Abstract
M-current-mediating KCNQ (Kv7) channels play an important role in regulating the excitability of neuronal cells, as highlighted by mutations in Kcnq2 and Kcnq3 that underlie certain forms of epilepsy. In addition to their expression in brain, KCNQ2 and -3 are also found in the somatosensory system. We have now detected both KCNQ2 and KCNQ3 in a subset of dorsal root ganglia neurons that correspond to D-hair Aδ-fibers and demonstrate KCNQ3 expression in peripheral nerve endings of cutaneous D-hair follicles. Electrophysiological recordings from single D-hair afferents from Kcnq3−/− mice showed increased firing frequencies in response to mechanical ramp-and-hold stimuli. This effect was particularly pronounced at slow indentation velocities. Additional reduction of KCNQ2 expression further increased D-hair sensitivity. Together with previous work on the specific role of KCNQ4 in rapidly adapting skin mechanoreceptors, our results show that different KCNQ isoforms are specifically expressed in particular subsets of mechanosensory neurons and modulate their sensitivity directly in sensory nerve endings.
Keywords: electrophysiology, ion channel, mechanotransduction, mouse genetics, neurobiology, potassium channel, M-current, skin-nerve preparation
Introduction
KCNQ (Kv7) K+ channels are key regulators of cellular electrical activity that act by stabilizing and regulating the membrane-resting potential (1–3). KCNQ1 is mainly expressed in cardiac and epithelial cells, KCNQ2–5 are found in neuronal and primary sensory cells (1, 4), and several KCNQ channels are also found in vascular smooth muscle (5–7). The neuronal KCNQ channels (KCNQ2–5) show properties of the M-current (8–11), a slowly activating, non-inactivating outward rectifying potassium current that can be inhibited by muscarinic stimulation (12). Mutations in four of the KCNQ channel genes (KCNQ1-4) give rise to human genetic disorders (1, 4). KCNQ1 mutations lead to cardiac arrhythmias and deafness (13, 14), mutations in KCNQ2 and -3 cause BFNS (benign familial neonatal seizures) (also known as BFNC (benign familial neonatal convulsions)) (15–17). KCNQ2 mutations can also cause myokymia (18) and lead to epileptic encephalopathy (19), whereas mutations in KCNQ4, which is expressed in the cochlea and the vestibular organ (20, 21), entail progressive hearing loss (22). KCNQ5 has not yet been associated to human disease.
Recently, KCNQ4 was detected in a small subset of rapidly adapting low-threshold mechanoreceptors in dorsal root ganglia (DRG)2 and associated Meissner corpuscles, as well as in lanceolate endings and circular nerve fibers around hair follicles in the periphery (23). The mutational inactivation of KCNQ4 increased responses to low-frequency mechanical stimuli in mice and humans (23). Other neuronal KCNQ isoforms were also reported to be expressed in sensory neurons. Several studies have identified the M-current and their underlying molecular correlates in nociceptive DRG neurons (10, 24–29). Both KCNQ2 (26) and KCNQ5 (27) were reported to be predominant KCNQ subunits in small diameter nociceptive DRG neurons.
Skin mechanoreceptors express a variety of ionic currents that greatly influence their specific transduction properties, but our knowledge of the underlying channels and their role in shaping their responses is far from being complete (30). We reported previously that the KCNQ inhibitor linopirdine increased the firing rate of D-hair fibers in response to mechanical stimulation in skin-nerve preparations (23), suggesting a role of KCNQ channels in these mechanoreceptors. D-hairs are the most sensitive mechanoreceptors in the skin and are characterized by large receptive fields (31, 32). According to their intermediate conduction velocity and medium cell diameter, D-hairs are classified as Aδ-low-threshold mechanoreceptors (Aδ-LTMR). They form lanceolate endings around hair follicles of awl/auchene and zigzag hairs (33).
In this study, we investigated which KCNQ isoforms are expressed in D-hairs. We found that a subset of KCNQ2- or KCNQ3-positive neurons in DRG sections was co-labeled with D-hair markers. Immunohistochemistry revealed KCNQ3 expression in lanceolate endings around hair follicles in the skin where it may modulate mechanotransduction. Indeed, skin-nerve preparations showed that KCNQ channels dampen the response of D-hairs to mechanical stimuli. During the movement phase of a mechanical ramp-and-hold stimulus, the firing rate of D-hair fibers from Kcnq3−/− and Kcnq2+/−/Kcnq3−/− mice was increased. We conclude that KCNQ3 and KCNQ2 directly modulate the mechanosensitivity of D-hairs at their peripheral nerve endings.
Materials and Methods
Kcnq Mouse Models
All mice were housed in the animal facility of the MDC Berlin according to institutional guidelines, and all animal experiments were done according to the German Animal Protection Law. Kcnq2tm1Dgen/Kcnq2+ (Kcnq2+/−) mice were obtained from The Jackson laboratory, Bar Harbor, ME, and contain a deletion in the Kcnq2 gene from base 418 to 535 by insertion of a Lac0-SA-IRES-lacZ-Neo555G/Kan construct. Kcnq3−/− mice were newly generated. We first generated Kcnq3lox/lox mice in which the protein coding exon 3 was flanked by loxP sites. C57Bl/6J Kcnq3 genomic DNA encompassing exon 1 to exon 5 was obtained from a BAC clone (Sanger Institute). The DNA was inserted into the XhoI and AatII restriction sites of the pKO901-DTA vector (where DTA indicates diphtheria toxin A). A neomycin resistance (NEOR) cassette flanked by FRT/loxP sites was ligated into PacI site of Kcnq3 intron 3. The construct was sequenced and electroporated into R1 (129/SvJ) mouse ES cells, which were analyzed for correct targeting and injected into C57Bl/6J blastocysts that were implanted into foster mothers. Chimeric offspring was first crossed to FLPe-recombinase-expressing deleter mice (34) to excise the NEOR cassette and next to Cre-recombinase-expressing deleter mice (35) to remove exon 3 from the Kcnq3 locus, which leads to a frameshift. The inactivation of Kcnq3 and the loss of KCNQ3 protein were confirmed by Southern blotting, RT-PCR, and Western blotting analysis (data not shown). Kcnq2+/−/Kcnq3−/− double mutant mice were generated by breeding Kcnq2+/− and Kcnq3−/− mice.
Antibodies
The following primary antibodies were used: guinea pig anti-KCNQ2 (raised against the peptide VQKSRNGGVYPGTSGEKKL that was coupled by a C-terminally added cysteine to Keyhole Limpet Hemocyanin; named Q2Agp1), 1:100; rabbit anti-KCNQ3 (named Q3Crb1 (36)), 1:200; goat anti-TrkB (R&D Systems, AF1494), 1:500; mouse anti-NF200 (Sigma, N0142), 1:1000; and mouse anti-ankyrin G (Zymed Laboratories Inc., 33–8800), 1:50. Secondary antibodies were from Molecular Probes.
Immunohistochemistry
Mice were anesthetized with ketamine and xylazine and perfused through the heart with 1% (w/v) paraformaldehyde in PBS. DRGs and hairy skin were dissected and postfixed at 4 °C with 1% paraformaldehyde for 30 min or 1 h, respectively. Tissues were then incubated overnight in 30% (w/v) sucrose at 4 °C and embedded in Tissue-Tek O.C.T. (Sakura). Cryosections were cut at 8 μm (for DRGs) and 35 μm (for hairy skin) and were blocked in 2% (w/v) BSA and 0.5% (v/v) Nonidet P-40 in PBS for 2 h. Antibodies were diluted in PBS containing 1% BSA and 0.25% Nonidet P-40 and incubated for 1 h. Nuclei were stained with DAPI. All sections were imaged using a Zeiss LSM 510 confocal microscope. Image analysis and stack assembly were performed off-line with the ZEN 2009 light edition software (Zeiss) and Adobe Photoshop.
