Abstract
The secoiridoids are the main class of specialized metabolites present in olive (Olea europaea L.) fruit. In particular, the secoiridoid oleuropein strongly influences olive oil quality because of its bitterness, which is a desirable trait. In addition, oleuropein possesses a wide range of pharmacological properties, including antioxidant, anti-inflammatory, and anti-cancer activities. In accordance, obtaining high oleuropein varieties is a main goal of molecular breeding programs. Here we use a transcriptomic approach to identify candidate genes belonging to the secoiridoid pathway in olive. From these candidates, we have functionally characterized the olive homologue of iridoid synthase (OeISY), an unusual terpene cyclase that couples an NAD (P)H-dependent 1,4-reduction step with a subsequent cyclization, and we provide evidence that OeISY likely generates the monoterpene scaffold of oleuropein in olive fruits. OeISY, the first pathway gene characterized for this type of secoiridoid, is a potential target for breeding programs in a high value secoiridoid-accumulating species.
Keywords: biosynthesis, enzyme, plant, secondary metabolism, transcriptomics, Oleaceae, monoterpenes, phenolic compounds, secoiridoids
Introduction
Olive (Olea europaea L.) produces a range of secondary metabolites that strongly affect the taste and nutritional properties of olive oil and fruits. The most abundant of these secondary metabolites are the secoiridoids, monoterpenoids with a 3,4-dihydropyran skeleton. These compounds are present as oleosidic secoiridoids or oleosides that have an exocyclic olefinic functionality (1) and possess a tyrosine-derived component (see Fig. 1A). Secoiridoids, which are recovered in virgin olive oil in small amounts, strongly influence olive oil taste, being responsible for the bitterness and pungency sensory notes (2), which are desirable traits for high quality olive oil.
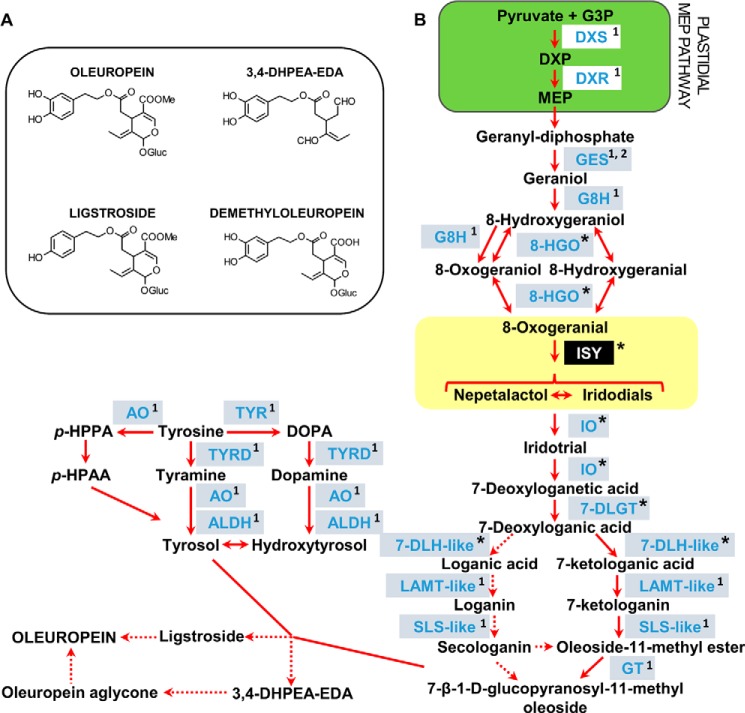
FIGURE 1.
Secoiridoid pathway. A, chemical structures of major secoiridoids of olive fruits. B, schematic representation of olive secoiridoid pathway. Iridoid synthase reaction is indicated in the yellow box. 7-DLH-like, LAMT-like, SLS-like, might convert the same intermediates as in the C. roseus pathway or the keto-analogues previously proposed for olive (24). G3P, glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose-5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; DXS, DXP synthase; DXR, DXP reductoisomerase; GES, geraniol synthase; G8H, geraniol 8-hydroxylase; 8-HGO, 8-hydroxygeraniol oxidoreductase; ISY, iridoid synthase; IO, iridoid oxidase; 7-DLGT, 7-deoxyloganetic acid-O-glucosyl transferase; 7-DLH-like, 7-deoxyloganic acid hydroxylase-like; LAMT-like, loganic acid methyltransferase-like; SLS-like, secologanin synthase-like; GT, glucosyltransferase; AO, amine oxidase; TYR, tyrosinase; TYRD, tyrosine/dopa decarboxylase; ALDH, alcohol dehydrogenase; p-HPPA, p-hydroxyphenylpyruvic acid; p-HPAA, p-hydroxyphenylacetic acid; p-HPEA, p-hydroxyphenyl ethanol; p-HPEA-EDA, elenolic acid linked to p-HPEA; 3,4-DHPEA-EDA, elenolic acid linked to 3,4-dihydroxyphenyl ethanol (3,4-DHPEA). *, candidate genes identified in this study; 1, candidate gene previously identified by Alagna et al. (3); 2, candidate genes previously identified by Vezzaro et al. (15).
Oleuropein is the most abundant olive oleoside. It can represent up to 90% of total fruit secoiridoids (3), although other structurally related secoiridoids (ligstroside, demethyloleuropein, 3,4-DHPEA-EDA (the dialdehydic form of decarboxymethyl elenolic acid linked to 3,4-dihydroxyphenyl ethanol), p-HPEA-EDA (the dialdehydic form of decarboxymethyl elenolic acid linked to p-hydroxyphenyl ethanol), oleuropein aglycon, 3,4-DHPEA-EA (the isomer of oleuropein aglycon), and ligstroside aglycon) are present in olive oil (4).
Oleuropein is strongly associated with the beneficial properties of olive oil for human health (2, 5). In particular, oleuropein exhibits antioxidant, anti-inflammatory, anti-atherogenic, anti-cancer, antimicrobial, and antiviral activities, and it has hypolipidemic and hypoglycemic effects (6). For instance, it plays a role in prevention of atherosclerosis and inhibition of low density lipoprotein peroxidation (7). It also exhibits cancer preventive activities (8) and can contribute to the nutritional prevention of osteoporosis (9). Additionally, this compound has been implicated in plant defense. Indeed, β-glucosidases released from herbivore-attacked tissues can convert oleuropein into a strong protein denaturant that has protein-cross-linking and lysine-alkylating activities (10, 11). Oleuropein content differs markedly among different genotypes (3). However, high oleuropein varieties are desirable, and this trait is considered a target for olive breeding programs.
Oleuropein is present in all constituent parts of the plant but accumulates at higher levels in the fruits and leaves (3, 12–14). In olive fruit, oleuropein is present at highest amounts in small unripe fruits (45 days after flowering) and then dramatically decreases during fruit development and ripening (3).
Considering the high value of oleuropein, the identification of the genes and enzymes required for its synthesis is particularly important to facilitate the development of high oleuropein varieties and to develop synthetic pathways in microbial or plant hosts using metabolic engineering approaches. Until now, only a few candidate genes of the secoiridoid pathway have been proposed in olive (3, 15). Recently, the iridoid pathway for the secoiridoid secologanin has been completely elucidated in Madagascar periwinkle (Catharanthus roseus), where the pathway feeds directly into the monoterpene indole alkaloid pathway (16–20) (see Fig. 1B). Because the secoiridoids in the Oleaceae family are derived from secologanin or a secologanin precursor (21–24), it is likely that the Oleaceae contain homologues of these C. roseus biosynthetic genes that participate in secoiridoid biosynthesis.
Iridoid biosynthesis is initiated from geranyl-pyrophosphate, which is then converted to secologanin by a series of reactions that include oxidations, reductions, glycosylations, and methylations. Secoiridoids are derived from iridoids by opening of the cyclopentane ring, and in the Oleaceae family, the resulting carbonyl group is oxidized and conjugated with a phenolic moiety. These Olea-specific reactions have not yet been resolved, although a putative pathway has been proposed (25) (see Fig. 2).
FIGURE 2.
Putative downstream reactions of oleuropein biosynthesis. The intermediates from 7-deoxyloganic acid to oleuropein are reported as previously hypothesized (24, 25). Olive enzymes similar to C. roseus 7-deoxyloganic acid hydroxylase-like (7-DLH-like), loganic acid methyltransferase-like (LAMT-like), and secologanin synthase-like (SLS-like) (23, 57) might catalyze the indicated steps.
Integrated approaches coupling co-expression analyses of transcriptomic data with functional characterization studies have been used successfully for the investigation of numerous plant specialized metabolic pathways (26–28). Moreover, clustering of transcript and metabolite profiles is becoming a powerful technique for identifying candidate genes (29–32). Large expressed sequence tag collections from high secoiridoid-accumulating olive fruits and leaves (33–36) are a valuable resource for the identification of candidate transcripts in secoiridoid biosynthesis. With the exception of a geraniol synthase involved in the synthesis of geraniol from geranyl-diphosphate (15), up to now no secoiridoid biosynthetic genes have been biochemically characterized in olive.
We used these olive transcriptomic data sets to identify putative genes of the secoiridoid pathway. Homologues of C. roseus pathway enzymes (iridoid synthase (ISY), 8-hydroxygeraniol oxidoreductase (8HGO), iridoid oxidase (IO), 7-deoxyloganetic acid-O-glucosyl transferase (7-DLGT), and 7-deoxyloganic acid hydroxylase (7-DLH)) were identified.
