Abstract
The EGF receptor can bind seven different agonist ligands. Although each agonist appears to stimulate the same suite of downstream signaling proteins, different agonists are capable of inducing distinct responses in the same cell. To determine the basis for these differences, we used luciferase fragment complementation imaging to monitor the recruitment of Cbl, CrkL, Gab1, Grb2, PI3K, p52 Shc, p66 Shc, and Shp2 to the EGF receptor when stimulated by the seven EGF receptor ligands. Recruitment of all eight proteins was rapid, dose-dependent, and inhibited by erlotinib and lapatinib, although to differing extents. Comparison of the time course of recruitment of the eight proteins in response to a fixed concentration of each growth factor revealed differences among the growth factors that could contribute to their differing biological effects. Principal component analysis of the resulting data set confirmed that the recruitment of these proteins differed between agonists and also between different doses of the same agonist. Ensemble clustering of the overall response to the different growth factors suggests that these EGF receptor ligands fall into two major groups as follows: (i) EGF, amphiregulin, and EPR; and (ii) betacellulin, TGFα, and epigen. Heparin-binding EGF is distantly related to both clusters. Our data identify differences in network utilization by different EGF receptor agonists and highlight the need to characterize network interactions under conditions other than high dose EGF.
Keywords: cancer biology, epidermal growth factor (EGF), epidermal growth factor receptor (EGFR), growth factor, receptor tyrosine kinase, systems biology
Introduction
The EGF receptor is an intrinsic membrane protein composed of an extracellular ligand-binding domain connected to an intracellular tyrosine kinase domain by a single transmembrane α-helix. In the absence of ligand, the EGF receptor is thought to exist as a monomer, although inactive “pre-dimers” are known to form (1–5). Upon binding an agonist ligand, the EGF receptor dimerizes leading to the activation of its tyrosine kinase and the phosphorylation of tyrosine residues in the C-terminal tail of the receptor (6–8).
The phosphorylated tyrosines on the EGF receptor serve as binding sites for a large number of signaling proteins that contain SH2 and/or phosphotyrosine-binding domains (9, 10). Some of these proteins, such as Cbl, possess an enzymatic activity (11). Others, such as Grb2 or Shc, serve as adapter proteins that bring other proteins into the EGF receptor-containing complex. For example, Grb2 recruits the scaffolding protein, Gab1, to the EGF receptor (12). Phosphorylation of Gab1 by the EGF receptor allows Gab1 to recruit additional proteins, such as Shp2 or PI3K-R1, to the signaling complex (13–16). The recruitment of these signaling proteins to the receptor ultimately triggers the activation of a variety of downstream signaling pathways, thereby mediating the intracellular effects of growth factor binding.
The EGF receptor binds seven different agonist ligands, including some of high affinity (EGF, TGFα, BTC,6 and HB-EGF) and some of low affinity (AREG, EPG, and EPR) (17). It has been reported that different EGF receptor ligands induce different responses when binding to the same cell line (18–21). Given that these agonists bind to the same receptor and stimulate similar downstream signaling molecules, it is difficult to explain how these divergent responses are achieved.
We have previously used a luciferase fragment complementation system to assess the ability of EGF to induce dimerization of the EGF receptor (22–24). In this study, we use our luciferase fragment complementation assay to visualize the recruitment of a variety of signaling proteins to the EGF receptor. The fine temporal resolution and quantitative nature of the split luciferase complementation system allowed us to continuously monitor the association of Cbl, CrkL, Gab1, Grb2, PI3K-R1, p52 Shc, p66 Shc, and Shp2 with the EGF receptor in response to increasing concentrations of all seven different EGF receptor ligands. Principal component analysis was applied to this large dataset to determine how the response to these growth factors differed. The data demonstrate that each growth factor produces a unique signature for the recruitment of signaling proteins, and this signature differs at different doses of the same growth factor. This suggests that each growth factor utilizes the signaling network differently, preferentially promoting flux through some pathways over others, which could readily lead to a different net biological outcome.
Experimental Procedures
Materials
EGF was purchased from Biomedical Technologies. TGFα and amphiregulin were from Leinco. Betacellulin was from ProSpec. Heparin-binding EGF was from Sigma. Epigen and epiregulin were synthesized and purified in the laboratory of Dr. Mark Lemmon (University of Pennsylvania). FetalPlex was from Gemini Bioproducts. The anti-EGF receptor antibody was from Cell Signaling. The PY20 anti-phosphotyrosine antibody was from BD Transduction Laboratories.
DNA Constructs
Full-length cDNA constructs for the signaling proteins were obtained from Addgene (CrkL PI3K-R1 and Shp2), Source Bioscience (Gab1), Thermo Fisher (p52 Shc, p66 Shc, and Grb2), or Sino Biologicals (c-Cbl). The stop codon in each was removed, and an in-frame BsiWI site was inserted through site-directed mutagenesis. The cDNAs were cut with BsiWI and fused to the C-terminal fragment of luciferase (CLuc). The construct was moved into the pcDNA3.1 Zeo expression vector where expression of the fusion protein is driven off the constitutive CMV promoter.
Cell Lines
CHO cells stably expressing the tetracycline-inducible EGF receptor C-terminally fused to the N-terminal fragment of firefly luciferase (EGFR-NLuc) (24) were used as the starting parental line. These cells were transfected with the pcDNA3.1 Zeo plasmids encoding the CLuc fusion of each of the eight signaling proteins. Eight (double) stable cell lines were selected by growth in 5 mg/ml Zeocin. Quantitation of EGF receptor expression in each line by 125I-EGF saturation binding indicated that the number of cell surface EGF receptors expressed in each line is within ±20% of the average level of receptor expression (data not shown). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FetalPlex, 100 μg/ml G418, 100 μg/ml hygromycin, and 100 μg/ml Zeocin and maintained in an incubator at 37 °C in 5% CO2.