In Situ Hybridization
In situ hybridization was carried out as described previously (37, 38). Briefly, DRGs were freshly embedded into O.C.T. compound and cut into 16-μm cryosections. Frozen sections were acetylated and incubated overnight at 61 °C with 0.4 ng/μl DIG- and/or FITC-labeled in situ probes in a hybridization buffer. The following primers were used for synthesis of in situ hybridization probes from cDNA: KCNQ2: TGGGACCGGCTTTTACAGA (forward), and GTTTTACTAAGTGCCCAGGTCC (reverse); KCNQ3: AATGAACCATATGTAGCCAGGG (forward), and CACTGGGGCCAAATACATAATC (reverse); KCNQ4: AGCTGATCACCGCCTGGTACATCG (forward), and GGTGCGGTCATTGAAGCTCCAG (reverse); KCNQ5: ACGTCAGATAAGAAGAGCCGAG (forward), and GCAGGTGGTGACATCAGAAATA (reverse).
The PCR product was cloned into pGEMTeasy vector (Promega), linearized, and transcribed in vitro using DIG and FITC RNA labeling mix and Sp6 and T7 RNA polymerases (Roche Diagnostics). The in situ probes for TrkB and Cav3.2 (39) were a kind gift from C. Birchmeier (MDC Berlin).
After hybridization, DRG sections were treated with RNase A (Sigma-Aldrich) for 15 min at 37 °C. For single in situ hybridization, sections were incubated in 1% blocking reagent with alkaline phosphatase-conjugated sheep anti-DIG antibody (Roche Diagnostics) overnight. Alkaline phosphatase activity was visualized with nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate toluidine salt solution (NBT/BCIP, Roche Diagnostics).
For double in situ hybridization experiments, sections were incubated with anti-FITC-POD (PerkinElmer) overnight at 4 °C and treated with Tyramide Signal Amplification (TSATM Plus) Biotin Kit (PerkinElmer). After quenching the first POD with 4% paraformaldehyde for 2 h and 3% H2O2 for 30 min, anti-DIG-POD (PerkinElmer) was added overnight at 4 °C and subsequently treated with TSATM Plus Cyanine3 System (PerkinElmer). The biotin conjugate was then visualized with streptavidin Alexa Fluor 488 conjugate (Life Technologies).
For combined immunostaining/in situ hybridization, hybridized sections were incubated overnight with anti-DIG-POD conjugate and NF200 or TrkB antibody. The RNA signal was detected with the TSATM Plus Biotin System and streptavidin Alexa Fluor 488 conjugate. Cell sizes were determined using the ImageJ software.
In Vitro Skin-Nerve Preparation
The skin-nerve preparation was used as described previously (23, 40). Mice were sacrificed, and the saphenous nerve and the skin of the hind limb were dissected free. The skin was placed corium side up into an organ bath, where it was superfused by 32 °C warm oxygen-saturated synthetic interstitial fluid, consisting of (in mm) 123 NaCl; 3.5 KCl; 0.7 MgSO4; 1.7 NaH2PO4; 2.0 CaCl2; 9.5 sodium gluconate; 5.5 glucose; 7.5 sucrose; 10 HEPES at a pH of 7.4. The saphenous nerve was pulled through a gap into an adjacent chamber with synthetic interstitial fluid solution that was overlaid by mineral oil. Filaments were teased from the desheathed nerve and placed at the electrode. Single units were identified by a mechanical search stimulus that was applied with a glass rod and classified according to their conduction velocity and von Frey hair thresholds. A computer-controlled nanomotor (Kleindiek Nanotechnik) was used to apply mechanical ramp-and-hold stimuli with a stainless steel metal rod with a flat circular contact area of 0.8 mm2. The PowerLab 8/30 system and LabChart 7.3.5 software (ADInstruments) were used to record raw electrophysiological data and the signal for the movement of the mechanical stimulator.
Dorsal Root Ganglion Neuronal Culture
Primary cultures of DRG neurons were established as described previously (41). Briefly, mice were sacrificed and DRGs were quickly dissected and collected in DRG medium (Ham's F12 medium (PAN Biotech), 10% FBS, 1% penicillin/streptomycin). DRGs were first digested in 1 mg/ml collagenase IV (Gibco) at 37 °C for 60 min, followed by incubation in 0.05% trypsin (Gibco) in PBS at 37 °C for 10 min. After removal of trypsin, DRGs were dissociated in 1 ml of DRG medium until all DRGs had broken up, and then were pipetted through a cell strainer. DRGs were loaded onto a 2-ml BSA pillow (15% BSA in DRG medium) and centrifuged at 900 rpm for 10 min to separate the myelin and debris. The pellet was resuspended in 150 μl of DRG medium and distributed onto poly-d-lysine- (100 μg/ml) and laminin- (10 μg/ml) coated coverslips and incubated at 37 °C overnight. Coverslips were then incubated with fresh medium containing goat anti-TrkB antibody for 2 h at 4 °C, washed with DRG medium three times, and then incubated with donkey anti-goat Alexa Fluor 488 for 1 h at 4 °C.
Whole-cell Electrophysiology in Cultured DRG Neurons
An upright microscope (Olympus BX51WI) equipped with epifluorescence was used to identify large (300–500 μm2) and healthy DRG cells (1–2 days in vitro) with specific fluorescence labeling seen at the plasma membrane and at intracellular endocytic compartments. Whole-cell membrane currents from such identified cells were recorded at room temperature using the conventional patch clamp configuration. Signals were acquired with a MultiClamp 700B amplifier, (Molecular Devices), digitized at 10 kHz, and filtered at 2 kHz (Clampex 10.4, Molecular Devices). Patch pipettes were filled with a solution containing (in mm): 80 KOAc, 30 KCl, 40 HEPES, 3 MgCl2, 3 EGTA, 1 CaCl2, adjusted to pH 7.4 and 290 mosm/kg, and typically registered resistances of 3–4 megaohms. The bath solution consisted of (in mm): 144 NaCl, 2.5 KCl, 2 CaCl2, 2 MgCl2, 5 HEPES, 10 glucose, adjusted to pH 7.4 and 325 mosm/kg. Access resistances were typically <15 megaohms. M-current deactivation was recorded with 1-s hyperpolarizing pulses (−40 mV, −60 mV, −80 mV) from a holding potential of −20 mV. Tail currents were measured off-line using ClampFit 10.4. Patched cells were photographed, and the area of the soma was measured with ImageJ.
Statistical Analysis
Statistical significance was determined using GraphPad Prism 5.0 software. All data shown are the mean ± S.E.
