially decreased the Dbs-GEF-catalyzed nucleotide exchange compared with the reaction without RhoGDIα (Fig. 6B). All complexes with acetylated RhoGDIα, except for Ac-Lys-52G, showed exchange kinetics similar to non-acetylated RhoGDIαWT. However, for Ac-Lys-52G, the nucleotide exchange rates approached values similar to the reactions in the absence of RhoGDIα. The fact, that they are not exactly the same can be explained by the observed residual binding of Ac-Lys-52G to RhoA-F (Table 1). Structurally, Lys-52G forms a hydrogen bond with Tyr-63R (superscript “R” for RhoA) and is in intramolecular hydrogen bond distance to the main chain carbonyl oxygen of Leu-41G (Fig. 6C). Acetylation of Lys-52G might sterically interfere with the formation of the N-terminal helix-loop-helix conformation contacting RhoA switch I and switch II, thereby abolishing RhoGDIα-binding. These data suggest that acetylation at Lys-52G could act as a displacement factor for the high affinity RhoA·GDP·RhoGDIα complex.
CBP, p300, and pCAF Lysine-acetylate RhoGDIα
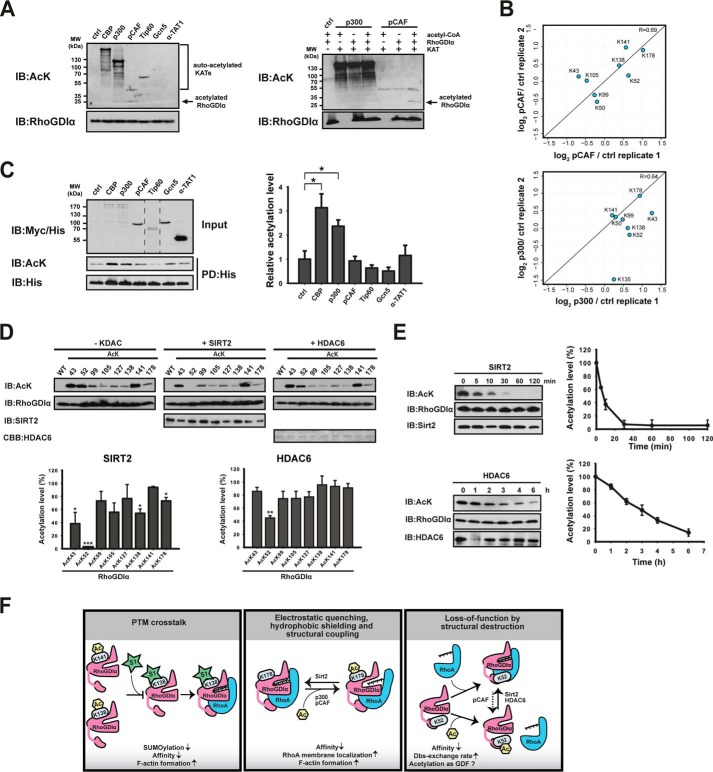
As shown above, RhoGDIα was acetylated at a basal level in HEK293T cells also in the non-KDAC inhibitor-treated samples, indicating KAT-catalyzed acetylation of RhoGDIα (Fig. 1, B and C). To identify RhoGDIα-specific KATs, we first used an in vitro approach. We selected six major KATs (CBP, p300, pCAF, Gcn5, Tip60, and α-TAT1) and used these active enzymes and purified RhoGDIα as a substrate for in vitro acetylation. By immunoblotting, we identified p300 and pCAF as KATs for RhoGDIα (Fig. 6A). The acetylation was dependent on the presence of acetyl-CoA, KAT enzyme, and RhoGDIα. We did not obtain a signal with acetyl-CoA alone or in the absence of RhoGDIα, excluding the possibility that the signal derives from non-enzymatic acetylation or KAT-autoacetylation activity, respectively (Fig. 7A, left and right (control)).
FIGURE 7.