We focused on iridoid synthase (CrISY), which is a key enzyme that catalyzes the formation of the iridoid scaffold (16). CrISY, which has high similarity to progesterone 5-β-reductase (P5βR)2 genes involved in cardenolide biosynthesis (37–39), reductively cyclizes 8-oxogeranial to form nepetalactol, the common biosynthetic precursor of all iridoids. Hence, ISY defines a new class of monoterpene cyclase and is mechanistically distinct from canonical terpene cyclases that operate via cationic intermediates. CrISY apparently couples an initial NAD(P)H-dependent 1,4-reduction step with a subsequent Michael-type cyclization (16, 40, 41).
In this study, three olive homologues of ISY were identified from the olive transcriptomic data. We discovered one gene that showed high similarity to CrISY and confirmed CrISY-like iridoid synthase activity with NADPH consumption assays and product characterization by GC-MS. Moreover, this gene showed a very similar co-expression profile to other putative iridoid biosynthetic genes, suggesting that this enzyme plays a physiological role in oleuropein biosynthesis. The biochemical functions of two other ISY homologues were also investigated, and their roles in secoiridoid production are discussed. Our data shed new light on oleuropein biosynthesis and more broadly on the evolutionary origin of secoiridoids and iridoids in Asterid families.
Experimental Procedures
Plant Material
Samples were harvested from field plants of cv. Leccino from the Olive Cultivar Collection of CNR-Institute of Biosciences and Bio-resources (Perugia, Italy). Fruits (mesocarp and exocarp) at different developmental stages (45, 75, 105, and 135 days after flowering (DAF)), flowers at anthesis stage, and young leaves were collected in three biological replicates. Immediately after harvest, samples were frozen in liquid nitrogen and stored at −80 °C.
Identification of Candidate Genes
The amino acid sequences from Madagascar periwinkle (C. roseus) iridoid synthase (ISY), 8-hydroxygeraniol oxidoreductase (8HGO), iridoid oxidase (IO), 7-deoxyloganetic acid-O-glucosyl transferase (7-DLGT), and 7-deoxyloganic acid hydroxylase (7-DLH), recently shown to be involved in monoterpene indole alkaloid biosynthesis (16, 17, 19, 20), were used to search olive-expressed sequence tags by basic local alignment (BLAST) in olive transcriptomic data of olive fruit (33) and other tissues (35). The sequences of candidate genes were assembled in silico using preliminary genomic sequences. The closest CrISY homologue, named OeISY, was selected for further analyses together with two other genes that showed high similarity to the Digitalis lanata progesterone-5β-reductase 1 (DlP5βR1) gene and were also expressed in fruit tissues. These genes were named Oe1,4-R1 and Oe1,4-R3. Specific primers were designed to resequence the homologues from fruit cDNA of cv. Leccino and to obtain the entire coding regions by rapid amplification of cDNA ends-PCR.
Co-expression Analyses
The analyses were performed by using a 454 transcriptomic data set from fruit (26,500 transcripts) designed for the identification of genes involved in the metabolism of phenolic and secoiridoid compounds (33). To reduce the transcriptomic data set to a workable size, we selected genes exhibiting high expression in fruits (1258 transcripts with total rpkm values over 130, calculated considering all fruit samples). In addition, we selected those genes that were differentially expressed between 45 and 135 DAF (746 transcripts) applying the statistical R test (42) (R > 10). Hierarchical clustering analysis of this filtered data set using Cluster 3.0 (43) allowed the identification of a set of co-expressed genes.
Heterologous Protein Expression and Purification
The entire coding sequences were amplified from fruit cDNA of cv. Leccino using gene specific primers for OeISY (5′-ATGAGCTGGTGGTTCAACAGATCT-3′, 5′-TCAAGGAATAAACCTATAAGCCCTC-3′), Oe1,4-R1 (5′-ATGAGTTGGTGGTGGAAAGGTGC-3′, 5′-TCAAGGAACAATCTTGTGTGATTTCA-3′), and Oe1,4-R3 (5′-ATGAGTTGGTGGTGGGCCGGAG-3′, 5′-TTAAGGGACGATCTTGTAAGCTTTCA-3′) and cloned into the Gateway vector pCR8-GWTOPO-T/A (Invitrogen). Subcloning into the vector pDEST17 using Gateway LR cloning yielded the Escherichia coli expression construct in frame with an N-terminal His tag. BL21 star cells (Life Technologies) harboring the desired plasmid were grown at 37 °C, induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at A600 nm of 0.8, and then cultured at 22 °C for 5 h. The cells were lysed by lysozyme treatment. For the purification, the soluble portion of the lysate was equilibrated with nickel-nitrilotriacetic acid-agarose resin (Qiagen). The proteins were eluted via a stepwise imidazole gradient (50, 100, 150, and 300 mm), where the enzymes of interest eluted in the 100 mm imidazole fraction. These fractions were then concentrated and buffer-exchanged into a storage buffer of 20 mm MOPS, pH 7.0. Proteins were quantified by Bradford assay using BSA protein as standard. Proteins were separated by SDS-PAGE by using 12% polyacrylamide gel and then transferred onto a PVDF membrane by electroblotting. After blocking with 5% milk in TBS buffer, the membrane was incubated with mouse 1:1000 anti-His polyclonal antibody (Roche) in TBSTT buffer. Anti-mouse IgG antibody (diluted 1:5000 in TBSTT) conjugated with alkaline phosphatase (Sigma-Aldrich) was used as detection antibody.
Chemical Synthesis
The synthesis of 8-oxoneral was performed as described previously for 8-oxogeranial (16) but starting with nerol instead of geraniol. The final purification step on a kugelrohr was omitted, because the washes in 15% ether in hexanes afforded sufficiently pure material in comparable yields. All other substrates and standards used in this study were either commercially available or had been synthesized as described previously (16).
GC-MS-based Assays
The enzyme assays were carried out essentially as described previously (16), using 20 mm MOPS, pH 7.0, as buffer. Substrates were kept as 50 mm stocks in tetrahydrofuran except progesterone, which was kept as a 50 mm stock in ethanol. The reactions (50 μl) were set up in glass vials using 200 μm of substrate, 400 μm of NADPH, and 1 μg of purified protein and terminated after 1 h by adding 120 μl of CH2Cl2. If the substrate conversion was incomplete, longer incubation (3 h and 24 h) with substrate was carried out. Enzymatic assays with progesterone were carried out at 30 °C, and the reactions were terminated after 3 h. The organic phase was used directly for GC-MS.
GC-MS analyses were carried out on an Agilent 6890N GC system coupled to an Agilent 5973 MS detector. For the initial assays with 8-oxogeranial as substrate, all nonchiral separations were performed with a Zebron ZB-5 HT column (30 m × 0.25 mm × 0.10 μm) using helium as carrier gas at 1 ml min−1 and with an injector temperature of 220 °C. The program used was the following: 5 min isothermal at 60 °C, 20 °C min−1 gradient up to 150 °C, 45 °C min−1 gradient up to 280 °C, and 4 min isothermal at 280 °C (run time = 16.39 min). GC-MS assays using progesterone as substrate were performed with a Zebron ZB-5 HT column (35 m × 0.25 mm × 0.10 μm) using helium as carrier gas at 1 ml min−1 and with an injector temperature of 250 °C. The program used was the following: 5 °C min−1 gradient up to 280 °C, 10 min isothermal at 280 °C (run time = 36.00 min). GC-MS assays with all the other substrates [8-oxoneral, (S)-8-oxocitronellal, citral, cinnamaldehyde, α-methylcinnamaldehyde] were carried out by using a DB-1 column (15 m × 0.25 mm × 0.10 μm) using helium as carrier gas at 1 ml min−1 and with an injector temperature of 220 °C. The program used was the following: 2 min isothermal at 60 °C, 12 °C min−1 gradient up to 150 °C, 45 °C min−1 gradient up to 280 °C, 2 min isothermal at 280 °C (run time = 14.39 min). All the analyses were repeated three times with similar results. Where possible, mass spectra were compared with those of authentic standards.
Spectrophotometry-based Assays for Kinetic Studies
For the determination of Michaelis-Menten parameters with 8-oxogeranial, kinetics of NADPH consumption were determined spectrophotometrically at 340 nm in cuvettes with 1-cm path length. The reactions contained 50 μm NADPH, 200 mm MOPS buffer, pH 7.0, 100 mm sodium chloride, and 2 nm (OeISY) or 40 nm (Oe1,4-P3) enzyme in a total volume of 800 μl. Substrate was added from a 50 mm stock solution in tetrahydrofuran resulting in a final tetrahydrofuran concentration of less than 0.006% (OeISY) or 2% (Oe1,4-P3). The reaction was equilibrated at room temperature (22 °C) and started by addition of enzyme. Plots of initial velocities versus substrate concentration were nonlinearly fit to the Michaelis-Menten equation in SigmaPlot 12.5 to obtain values of kcat and Km. Assays with Oe1,4-P3 and progesterone as a substrate were carried out at 40 °C in 200 μl of assay volume in 96-well plates. Each well contained 60 nm of enzyme. The data were collected for 20 min in 30-s intervals. NADPH consumption rates were determined taking into account background NADPH decay.
cDNA Synthesis and Gene Expression Analysis
Total RNA was extracted from 0.2 g of olive tissue with the RNeasy plant mini kit (Qiagen) and treated with DNase I (Ambion, Austin, TX). Reverse transcription of 2 μg of RNA was performed using oligo(dT)18 and the SuperScript III Reverse Transcriptase kit (Invitrogen) according to the manufacturer's instructions.