Results
KCNQ2 and -3 Are Expressed in D-hair Aδ Mechanoreceptors
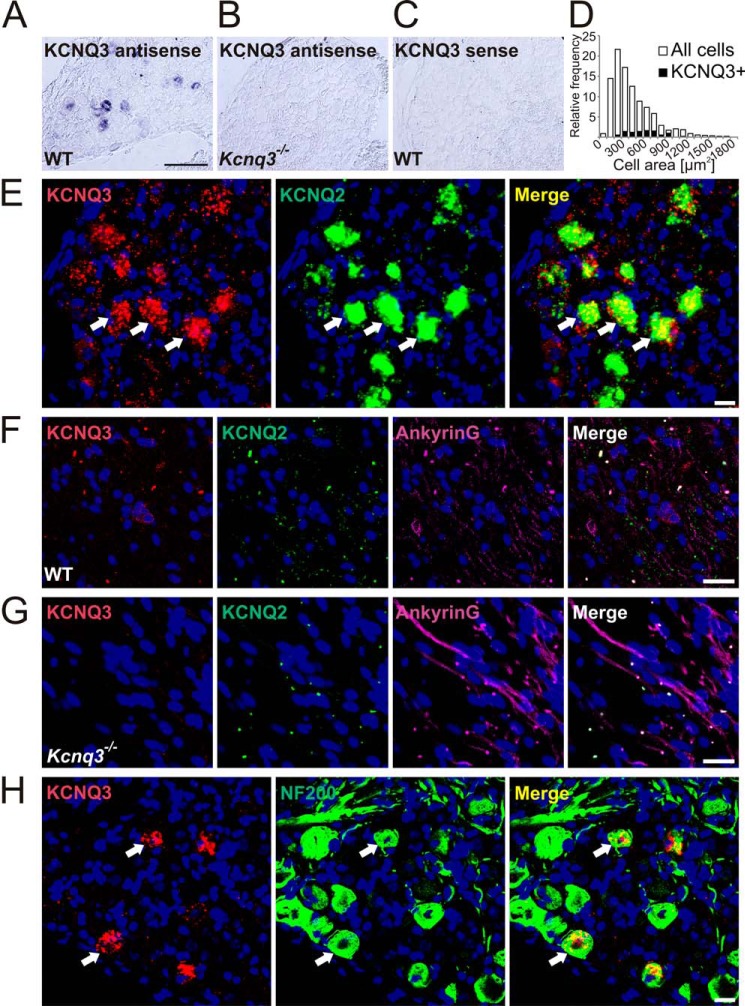
Pharmacological experiments with skin-nerve preparations have suggested that KCNQ channels are expressed in low-threshold D-hair mechanoreceptors (23). To identify KCNQ channel subunits in specific subsets of DRG neurons, we used in situ hybridization because KCNQ2 and KCNQ3 proteins are predominantly localized to axon initial segments and nodes of Ranvier rather than to neuronal somata. This severely limits their co-localization with neuron-specific markers by immunohistochemistry. We hybridized DRG sections with probes for each of the neuronal KCNQ isoforms together with a probe for the neurotrophin receptor TrkB, which in the DRG serves as a D-hair specific marker (42, 43). In these double in situ hybridization experiments (Fig. 1), ∼30% of KCNQ2-positive cells and 44% of KCNQ3-labeled cells were found to be positive for TrkB (Fig. 1, A and B; Table 1 for summary of co-localization results), suggesting that both KCNQ2 and KCNQ3 are found in D-hairs. In addition, KCNQ3 in situ hybridization experiments were combined with TrkB immunostainings, showing ∼45% co-localization of KCNQ3 with TrkB (data not shown). Approximately 35% of KCNQ3-positive neurons co-expressed the receptor protein-tyrosine kinase c-Ret and 48% co-expressed the T-type calcium channel Cav3.2 (Fig. 1, E and F). c-Ret has been reported to be a marker for LTMRs (44, 45), whereas Cav3.2 is specific for D-hair receptors (46, 47). In contrast, KCNQ4- and -5-expressing DRG neurons were not labeled with the D-hair markers TrkB and Cav3.2 (Fig. 1, C, D, and G). Approximately 10% of neuronal somata were labeled with the KCNQ3 probe, whereas labeling was absent in Kcnq3−/− mice and with the control sense probe (Fig. 2, A–C). The specificity of KCNQ3, -4, and -5 in situ hybridization was confirmed using DRG sections from the respective knock-out mice (Fig. 2B and data not shown). When compared with KCNQ4, which is expressed in large cells with an average area of 811 ± 23 μm2 (23), KCNQ3 was mainly detected in cells with medium cell size (569 ± 21 μm2; n = 84; Fig. 2D). Double in situ hybridization revealed that expression of Kcnq2 and Kcnq3 largely overlapped in DRG somata (Fig. 2E). Agreeing with previous results (48–50), KCNQ2 and -3 were detected at ankyrin-G-positive nodes of Ranvier (Fig. 2F). No KCNQ3 labeling of nodes of Ranvier was detected in Kcnq3−/− mutant mice (Fig. 2G). The localization at nodes of Ranvier indicates that KCNQ2 and KCNQ3 are expressed in myelinated neurons. Indeed, almost all KCNQ3-positive neurons co-expressed neurofilament 200 (NF200) (Fig. 2H), a marker for myelinated neurons.
FIGURE 1.
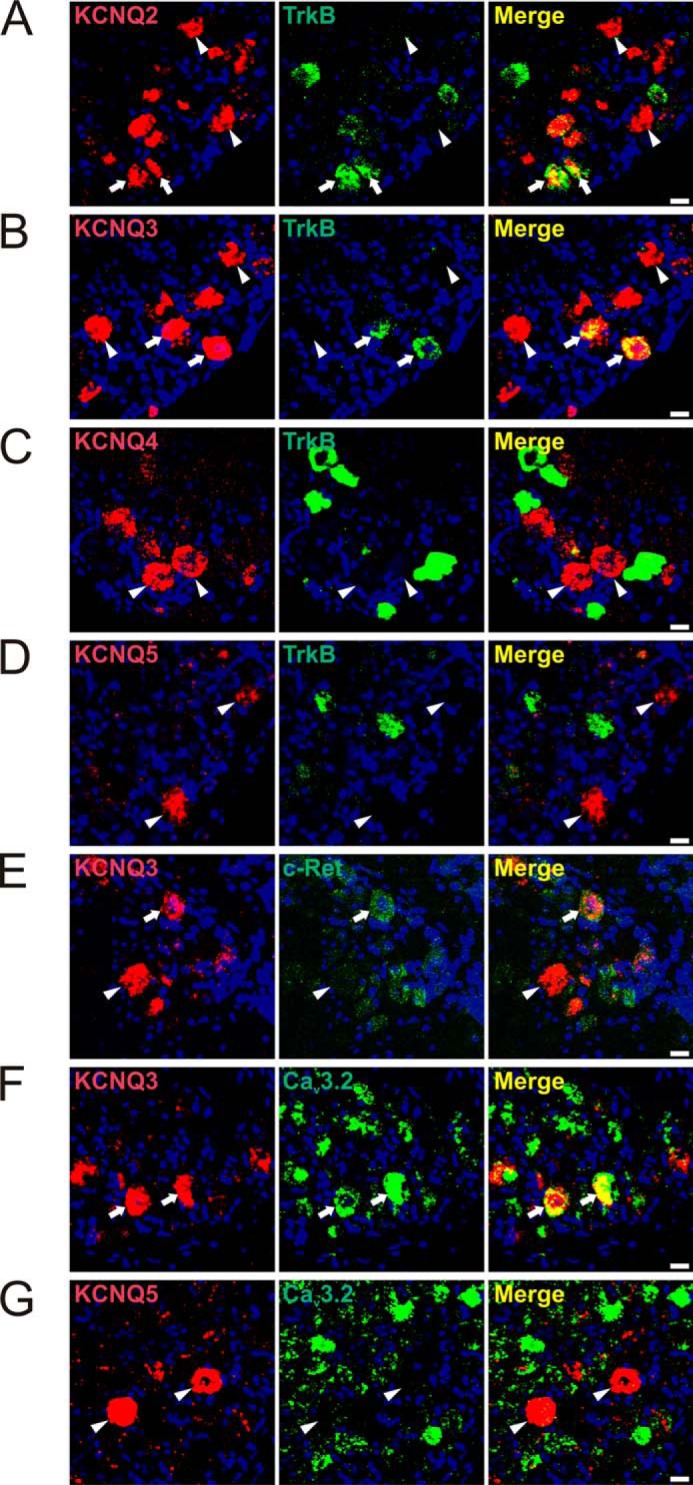
KCNQ2 and -3, but not KCNQ4 and -5, are expressed in a subset of TrkB-positive, low-threshold D-hair fibers. Expression of different KCNQ channels in DRG sections was revealed by double in situ hybridization. A and B, co-expression of KCNQ2 and KCNQ3 with TrkB, respectively (arrows). A, 27 out of 102 KCNQ2-positive and of 53 TrkB-positive cells were co-labeled for both proteins. B, out of 190 KCNQ3-positive neurons and 84 TrkB-positive cells, 71 were labeled for both TrkB and KCNQ3. C, KCNQ4-expressing neurons were not labeled for TrkB (n = 89). D, TrkB-labeling is also absent in KCNQ5-positive cells (n = 71). E, 26 out of 72 KCNQ3-positive cells were labeled for c-Ret (arrows). F, co-expression of KCNQ3 and Cav3.2. 35 out of 71 KCNQ3-positive neurons were labeled for Cav3.2 (arrows). G, 114 out of 131 KCNQ5-positive neurons were not labeled for Cav3.2. Arrowheads, KCNQ2-, -3-, -4-, and -5-positive DRG neurons, which were not labeled for TrkB (A–D), c-Ret (E), or Cav3.2 (G), respectively. Nuclei were labeled with DAPI (blue). Scale bars: 20 μm.