Regulation of RhoGDIα acetylation by KDACs and KATs. A, RhoGDIα is acetylated by p300 and pCAF in vitro (left). As a control for the specificity of RhoGDIα acetylation catalyzed by p300 and pCAF and to exclude non-enzymatic acetylation, reactions were performed in the absence of KAT, RhoGDIα, or acetyl-CoA (right). Acetylation was visualized by IB using an anti-Ac-Lys antibody. B, correlation scatter plot of RhoGDIα acetylation sites identified by mass spectrometry. Plotted are the log2 results of two replicates for pCAF (top) and p300 (bottom). Shown are the ratios of the KAT-treated versus control samples. C, in vivo KAT assay. His6-RhoGDIα was cotransfected with expression constructs of various KATs (CBP, p300, pCAF, Tip60, Gcn5, and α-TAT1), and acetylation was assessed by Ni2+-NTA pull-down (PD) and IB (left). Quantifications were done using ImageJ software by normalizing the acetylation signal to the amount of overexpressed RhoGDIα. Shown is the mean ± S.E. from five independent experiments. Only for p300 and CBP do we observe a statistically significant increase in RhoGDIα acetylation compared with the non-enzyme control. *, p < 0.01 for the indicated comparison (right). D, Sirt2 and HDAC6 are RhoGDIα deacetylases. 2 μg of acetylated RhoGDIα proteins were incubated with 0.5 μg of Sirt2 or HDAC6 for 4 h at room temperature. Reaction products were analyzed by IB (top panels). For Sirt2, we observed a statistically significant deacetylation for Ac-Lys-52G, Ac-Lys-138G, and Ac-Lys-178G, and for HDAC6, we observed this only for Ac-Lys-52G. The quantification was done using ImageJ by normalizing to the samples without KDACs (bottom panels). Values represent the mean ± S.E. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.001, statistically significant difference from samples without KDACs. E, kinetics for RhoGDIα Ac-Lys-52G deacetylation by Sirt2 and HDAC6. 2 μg of Ac-Lys-52G were incubated with either Sirt2 (0.25 μg) or HDAC6 (0.5 μg). Quantification of the acetylation level was done using ImageJ software and normalizing it to the band intensity at t = 0 min. Shown is the mean ± S.D. (error bars) from three independent experiments. F, working model for the regulation of RhoGDIα function by lysine acetylation. Left, indirect cross-talk of Lys-141G acetylation and Lys-138G SUMOylation and direct cross-talk by acetylation of Lys-138G affecting RhoA-affinity; middle, electrostatic quenching, hydrophobic shielding, and structural coupling of Ac-Lys-178G, decreasing RhoA affinity and thereby increasing cellular F-actin content; right, acetylation of Lys-52G in the RhoGDIα N-terminal domain results in loss of function by structural destruction.
To identify the RhoGDIα acetylation sites, we performed mass spectrometric analyses (Fig. 7B). We found that acetylation of Lys-178G was most strongly up-regulated. Supporting our immunoblotting results, pCAF and p300 both led to a 2-fold increase in Lys-178G acetylation. Additionally, pCAF acetylated RhoGDIα on Lys-52, Lys-138, and Lys-141, whereas p300 catalyzed Lys-43 acetylation. Thus, for all acetylation sites affecting RhoGDIα-function (Lys-52, Lys-138, Lys-141, and Lys-178), we identified a specific acetyltransferase.
To confirm our results, we also used an in vivo approach by transiently coexpressing His6-RhoGDIα together with Myc-tagged (CBP, p300, pCAF, Gcn5, and α-TAT1) or His6-tagged (Tip60) KATs in HEK293T cells. We pulled down His6-RhoGDIα and analyzed the amount of acetylated RhoGDIα by immunoblotting. As shown in Fig. 7C, overexpression of both p300 and CBP resulted in a significant increase of acetylated RhoGDIα, supporting our in vitro data. Notably, the overexpression of pCAF did not increase the acetylation level of RhoGDIα, indicating that different mechanisms operate to acetylate RhoGDIα in vitro and in vivo.
Sirt2 and HDAC6 Act as RhoGDIα Deacetylases
Treatment of cells with KDAC inhibitors resulted in an increase in the acetylation level of endogenous and transiently expressed RhoGDIα (Fig. 1, B and C). To identify the KDACs responsible for RhoGDIα deacetylation, we performed an in vitro dot blot screen with all mammalian deacetylases (data not shown). We discovered the cytosolic enzymes Sirt2 and HDAC6 to act as RhoGDIα deacetylases. To verify these results, we determined the extent of RhoGDIα deacetylation by immunoblotting (Fig. 7D). Apparently, compared with the non-enzyme control, RhoGDIα Ac-Lys-43, Ac-Lys-52, Ac-Lys-138, and Ac-Lys-178 were significantly deacetylated by Sirt2 (Fig. 7D, bottom left). For HDAC6, Ac-Lys-52G again is the preferred substrate becoming significantly deacetylated (Fig. 7D, bottom right). The other acetylation sites were not significantly deacetylated by HDAC6. Time course experiments with Ac-Lys-52G revealed a nearly complete deacetylation by Sirt2 after 30 min, whereas even using double the amount of HDAC6 did not show complete deacetylation after 6 h (Fig. 7E).