Luciferase Assays
Double stable CHO cells were plated into 96-well black-walled dishes 2 days prior to use in medium containing 1.5 μg/ml doxycycline to induce expression of the EGFR-NLuc fusion protein. For assay, cells were transferred into Dulbecco's phosphate-buffered saline supplemented with 5 mg/ml BSA and 20 mm MOPS, pH 7.2. Cells were incubated with 0.9 mg/ml d-luciferin for 30 min at 37 °C prior to the addition of growth factor and the start of imaging. Cell radiance (photons/s/cm2/steradian) was measured every 30 s for 25 min using a cooled charge-coupled device camera in the IVIS50 or IVIS Lumina imaging system. Assays were performed in hextuplicate. The lines through the data were drawn using Equation 1, which represents the sum of a logistics association equation and an exponential dissociation equation.
 |
where Y = photons/s at time (t). k1 represents the association rate constant, and k2 is the dissociation rate constant. This curve drawing was not part of the principal component analysis and was used only for visual presentation of the dose-response curves.
Western Blotting
CHO cells expressing the wild type EGF receptor were treated without or with 5 μm erlotinib or 10 μm lapatinib for 1 h and then stimulated with the indicated concentrations of EGF for 5 min. Lysates were prepared, and Western blotting with anti-EGF receptor and anti-phosphotyrosine antibodies were performed as described previously (23).
PCA and Enrichment Analysis
Computational analysis was performed using the Python programming language. PCA utilized the scikit-learn package (25). PCA was performed on a 280 × 44 matrix, with 280 unique combinations of protein, growth factor, and dose, each with 44 time points, normalized to the maximal response elicited for that agonist/protein pair. For PCA, a subset of five (out of seven) doses was chosen for each growth factor to bracket the EC50 value for the recruited signaling proteins as follows: for BTC and EGF, the doses ranged from 0.03 to 3 nm; for TGF, the doses ranged from 0.1 to 10 nm; for HB-EGF, the doses ranged from 0.3 to 30 nm; and for AREG, EPG, and EPR, the doses ranged from 3 to 300 nm. References to “low” doses of growth factor (as used in Figs. 4 and 7) represent the second dose in the five-dose series, and references to “high” doses (as used for Fig. 7) represent the fourth dose in the five-dose series. Distances between protein pairs were calculated using Euclidean distance between the five-dimensional vector across doses in PC space. Top- and bottom-quartile enrichment was calculated using the hypergeometric test and Bonferroni-corrected.
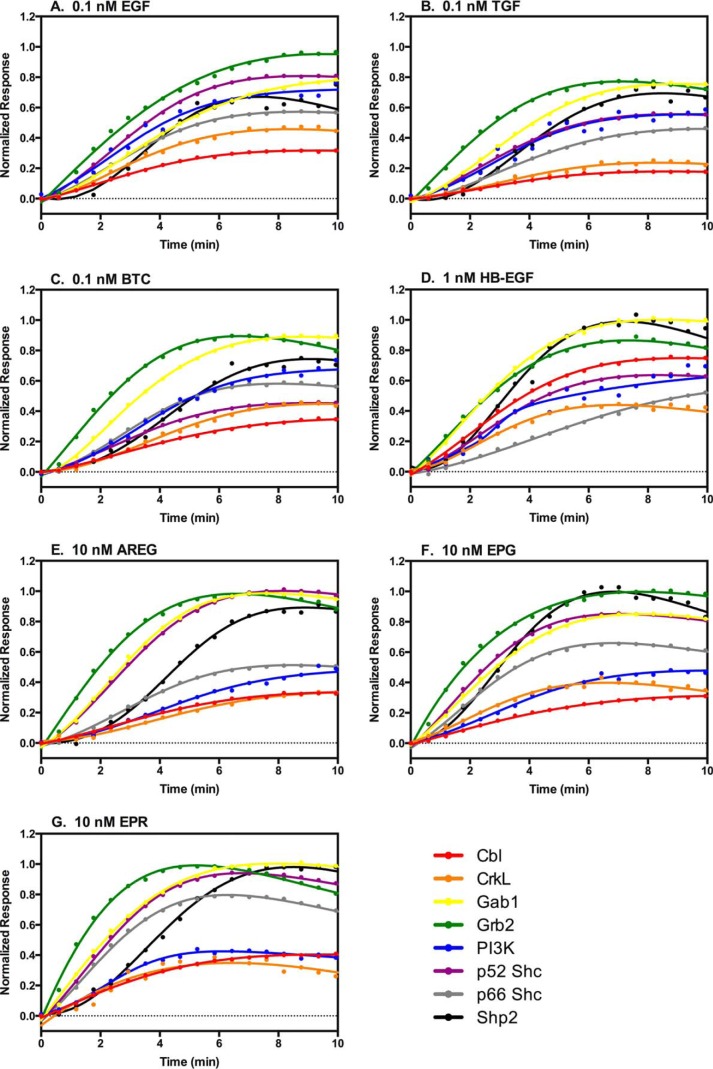
FIGURE 4.
Relative response times for the recruitment of the eight signaling proteins by comparable low doses of each of the EGF receptor ligands. The response to the indicated low concentration of each of the seven agonists was normalized to the maximal response elicited by that agonist for that EGF receptor/protein pair. The normalized responses for all signaling proteins stimulated by a single agonist were then plotted on the same graph.
FIGURE 7.

Signaling protein response varies by growth factor dose. To quantify differences in protein response, Euclidean distances were calculated between proteins for each growth factor at both a low and high dose and visualized as a heat map. The response pattern for each growth factor at low dose is in the left column, and the response pattern for a high dose is in the right column.