TABLE 1.
Summary of co-localization of KCNQ subunits with somatosensory markers
Fractions signify the total number of cells positive for proteins designated by row which cells co-express proteins designated by column. For example, of 89 KCNQ3-positive cells, 71 co-express KCNQ2.
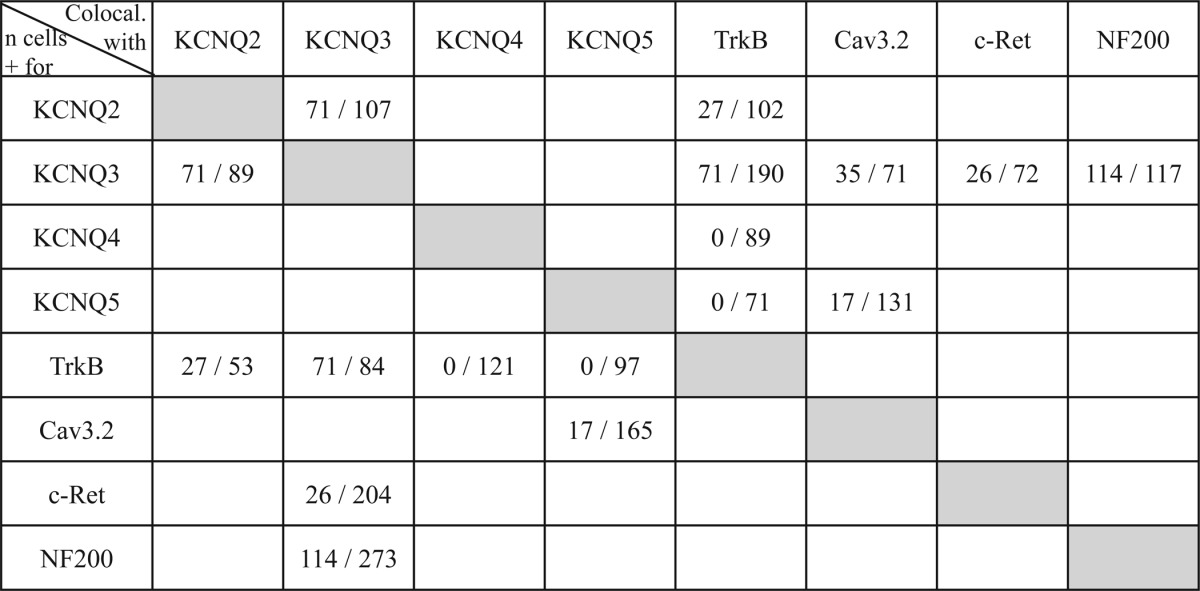
FIGURE 2.
KCNQ3 is expressed in medium diameter myelinated DRG neurons and co-localizes with KCNQ2. A–C, DRG sections from WT (A and C) and Kcnq3−/− (B) mice were labeled with antisense (A and B) and negative control sense (C) in situ hybridization probes for KCNQ3. A, of 924 DRG neurons from six sections, 87 were labeled for KCNQ3. KCNQ3 labeling is absent in Kcnq3−/− mice (B) and with the sense in situ probe (C). D, quantification of A with distribution of KCNQ3 cell size over cell sizes. E, double in situ hybridization for KCNQ2 and -3 demonstrates co-expression (arrows show examples). Of 107 KCNQ2-positive and 89 KCNQ3-positive DRG neurons, 71 were labeled for both KCNQ2 and KCNQ3. F and G, nodes of Ranvier were identified by ankyrin-G antibody staining. F, of 848 KCNQ2-positive nodes of Ranvier in WT mice, 722 were co-stained with KCNQ3 antibody. G, KCNQ3 antibody labeling is absent in Kcnq3−/− mice (n = 588). H, co-expression of KCNQ3 (detected by in situ hybridization) with NF200 (antibody labeling). Of 273 NF200-positive and 117 KCNQ3-positive DRG neurons, 114 were co-labeled for these markers (arrows show examples). E–H, nuclei were labeled with DAPI (blue). Scale bars: 100 μm (A) and 20 μm (E–H).
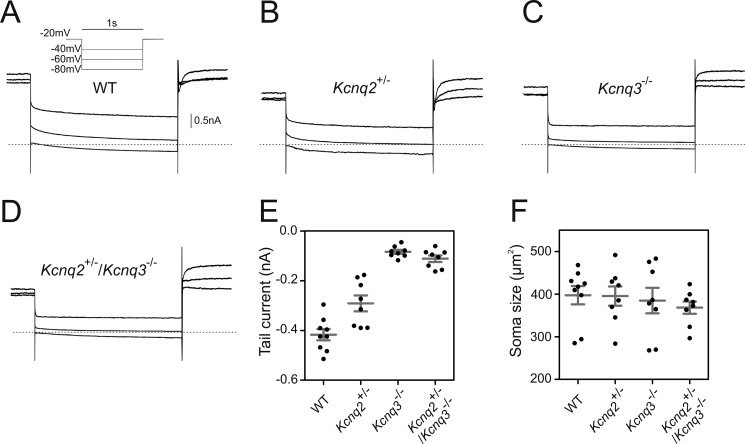
To confirm that a subset of DRG neurons expresses KCNQ2 and KCNQ3, membrane currents were measured by patch-clamping primary DRG neurons from WT and genetically modified mice (Fig. 3). Large cells labeled with TrkB antibody were selected and revealed a slowly deactivating current upon membrane hyperpolarizing from −20 to −40 mV when derived from WT mice (Fig. 3, A and E). This slow tail current, typical of M-currents, was inhibited by XE991 (data not shown). The M-current component was about halved in Kcnq2+/− mice (Fig. 3, B and E) consistent with previous results (51), and virtually abolished in Kcnq3−/− and Kcnq2+/−/Kcnq3−/− double mutant mice (Fig. 3, C–E). These results confirm the conclusion from our in situ hybridization that both KCNQ2 and KCNQ3 are expressed in TrkB-positive DRG neurons and suggest that they predominantly form heteromeric channels when studied at somata of cultured DRG neurons.
FIGURE 3.