Quantitative real time PCR was performed on a PCR real time CFX96, C1000 Touch Thermal Cycler (Bio-Rad) according to the manufacturer's protocol using SYBR Green jump start reaction mixtures (Sigma-Aldrich) and gene-specific primers for OeISY (5′-AAGGATAAGGACTCCGTGTGG-3′, 5′-GCTCAGCACATTCTCAAGACAA-3′), Oe1,4-R1.1 (5′-CACAGCCACCGTGGAATGC-3′, 5′-CAGTTAGAGGAGAAAGCTTGGC-3′), Oe1,4-R1.2 (5′-CGCAGCCATCATGGAACGC-3′, 5′-CAGTAAGAGGAGAAAGCTTGGC-3′), and Oe1,4-R3 (5′-GGGAGTGAAATTGCCTGGGA-3′, 5′-TAGGGATCCACTGCAGACCA-3′). Primer efficiency was assessed by measuring a standard curve for each gene with six dilution points, each one replicated three times. Only primer pairs that produced the expected amplicon and showed similar PCR efficiency were selected for use. All reactions were performed in triplicate. After each assay, a dissociation kinetics analysis was performed to verify the specificity of the amplification products. Relative amounts of all mRNAs were calculated using the 2−ΔΔCt method (44), where ΔCt = Ct(target gene) − Ct(reference gene). The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH2) was used as an endogenous reference for normalization (45). Whether differences were statistically significant was determined by t tests on three biological replicates (*, p < 0.05; **, p < 0.01; n = 3).
Phylogenetic Analysis
Amino acid sequences of progesterone 5β-reductase homologues from a variety of plant species were aligned with ClustalW using Bioedit Sequence Alignment Editor (version 7.0). The phylogenetic tree was built with the Maximum Likelihood method, using MEGA program (version 6.0) (46) with 1000 bootstrap replicates. The evolutionary distances were computed with the Jones-Taylor-Thornton model, and all positions containing gaps and missing data were eliminated from the data set. Physcomitrella patens (GenBankTM accession number EDQ81106.1) was used as outgroup. The analyses include also the characterized P5βR of D. lanata (P5βR1 (GenBankTM accession number AAS93804.1), P5βR2 (GenBankTM accession number ADL28122.1)) and Digitalis purpurea (P5βR1 (GenBankTM accession number AAS93805.1), P5βR2 (GenBankTM accession number ACZ66261.1)), CrISY (GenBankTM accession number AFW98981.1), other P5βR-like proteins from C. roseus (GenBankTM accession numbers KJ873882 (CrP5βR1), KJ873883 (CrP5βR2), KJ873884 (CrP5βR3), KJ873885 (CrP5βR4), KJ873886 (CrP5βR5), and KJ873887 (CrP5βR6)) and Medicago truncatula (GenBankTM accession numbers KJ873888 (MtP5βR1), KJ873889 (MtP5βR2), KJ873890 (MtP5βR3), and KJ873891 (MtP5βR4)), recently biochemically characterized (19).
Accession Numbers
Sequence data from this article were submitted to the GenBankTM database under accession numbers KT954038 (OeISY), KT954039 (Oe1,4-R1.1a), KT954040 (Oe1,4-R1.1b), KT954041 (Oe1,4-R1.2a), KT954042 (Oe1,4-R1.2b), KT954043 (Oe1,4-R2), KT954044 (Oe1,4-R3), KT954045 (Oe8HGO), KT954046 (OeIO), KT954047 (Oe7-DLGT1), KT954048 (Oe7-DLGT2), and KT954049 (Oe7-DLH-like).
Results
Identification of Candidate Genes for the Olive Secoiridoid Pathway
Based on homology with known iridoid biosynthetic genes, we identified a group of olive genes putatively involved in secoiridoid biosynthesis to be used in co-expression analyses. These candidates were identified in olive transcriptomic data (33, 35) by using the local alignment search tool tBLASTn with five C. roseus enzymes involved in iridoid biosynthesis (16, 17, 19, 20) as query sequences. We identified a putative iridoid synthase (OeISY), 8-hydroxygeraniol oxidoreductase (Oe8HGO),iridoid oxidase (OeIO), 7-deoxyloganetic acid-O-glucosyl transferase (Oe7-DLGT1 and Oe7-DLGT2), and 7-deoxyloganic acid hydroxylase-like (Oe7-DLH-like) genes (Table 1 and Figs. 1B and 2).
TABLE 1.
Similarity of olive candidates to Catharanthus roseus proteins
| Transcript | GenBankTM accession no.a | Length | Similarityb | Blast resultsc |
|---|---|---|---|---|
| bp | ||||
| OeISY | KT954038 | 1179 | Iridoid synthase | 0/72% (AFW98981.1) |
| Oe1,4-R1.1a | KT954039 | 1173 | Progesterone-5-β-reductase 4 | 0/80% (AIW09146.1) |
| Oe1,4-R1.1b | KT954040 | 1173 | Progesterone-5-β-reductase 4 | 0/80% (AIW09146.1) |
| Oe1,4-R1.2a | KT954041 | 1173 | Progesterone-5-β-reductase 4 | 0/80% (AIW09146.1) |
| Oe1,4-R1.2b | KT954042 | 1173 | Progesterone-5-β-reductase 4 | 0/80% (AIW09146.1) |
| Oe1,4-R2 | KT954043 | 1170 | Progesterone-5-β-reductase 2 | 0/72% (AIW09144.1) |
| Oe1,4-R3 | KT954044 | 1170 | Progesterone-5-β-reductase 2 | 0/77% (AIW09144.1) |
| Oe8HGO | KT954045 | 1128 | 8-Hydroxygeraniol oxidoreductase | 0/78% (AHK60836.1) |
| OeIO | KT954046 | 1365 | Iridoid oxidase | 0/84% (AHK60833.1) |
| Oe7-DLGT1 | KT954047 | 1461 | 7-Deoxyloganetic acid-O-glucosyl transferase | 0/77% (AGX93065) |
| Oe7-DLGT2 | KT954048 | 1266 | 7-Deoxyloganetic acid-O-glucosyl transferase | 0/63% (AGX93065) |
| Oe7-DLH-like | KT954049 | 1431 | 7-Deoxyloganic acid 7-hydroxylase | 1e-180/51% (AGX93062.1) |
a GenBankTM accession numbers of the transcripts.
b Enzymatic functions.
c E value and percentage of identity of the BLAST search. These values were used to indicate the significance of sequence similarity. The text in parentheses indicates the GenBankTM accession number of C. roseus proteins.
OeISY shared 72% of amino acid identity with CrISY(GenBankTM accession number AFW98981.1). We hypothesized that this gene encodes the iridoid synthase required for secoiridoid biosynthesis in olive. Three other putative 1,4-reductases (∼60% amino acid identity to CrISY) were identified and named Oe1,4-R1, Oe1,4-R2, and Oe1,4-R3. Oe8HGO shared 78% of amino acid identity with the C. roseus 8-hydroxygeraniol oxidoreductase (CrHGO, GenBankTM accession number AHK60836.1), an enzyme able to oxidize both hydroxyl groups of 8-hydroxygeraniol. OeIO showed 84% amino acid identity to CrIO (GenBankTM accession number AHK60833.1), which converts iridodial into 7-deoxyloganetic acid in a two-step oxidation. Oe7-DLGT1 and Oe7-DLGT2 shared, respectively, 77 and 63% of amino acid identity with Cr7-DLGT (GenBankTM accession number AGX93065), which catalyzes the glucosylation of 7-deoxyloganetic acid to form 7-deoxyloganic acid. Oe7-DLH-like, which shared 50% amino acid identity with Cr7-DLH (GenBankTM accession number AGX93062), putatively hydroxylates 7-deoxyloganic acid to yield loganic acid or its isomer 7-ketologanic acid.
Secoiridoid Genes are Co-expressed
To find sets of co-expressed genes possibly involved in secoiridoid biosynthesis, we analyzed transcriptomic data (33) from four cDNA libraries obtained from fruits of two olive varieties (Coratina and Tendellone) at two developmental stages (45 and 165 DAF). Coratina has a higher secoiridoid content compared with Tendellone, and both varieties are richer in secoiridoids at 45 DAF than at 165 DAF. To reduce the transcriptomic data set to a workable size, we considered all genes exhibiting high expression in fruits and selected those differentially expressed among developmental stages (R > 10). Hierarchical clustering analysis of the normalized data revealed that most of the candidate genes of secoiridoid biosynthesis that we had identified grouped within the same cluster (Fig. 3). In particular, OeISY and Oe1,4-R1 grouped together with Oe8HGO, OeIO, and other candidate transcripts of the secoiridoid pathway that we had previously identified (3) (1-deoxy-d-xylulose 5-phosphate reductoisomerase (OeDXR), geraniol 8-hydroxylase (OeG8H), and secologanin synthase-like (OeSLS-like3)), indicating that this set of genes is co-expressed (p = 0.966). In addition, a transcript (OeCYP76A1) putatively encoding for an uncharacterized CYP76A1 also grouped with these genes. This CYP450 enzyme might carry out an oxidation reaction later in the oleuropein pathway. Other candidate genes for the biosynthesis of the terpenic moiety (1-deoxy-d-xylulose 5-phosphate synthase (OeDXS), geraniol synthase (OeGES), Oe7-DLGT2, loganic acid methyltransferase-like (OeLAMT), and secologanin synthases-like (OeSLS-like2 and OeSLS-like4)) and the phenolic moiety (tyrosine decarboxylase (OeTYRD)) of the secoiridoids group together (p = 0.832) and with a lower score (p = 0.659) also with OeISY and Oe1,4-R1. In contrast, other potential candidates such as Oe7-DLGT1, Oe7-DLH-like, OeSLS-like1 and the other 1,4-reductases that we identified (1,4-R2 and 1,4-R3) had a different expression pattern and do not group with the other secoiridoid candidates.