For clustering of the growth factors based on protein recruitment across all doses, pairwise protein distances for each ligand were converted to a one-dimensional vector. The vectors for each ligand were then clustered using hierarchical clustering. An ensemble of 35 clustering results was generated by varying linkages (single, complete, average, and weighted) and distance metrics (Euclidean, Pearson correlation, city block, cosine, Bray-Curtis, Canberra, Chebyshev, and square Euclidean). The Euclidean metric was also used with median, centroid, and Ward linkage. The results for each ligand were assembled into a matrix and hierarchically clustered using single linkage and Euclidean distance. p66 Shc was not included in this analysis so as not to over-weight the results toward the contribution of Shc isoforms.
Results
Generation and Characterization of Stable Cell Lines
The split luciferase complementation assay utilizes an N-terminal (NLuc) and C-terminal (CLuc) fragment of firefly luciferase (26). Individually, the fragments have no enzymatic activity. However, when they are brought into proximity, they complement each other forming a catalytically active luciferase that produces light upon oxidation of luciferin. For our luciferase complementation assays, each of eight signaling proteins (Cbl, CrkL, Gab1, Grb2, PI3K-R1, p52 Shc, p66 Shc, and Shp2) was C-terminally fused to the CLuc fragment via a 16-amino acid flexible linker. The cDNA for the fusion protein was then transfected into a CHO cell line that stably expressed the EGF receptor C-terminally fused to the NLuc fragment (EGFR-NLuc) off a tetracycline-inducible promoter. Double stable cell lines were selected for use in these experiments. For assay, the CHO cells were cultured for 24 h in 1.5 μg/ml doxycycline to induce expression of the EGFR-NLuc fusion protein. The signaling proteins were constitutively expressed from a CMV promoter.
Luciferase Complementation between the EGF Receptor and Signaling Proteins
All eight signaling proteins yielded an EGF-stimulated increase in luciferase activity when co-expressed in cells with EGFR-NLuc (Fig. 1). EGF-stimulated complementation between the EGF receptor and these signaling proteins was seen as early as 30 s after the addition of EGF. At low concentrations of EGF, essentially all of the pairings exhibited a rapid rise in luciferase activity, which peaked by ∼5–8 min. For some pairings, such as the EGF receptor and PI3K-R1 (Fig. 1E), this level of complementation was maintained over the entire time course at all doses. In other pairings, such as Cbl (Fig. 1A) or CrkL (Fig. 1B), complementation plateaued at low concentrations of growth factor but declined after an early peak at high concentrations of EGF. Still other proteins demonstrated a bimodal response across doses. For example, for Grb2 (Fig. 1D) and Shp2 (Fig. 1H), the maximum complementation occurred at a relatively low dose of EGF, with higher doses of growth factor resulting in lower peak responses and a marked decrease at longer times.
FIGURE 1.
EGF-stimulated association of eight signaling proteins with the EGF receptor measured using luciferase fragment complementation imaging. CHO cells stably co-expressing EGFR-NLuc and the CLuc-fused version of one of eight signaling proteins were assayed for EGF-stimulated light production in the presence of luciferin. Cells were stimulated with the indicated concentration of EGF at time t = 0, and light production was monitored for 25 min.
In the EGF receptor/Shp2 pairing (Fig. 1H), the luciferase activity observed at the highest concentrations of EGF actually fell below the basal level after about 15 min. These data imply that Shp2 associates with the EGF receptor under non-stimulated conditions. This association is apparently disrupted upon stimulation with high doses of growth factor.
If these signaling proteins are being recruited to the EGF receptor via phosphotyrosine-dependent interactions, then the associations visualized through luciferase complementation should be sensitive to inhibition of the EGF receptor tyrosine kinase. As shown in Fig. 2, A–H, treatment of cells with 5 μm erlotinib (green lines) effectively inhibited EGF-stimulated complementation between the EGF receptor and each of these eight signaling proteins. Inhibition was essentially complete for all pairings with the exception of p52 Shc and p66 Shc, for which the inhibition was ∼70%. The complementation between the EGF receptor and Shp2 actually showed an EGF-stimulated decline in luciferase activity, again consistent with there being a basal level of association between the two proteins, which is disrupted after ligand binding.
FIGURE 2.
Effect of erlotinib and lapatinib on EGF-stimulated association of eight signaling proteins with the EGF receptor. A–H, CHO cells stably co-expressing EGFR-NLuc and the CLuc-fused version of one of eight signaling proteins were treated with 5 μm erlotinib (green lines) or 10 μm lapatinib (red lines) for 60 min prior to stimulation without or with 0.3 nm EGF. EGF-stimulated light production was monitored for 25 min after addition of EGF. I, CHO cells expressing wild type EGF receptor were treated with 5 μm erlotinib or 10 μm lapatinib for 60 min prior to stimulation with 0.3 or 3.0 nm EGF. Lysates were prepared and equal amounts of protein analyzed by SDS-gel electrophoresis and Western blotting with an anti-phosphotyrosine antibody or an anti-EGF receptor antibody. Quantitation of anti-phosphotyrosine blot is shown.
Despite the fact that lapatinib appeared to inhibit EGF receptor autophosphorylation to the same extent as erlotinib (Fig. 2I), pretreatment of the cells with 10 μm lapatinib (red lines) was far less effective than pretreatment with erlotinib at blocking the association of these signaling proteins with the EGF receptor. Although lapatinib was able to completely block complementation between the EGF receptor and Cbl, CrkL, and Shp2, the other five signaling proteins all showed at least 20% residual EGF-stimulated luciferase activity in the presence of lapatinib. The association of Gab1 was particularly insensitive to lapatinib treatment. Thus, there is a significant difference between erlotinib and lapatinib in terms of their efficacy for inhibiting EGF-stimulated signaling complex assembly.
Recruitment Stimulated by Other EGF Receptor Ligands
The EGF receptor is activated by a family of homologous growth factors, including EGF TGFα, BTC, HB-EGF, AREG, EPG, and EPR (17). To quantify the similarities and differences in the response of cells to stimulation by each of these ligands, the luciferase complementation assay was used to monitor the recruitment of the eight different signaling proteins to the EGF receptor in response to each of these agonist ligands.