M-current in TrkB-expressing DRG neurons depends on KCNQ2 and KCNQ3. A–D, representative traces of M-current deactivation by patch clamp in cultured DRG neurons (1–2 days in vitro) labeled with TrkB antibody from WT (9 cells) (A), Kcnq2+/− (8 cells) (B), Kcnq3−/− (8 cells) (C), and Kcnq2+/−/Kcnq3−/− (8 cells) (D) mice. E, tail current measurements at membrane voltages clamped to −40 mV. Currents were statistically significantly reduced in Kcnq2+/− (p = 0.0061), Kcnq3−/− (p = 0.0006), and Kcnq2+/−/Kcnq3−/− (p = 0.0006) when compared with WT mice. F, soma sizes of cells measured in E. Cells were hyperpolarized for 1 s from a holding potential of −20 mV (see clamp protocol in A). Dotted line denotes 0 nA. Data are reported as mean (long bar) ± S.E. (short bars). Mann-Whitney U test was used to assess statistical significance.
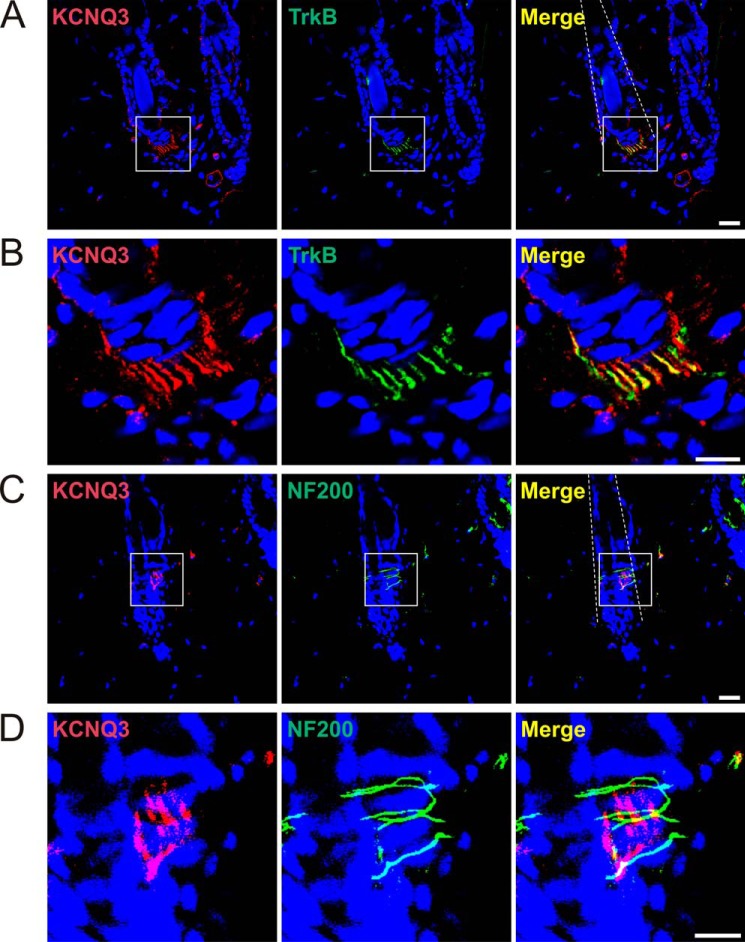
We next investigated KCNQ expression in the skin. Three types of major hair follicles of trunk hairy skin were described, called guard, awl/auchene, and zigzag hair (52). Each of them is innervated by a unique combination of C-LTMRs, Aδ-LTMRs, and rapidly adapting Aβ-LTMRs (33), which are neurochemically defined as type I, II, and III lanceolate endings, respectively (39). KCNQ3 staining was observed in type II endings (Fig. 4), which are defined as TrkB-positive (Fig. 4, A and B) and NF200-negative (Fig. 4, C and D) (39). KCNQ2 could not be detected in the skin with our antibody.
FIGURE 4.
KCNQ3 in mouse skin mechanoreceptors. A, immunohistochemistry of hairy skin sections shows KCNQ3 expression in TrkB-positive lanceolate endings. Dotted lines outline hair shafts. B, magnified views of boxes in A. C, KCNQ3 is not detected in NF200-positive circular nerve endings. Dotted lines outline hair shafts. D, magnified views of boxes in C. Nuclei were labeled with DAPI (blue). Scale bars: 20 μm (A and C) and 10 μm (B and D).
Mechanosensitivity of D-hair Receptors Is Increased in Kcnq3−/− and in Kcnq2+/−/Kcnq3−/− Mutant Mice
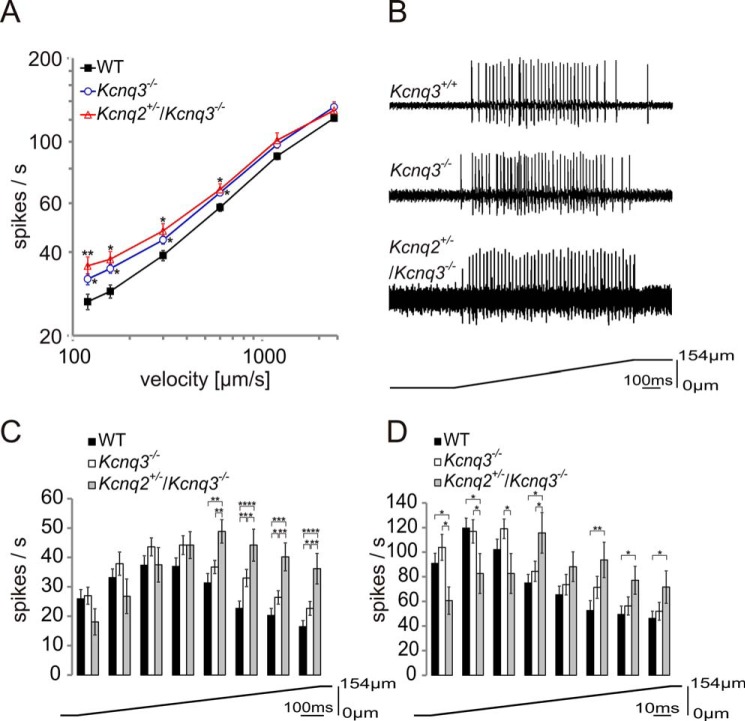
We used in vitro skin-nerve preparations of the mouse saphenous nerve to investigate the role of KCNQ3 in D-hair mechanoreceptors. D-hair fibers were classified according to their conduction velocity between 1 and 10 m/s and von Frey thresholds that are typically much lower than the force of the weakest von Frey hair used in the present study (0.35 millinewtons) (46, 53). Axonal conduction velocity and the force necessary to elicit the first action potential were not affected in our mouse models (data not shown). Receptive properties of D-hair mechanoreceptors were tested with repeated ramp-and-hold mechanical stimuli with constant displacement amplitudes of 154 μm and a range of ramp velocities varying from 120 to 2400 μm/s. The firing frequency of D-hairs during the ramp phase was consistently higher in Kcnq3−/− mice than in Kcnq3+/+ mice (Fig. 5, A and B), with a larger impact of Kcnq3 disruption at low indentation velocities (Fig. 5A).
FIGURE 5.
Mechanosensitivity of D-hair receptors in WT, Kcnq3−/−, and Kcnq2+/−/Kcnq3−/− mice. A, the response of D-hair receptors to a constant displacement stimulus of 154 μm with a range of ramp velocities from 120 to 2400 μm/s in WT (40 fibers), Kcnq3−/− (29 fibers), and Kcnq2+/−/Kcnq3−/− (12 fibers) mutant mice. B, example traces of electrical responses of D-hairs to 158 μm/s mechanical stimuli from WT, Kcnq3−/−, and Kcnq2+/−/Kcnq3−/− mice. C and D, detailed analysis of phasic firing frequency at velocities of 158 μm/s (C) and 1163 μm/s (D) of D-hairs from WT (158 μm/s, 40 fibers; 1163 μm/s, 38 fibers), Kcnq3−/− (158 μm/s, 29 fibers; 1163 μm/s, 29 fibers), and Kcnq2+/−/Kcnq3−/− (158 μm/s, 12 fibers; 1163 μm/s, 11 fibers) mutant mice. The ramp was divided into eight intervals (interval size 124.5 ms for C and 16.5 ms for D), and firing frequency was measured for each interval. Statistical significance for A, C, and D: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; Student's t test.