Discussion
RhoGDIα is a major regulator of RhoGNBP function modulating its subcellular distribution and turnover, thereby creating a cytosolic pool of inactive Rho easily activatable for a fast cellular response. Thus, RhoGDIα is essential for the precise spatial and temporal regulation of RhoGNBPs and affects their downstream signaling. How RhoGDIα is regulated to create a high specificity of action is only marginally understood. Here, we present the first functional and structural study describing how RhoGDIα function is controlled by post-translational lysine acetylation (Fig. 7F). Using the powerful genetic code expansion concept to produce site-specifically lysine-acetylated proteins allows us to study its real impact on protein function.
As we show here, RhoGDIα acetylation influences the membrane/cytosol and GTP/GDP cycle of Rho proteins, phenotypically observable by affecting the cellular actin cytoskeleton (Lys-52G, Lys-99G, Lys-141G, and Lys-178G). It furthermore interferes with binding to prenylated (Ac-Lys-52G, Ac-Lys-138G, and Ac-Lys-178G) RhoGNBPs. Lysine acetylation per se leads to an electrostatic quenching of the lysine side chain's positive charge and an increase of the hydrophobicity. Additionally, we observe that Ac-Lys-178G structurally couples β5, β9, and β10 of the Ig domain, most likely interfering with the inherent Ig flexibility (structural coupling). Moreover, we show structurally that the volume of the hydrophobic cavity of the Ig domain is increased, adopting the farnesyl group differently compared with the geranylgeranyl group (PDB entry 4F38). These mechanisms might contribute to the observed reduced RhoA-F binding. Ac-Lys-52G is a loss-of-function modification completely abolishing RhoGDIα function, most likely by interfering with the transition of the intrinsically unfolded N-terminal domain to adopt the helix-loop-helix conformation upon binding to the Rho protein (structural destruction). Thereby, the cell has a system to quickly switch on/off RhoGDIα function. We observed that acetylation at the solvent-exposed Lys-138G also reduced the binding toward RhoA-F. The exact molecular mechanisms need further examination but might also include hydrophobic shielding from the polar solvent as well as structural coupling. RhoGDIα acetylation interferes directly (Ac-Lys-138G) and indirectly (Ac-Lys-141G) with Lys-138G SUMOylation, reported to increase the RhoA affinity (PTM cross-talk). By impairing SUMOylation, Lys-141G acetylation might therefore indirectly affect the observed increase of RhoA at the membrane and cellular F-actin content. Using these mechanisms, RhoGDIα lysine acetylation is a powerful cellular system to tightly regulate the RhoGNBP turnover and residence time at the membrane, finally resulting in a modulation of RhoGNBP signaling.
We have discovered that acetylation at Lys-178G lowers the affinity toward RhoA-F, resulting in more RhoA located at the plasma membrane. Consistent with an increased Rho signaling, we found more filamentous actin in cells expressing the respective acetylation mimicking mutant.
RhoGDIα Lys-52 acetylation completely blocks the interaction with RhoA, drastically reduces RhoA-F binding (4 orders of magnitude) and the membrane extraction capability. This is supported by previous results showing that the N-terminal domain of RhoGDIα is essential for membrane extraction as well as delivery of Rho proteins. Consistently, the expression of K52QG in the N-terminal domain resulted in a cellular F-actin content comparable with mock control, indicating that acetylation at Lys-52G leads to a complete loss of function. Notably, Lys-52G acetylation restores the GEF-catalyzed nucleotide exchange rates, nearly approaching values observed for uncomplexed RhoA. This shows that it might act as a bona fide RhoGDIα displacement factor.
An open question for many proteins identified by quantitative mass spectrometry to be lysine-acetylated is the biological significance of the modification. Many acetylation sites are of low stoichiometry, for which only a gain of function might confer an additional property. Technical progress to determine absolute quantities of acetylation on a whole proteome scale will help to identify biologically important sites (52, 53). Notably, in contrast to many reported studies, for RhoGDIα, we were able to find endogenously acetylated protein in several human cell lines, showing that a significant amount is lysine-acetylated and suggesting that acetylation is of biological importance to control RhoGDIα function. However, the exact absolute quantities could vary and might depend on the cellular metabolic, physiologic, and cell cycle state. Additionally, the expression level and activity of KDACs and KATs might play an important role. Here, we show that RhoGDIα is targeted by the KATs CBP, p300, and pCAF. The KATs p300 and pCAF acetylate RhoGDIα at Lys-52, -138, -141, and -178, the sites being important to control RhoGDIα function as shown here. Moreover, RhoGDIα is differentially deacetylated by Sirt2 and HDAC6 at these sites.