FIGURE 3.
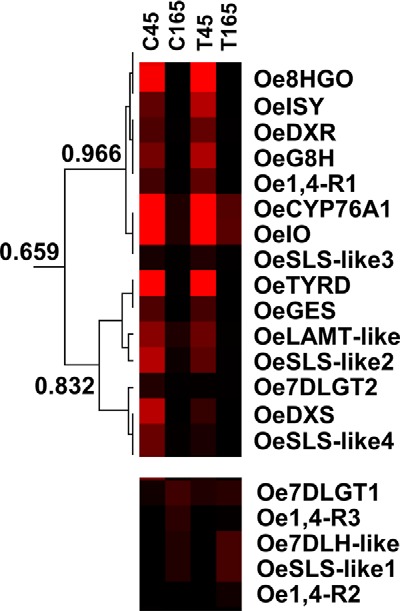
Hierarchical clustering analysis of secoiridoid gene expression in olive fruits. Transcriptomic data from Alagna et al. (33) were used for the analysis. Colors indicate transcriptional activation (red) or repression (black). Top panel, transcripts co-expressed as indicated by Pearson correlation coefficient. Bottom panel, not co-expressed. C45, cv. Coratina at 45 DAF; C135, cv. Coratina at 135 DAF; T45, cv. Tendellone at 45 DAF; T135, cv. Tendellone 135 DAF. Oe8-HGO (8-hydroxygeraniol oxidoreductase), OeISY (iridoid synthase), Oe1,4-R (related 1,4-reductase genes numbered 1–4), OeCYP76A1 (cytochrome P450 76A1), OeIO (iridoid oxidase), Oe7-DLGT (7-deoxyloganetic acid-O-glucosyl transferase), and Oe7-DLH-like (7-deoxyloganic acid hydroxylase-like) expression data refer to the candidate genes identified in this study. OeDXR (DXP reductoisomerase, GenBankTM accession number JX266164), OeG8H (geraniol 8-hydroxylase, GenBankTM accession number JX266182) OeSLS-like (secologanin synthase-like; GenBankTM accession numbers JX266184 (OeSLS-like1), JX266186 (OeSLS-like2), JX266188 (OeSLS-like3), JX266190 (OeSLS-like4)), OeTYRD (tyrosine/dopa decarboxylase, GenBankTM accession number JX266195), OeGES (geraniol synthase; GenBankTM accession number JX266180), OeLAMT-like (loganic acid methyltransferase-like; GenBankTM accession number JX266191), and OeDXS (DXP synthase, GenBankTM accession number JX266162) expression data refer to the candidate genes identified by Alagna et al. (3).
These data indicate that most secoiridoid pathway candidates are co-expressed and have an expression profile consistent with the oleuropein content in the fruit that is higher at 45 DAF compared with 165 DAF (3). These results support the involvement of OeISY, Oe1,4-R1, Oe8HGO, OeIO, and Oe7-DLGT2 in the biosynthesis of olive secoiridoids.
OeISY, Oe1,4-R1, and Oe1,4-R3 Encode for 1,4-Reductases with Different Substrate Specificities
Considering the key role of ISY in the formation of the iridoid scaffold, we selected OeISY and Oe1,4-R1 (both potential iridoid synthases expressed in fruit) for heterologous expression and biochemical characterization. In addition, we selected Oe1,4-R3, which does not cluster with these iridoid biosynthetic genes, as a negative control that may potentially be involved in a different pathway. Oe1,4-R2 was not included in this analysis because it was expressed at low levels in fruits, and as with Oe1,4-R3, it did not cluster with the other candidate genes.
The entire coding regions of OeISY, Oe1,4-R1, and Oe1,4-R3 were obtained by rapid amplification of cDNA ends-PCR from the cv. Leccino. One single allele was obtained for OeISY and Oe1,4-R3, whereas four allelic variants (Oe1,4-R1.1a, Oe1,4-R1.1b, Oe1,4-R1.2a, and Oe1,4-R1.2b) were obtained for Oe1,4-R1, indicating at least two loci for this gene. Oe1,4-R1.1 and Oe1,4-R1.2 transcripts shared 98% of nucleotide identity. Most of the mutations identified in Oe1,4-R1 alleles were synonymous, resulting in marginal differences in protein sequence; in fact, alleles 2a and 2b are predicted to encode the same protein, and the 1a and 1b predicted proteins differ by only one amino acid. For this reason, only the 1a and 2a alleles (99% amino acid identity) were selected for functional characterization.
The entire coding regions of OeISY, Oe1,4-R1.1a, Oe1,4-R1.2a, and Oe1,4-R3 genes were cloned into vectors for expression in E. coli. The proteins were expressed, purified, and then subjected to in vitro biochemical assays. We were not able to detect differences between the activities of Oe1,4-R1.1a and those of Oe1,4-R1.2a, and therefore, these two enzymes will be referred to generically as Oe1,4-R1. Seven compounds (8-oxogeranial, 8-oxoneral, (S)-8-oxocitronellal, progesterone, citral, cinnamaldehyde, and α-methylcinnamaldehyde) were used as substrates for the enzyme assays. The enzymes were incubated with substrates in the presence or absence (negative control) of NADPH. GC-MS analyses revealed that OeISY recombinant enzyme was able to reduce and cyclize 8-oxogeranial in the presence of NADPH. Nepetalactol and its open dialdehyde forms (iridodials) were obtained as products, as previously reported for CrISY (16) (Fig. 4, A–C). The observed products do not readily interconvert under the assay conditions used (40), and the existence of a product mixture is therefore suggestive of a Michael reaction mechanism, as proposed previously for C. roseus CrISY (40). The steady-state kinetic constants of the reaction were determined using spectrophotometric NADPH consumption assays (kcat = 3.8 ± 0.2 s−1, Km = 0.6 ± 0.1 μm, kcat/Km = 6.3 μm−1 s−1; all data are means ± error of fit) (Fig. 4D). These catalytic parameters are similar to those of CrISY (16), demonstrating high catalytic efficiency of OeISY for 8-oxogeranial and supporting a role in secoiridoid biosynthesis.
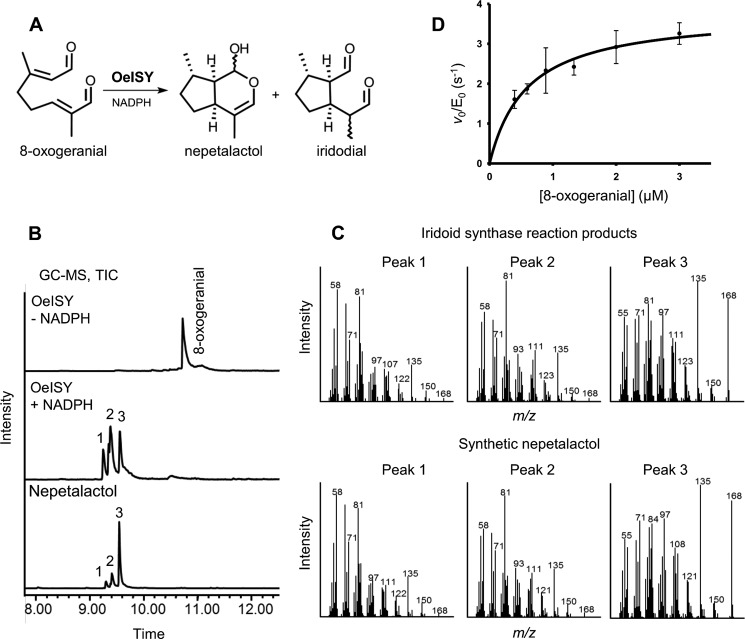
FIGURE 4.
Functional characterization of OeISY. A, iridoid synthase reaction. B, analysis of ISY reaction products by GC-MS after 1 h of incubation with 200 μm 8-oxogeranial. Total ion chromatograms of CH2Cl2 extracts of the iridoid synthase reaction (plus NADPH) and of a negative control (minus NADPH) are compared with the TICs of a cis-trans-nepetalactol standard. In the presence of NADPH, OeISY converted 8-oxogeranial to cis-trans-nepetalactol (peak 3) and to its open dialdehyde forms (cis-trans-iridodials, peaks 1 and 2), as previously observed for CrISY (16, 40). GC-MS assays were performed with a Zebron ZB-5 HT column (35 m × 0.25 mm × 0.10 μm). C, MS spectra of the products. D, steady-state kinetic analysis of the iridoid synthase reaction. The reaction rates were measured spectrophotometrically monitoring NADPH consumption at 340 nm. Individual data points are the average of four replicates ± error of fit.