Because KCNQ2 and KCNQ3 were largely co-expressed in DRG neurons and as the fraction of TrkB-labeled cells was similar for KCNQ2- and KCNQ3-positive neurons, we also addressed the role of KCNQ2. Because Kcnq2−/− mutant mice die within a few hours after birth due to pulmonary atelectasis (54) and as we lack conditional Kcnq2 knock-out mice, we examined whether a reduction of KCNQ2 levels would enhance the effect of KCNQ3 deletion on D-hair excitability. Moderate loss of KCNQ2 function may already have significant effects, as evident from benign familial neonatal seizures where loss of KCNQ2 on only one allele already leads to epilepsy (15, 16, 55). Heterozygous Kcnq2+/− animals are viable, express reduced levels of the KCNQ2 protein, and show increased sensitivity to seizure-inducing drugs (54). We therefore crossed Kcnq2+/− mice with Kcnq3−/− animals to yield Kcnq2+/−/Kcnq3−/− double mutant mice. The additional decrease in KCNQ2 protein levels did not lead to a significant further increase of the firing frequency when measured over the entire duration of the ramp (Fig. 5A). Agreeing with previous results (46), control D-hair fibers displayed their highest firing frequencies during the first half of the ramp and reached their maximal firing frequencies after about 375 ms with a slow ramp (158 μm/s) and within the first 35 ms with a fast ramp (1163 μm/s) before decreasing below initial rates (Fig. 5, C and D). The increased sensitivity of Kcnq3−/− and Kcnq2+/−/Kcnq3−/− D-hairs was mainly due to increased firing rates toward the end of the ramp (Fig. 5, C and D). In particular at low indentation velocities, a significant effect of the additional heterozygous disruption of Kcnq2 became visible (Fig. 5C).
Discussion
We have detected KCNQ2 and -3, but not KCNQ4 and -5, in murine D-hair Aδ-fibers. KCNQ3 was targeted to peripheral nerve endings of defined mechanoreceptors in the skin, but the KCNQ2 antibody failed to label nerve endings around hair follicles. Loss of KCNQ3 enhanced mechanoreceptor sensitivity in low-threshold D-hair fibers in response to mechanical ramp-and-hold stimuli in electrophysiological skin-nerve measurements.
Low-threshold D-hair fibers were first described in 1967 in the cat (31). We used several criteria to identify KCNQ3 expression in D-hair mechanosensitive neurons. In morphological studies, we found that KCNQ3 co-localized with NF200 and TrkB in DRG somata and that the channel was expressed in TrkB-positive and NF200-negative peripheral longitudinal lanceolate nerve endings that are typical of D-hairs. Although NF200 is a general marker for myelinated neurons, TrkB is widely accepted as a marker for D-hairs, both in DRGs and in longitudinal lanceolate endings in the skin (33, 39). The TrkB tyrosine kinase receptor binds neurotrophin-4 (NT4) (56), which is essential for the maintenance and survival of adult D-hairs (43). Furthermore, KCNQ3 partially co-localized with c-Ret, a marker for LTMRs (44, 45). Some KCNQ3-positive neurons were also co-labeled for Cav3.2 T-type Ca2+ channels that are considered specific for D-hairs (46, 47) and that amplify mechanoreceptor sensitivity (46). Finally, our functional analysis revealed that Kcnq3 disruption affected the sensitivity of mechanosensitive fibers that displayed typical characteristics of D-hairs.
Three different hair types can be differentiated in murine fur, called guard, awl/auchene, and zigzag hair. Each of these hair types is associated with a unique combination of distinct LTMR endings (33). Guard hair follicles are innervated by rapidly adapting Aβ-LTMRs and associate with Merkel cell clusters in touch domes, which are innervated by Aβ SA1-LTMRs. Awl/auchene hairs are triply innervated by rapidly adapting Aβ-LTMRs, Aδ-LTMRs, and C-LTMRs, whereas zigzag hair follicles are associated with Aδ-LTMRs and C-LTMRs (33). Each of these fibers can be uniquely characterized by a different set of neurochemical markers (39). Aβ-LTMRs are characterized as S100+ (a Ca2+-binding protein), NF200+, calbindin+, and TrkBlow, Aδ-LTMRs are identified by a S100+, NF200−, calbindin−, and TrkBhigh expression profile, and C-LTMR endings are solely positive for S100 and lack expression of NF200, calbindin, and TrkB. This work showed that KCNQ3 localizes to TrkB-positive and NF200-negative hair follicle-associated nerve endings that most likely represent Aδ D-hair fibers around awl/auchene and zigzag hairs.
However, KCNQ3 is not specific for D-hairs. Less than 50% of KCNQ2-and -3-expressing DRG neurons were positive for the D-hair markers TrkB or Cav3.2, indicating that these KCNQ subunits are found in various sensory subpopulations. Indeed, KCNQ2 and -3 were previously reported to be expressed by both small and large diameter DRG neurons (10). M-current was found in bradykinin- (25, 57) and capsaicin- (10) sensitive nociceptive neurons, in mas-related G-protein-coupled receptor member D (MrgD)- (58) and protease-activated receptor 2 (PAR-2)- (24) positive nociceptors, as well as in transient receptor potential cation channel subfamily M member 8 (TRPM8)-expressing cold nociceptors (29). Both KCNQ2 (26, 28, 29) and KCNQ5 (27) were postulated to be the predominant KCNQ subunit in these nociceptive neurons.
As we and others (48, 50) have shown, KCNQ2 strongly co-localizes with KCNQ3 in DRGs. Both isoforms partially co-localized with TrkB, suggesting that KCNQ2/3 heteromeric channels may not only be found at initial axon segments or nodes of Ranvier, but possibly also in D-hair nerve endings around hair follicles. This issue seems particularly important because KCNQ2/KCNQ3 heteromers yield larger currents than either KCNQ2 or KCNQ3 homomeric channels (8, 55, 59) and because the loss of KCNQ2 generally affects neuronal excitability more than a loss of KCNQ3 (60). Although our inability to detect KCNQ2 in D-hair nerve endings might be because of the low sensitivity of the KCNQ2 antibody, it is possible that only KCNQ3 homo-oligomeric channels, but not KCNQ2/3 heteromers, are targeted to these nerve endings. The presence of KCNQ2/3 heteromers in WT D-hair nerve endings, however, is supported by the increase in D-hair sensitivity of Kcnq2+/−/Kcnq3−/− mice when compared with Kcnq3−/− mice that became apparent in the second half of slowly indenting ramps (Fig. 5C). This difference in sensitivity is compatible with the formation of KCNQ2 homo-oligomeric channels in D-hair nerve endings of Kcnq3−/− mice. The formation of KCNQ2/KCNQ3 heteromeric channels in WT D-hairs is further supported by the detection of both KCNQ2 and KCNQ3 in TrkB-positive DRG neurons and the ∼50% reduction of KCNQ3-dependent M-currents in cultured bona fide D-hair DRG neurons from Kcnq2+/− mice (Fig. 3).
Unfortunately, we could not study the effect of a complete lack of KCNQ2 because Kcnq2−/− mice show early postnatal lethality (54). Recently, Kcnq2 has been disrupted in sensory neurons using paired box 3 (Pax3)-driven Cre-recombinase and Kcnq2lox/lox mice (61). As expected from the role of M-currents in dampening neuronal excitation, DRG neurons from these mice showed increased excitability and reduced spike frequency adaptation. Consistent with a role of KCNQ2 in a wide range of sensory neurons, these mice displayed thermal hyperalgesia and mechanical allodynia (61). The role of KCNQ2 in specific mechanoreceptors, however, was not investigated.