Supplemental Figs. 1–6 show the time courses of the recruitment of these eight signaling proteins to the EGF receptor in response to increasing doses of each of these additional six growth factors. Like EGF, all of these growth factors stimulated the recruitment of all eight signaling proteins in a dose-dependent manner. However, the patterns of the dose response curves for all seven growth factors for each individual pairing were similar. For example, for all growth factors, PI3K-R1 recruitment plateaued early, and the level of signal was maintained over the entire time course. Similarly, the bimodal response for the recruitment of Grb2 and Shp2 was observed for all growth factors.
Table 1 reports the estimated EC50 values for each ligand stimulating the recruitment of each protein. As expected from their low binding affinities, AREG, EPG, and EPR required ∼30–100-fold greater concentrations of ligand to stimulate a maximal response than did EGF, TGFα, BTC, or HB-EGF. Surprisingly, the EC50 values for a given growth factor for stimulating the recruitment of the different signaling proteins differed up to 18-fold.
TABLE 1.
EC50 values for agonist-stimulated EGF receptor/protein association
Table compares the EC50 values for each growth factor for stimulating the recruitment of the eight signaling proteins. These values were estimated based on the response to each dose of growth factor at t = 2.5 min. This largely eliminates the effects of the declines in signal at longer times and means that these values reflect mainly the initial association of the two proteins. The EC50 values differed for the recruitment of different proteins by the same growth factor. So, for example, EGF exhibited an EC50 of ∼0.03 nm for recruiting Grb2 and PI3K-R1 but an EC50 about 10-fold higher for recruiting Cbl. EPG exhibited the widest range of EC50 values (∼18-fold difference), whereas HB-EGF showed the smallest range of EC50 values (∼3-fold).
| EC50 | EGF | TGF | BTC | HB-EGF | AREG | EPG | EPR |
|---|---|---|---|---|---|---|---|
| nm | |||||||
| Cbl | 0.31 | 0.73 | 0.40 | 0.85 | 36.0 | 33.0 | 21.0 |
| CrkL | 0.14 | 0.40 | 0.25 | 1.5 | 26.0 | 19.0 | 21.0 |
| Gab1 | 0.08 | 0.11 | 0.06 | 0.47 | 5.9 | 4.8 | 2.7 |
| Grb2 | 0.03 | 0.06 | 0.04 | 0.64 | 4.0 | 1.8 | 2.7 |
| PI3K-RI | 0.03 | 0.24 | 0.11 | 1.2 | 21.0 | 22.0 | 18.0 |
| p52 Shc | 0.06 | 0.12 | 0.15 | 0.85 | 3.7 | 3.7 | 2.6 |
| p66 Shc | 0.10 | 0.21 | 0.05 | 1.7 | 13.0 | 6.1 | 4.0 |
| Shp2 | 0.09 | 0.11 | 0.09 | 0.50 | 13.0 | 3.3 | 4.0 |
Fig. 3 compares the extent of recruitment of the eight signaling proteins in response to an optimal concentration of each of the seven growth factors. The concentrations compared were those that gave the maximal peak response for that particular pairing (Fig. 1 and supplemental Figs. 1–6). For most of the pairs, all seven ligands stimulated a similar maximal response. However, HB-EGF routinely elicited a slightly lower response than the other growth factors. The greatest difference in response was observed for the recruitment of Grb2 for which the response to EPG and EPR was ∼30% higher than that to EGF, while the response to HB-EGF was ∼30% lower than that to EGF. Consequently, there was nearly a 2-fold difference in the relative extent of Grb2 recruitment between the high of EPG/EPR and the low of HB-EGF.
FIGURE 3.
Comparison of the association of eight signaling proteins with the EGF receptor stimulated by optimal concentrations of the seven EGF receptor agonists. CHO cells stably co-expressing EGFR-NLuc and the CLuc-fused version of one of eight signaling proteins were stimulated at t = 0 with the concentration of each growth factor that yielded maximal peak complementation for a given receptor/protein pair and light production monitored for 25 min.
Fig. 4 compares the ability of a fixed (comparable) low dose of each growth factor to stimulate the recruitment of all eight signaling proteins. The responses have been normalized to the maximal response observed for that EGFR/protein pair at the optimal dose of that growth factor. For all growth factors, Grb2 appears to have the fastest relative response time. Cbl and CrkL most frequently have the slowest relative response time. The recruitment of PI3K-R1 shows the most variability being similar to Cbl and CrkL for the low affinity ligands but closer to Grb2 and Gab1 for the high affinity ligands. Interestingly, the recruitment of p52 Shc and p66 Shc differs noticeably from each other. In many cases, p52 Shc shows a shorter relative response time than p66 Shc, often significantly shorter, as for AREG and HB-EGF. However, this order is reversed for BTC where p66 Shc is recruited more rapidly than p52 Shc. Thus, at the earliest times of signal transduction, differences in response to the different the growth factors can be identified and would contribute to a different biological outcome.
Global Behaviors Observed via Reduced Dimensionality
The foregoing data represent an extremely rich set of measurements of the recruitment of eight different signaling proteins by the EGF receptor. Within this dataset, discovering relationships among the proteins and growth factors is difficult due to the high dimensionality of the problem. To reduce the dimensionality of the dataset, while keeping the relationships within the data intact, we used PCA.
For this analysis, a fixed subset of five (out of the seven) doses of each growth factor was used (Supplemental Table 1). The subset of doses was chosen so that we captured a comparable range of response above and below the EC50 values for each of the different growth factors. As a result, the doses that were not included in the analysis were either the very lowest concentrations that elicited a weak or no response or the very highest concentrations that were super-saturating. This approach allows us to compare the behavior of the same dose of a single growth factor across all eight signaling proteins and to compare the behavior of a single signaling protein at comparable concentrations across the different growth factors.