GC-MS analyses with the other substrates indicated that OeISY accepts other compounds closely related to 8-oxogeranial as a substrate (Table 2). OeISY was able to cyclize 8-oxoneral and reduce citral, whereas it was unable to metabolize progesterone, cinnamaldehyde, α-methylcinnamaldehyde, and (S)-8-oxocitronellal. The inability to reduce (S)-8-oxocitronellal indicates that OeISY specifically reduces the double bond at position C2. These data suggest that 8-oxogeranial and its isomer 8-oxoneral are the preferred substrates of OeISY.
TABLE 2.
Enzymatic activity of olive 1,4-reductases on different substrates
n.d.a. = no detectable activity; incomplete: conversion of substrate (200 μm) to product was incomplete after 24 h.

According to GC-MS assays, Oe1,4-R1 was unable to metabolize 8-oxogeranial (Table 2). However, Oe1,4-R1 reduced citral (Fig. 5A) and inefficiently cyclized 8-oxoneral (Fig. 5B). The activity with 8-oxoneral was observed only after long incubation (24 h) with the substrate, and most of the substrate was not converted. These results indicate that Oe1,4-R1 is not involved in the iridoid synthase step of secoiridoid biosynthesis. The low efficiency of conversion observed with all the substrates tested probably indicates that we have not identified the native substrate for this enzyme. However, the strong co-expression of Oe1,4-R1 with OeISY and with the other candidate genes of secoiridoid biosynthesis suggests that it might be involved in another step of this pathway.
FIGURE 5.
Functional characterization of Oe1,4-R1. Analysis of the reaction products by GC-MS. Total ion chromatograms of CH2Cl2 extracts and MS spectra are reported. GC-MS assays were carried out by using a DB-1 column (15 m × 0.25 mm × 0.10 μm). Purified enzyme was incubated for 3 h with 200 μm citral and NADPH (A). The conversion of 200 μm 8-oxoneral (plus NADPH) was incomplete after 24 h (B). As negative control, the same reactions were prepared without NADPH.
GC-MS analyses of Oe1,4-R3 biochemical assays revealed a wide activity range for this enzyme, because it was able to reduce all the tested substrates except cinnamaldehyde and α-methylcinnamaldehyde (Table 2). This enzyme reduced progesterone (Fig. 6A) and cyclized 8-oxogeranial (Fig. 6B) and 8-oxoneral. The ability to reduce (S)-8-oxocitronellal indicated that this enzyme, like Oe1,4-R1, does not specifically reduce the double bond at position C2. The steady-state kinetic constants of the reaction were determined through spectrophotometric NADPH consumption assays for progesterone (kcat = 3.2 ± 0.3 s−1, Km = 430 ± 60 μm, kcat/Km = 0.0074 μm−1 s−1; all data are means ± error of fit) and 8-oxogeranial (kcat = 0.90 ± 0.08 s−1, Km = 2900 ± 400 μm, kcat/Km = 0.0003 μm−1 s−1; all data are means ± error of fit). These results indicate low catalytic efficiencies for both substrates compared with OeISY with 8-oxogeranial but higher efficiency for progesterone compared with 8-oxogeranial (Fig. 6C). As hypothesized based on the expression analyses, Oe1,4-R3 appears not to be involved in the secoiridoid pathway.
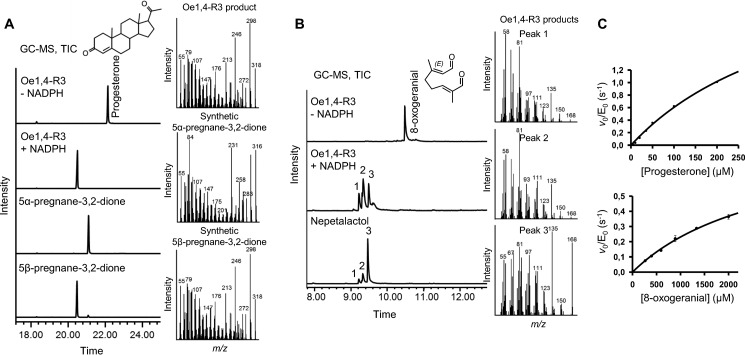
FIGURE 6.
Functional characterization of Oe1,4-R3. A and B, analysis of the reaction products by GC-MS after 3 h of incubation with 200 μm of progesterone (A) or after 1 h of incubation with 200 μm of 8-oxogeranial (B). Total ion chromatograms and MS spectra of CH2Cl2 extracts of the enzymatic reaction (plus NADPH) and of a negative control (minus NADPH) are compared with the TICs of 5α-pregnane-3,20-dione and 5β-pregnane-3,20-dione (A) or a cis-trans-nepetalactol standard (B). GC-MS assays were performed with a Zebron ZB-5 HT column (35 m × 0.25 mm × 0.10 μm). C, steady-state kinetic analysis with progesterone (left panel) or 8-oxogeranial (right panel) as substrate. The reaction rates were measured spectrophotometrically monitoring NADPH consumption at 340 nm. Individual data points are the averages of four replicates ± error of fit.
The Expression Patterns of OeISY and Oe1,4-R1 Are Consistent with a Role in Secoiridoid Biosynthesis
Even though secoiridoids accumulate in all the organs of the olive plant, the highest content is found in leaf and fruit tissues. We conducted an expression analysis of different tissues using real time, quantitative PCR and found that OeISY and Oe1,4-R1 mRNA levels were several hundred-fold higher in fruits and leaves compared with roots and flowers (Fig. 7A). In contrast, Oe1,4-R3 was more highly expressed in the roots, where its mRNA levels were about 4-fold higher than in leaves, the tissue characterized by the lowest relative expression (Fig. 7A).
FIGURE 7.
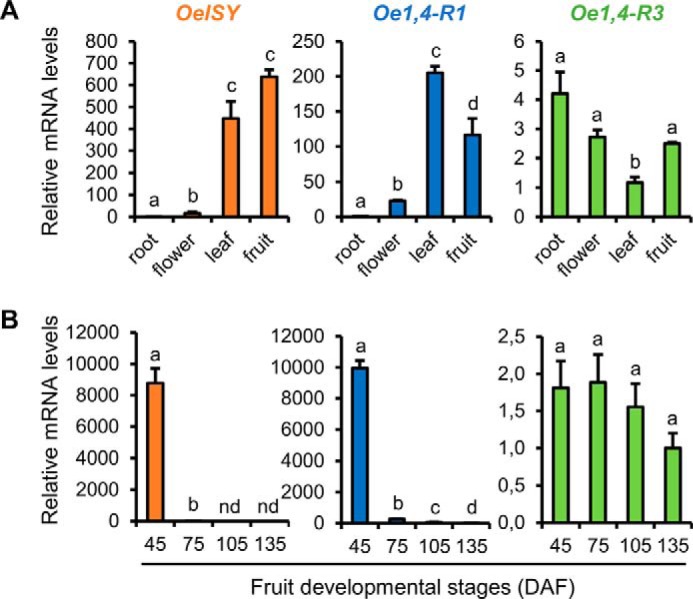
Relative mRNA levels of OeISY, Oe1,4-R1, and Oe1,4-R3 in olive tissues (A) and fruits during development (B). The values are expressed as ΔΔCt. Bars, ± S.E., n = 3. Different letters indicate significant differences between samples as determined using analysis of variance (Bonferroni's post hoc tests, p < 0.05).
Previous analysis of the secoiridoid content of olive fruit during development showed that, after a peak at 45 DAF (about 120 mg/g dry weight for cultivar Leccino), the levels decrease until 165 DAF, where they reach the lowest content (about 48 mg/g dry weight for the same cultivar) (3). We included four fruit developmental stages in our expression analysis by quantitative PCR. In accordance with the reported trends in secoiridoid accumulation, we found that OeISY and Oe1,4-R1 mRNA levels were highest at 45 DAF and then decreased in the subsequent stages (Fig. 7B). In particular, at 45 DAF OeISY reached mRNA levels up to 8000-fold higher compared with 75 DAF, whereas at 105 and 135 DAF, it was not detected. Oe1,4-R1 relative mRNA levels were about 10,000-fold higher at 45 DAF compared with 135 DAF, which was the stage characterized by the lowest expression. In contrast, Oe1,4-R3 mRNA levels did not change significantly during fruit development (Fig. 7B). These results indicate that OeISY and Oe1,4-R1 expression patterns correlate well with the secoiridoid content of olive tissues. In contrast, the expression profile of Oe-1,4-R3 is unrelated to the secoiridoid content, suggesting that this gene may be involved in a different metabolic pathway.
Conserved Domains and Phylogenetic Analyses of Olive 1,4-Reductases
We classified OeISY, Oe1,4-R1, and Oe1,4-R3 as 1,4-reductases according to their ability to perform 1,4-reductions and showed that the sequences of the predicted proteins, like CrISY, are highly similar to P5βRs. Amino acid sequence comparisons revealed that, like P5βRs, they belong to a class of short chain dehydrogenases/reductases (SDRs) structurally characterized by Thorn et al. (47). This class of SDRs is defined by eight conserved motifs and two conserved active site residues (Tyr-179 and Lys-147) (Fig. 8). Residue numbering refers to the crystal structure of DlP5βR (Protein Data Bank code 2V6G) (47).