The increased mechanical sensitivity of Kcnq3−/− and Kcnq2+/−/Kcnq3−/− D-hairs observed here fits with a role of KCNQ channels in dampening neuronal excitability. Compatible with the role of the M-current in spike frequency adaptation (1, 2), the effect of KCNQ2/3 on D-hair firing was most obvious in the later phases of mechanical indentation. Analogous to results for KCNQ4, which influences the sensitivity of rapidly adapting mechanosensors at low, but not at high vibration frequencies (23), the effect of KCNQ2/3 on D-hair sensitivity was largest at low indentation speeds. Similar to KCNQ4, which localizes to circular and lanceolate endings of rapidly adapting mechanoreceptors (23), KCNQ3 is strategically localized to lanceolate endings of D-hairs where it can directly influence sensitivity by shunting depolarizing receptor currents upstream of action potential generation. The overall impact of KCNQ2/3 on D-hair sensitivity was modest and did not result in any obvious behavioral phenotype, but a larger effect might be seen with a complete loss of KCNQ2 in D-hairs or with more physiological stimuli (62). Furthermore, we cannot exclude that D-hairs adapted to the loss of KCNQ channel subunits in our constitutive knock-out mouse model. Importantly, the present study, together with our previous study on KCNQ4 (23), suggests a more general role of KCNQ M-type channels in regulating neuronal sensitivity directly at sensory nerve endings.
Author Contributions
S. S. and I. J. O. planned, performed, and analyzed experiments and wrote the paper. T. J. J. planned and analyzed experiments and wrote the paper.
Acknowledgments
We thank P. Seidler for technical support and J. Walcher (Lewin group, MDC) for sharing skin-nerve preparation experiences.
This work was supported by a Senatsausschuss Wettbewerb (SAW) grant of the Leibniz Gemeinschaft and by the Deutsche Forschungsgemeinschaft (DFG Exc 257 'Neurocure') (to T. J. J). The authors declare that they have no conflicts of interest with the contents of this article.
- DRG
- dorsal root ganglion
- Cav3.2
- voltage-gated calcium channel 3.2
- D-hair
- down-hair
- DIG
- digoxigenin
- LTMR
- low-threshold mechanoreceptor
- NF200
- neurofilament 200
- POD
- peroxidase
- S100
- a family of low molecular weight Ca2+-binding proteins
- TrkB
- tyrosine kinase receptor type B.
References
- 1. Jentsch T. J. (2000) Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Rev. Neurosci. 1, 21–30 [DOI] [PubMed] [Google Scholar]
- 2. Delmas P., and Brown D. A. (2005) Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat. Rev. Neurosci. 6, 850–862 [DOI] [PubMed] [Google Scholar]
- 3. Brown D. A., and Passmore G. M. (2009) Neural KCNQ (Kv7) channels. Br. J. Pharmacol. 156, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soldovieri M. V., Miceli F., and Taglialatela M. (2011) Driving with no brakes: molecular pathophysiology of Kv7 potassium channels. Physiology (Bethesda) 26, 365–376 [DOI] [PubMed] [Google Scholar]
- 5. Zhong X. Z., Harhun M. I., Olesen S. P., Ohya S., Moffatt J. D., Cole W. C., and Greenwood I. A. (2010) Participation of KCNQ (Kv7) potassium channels in myogenic control of cerebral arterial diameter. J. Physiol. 588, 3277–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ng F. L., Davis A. J., Jepps T. A., Harhun M. I., Yeung S. Y., Wan A., Reddy M., Melville D., Nardi A., Khong T. K., and Greenwood I. A. (2011) Expression and function of the K+ channel KCNQ genes in human arteries. Br. J. Pharmacol. 162, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schleifenbaum J., Kassmann M., Szijártó I. A., Hercule H. C., Tano J.-Y., Weinert S., Heidenreich M., Pathan A. R., Anistan Y.-M., Alenina N., Rusch N. J., Bader M., Jentsch T. J., and Gollasch M. (2014) Stretch-activation of angiotensin II type 1a receptors contributes to the myogenic response of mouse mesenteric and renal arteries. Circ. Res. 115, 263–272 [DOI] [PubMed] [Google Scholar]
- 8. Wang H. S., Pan Z., Shi W., Brown B. S., Wymore R. S., Cohen I. S., Dixon J. E., and McKinnon D. (1998) KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 9. Shah M., Mistry M., Marsh S. J., Brown D. A., and Delmas P. (2002) Molecular correlates of the M-current in cultured rat hippocampal neurons. J. Physiol. 544, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Passmore G. M., Selyanko A. A., Mistry M., Al-Qatari M., Marsh S. J., Matthews E. A., Dickenson A. H., Brown T. A., Burbidge S. A., Main M., and Brown D. A. (2003) KCNQ/M currents in sensory neurons: significance for pain therapy. J. Neurosci. 23, 7227–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schroeder B. C., Hechenberger M., Weinreich F., Kubisch C., and Jentsch T. J. (2000) KCNQ5, a novel potassium channel broadly expressed in brain, mediates M-type currents. J. Biol. Chem. 275, 24089–24095 [DOI] [PubMed] [Google Scholar]
- 12. Brown D. A., and Adams P. R. (1980) Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature 283, 673–676 [DOI] [PubMed] [Google Scholar]
- 13. Neyroud N., Tesson F., Denjoy I., Leibovici M., Donger C., Barhanin J., Fauré S., Gary F., Coumel P., Petit C., Schwartz K., and Guicheney P. (1997) A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat. Genet. 15, 186–189 [DOI] [PubMed] [Google Scholar]
- 14. Wang Q., Curran M. E., Splawski I., Burn T. C., Millholland J. M., VanRaay T. J., Shen J., Timothy K. W., Vincent G. M., de Jager T., Schwartz P. J., Toubin J. A., Moss A. J., Atkinson D. L., Landes G. M., Connors T. D., and Keating M. T. (1996) Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat. Genet. 12, 17–23 [DOI] [PubMed] [Google Scholar]
- 15. Biervert C., Schroeder B. C., Kubisch C., Berkovic S. F., Propping P., Jentsch T. J., and Steinlein O. K. (1998) A potassium channel mutation in neonatal human epilepsy. Science 279, 403–406 [DOI] [PubMed] [Google Scholar]
- 16. Singh N. A., Charlier C., Stauffer D., DuPont B. R., Leach R. J., Melis R., Ronen G. M., Bjerre I., Quattlebaum T., Murphy J. V., McHarg M. L., Gagnon D., Rosales T. O., Peiffer A., Anderson V. E., and Leppert M. (1998) A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat. Genet. 18, 25–29 [DOI] [PubMed] [Google Scholar]
- 17. Charlier C., Singh N. A., Ryan S. G., Lewis T. B., Reus B. E., Leach R. J., and Leppert M. (1998) A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat. Genet. 18, 53–55 [DOI] [PubMed] [Google Scholar]
- 18. Dedek K., Kunath B., Kananura C., Reuner U., Jentsch T. J., and Steinlein O. K. (2001) Myokymia and neonatal epilepsy caused by a mutation in the voltage sensor of the KCNQ2 K+ channel. Proc. Natl. Acad. Sci. U.S.A. 98, 12272–12277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orhan G., Bock M., Schepers D., Ilina E. I., Reichel S. N., Löffler H., Jezutkovic N., Weckhuysen S., Mandelstam S., Suls A., Danker T., Guenther E., Scheffer I. E., De Jonghe P., Lerche H., and Maljevic S. (2014) Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Ann. Neurol. 75, 382–394 [DOI] [PubMed] [Google Scholar]