We could account for 97.0% of the co-variation within the entire dataset by projecting the original data into the first two dimensions of the principal component space. Principal component 1 (PC1) captures 87.6% of the variance, whereas PC2 captures 9.4% of the variance. As a result, each time series for one growth factor dose and signaling protein response can be plotted as a single point in two-dimensional PC space while retaining almost all of the variation that exists in the original 44 dimensions (i.e. the 44 time points per curve). The loading plots (Fig. 5A) indicate that PC1 represents a positive integration of information across most time points. By contrast, PC2 negatively weights the earliest time points while positively weighting the latter half of the time course.
FIGURE 5.
Dimensionality can be reduced using principal component analysis. A, plot of the loadings of the first two principal components, which accounts for 97.0% of the covariance in the dataset (PC1 accounts for 87.6%; PC2 accounts for 9.4%). B, correlation of PC1 with maximum peak magnitude and PC2 with peak time was calculated across five doses for each protein-growth factor pair using Pearson correlation. Mean correlation for PC1 with maximum peak magnitude was 0.97, whereas mean correlation for PC2 with peak time was 0.75. C, dataset is plotted based on projections onto the first two principal components, capturing 97.0% of the variance. Individual points are colored according to the signaling protein measured.
Based on our observations of the data, we thought the latent dimensions of the principal component analysis might describe physical features of the data, specifically information about the relative maximum signal achieved and the rate at which this signal was achieved. To test this hypothesis, for each protein-ligand pair we determined the correlation between the dose-response vector in PC1 and the magnitude of the peak for each dose in the original normalized data. We also determined the correlation between the PC2 dose-response vector and the time of peak signal for each dose. As shown in Fig. 5B, there is an extremely high correlation between the PC1 value and the relative magnitude of the peak response (mean r = 0.97). Similarly, with the exception of a few outliers in PC2 space, there is a high correlation between the value in PC2 space and the time to peak response in the original data (mean r = 0.76). The high correlation indicates that we can ascribe physical meaning to our principal component axes. Specifically, higher values along PC1 indicate that the signal achieves a higher relative maximum value. Higher values in PC2 space indicate that the signal achieves its maximum value at a later time. Lower values in PC2 space indicate that the signal achieves its maximum value at an earlier time.
Fig. 5C shows the entire dataset reduced to the first two dimensions of PCA space. Each point represents a time course for a particular signaling protein at a single dose of a single growth factor. Points are colored to indicate the signaling protein being recruited to the EGF receptor. Points close together in PCA space represent responses that are similar to each other across the entire time course. The responses of Shp2 and PI3K-R1 are the most separated in both PCs indicating that they are the most different. The remaining points are densely packed in the intermediate region between the extremes of the PI3K-R1 and Shp2 signals.
To identify trends in the data, the measurements were organized into a dose series for each ligand/protein pair. This was visualized by connecting the PCA point representing the curve at the lowest dose of one growth factor to the point representing the curve at the next higher dose of that same growth factor with a directed arrow, continuing on for the five doses of each growth factor (see supplemental Fig. 7 for an example). This approach reveals significant trends in the evolution of signals across the dose range, despite the density initially observed in PC space (Fig. 6). Overall, the major mode of behavior for a given signaling protein is dominated by the identity of the protein rather than the identity of the growth factor. Therefore, the curves describing the recruitment of the same signaling protein stimulated by any of the seven ligands (Fig. 6A) are more similar to each other than they are to the curves that describe the recruitment of a different signaling protein stimulated by the same growth factor (Fig. 6B). As is apparent from Fig. 6B, there are differences in how the individual protein responses evolve based on the stimulatory ligand.
FIGURE 6.
Global trends based on dose response. The individual points in two-dimensional PC space representing a protein-ligand pair at a particular dose were organized into a dose-response series for the five chosen doses, by connecting the response at lower doses to the next higher dose using a directed arrow. A, resulting vectors are grouped by signaling protein and colored according to the growth factor. B, resulting vectors are grouped by growth factor and colored according to signaling protein.
Aside from these general observations, each PC shows two contrasting trends in a subset of proteins. First, for Cbl, CrkL, PI3K-RI, p52 Shc, and p66 Shc, there is a monotonic increase in PC1 (relative maximal signal) as the dose of growth factor increases. This is what is expected in a traditional dose-response curve, i.e. the signal increases with increasing dose. By contrast, Gab1, Grb2, and Shp2 show a bimodal response in PC1 space, reflecting an initial increase in response followed by a marked decrease in peak signal at the highest doses of most of the growth factors.
A second trend is that for most of the signals there is a monotonic progression down the PC2 axis. This indicates that the peak response is achieved more rapidly at higher concentrations of growth factor. An exception to this rule is the recruitment of p52 Shc in response to EGF, BTC, and HB-EGF, where there is little change in the time to peak response over the entire dose range tested.
Pairwise Interactions
The proteins chosen for this study were selected because they are involved in well documented interactions with the EGF receptor and with each other. Therefore, we would expect that the behaviors of some of these proteins should correlate in PC space. To quantify these relationships, we calculated the distances between interacting protein pairs in PC space for each dose of a single growth factor. Fig. 7 shows heat maps of protein-pair distances for a low dose and a high dose for each growth factor. Two important features are immediately apparent from these heat maps. First, the patterns seen for the low and high doses of the same growth factor are distinctly different. This suggests that the same growth factor utilizes the network differently when applied at different concentrations. Second, the heat maps for each growth factor are very different, suggesting that the different growth factors activate the network in a manner that is specific to that growth factor.
To evaluate the similarity of protein recruitment dynamics across the different growth factors, distances were calculated between protein pairs in PC space across the five doses of each individual growth factor. The complete set of these cumulative distances was then rank-ordered, and both the top and bottom quartiles were probed for statistical enrichment for individual proteins or specific protein pairs (supplemental Table 2).