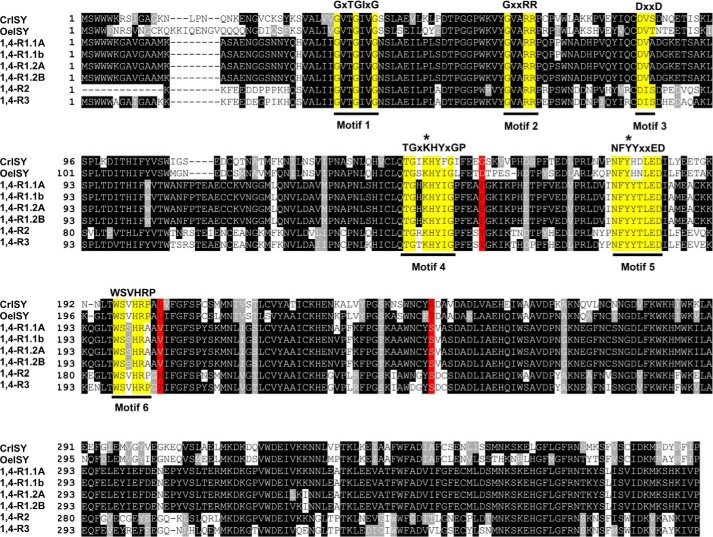
FIGURE 8.
Alignment of the deduced amino acid sequences of olive 1,4-reductases. The conserved motifs of the P5βR subclass of SDRs (47) are shown with bars. Conserved amino acids in the motifs are highlighted in yellow. In the motifs, x denotes any residue. Motif 1, coenzyme binding (recognition of pyrophosphate and ribose linked to the adenine ring); Motif 2, O2′-phosphate binding, discriminates between NADH and NADPH; Motif 3, involved in the interaction with the adenine ring of NADPH; Motif 4, contains active site residue Lys-147 (indicated by an asterisk); Motif 5, contains active site residue Tyr-179 (indicated by an asterisk); Motif 6, part of β-sheet, it helps to position the catalytically important motifs 4 and 5, proline (P) packs against the nicotinamide ring of NADPH. Three hot spots at positions 156, 205, and 248, in close proximity of the substrate-binding pocket and important for the enzymatic activity (48), are highlighted in red.
Some amino acid substitutions were observed in the conserved motifs of the olive 1,4-reductases, possibly controlling the enzymatic activity and the substrate specificity. OeISY, similarly to CrISY, showed substitution of Tyr-180 for histidine in motif 5, a motif that harbors the Tyr-179 residue that is important for catalytic activity. Oe1,4-R1 showed a substitution of Val-200 for serine in the motif 6 involved in NADPH binding. Moreover, OeISY and Oe1,4-R1 differ in three hot spots that, according to Bauer et al. (48), affect P5βR-like enzymatic activity. In particular, OeISY compared with DlP5βR, showed a substitution of Tyr-156 for aspartic acid and, similarly to CrISY, a substitution of Ser-248 for an apolar amino acid (valine). OeISY, as CrISY, and Oe1,4-R1 presented a substitution of Asn-205 for an apolar amino acid (leucine or valine) (Fig. 8).
We carried out a phylogenetic analysis using the amino acid sequences of olive 1,4-reductases and their homologues from other plants (Fig. 9). Along with a number of predicted sequences, this analysis included CrISY, the progesterone reductases P5βR1 and P5βR2 of D. lanata and D. purpurea (38, 39), and, in addition, some P5βR homologues from C. roseus (CrP5βR1–6) and M. truncatula (MtP5βR1–4) whose exact role in plant metabolism remains unclear (49). The analysis highlights the fact that proteins sharing homology with P5βR are very common in the plant kingdom, being present in different families of both monocots and dicots. Two main clades (A and B) seem to have originated from a common ancestor. All the proteins included in clade A are of unknown function and include only dicot sequences. Clade B gives rise to three different subclades: c and d, both specific to dicots, and e, which contains only monocot sequences. Oe1,4-R1, Oe1,4-R2, and Oe1,4-R3 group in the subclade c, the largest subclade, which includes representatives that are widely dispersed across numerous dicot families, whereas OeISY groups together with CrISY and with DlP5βR2 and DpP5βR2 in subclade d, which contains only proteins belonging to the lamiids. These data suggest that iridoid synthases might have originated from an ancestor exclusively common to Asterids, the subclass that contains the iridoid and secoiridoid synthesizing species (50), shedding new light on their possible differentiation from other SDRs.
FIGURE 9.
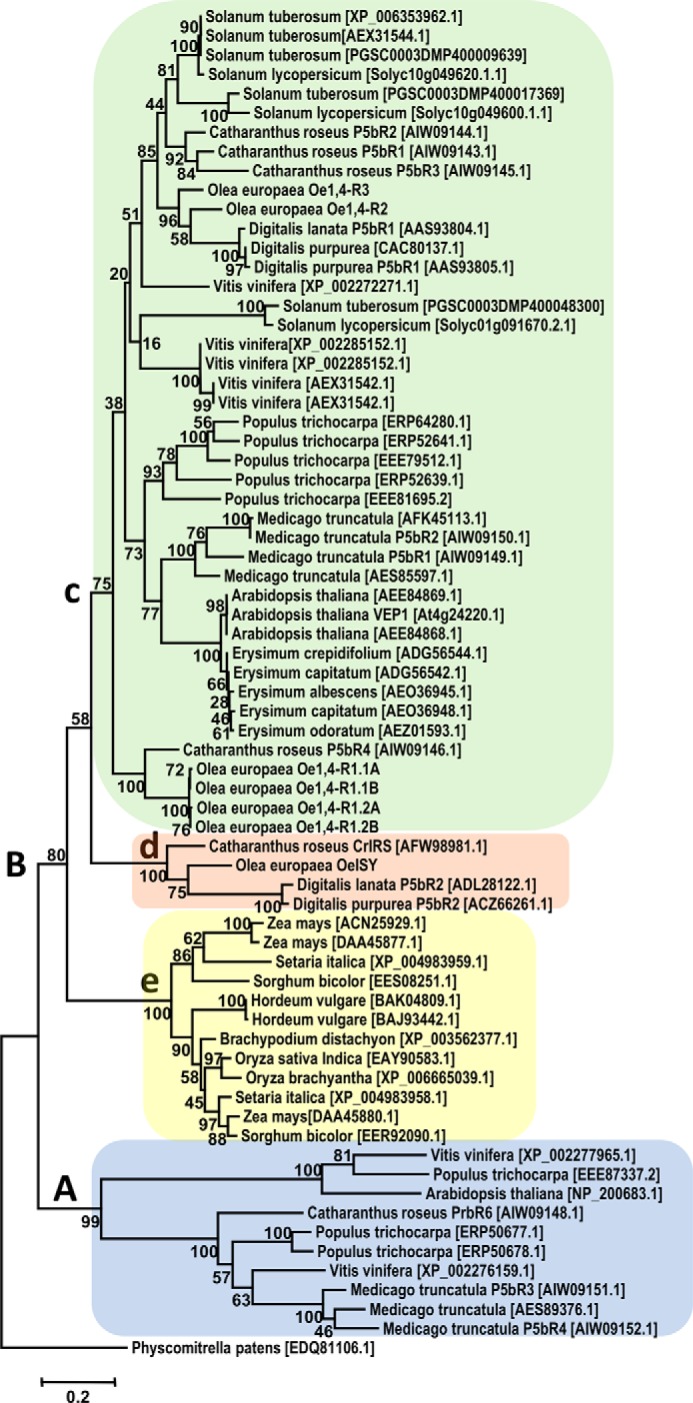
Phylogenetic tree of olive 1,4-reductases. Predicted amino acid sequences from monocots and dicots were aligned with the biochemically characterized enzymes from olive. The phylogenetic tree was drawn by maximum likelihood method with 1000 bootstrap replicates (percentage values shown at branch points), using P. patens (GenBankTM accession number EDQ81106.1) as outgroup.
Discussion
Secoiridoids are responsible for many of the health promoting effects of olive oil and positively affect its organoleptic properties (5, 6, 51). Hence, high secoiridoid content in olive fruits is a desirable trait in olive cultivation. Based on sequence and transcriptomic data mining, we have identified five genes (OeISY, Oe1,4-R1, Oe8HGO, OeIO, and Oe7-DLGT2) that are potentially involved in the biosynthesis of these economically important natural products. The genes of the secoiridoid pathway can potentially be used as targets for the identification of molecular markers in olive breeding programs aiming to increase oleuropein production. Moreover, the identification of the biosynthetic genes have also applications for metabolic engineering in microbial or plant hosts. They can be used to develop synthetic pathways for a large scale production of such valuable compounds.
Oleuropein is not restricted to the Olea genus but is also found in other important genera belonging to the Oleaceae family, including ash (Fraxinus excelsior and Fraxinus chinensis), lilac (Syringa josikaea, Syringa vulgaris, and Syringa dilatata), privet (Ligustrum ovalifolium), and jasmine (Jasminum polyanthum) (21, 52–55). Therefore, our discovery in olive will facilitate the elucidation of secoiridoid pathways in these closely related species.