Sirt2 and HDAC6 are the main cytosolic deacetylases known to physically interact, to work synergistically, to colocalize with microtubules, and to deacetylate many proteins involved in cytoskeleton regulation, such as cortactin, mDia2, and α-tubulin (54–56). Down-regulation or inhibition of Sirt2 and HDAC6 were both found to inhibit cell motility as well as cell migration (57). Sirt2 deacetylates and thereby inactivates p300, which in turn is able to inactivate both Sirt2 and HDAC6, constituting a feedback loop (58, 59). The balance between acetylation/deacetylation of RhoGDIα might interfere with cell adhesion, cell migration, and cell motility by directly affecting Rho signal transduction pathways (60).
In summary, the presence of endogenously acetylated RhoGDIα in mammalian cells, site-specific KATs, and KDACs suggests that acetylation is a physiologically important post-translational modification to control RhoGDIα function. Depending on when and where RhoGDIα acetylation takes place in vivo, the cell is able to directly control the Rho protein's lifetime and turnover, its residence time on the membrane, and its subcellular distribution. All of these processes strongly contribute to the initiation and termination of Rho signal transduction pathways, ultimately balancing cell resting and cell motility. A dysfunction in its regulation might therefore lead to severe cellular defects supporting tumorigenesis and neurodegeneration. Because there are just three RhoGDIs in mammalian cells, modification of RhoGDIα by post-translational lysine acetylation can create a sophisticated system to spatially and temporally balance RhoGNBP signal transduction pathways. Tackling the RhoGDIα lysine acetylation machinery might thus be a promising yet underestimated therapeutic approach.
Author Contributions
N. K., S. W. L. C. J., J. W. C., M. K., H. N., A. Z., G. P., and M. L. designed the experiments, discussed data, generated the data, and wrote the manuscript. L. S. and L. B. provided technical assistance with the project. S. W., U. B., and M. L. performed structural analysis. M. S., S. W., and M. L. took x-ray data sets. S. d. B., P. K., and J. B. performed experiments and discussed data.
Acknowledgments
We thank Dr. Astrid Schauss, Dr. Nikolay Kladt, and Dr. Christian Jüngst (CECAD Imaging Facility) for discussions of the cell biological experiments; Dr. Tobias Lamkemeyer for discussion of the proteomic experiments; and Astrid Wilbrand-Hennes and René Grandjean for technical assistance (CECAD Proteomics Facility). We thank Linda Baldus and Lukas Scislowski for expert technical assistance. We thank Prof. Dr. A. Wittinghofer for discussions and experimental materials. We thank the beamline groups at the SLS Villingen/Switzerland and at the Diamond/Oxford UK, whose outstanding efforts have made these experiments possible.
This work was supported by Emmy Noether Grant LA2984-1/1, Sonderforschungsbereich 635 (SFB635; Post-translational Control of Protein Function), and Priority Programs SPP1365 and SPP1580, all from Deutsche Forschungsgemeinschaft. This work was also supported by the European BIOSTRUCTX_5870. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5FR2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GNBP
- guanine nucleotide-binding protein
- RhoGDI
- Rho guanine-nucleotide dissociation inhibitor
- GEF
- guanine nucleotide exchange factor
- mantGDP
- 2/3′-O-(N-methyl-anthraniloyl)-guanosine-5′-diphosphate
- PDB
- Protein Data Bank
- PTM
- post-translational modification
- NTA
- nitrilotriacetic acid
- aa
- amino acid(s)
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- KAT
- lysine acetyltransferase
- ITC
- isothermal titration calorimetry
- KDAC
- lysine deacetylase
- ESI
- electrospray ionization
- RhoA-G
- geranylgeranylated RhoA
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- EGFP
- enhanced GFP.