Several specific protein pairs were strongly represented in the top quartile. The pairwise distances of Cbl with CrkL and p52 Shc with p66 Shc were the most significantly enriched protein pairs in the top quartile (p < 0.005, Bonferroni-corrected), whereas the interaction of Gab1 with p52 Shc was also significantly enriched (p < 0.05, Bonferroni-corrected).
Enrichment in the bottom quartile was also calculated to identify proteins and protein pairs that rarely exhibited similar dynamics. Shp2-based interactions as a group were identified as being enriched in this quartile (p < 0.0005, Bonferroni-corrected).
To compare the global response of the network to each of the seven different growth factors, we performed ensemble clustering on the pairwise distances between proteins, as described under “Experimental Procedures.” The percentage of time each growth factor clustered with another growth factor in the ensemble of clustering solutions is visualized in Fig. 8 as a heat map. The growth factors were then hierarchically clustered. As can be seen from this figure, BTC, EPG, and TGFα form a strong cluster (the BTC cluster) because they cluster together in every clustering solution in the ensemble. AREG and EPR form a second strong cluster (the AREG cluster). EGF clusters most frequently with the AREG cluster (77%) but shares some membership in the BTC cluster (23%). HB-EGF is rather unique and is far from both the BTC and AREG clusters.
FIGURE 8.

Heat map and dendrogram showing the results of clustering of the responses to the seven growth factors. The pairwise protein distances for each ligand were converted to a vector, and the vectors for each ligand were hierarchically clustered via the ensemble approach described under “Experimental Procedures.” The results are visualized via a heat map displaying the fraction of time each ligand pair clustered together across the ensemble.
Discussion
We report here on the use of luciferase fragment complementation to study the association of downstream signaling proteins with the EGF receptor. The advantages of this system include the ease of assay and the fact that it can be done in live cells with continuous monitoring. In addition, the signals generated from the eight signaling proteins examined here were robust, allowing detection of differences associated with changes in the concentration of growth factor. Finally, the approach is scalable and useful for screening applications.
Using this system, we found that all eight of the selected signaling proteins are rapidly recruited to an EGF receptor-containing complex, with association being apparent by 30 s. The peak extent of association occurred between 5 and 7 min, depending on the pairing. This is consistent with the time course of assembly of Shc-containing complexes after stimulation of cells with EGF, as documented through quantitative mass spectrometry (27).
In most of the pairings, the luciferase signal decreased slowly over time particularly at the higher doses of growth factor. As internalization of the EGF receptor begins almost immediately after addition of growth factor (28), it seems likely that at least part of the decrease in signal at longer times is due to internalization and degradation of the receptor and its associated signaling proteins. Nevertheless, at least some fraction of the agonist-induced increase in luciferase activity is maintained for as long as 25 min after the addition of EGF. These data imply that these signaling proteins remain associated with the EGF receptor even after it has been internalized. Thus, some aspects of signaling probably continue to occur well after the receptor has been removed from the cell surface.
Receptor internalization is unlikely to account for the decrease in peak signal observed for the recruitment of Gab1, Grb2, and Shp2 at high concentrations of all the growth factors. This decrease could reflect increased competition between signaling proteins for binding to sites on the EGF receptor when the signal is strong. It could also arise from depletion of a common pool of adapter or scaffold proteins when the stimulatory signal is maximal. Finally, it is possible that there is steric interference with luciferase complementation when the signal is strong, and Gab1, Grb2, and Shp2 bind to the EGF receptor in a multiprotein complex.
All EGF receptor/signaling protein pairs showed a dosedependence on the concentration of growth factor. However, the EC50 for any given growth factor varied as much as 18-fold for the recruitment of different proteins. Knudsen et al. (29) reported similar differences in the EC50 values of four EGF receptor ligands for inducing the phosphorylation of the EGF receptor and several signaling proteins. The molecular basis for this observation is not known, but it may reflect differences in the order or extent of phosphorylation of sites in response to these seven agonists (21, 30–32).
Surprisingly, there were significant differences in the ability of saturating concentrations of erlotinib and lapatinib to inhibit the recruitment of these signaling proteins. This is likely due to differences in residual phosphorylation of the EGF receptor. These findings clearly identify erlotinib as a more effective inhibitor of signaling in this system than lapatinib and suggest that these complementation assays may be useful for identifying residual signaling pathways that could be targeted for therapeutic benefit.
The overarching message from the principal component analysis is that there are significant differences in signaling protein recruitment depending on both ligand and dose. These variable responses likely reflect different signaling protein recruitment strategies employed by the individual ligands over their entire dose range. Although the observed differences are subtle at the level of individual proteins recruited, collectively they could readily give rise to a distinctly different biological outcome for each of the agonist ligands.
In vivo levels of EGF and other ErbB family growth factors vary widely from tissue to tissue, being low in plasma but 10–100-fold higher in secretions such as saliva and tears (33–35). Given the differences in network behavior identified here, our data imply that therapeutic agents that target one particular node in the signaling pathway could be efficacious in one tissue but not in another, simply because of differences in network utilization based on the identity of the stimulating growth factor and/or the dose involved. This underscores the need to understand the signaling network at all doses of growth factor, as different tissues will likely be responding to vastly different doses of EGF or other EGF receptor agonists. Because many of the experiments that have defined our understanding of this network have been carried out using high dose EGF (36–40), our current appreciation of the network may not reflect the actual flux through the pathways under all physiological conditions.
Ensemble clustering of the responses to the growth factors demonstrated that the seven different EGF receptor ligands basically cluster into three groups as follows: (i) BTC, TGFα, and EPG; and (ii) EGF, AREG, and EPR. HB-EGF is distantly related to both clusters. Thus, based on their ability to recruit these signaling proteins to the EGF receptor, these ligands do not fall neatly into groups defined by high versus low affinity nor do they fall into groups based on whether they bind only to the EGF receptor or to both the EGF receptor and ErbB4 (17). Whether there is some specific functional difference that distinguishes the two main groups of EGF receptor agonists, such as temporal or spatial differences in expression, remains to be determined.