The co-expression observed for the genes of the putative olive secoiridoid pathway has also been observed for genes involved in iridoid biosynthesis in C. roseus (19, 56). Conservation of iridoid biosynthesis genes in C. roseus and olive suggests common biosynthetic intermediates until the formation of 7-deoxyloganic acid (Fig. 1B). However, the downstream biosynthetic steps after this point still need to be clarified in the olive pathway. It has been hypothesized (22–24) that oleoside-11-methyl ester is an intermediate of the pathway, and it is presumably synthesized from 7-deoxyloganic acid via 7-ketologanic acid and 7-ketologanin (Fig. 2), but more experimental evidence for this prediction is required. These enzymatic reactions, which include oxidoreduction, methylation, and cleavage of the cyclopentane ring, are similar to those identified in the secologanin biosynthesis of monoterpene indole alkaloids (19). Therefore, we hypothesize that, in olive, genes similar to CrDLH, CrLAMT, and CrSLS might work on these compounds.
Among the candidate genes involved in secoiridoid biosynthesis, we focused our attention on the functional characterization of iridoid synthase, an enzyme that is key for the formation of the iridoid scaffold and belongs to a new and unexplored class of enzymes. The high specificity of OeISY for 8-oxogeranial, its co-expression with other candidate genes of the pathway (3), and the correlation of its expression levels with secoiridoid content all strongly suggest that OeISY synthesizes the iridoid scaffold in olive. Although iridoids are very common in flowering plants, only in very few cases has iridoid synthase activity been demonstrated in vitro (16, 49). Similar to CrISY (16, 40), OeISY gave a mixture of nepetalactol and iridodials as enzymatic products. It remains unclear how this mixture of products is channeled into secoiridoid biosynthesis. However, it has been shown in C. roseus that the enzyme that acts directly downstream of ISY in the pathway, iridoid oxidase, can convert both nepetalactol and iridodials in this mixture into 7-deoxyloganetic acid, although nepetalactol was consumed faster (19).
Oe1,4-R1 was co-expressed with OeISY and other candidates of secoiridoid pathway, but it did not show iridoid synthase activity in vitro. The strong co-expression profile with the iridoid synthase gene suggests a role in this pathway, but the function remains to be identified. Based on the current knowledge on oleuropein biosynthesis, we are unable to suggest a role for Oe1,4-R1. Many reactions in oleuropein biosynthesis are still uncertain or hypothetical, particularly the downstream reactions, and Oe1,4-R1 may have a role in an unpredicted step. Moreover, we cannot exclude the existence of alternative routes to oleuropein biosynthesis.
All ISY proteins show high sequence similarity to P5βRs, which convert progesterone to 5β-pregnane-3,20-dione in cardenolide-producing plants (i.e. D. lanata, D. purpurea, and Erysimum crepidifolium) but, with unknown functions, also occur in numerous cardenolide-free plants such as Arabidopsis thaliana (48), C. roseus, and M. truncatula (49). A large number of P5βR homologues from many species, including cardenolide- and iridoid-free plants, indicate a large and heterogeneous class of enzymes that is not fully explored in terms of biological function and biocatalytic potential.
The high similarity of OeISY and CrISY in terms of sequence and biological function may indicate that they are orthologues derived from a common ancestor belonging to the P5βR family (Fig. 9). This hypothesis was substantiated by our phylogenetic analysis, which indicates that proteins similar to Oe1,4-R1 and Oe1,4-R3 are almost ubiquitous in dicot families, whereas ISYs group in a small subclade which includes only proteins belonging to the Asterid clade. Most likely, a common ISY ancestor in this subclass can account for the chemical diversity of iridoids and secoiridoids (50) synthesized by various species in this clade. As more ISY sequences from this subclass become available, it may be possible to elucidate how ISYs have evolutionarily diverged from other SDRs.
The enzymatic cyclization of 8-oxogeranial performed by iridoid synthases is a mechanistically remarkable reaction performed by a large number of plants. By adding new members, like OeISY, to this recently discovered class of catalysts, we can better understand how the iridoid biosynthetic pathway evolved and which molecular features are essential for the iridoid synthase reaction. Knowledge of the enzymes performing secoiridoid synthesis in olives can lay the basis for enhancing the health promoting features of this important food crop.
Author Contributions
F. A., A. O., S. E. O., and L. B. conceived the work. F. A. performed most of the experiments. F. G.-F. synthesized 8-oxoneral and interpreted the GC-MS data. H. K. performed the kinetic experiments. L. B. provided the plant samples and the sequence data. F. A. and F. P. analyzed the transcriptomic data. F. A., F. P., A. O., and S. E. O. wrote the manuscript.
Acknowledgments
We are grateful to Dr. Jo Dicks and Dr. Thomas Loveau for suggestions on methodology.
This work was supported by the I-MOVE FP7 Marie Curie COFUND Program funded by the European Union and Regione Umbria (Italy) and also by the Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BB/J004561/1 (Understanding and Exploiting Plant and Microbial Metabolism) and by the John Innes Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
- P5βR
- progesterone 5-β-reductase
- DAF
- days after flowering
- SDR
- short chain dehydrogenase/reductase.
References
- 1. Obied H. K., Prenzler P. D., Ryan D., Servili M., Taticchi A., Esposto S., and Robards K. (2008) Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat. Prod. Rep. 25, 1167–1179 [DOI] [PubMed] [Google Scholar]
- 2. Servili M., and Montedoro G. F. (2002) Contribution of phenolic compounds to virgin olive oil quality. Eur. J. Lipid Sci. Tech. 104, 602–613 [Google Scholar]
- 3. Alagna F., Mariotti R., Panara F., Caporali S., Urbani S., Veneziani G., Esposto S., Taticchi A., Rosati A., Rao R., Perrotta G., Servili M., and Baldoni L. (2012) Olive phenolic compounds: metabolic and transcriptional profiling during fruit development. BMC Plant Biol. 12, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Servili M., Selvaggini R., Esposto S., Taticchi A., Montedoro G., and Morozzi G. (2004) Health and sensory properties of virgin olive oil hydrophilic phenols: agronomic and technological aspects of production that affect their occurrence in the oil. J. Chromatogr. A 1054, 113–127 [PubMed] [Google Scholar]
- 5. Hassen I., Casabianca H., and Hosni K. (2015) Biological activities of the natural antioxidant oleuropein: Exceeding the expectation: A mini-review. J. Funct. Foods 18, 926–940 [Google Scholar]
- 6. Omar S. H. (2010) Oleuropein in olive and its pharmacological effects. Sci. Pharm. 78, 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waterman E., and Lockwood B. (2007) Active components and clinical applications of olive oil. Altern. Med. Rev. 12, 331–342 [PubMed] [Google Scholar]
- 8. Owen R. W., Haubner R., Mier W., Giacosa A., Hull W. E., Spiegelhalder B., and Bartsch H. (2003) Isolation, structure elucidation and antioxidant potential of the major phenolic and flavonoid compounds in brined olive drupes. Food Chem. Toxicol. 41, 703–717 [DOI] [PubMed] [Google Scholar]
- 9. Puel C., Mathey J., Agalias A., Kati-Coulibaly S., Mardon J., Obled C., Davicco M. J., Lebecque P., Horcajada M. N., Skaltsounis A. L., and Coxam V. (2006) Dose-response study of effect of oleuropein, an olive oil polyphenol, in an ovariectomy/inflammation experimental model of bone loss in the rat. Clin. Nutr. 25, 859–868 [DOI] [PubMed] [Google Scholar]
- 10. Kubo I., Matsumoto A., and Takase I. (1985) A multichemical defense mechanism of bitter olive Olea europaea (Oleaceae). Is oleuropein a phytoalexin precursor? J. Chem. Ecol. 11, 251–263 [DOI] [PubMed] [Google Scholar]
- 11. Konno K., Hirayama C., Yasui H., and Nakamura M. (1999) Enzymatic activation of oleuropein: a protein crosslinker used as a chemical defense in the privet tree. Proc. Natl. Acad. Sci. U.S.A. 96, 9159–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malik N. S., and Bradford J. M. (2008) Recovery and stability of oleuropein and other phenolic compounds during extraction and processing of olive (Olea europaea L.) leaves. J. Food Agric. Environ. 6, 8–13 [Google Scholar]
- 13. Ortega-García F., and Peragón J. (2010) Phenol metabolism in the leaves of the olive tree (Olea europaea L.) cv. Picual, Verdial, Arbequina, and Frantoio during ripening. J. Agric. Food Chem. 58, 12440–12448 [DOI] [PubMed] [Google Scholar]
- 14. Soler-Rivas C., Espín J. C., and Wichers H. J. (2000) Oleuropein and related compounds. J. Sci. Food Agric. 80, 1013–1023 [Google Scholar]
- 15. Vezzaro A., Krause S. T., Nonis A., Ramina A., Degenhardt J., and Ruperti B. (2012) Isolation and characterization of terpene synthases potentially involved in flavor development of ripening olive (Olea europaea) fruits. J. Plant. Physiol. 169, 908–914 [DOI] [PubMed] [Google Scholar]