References
- 1. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 2. Gundersen G. G., Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., and Gomes E. R. (2005) Regulation of microtubules by Rho GTPases in migrating cells. Novartis Found. Symp. 269, 106–116; discussion 116–126, 223–230 [PubMed] [Google Scholar]
- 3. Jaffe A. B., and Hall A. (2005) Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 4. Vetter I. R., and Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 5. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 6. Geyer M., and Wittinghofer A. (1997) GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr. Opin. Struct. Biol. 7, 786–792 [DOI] [PubMed] [Google Scholar]
- 7. Dovas A., and Couchman J. R. (2005) RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Toledo M., Senic-Matuglia F., Salamero J., Uze G., Comunale F., Fort P., and Blangy A. (2003) The GTP/GDP cycling of rho GTPase TCL is an essential regulator of the early endocytic pathway. Mol. Biol. Cell 14, 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunet N., Morin A., and Olofsson B. (2002) RhoGDI-3 regulates RhoG and targets this protein to the Golgi complex through its unique N-terminal domain. Traffic 3, 342–357 [DOI] [PubMed] [Google Scholar]
- 10. DerMardirossian C., Rocklin G., Seo J. Y., and Bokoch G. M. (2006) Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol. Biol. Cell 17, 4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheffzek K., Stephan I., Jensen O. N., Illenberger D., and Gierschik P. (2000) The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 7, 122–126 [DOI] [PubMed] [Google Scholar]
- 12. Grizot S., Fauré J., Fieschi F., Vignais P. V., Dagher M. C., and Pebay-Peyroula E. (2001) Crystal structure of the Rac1-RhoGDI complex involved in NADPH oxidase activation. Biochemistry 40, 10007–10013 [DOI] [PubMed] [Google Scholar]
- 13. Hoffman G. R., Nassar N., and Cerione R. A. (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 [DOI] [PubMed] [Google Scholar]
- 14. Tnimov Z., Guo Z., Gambin Y., Nguyen U. T., Wu Y. W., Abankwa D., Stigter A., Collins B. M., Waldmann H., Goody R. S., and Alexandrov K. (2012) Quantitative analysis of prenylated RhoA interaction with its chaperone, RhoGDI. J. Biol. Chem. 287, 26549–26562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dransart E., Morin A., Cherfils J., and Olofsson B. (2005) RhoGDI-3, a promising system to investigate the regulatory function of rhoGDIs: uncoupling of inhibitory and shuttling functions of rhoGDIs. Biochem. Soc. Trans. 33, 623–626 [DOI] [PubMed] [Google Scholar]
- 16. Takahashi K., Sasaki T., Mammoto A., Takaishi K., Kameyama T., Tsukita S., and Takai Y. (1997) Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem. 272, 23371–23375 [DOI] [PubMed] [Google Scholar]
- 17. DerMardirossian C., Schnelzer A., and Bokoch G. M. (2004) Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 15, 117–127 [DOI] [PubMed] [Google Scholar]
- 18. DerMardirossian C. M., and Bokoch G. M. (2006) Phosphorylation of RhoGDI by p21-activated kinase 1. Methods Enzymol. 406, 80–90 [DOI] [PubMed] [Google Scholar]
- 19. Gorvel J. P., Chang T. C., Boretto J., Azuma T., and Chavrier P. (1998) Differential properties of D4/LyGDI versus RhoGDI: phosphorylation and rho GTPase selectivity. FEBS Lett. 422, 269–273 [DOI] [PubMed] [Google Scholar]
- 20. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 21. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., and Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 22. Boulter E., and Garcia-Mata R. (2010) RhoGDI: A rheostat for the Rho switch. Small GTPases 1, 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sawada N., Itoh H., Miyashita K., Tsujimoto H., Sone M., Yamahara K., Arany Z. P., Hofmann F., and Nakao K. (2009) Cyclic GMP kinase and RhoA Ser188 phosphorylation integrate pro- and antifibrotic signals in blood vessels. Mol. Cell Biol. 29, 6018–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rolli-Derkinderen M., Sauzeau V., Boyer L., Lemichez E., Baron C., Henrion D., Loirand G., and Pacaud P. (2005) Phosphorylation of serine 188 protects RhoA from ubiquitin/proteasome-mediated degradation in vascular smooth muscle cells. Circ. Res. 96, 1152–1160 [DOI] [PubMed] [Google Scholar]