With respect to similarities in the utilization of the network by the different growth factors, our analysis identified a strong correlation between the recruitment of Cbl and the recruitment of CrkL to the EGF receptor. As CrkL is known to bind directly to Cbl (41, 42), the detection of a correlation between Cbl and CrkL binding to the EGF receptor suggests that the primary mechanism through which CrkL associates with the EGF receptor may be through binding to tyrosine-phosphorylated Cbl. The fact that this relationship is clearly observed in our dataset suggests that this analysis is capable of identifying interactions between proteins that associate within this signaling network. Viewed in this light, the significant correlation between p52 Shc and Gab1 suggests that this also represents a preferred interaction in this network. The direct binding of p52 Shc to Gab1 has been reported (43, 44).
The finding that other canonical network interactions, such as Grb2/Shc or Grb2-Gab1, were not detected in this analysis likely reflects the complex and dynamic behavior of the network. Grb2 is an adapter protein that recruits a number of proteins, including Cbl, Gab1, and Shp2, to the EGF receptor. It can bind directly to the EGF receptor or indirectly through Shc. As a result, the interaction of Grb2 with the EGF receptor represents the summation of a multiplicity of different binding events. Variation in the dynamics of the different binding events, such as Grb2-Cbl versus Grb2-Gab1, could easily obscure any correlations between the binding of the individual partners in the protein pairs. Thus, it will be necessary to assess these interactions more directly to determine whether their association is differentially affected by the seven EGF receptor agonists.
Ultimately, we would like to be able to determine which path through the network is used to recruit a particular protein to the EGF receptor signaling complex by a particular growth factor at a particular dose. Prediction on this level is likely to require careful modeling of network behavior. To this end, these data can be used, in conjunction with other information, to build mechanistic models of the network interactions to determine the dose-dependent network paths of a given signaling protein.
Author Contributions
L. J. P. conceived and designed the study, oversaw all the work, and wrote the manuscript. J. L. M. and L. H. performed the experiments. N. J. B. synthesized and characterized the epigen and epiregulin. T. R. and K. M. N. performed and interpreted the principal component analysis and ensemble clustering, and wrote the manuscript.
Supplementary Material
This work was supported in part by National Institutes of Health Grant RO1GM099695 (to L. J. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.

This article contains supplemental Tables S1 and S2 and Figs. S1–S7.
- BTC
- betacellulin
- AREG
- amphiregulin
- EPG
- epigen
- EPR
- epiregulin
- HB-EGF
- heparin-binding EGF
- EGFR
- EGF receptor
- PCA
- principal component analysis
- PC
- principal component.
References
- 1. Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., and Burgess A. W. (2005) Ligand-induced dimer-tetramer transition during the activation of cell surface epidermal growth factor receptor. A multidimensional microscopy analysis. J. Biol. Chem. 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 2. Hofman E. G., Bader A. N., Voortman J., van den Heuvel D. J., Sigismund S., Verkleij A. J., Gerritsen H. C., and van Bergen en Henegouwen P. M. (2010) Ligand-induced epidermal growth factor receptor (EGFR) oligomerization is kinase-dependent and enhances internalization. J. Biol. Chem. 285, 39481–39489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moriki T., Maruyama H., and Maruyama I. N. (2001) Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 4. Saffarian S., Li Y., Elson E. L., and Pike L. J. (2007) Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys. J. 93, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu X., Sharma K. D., Takahashi T., Iwamoto R., and Mekada E. (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H.-J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., and Ward C. W. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 7. Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J.-H., Saito K., Sakamoto A., Inoue M., Shirouzu M., and Yokoyama S. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Gureasko J., Shen K., Cole P. A., and Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 9. Citri A., and Yarden Y. (2006) EGF-ErbB signalling: toward the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 10. Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 11. Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., and Yarden Y. (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 12. Holgado-Madruga M., Emlet D. R., Moscatello D. K., Godwin A. K., and Wong A. J. (1996) A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature 379, 560–564 [DOI] [PubMed] [Google Scholar]
- 13. Gu H., and Neel B. G. (2003) The ‘Gab’ in signal transduction. Trends Cell Biol. 13, 122–130 [DOI] [PubMed] [Google Scholar]
- 14. Rodrigues G. A., Falasca M., Zhang Z., Ong S. H., and Schlessinger J. (2000) A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 20, 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simister P. C., and Feller S. M. (2012) Order and disorder in large multi-site docking proteins of the Gab family–implications for signalling complex formation and inhibitor design strategies. Mol. Biosyst. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 16. Wang W., Xu S., Yin M., and Jin Z. G. (2015) Essential roles of Gab1 tyrosine phosphorylation in growth factor-mediated signaling and angiogenesis. Int. J. Cardiol. 181, 180–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson K. J., Gilmore J. L., Foley J., Lemmon M. A., and Riese D. J. 2nd (2009) Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol. Ther. 122, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito T., Okada S., Ohshima K., Yamada E., Sato M., Uehara Y., Shimizu H., Pessin J. E., and Mori M. (2004) Differential activation of epidermal growth factor (EGF) receptor downstream signaling pathways by betacellulin and EGF. Endocrinology 145, 4232–4243 [DOI] [PubMed] [Google Scholar]