- 16. Geu-Flores F., Sherden N. H., Courdavault V., Burlat V., Glenn W. S., Wu C., Nims E., Cui Y., and O'Connor S. E. (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492, 138–142 [DOI] [PubMed] [Google Scholar]
- 17. Asada K., Salim V., Masada-Atsumi S., Edmunds E., Nagatoshi M., Terasaka K., Mizukami H., and De Luca V. (2013) A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar periwinkle. Plant Cell 25, 4123–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salim V., Wiens B., Masada-Atsumi S., Yu F., and De Luca V. (2014) 7-deoxyloganetic acid synthase catalyzes a key 3 step oxidation to form 7-deoxyloganetic acid in Catharanthus roseus iridoid biosynthesis. Phytochemistry 101, 23–31 [DOI] [PubMed] [Google Scholar]
- 19. Miettinen K., Dong L., Navrot N., Schneider T., Burlat V., Pollier J., Woittiez L., van der Krol S., Lugan R., Ilc T., Verpoorte R., Oksman-Caldentey K. M., Martinoia E., Bouwmeester H., Goossens A., Memelink J., and Werck-Reichhart D. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5, 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salim V., Yu F., Altarejos J., and De Luca V. (2013) Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 76, 754–765 [DOI] [PubMed] [Google Scholar]
- 21. Damtoft S., Franzyk H., and Jensen S. R. (1993) Biosynthesis of secoiridoid glucosides in Oleaceae. Phytochemistry 34, 1291–1299 [Google Scholar]
- 22. Damtoft S., Franzyk H., and Jensen S. R. (1995) Biosynthesis of iridoids in Syringa and Fraxinus: carbocyclic iridoid precursors. Phytochemistry 40, 785–792 [Google Scholar]
- 23. Damtoft S., Franzyk H., and Jensen S. R. (1995) Biosynthesis of iridoids in Syringa and Fraxinus: secoiridoid precursors. Phytochemistry 40, 773–784 [Google Scholar]
- 24. Jensen S. R., Franzyk H., and Wallander E. (2002) Chemotaxonomy of the Oleaceae: iridoids as taxonomic markers. Phytochemistry 60, 213–231 [DOI] [PubMed] [Google Scholar]
- 25. Ryan D., Antolovich M., Herlt T., Prenzler P. D., Lavee S., and Robards K. (2002) Identification of phenolic compounds in tissues of the novel olive cultivar hardy's mammoth. J. Agric. Food Chem. 50, 6716–6724 [DOI] [PubMed] [Google Scholar]
- 26. Field B., Fiston-Lavier A. S., Kemen A., Geisler K., Quesneville H., and Osbourn A. E. (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. U.S.A. 108, 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naoumkina M. A., Modolo L. V., Huhman D. V., Urbanczyk-Wochniak E., Tang Y., Sumner L. W., and Dixon R. A. (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22, 850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schilmiller A. L., Pichersky E., and Last R. L. (2012) Taming the hydra of specialized metabolism: how systems biology and comparative approaches are revolutionizing plant biochemistry. Curr. Opin. Plant Biol. 15, 338–344 [DOI] [PubMed] [Google Scholar]
- 29. Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., and Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yonekura-Sakakibara K., Tohge T., Niida R., and Saito K. (2007) Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282, 14932–14941 [DOI] [PubMed] [Google Scholar]
- 31. Saito K., Hirai M. Y., and Yonekura-Sakakibara K. (2008) Decoding genes with coexpression networks and metabolomics: “majority report by precogs.” Trends Plant Sci. 13, 36–43 [DOI] [PubMed] [Google Scholar]
- 32. Shulaev V., Cortes D., Miller G., and Mittler R. (2008) Metabolomics for plant stress response. Physiol. Plant. 132, 199–208 [DOI] [PubMed] [Google Scholar]
- 33. Alagna F., D'Agostino N., Torchia L., Servili M., Rao R., Pietrella M., Giuliano G., Chiusano M. L., Baldoni L., and Perrotta G. (2009) Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. BMC Genomics 10, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galla G., Barcaccia G., Ramina A., Collani S., Alagna F., Baldoni L., Cultrera N. G., Martinelli F., Sebastiani L., and Tonutti P. (2009) Computational annotation of genes differentially expressed along olive fruit development. BMC Plant Biol. 9, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muñoz-Mérida A., González-Plaza J. J., Cañada A., Blanco A. M., García-López Mdel C., Rodríguez J. M., Pedrola L., Sicardo M. D., Hernández M. L., De la Rosa R., Belaj A., Gil-Borja M., Luque F., Martínez-Rivas J. M., Pisano D. G., Trelles O., Valpuesta V., and Beuzón C. R. (2013) De novo assembly and functional annotation of the olive (Olea europaea) transcriptome. DNA Res. 20, 93–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ozdemir Ozgenturk N., Oruç F., Sezerman U., Kuçukural A., Vural Korkut S., Toksoz F., and Un C. (2010) Generation and Analysis of Expressed Sequence Tags from Olea europaea L. Comp. Funct. Genomics 2010, 757512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bauer P., Munkert J., Brydziun M., Burda E., Müller-Uri F., Gröger H., Muller Y. A., and Kreis W. (2010) Highly conserved progesterone 5β-reductase genes (P5βR) from 5β-cardenolide-free and 5β-cardenolide-producing angiosperms. Phytochemistry 71, 1495–1505 [DOI] [PubMed] [Google Scholar]
- 38. Pérez-Bermúdez P., García A. A., Tuñón I., and Gavidia I. (2010) Digitalis purpurea P5βR2, encoding steroid 5β-reductase, is a novel defense-related gene involved in cardenolide biosynthesis. New Phytol. 185, 687–700 [DOI] [PubMed] [Google Scholar]
- 39. Herl V., Fischer G., Müller-Uri F., and Kreis W. (2006) Molecular cloning and heterologous expression of progesterone 5β-reductase from Digitalis lanata Ehrh. Phytochemistry 67, 225–231 [DOI] [PubMed] [Google Scholar]
- 40. Lindner S., Geu-Flores F., Bräse S., Sherden N. H., and O'Connor S. E. (2014) Conversion of substrate analogs suggests a Michael cyclization in iridoid biosynthesis. Chem. Biol. 21, 1452–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kries H., Caputi L., Stevenson C. E., Kamileen M. O., Sherden N. H., Geu-Flores F., Lawson D. M., and O'Connor S. E. (2016) Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 12, 6–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stekel D. J., Git Y., and Falciani F. (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res. 10, 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eisen M. B., Spellman P. T., Brown P. O., and Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 45. Nonis A., Vezzaro A., and Ruperti B. (2012) Evaluation of RNA extraction methods and identification of putative reference genes for real-time quantitative polymerase chain reaction expression studies on olive (Olea europaea L.) fruits. J. Agric. Food Chem. 60, 6855–6865 [DOI] [PubMed] [Google Scholar]
- 46. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thorn A., Egerer-Sieber C., Jäger C. M., Herl V., Müller-Uri F., Kreis W., and Muller Y. A. (2008) The crystal structure of progesterone 5β-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J. Biol. Chem. 283, 17260–17269 [DOI] [PubMed] [Google Scholar]
- 48. Bauer P., Rudolph K., Müller-Uri F., and Kreis W. (2012) Vein patterning 1-encoded progesterone 5β-reductase: activity-guided improvement of catalytic efficiency. Phytochemistry 77, 53–59 [DOI] [PubMed] [Google Scholar]
- 49. Munkert J., Pollier J., Miettinen K., Van Moerkercke A., Payne R., Müller-Uri F., Burlat V., O'Connor S. E., Memelink J., Kreis W., and Goossens A. (2015) Iridoid synthase activity is common among the plant progesterone 5β-reductase family. Mol. Plant. 8, 136–152 [DOI] [PubMed] [Google Scholar]
- 50. Jensen S. R. (1991) Plant iridoids, their biosynthesis and distribution in angiosperms. In Proceedings of the Phytochemical Society of Europe: Ecological Chemistry and Biochemistry of Plant Terpenoids (Harborne J. B., and Tomas-Barberan F. A., eds) pp. 133–158, Clarendon Press Oxford [Google Scholar]
- 51. Buckland G., and Gonzalez C. A. (2015) The role of olive oil in disease prevention: a focus on the recent epidemiological evidence from cohort studies and dietary intervention trials. Br. J. Nutr. 113, S94–101 [DOI] [PubMed] [Google Scholar]
- 52. Kuwajima H., Uemura T., Takaishi K., Inoue K., and Inouyet H. (1988) A secoiridoid glucoside from Olea europaea. Phytochemistry 27, 1757–1759 [Google Scholar]
- 53. Oh H., Ko E.-K., Kim D.-H., Jang K.-K., Park S.-E., Lee H.-S., and Kim Y.-C. (2003) Secoiridoid glucosides with free radical scavenging activity from the leaves of Syringa dilatata. Phytother. Res. 17, 417–419 [DOI] [PubMed] [Google Scholar]
- 54. Tanahashi T., Takenaka Y., and Nagakura N. (1996) Two dimeric secoiridoid glucosides from Jasminum polyanthum. Phytochemistry 41, 1341–1345 [Google Scholar]
- 55. Sugiyama M., Machida K., Matsuda N., and Kikuchi M. (1993) A secoiridoid glycoside from Osmanthus asiaticus. Phytochemistry 34, 1169–1170 [DOI] [PubMed] [Google Scholar]
- 56. Van Moerkercke A., Fabris M., Pollier J., Baart G. J., Rombauts S., Hasnain G., Rischer H., Memelink J., Oksman-Caldentey K. M., and Goossens A. (2013) CathaCyc, a metabolic pathway database built from Catharanthus roseus RNA-Seq data. Plant Cell Physiol. 54, 673–685 [DOI] [PubMed] [Google Scholar]
- 57. Dewick P. M. (2002) The biosynthesis of C5-C25 terpenoid compounds. Nat. Prod. Rep. 19, 181–222 [DOI] [PubMed] [Google Scholar]