- 25. Yu J., Zhang D., Liu J., Li J., Yu Y., Wu X. R., and Huang C. (2012) RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J. Biol. Chem. 287, 13752–13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park J., Chen Y., Tishkoff D. X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B. M., Skinner M. E., Lombard D. B., and Zhao Y. (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y., Zhao W., Yang J. S., Cheng Z., Luo H., Lu Z., Tan M., Gu W., and Zhao Y. (2012) Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol. Cell Proteomics 11, 1048–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lammers M., Neumann H., Chin J. W., and James L. C. (2010) Acetylation regulates cyclophilin A catalysis, immunosuppression and HIV isomerization. Nat. Chem. Biol. 6, 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weisshaar S. R., Keusekotten K., Krause A., Horst C., Springer H. M., Göttsche K., Dohmen R. J., and Praefcke G. J. (2008) Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582, 3174–3178 [DOI] [PubMed] [Google Scholar]
- 31. Fres J. M., Müller S., and Praefcke G. J. (2010) Purification of the CaaX-modified, dynamin-related large GTPase hGBP1 by coexpression with farnesyltransferase. J. Lipid Res. 51, 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moriarty N. W., Grosse-Kunstleve R. W., and Adams P. D. (2009) Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeLano W. L. (2002) The PyMOL Molecular Graphics System, version 1.7.2.0, DeLano Scientific, San Carlos, CA [Google Scholar]
- 43. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 44. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 45. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 46. Neumann H., Peak-Chew S. Y., and Chin J. W. (2008) Genetically encoding Nϵ-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 [DOI] [PubMed] [Google Scholar]
- 47. de Boor S., Knyphausen P., Kuhlmann N., Wroblowski S., Brenig J., Scislowski L., Baldus L., Nolte H., Krüger M., and Lammers M. (2015) Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc. Natl. Acad. Sci. U.S.A. 112, E3679–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solski P. A., Helms W., Keely P. J., Su L., and Der C. J. (2002) RhoA biological activity is dependent on prenylation but independent of specific isoprenoid modification. Cell Growth Differ. 13, 363–373 [PMC free article] [PubMed] [Google Scholar]
- 49. Moissoglu K., McRoberts K. S., Meier J. A., Theodorescu D., and Schwartz M. A. (2009) Rho GDP dissociation inhibitor 2 suppresses metastasis via unconventional regulation of RhoGTPases. Cancer Res. 69, 2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Platko J. V., Leonard D. A., Adra C. N., Shaw R. J., Cerione R. A., and Lim B. (1995) A single residue can modify target-binding affinity and activity of the functional domain of the Rho-subfamily GDP dissociation inhibitors. Proc. Natl. Acad. Sci. U.S.A. 92, 2974–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Euden J., Mason S. A., Viero C., Thomas N. L., and Williams A. J. (2013) Investigations of the contribution of a putative glycine hinge to ryanodine receptor channel gating. J. Biol. Chem. 288, 16671–16679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baeza J., Dowell J. A., Smallegan M. J., Fan J., Amador-Noguez D., Khan Z., and Denu J. M. (2014) Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289, 21326–21338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinert B. T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., and Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 55. North B. J., Marshall B. L., Borra M. T., Denu J. M., and Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 56. Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., and Sánchez-Madrid F. (2008) HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 18, 291–297 [DOI] [PubMed] [Google Scholar]
- 57. Zuo Q., Wu W., Li X., Zhao L., and Chen W. (2012) HDAC6 and SIRT2 promote bladder cancer cell migration and invasion by targeting cortactin. Oncol. Rep. 27, 819–824 [DOI] [PubMed] [Google Scholar]
- 58. Han Y., Jin Y. H., Kim Y. J., Kang B. Y., Choi H. J., Kim D. W., Yeo C. Y., and Lee K. Y. (2008) Acetylation of Sirt2 by p300 attenuates its deacetylase activity. Biochem. Biophys. Res. Commun. 375, 576–580 [DOI] [PubMed] [Google Scholar]
- 59. Han Y., Jeong H. M., Jin Y. H., Kim Y. J., Jeong H. G., Yeo C. Y., and Lee K. Y. (2009) Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem. Biophys. Res. Commun. 383, 88–92 [DOI] [PubMed] [Google Scholar]
- 60. Garcia-Mata R., Boulter E., and Burridge K. (2011) The “invisible hand”: regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 12, 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wiseman T., Williston S., Brandts J. F., and Lin L. N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 62. Diederichs K., and Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275; Erratum (1997) Nat. Struct. Biol. 4, 592 [DOI] [PubMed] [Google Scholar]
- 63. Diederichs K., and Karplus P. A. (2013) Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 69, 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brünger A. T. (1997) Free R value: cross-validation in crystallography. Methods Enzymol. 277, 366–396 [DOI] [PubMed] [Google Scholar]