- 19. Shin H. S., Lee H. J., Nishida M., Lee M.-S., Tamura R., Yamashita S., Matsuzawa Y., Lee I.-K., and Koh G. Y. (2003) Betacellulin and amphiregulin induce upregulation of cyclin D1 and DNA synthesis activity through differential signaling pathways in vascular smooth muscle cells. Circ. Res. 93, 302–310 [DOI] [PubMed] [Google Scholar]
- 20. Streicher K. L., Willmarth N. E., Garcia J., Boerner J. L., Dewey T. G., and Ethier S. P. (2007) Activation of a nuclear factor-κB/interleukin-1 positive feedback loop by amphiregulin in human breast cancer cells. Mol. Cancer Res. 5, 847–861 [DOI] [PubMed] [Google Scholar]
- 21. Wilson K. J., Mill C., Lambert S., Buchman J., Wilson T. R., Hernandez-Gordillo V., Gallo R. M., Ades L. M., Settleman J., and Riese D. J. 2nd (2012) EGFR ligands exhibit functional differences in models of paracrine and autocrine signaling. Growth Factors 30, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Macdonald-Obermann J. L., Adak S., Landgraf R., Piwnica-Worms D., and Pike L. J. (2013) Dynamic analysis of the epidermal growth factor (EGF) receptor-ErbB2-ErbB3 protein network by luciferase fragment complementation imaging. J. Biol. Chem. 288, 30773–30784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macdonald-Obermann J. L., and Pike L. J. (2014) Different epidermal growth factor (EGF) ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J. Biol. Chem. 289, 26178–26188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang K. S., Ilagan M. X., Piwnica-Worms D., and Pike L. J. (2009) Luciferase fragment complementation imaging of conformational changes in the EGF receptor. J. Biol. Chem. 284, 7474–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., and Duchesnay E. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 [Google Scholar]
- 26. Luker K. E., Smith M. C., Luker G. D., Gammon S. T., Piwnica-Worms H., and Piwnica-Worms D. (2004) Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U.S.A. 101, 12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng Y., Zhang C., Croucher D. R., Soliman M. A., St.-Denis N., Pasculescu A., Taylor L., Tate S. A., Hardy W. R., Colwill K., Dai A. Y., Bagshaw R., Dennis J. W., Gingras A.-C., Daly R. J., and Pawson T. (2013) Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beguinot L., Lyall R. M., Willingham M. C., and Pastan I. (1984) Down-regulation of the epidermal growth factor receptor in KB cells is due to receptor internalization and subsequent degradation in lysosomes. Proc. Natl. Acad. Sci. U.S.A. 81, 2384–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knudsen S. L., Mac A. S., Henriksen L., van Deurs B., and Grøvdal L. M. (2014) EGFR signaling patterns are regulated by its different ligands. Growth Factors 32, 155–163 [DOI] [PubMed] [Google Scholar]
- 30. Gilmore J. L., Scott J. A., Bouizar Z., Robling A., Pitfield S. E., Riese D. J. 2nd, and Foley J. (2008) Amphiregulin-EGFR signaling regulates PTHrP gene expression in breast cancer cells. Breast Cancer Res. Treat. 110, 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Curran T. G., Zhang Y., Ma D. J., Sarkaria J. N., and White F. M. (2015) A multiplex method for absolute quantification of peptides and posttranslational modifications. Nat. Commun. 6, 5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo L., Kozlosky C. J., Ericsson L. H., Daniel T. O., Cerretti D. P., and Johnson R. S. (2003) Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J. Am. Soc. Mass Spectrom. 14, 1022–1031 [DOI] [PubMed] [Google Scholar]
- 33. Grayson L. S., Hansbrough J. F., Zapata-Sirvent R. L., Dore C. A., Morgan J. L., and Nicolson M. A. (1993) Quantitation of cytokine levels in skin graft donor site wound fluid. Burns 19, 401–405 [DOI] [PubMed] [Google Scholar]
- 34. Lemos-González Y., Rodríguez-Berrocal F. J., Cordero O. J., Gómez C., and Páez de la Cadena M. (2007) Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. Br. J. Cancer 96, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marti U., Burwen S. J., and Jones A. L. (1989) Biological effects of epidermal growth factor, with emphasis on the gastrointestinal tract and liver: an update. Hepatology 9, 126–138 [DOI] [PubMed] [Google Scholar]
- 36. Blagoev B., Ong S.-E., Kratchmarova I., and Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 37. Dengjel J., Akimov V., Olsen J. V., Bunkenborg J., Mann M., Blagoev B., and Andersen J. S. (2007) Quantitative proteomic assessment of very early cellular signaling events. Nat. Biotechnol. 25, 566–568 [DOI] [PubMed] [Google Scholar]
- 38. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 39. Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., and White F. M. (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 40. Steen H., Kuster B., Fernandez M., Pandey A., and Mann M. (2002) Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem. 277, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 41. Kyono W. T., de Jong R., Park R. K., Liu Y., Heisterkamp N., Groffen J., and Durden D. (1998) Differential interaction of CrkL with Cbl or C3G, Hef-1 and γ subunit immunoreceptor tyrosine-based activation motif in signaling of myeloid high affinity Fc receptor for IgG (FcγRI). J. Immunol. 161, 5555–5563 [PubMed] [Google Scholar]
- 42. Sattler M., Salgia R., Shrikhande G., Verma S., Uemura N., Law S. F., Golemis E. A., and Griffin J. D. (1997) Differential signaling after β1 integrin ligation is mediated through binding of CrkL to p120Cbl and p110HEF1. J. Biol. Chem. 272, 14320–14326 [DOI] [PubMed] [Google Scholar]
- 43. Ingham R. J., Holgado-Madruga M., Siu C., Wong A. J., and Gold M. R. (1998) The Gab1 protein is a docking site for multiple proteins involved in signaling by the B cell antigen receptor. J. Biol. Chem. 273, 30630–30637 [DOI] [PubMed] [Google Scholar]
- 44. Lecoq-Lafon C., Verdier F., Fichelson S., Chrétien S., Gisselbrecht S., Lacombe C., and Mayeux P. (1999) Erythropoietin induces the tyrosine phosphorylation of Gab1 and its association with Shc, Shp2, SHIP and phosphatidylinositol 3-kinase. Blood 93, 2578–2585 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.