- 20. Kharkovets T., Hardelin J. P., Safieddine S., Schweizer M., El-Amraoui A., Petit C., and Jentsch T. J. (2000) KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc. Natl. Acad. Sci. U.S.A. 97, 4333–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spitzmaul G., Tolosa L., Winkelman B. H. J., Heidenreich M., Frens M. A., Chabbert C., de Zeeuw C. I., and Jentsch T. J. (2013) Vestibular role of KCNQ4 and KCNQ5 K+ channels revealed by mouse models. J. Biol. Chem. 288, 9334–9344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kubisch C., Schroeder B. C., Friedrich T., Lütjohann B., El-Amraoui A., Marlin S., Petit C., and Jentsch T. J. (1999) KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell 96, 437–446 [DOI] [PubMed] [Google Scholar]
- 23. Heidenreich M., Lechner S. G., Vardanyan V., Wetzel C., Cremers C. W., De Leenheer E. M., Aránguez G., Moreno-Pelayo M. Á., Jentsch T. J., and Lewin G. R. (2012) KCNQ4 K+ channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat. Neurosci. 15, 138–145 [DOI] [PubMed] [Google Scholar]
- 24. Linley J. E., Rose K., Patil M., Robertson B., Akopian A. N., and Gamper N. (2008) Inhibition of M current in sensory neurons by exogenous proteases: a signaling pathway mediating inflammatory nociception. J. Neurosci. 28, 11240–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu B., Linley J. E., Du X., Zhang X., Ooi L., Zhang H., and Gamper N. (2010) The acute nociceptive signals induced by bradykinin in rat sensory neurons are mediated by inhibition of M-type K+ channels and activation of Ca2+-activated Cl− channels. J. Clin. Invest. 120, 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rose K., Ooi L., Dalle C., Robertson B., Wood I. C., and Gamper N. (2011) Transcriptional repression of the M channel subunit Kv7.2 in chronic nerve injury. Pain 152, 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King C. H., and Scherer S. S. (2012) Kv7.5 is the primary Kv7 subunit expressed in C-fibers. J. Comp. Neurol. 520, 1940–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Passmore G. M., Reilly J. M., Thakur M., Keasberry V. N., Marsh S. J., Dickenson A. H., and Brown D. A. (2012) Functional significance of M-type potassium channels in nociceptive cutaneous sensory endings. Front Mol Neurosci. 5, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vetter I., Hein A., Sattler S., Hessler S., Touska F., Bressan E., Parra A., Hager U., Leffler A., Boukalova S., Nissen M., Lewis R. J., Belmonte C., Alzheimer C., Huth T., Vlachova V., Reeh P. W., and Zimmermann K. (2013) Amplified cold transduction in native nociceptors by M-channel inhibition. J. Neurosci. 33, 16627–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hao J., Bonnet C., Amsalem M., Ruel J., and Delmas P. (2015) Transduction and encoding sensory information by skin mechanoreceptors. Pflügers Arch. 467, 109–119 [DOI] [PubMed] [Google Scholar]
- 31. Brown A. G., and Iggo A. (1967) A quantitative study of cutaneous receptors and afferent fibres in the cat and rabbit. J. Physiol. 193, 707–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lechner S. G., and Lewin G. R. (2013) Hairy sensation. Physiology 28, 142–150 [DOI] [PubMed] [Google Scholar]
- 33. Li L., Rutlin M., Abraira V. E., Cassidy C., Kus L., Gong S., Jankowski M. P., Luo W., Heintz N., Koerber H. R., Woodbury C. J., and Ginty D. D. (2011) The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 147, 1615–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farley F. W., Soriano P., Steffen L. S., and Dymecki S. M. (2000) Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 [PubMed] [Google Scholar]
- 35. Schwenk F., Baron U., and Rajewsky K. (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tzingounis A. V., Heidenreich M., Kharkovets T., Spitzmaul G., Jensen H. S., Nicoll R. A., and Jentsch T. J. (2010) The KCNQ5 potassium channel mediates a component of the afterhyperpolarization current in mouse hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 10232–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watakabe A., Ichinohe N., Ohsawa S., Hashikawa T., Komatsu Y., Rockland K. S., and Yamamori T. (2007) Comparative analysis of layer-specific genes in mammalian neocortex. Cereb. Cortex. 17, 1918–1933 [DOI] [PubMed] [Google Scholar]
- 38. Abe Y., Nawa H., and Namba H. (2009) Activation of epidermal growth factor receptor ErbB1 attenuates inhibitory synaptic development in mouse dentate gyrus. Neurosci. Res. 63, 138–148 [DOI] [PubMed] [Google Scholar]
- 39. Wende H., Lechner S. G., Cheret C., Bourane S., Kolanczyk M. E., Pattyn A., Reuter K., Munier F. L., Carroll P., Lewin G. R., and Birchmeier C. (2012) The transcription factor c-Maf controls touch receptor development and function. Science 335, 1373–1376 [DOI] [PubMed] [Google Scholar]
- 40. Milenkovic N., Wetzel C., Moshourab R., and Lewin G. R. (2008) Speed and temperature dependences of mechanotransduction in afferent fibers recorded from the mouse saphenous nerve. J. Neurophysiol. 100, 2771–2783 [DOI] [PubMed] [Google Scholar]
- 41. Frank J. A., Moroni M., Moshourab R., Sumser M., Lewin G. R., and Trauner D. (2015) Photoswitchable fatty acids enable optical control of TRPV1. Nat Commun. 6, 7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stucky C. L., Shin J.-B., and Lewin G. R. (2002) Neurotrophin-4: a survival factor for adult sensory neurons. Curr. Biol. 12, 1401–1404 [DOI] [PubMed] [Google Scholar]
- 43. Stucky C. L., DeChiara T., Lindsay R. M., Yancopoulos G. D., and Koltzenburg M. (1998) Neurotrophin 4 is required for the survival of a subclass of hair follicle receptors. J. Neurosci. 18, 7040–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bourane S., Garces A., Venteo S., Pattyn A., Hubert T., Fichard A., Puech S., Boukhaddaoui H., Baudet C., Takahashi S., Valmier J., and Carroll P. (2009) Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron. 64, 857–870 [DOI] [PubMed] [Google Scholar]
- 45. Luo W., Enomoto H., Rice F. L., Milbrandt J., and Ginty D. D. (2009) Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 64, 841–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang R., and Lewin G. R. (2011) The Cav3.2 T-type calcium channel regulates temporal coding in mouse mechanoreceptors. J. Physiol. 589, 2229–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin J.-B., Martinez-Salgado C., Heppenstall P. A., and Lewin G. R. (2003) A T-type calcium channel required for normal function of a mammalian mechanoreceptor. Nat. Neurosci. 6, 724–730 [DOI] [PubMed] [Google Scholar]
- 48. Pan Z., Kao T., Horvath Z., Lemos J., Sul J.-Y., Cranstoun S. D., Bennett V., Scherer S. S., and Cooper E. C. (2006) A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J. Neurosci. 26, 2599–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Devaux J. J., Kleopa K. A., Cooper E. C., and Scherer S. S. (2004) KCNQ2 is a nodal K+ channel. J. Neurosci. 24, 1236–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cooper E. C. (2011) Made for “anchorin”: Kv7.2/7.3 (KCNQ2/KCNQ3) channels and the modulation of neuronal excitability in vertebrate axons. Semin. Cell Dev. Biol. 22, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robbins J., Passmore G. M., Abogadie F. C., Reilly J. M., and Brown D. A. (2013) Effects of KCNQ2 gene truncation on M-type Kv7 potassium currents. PLoS ONE 8, e71809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dry J. W. (1926) The coat of the mouse (Mus musculus). J Genet. 16, 287–340 [Google Scholar]
- 53. Koltzenburg M., Stucky C. L., and Lewin G. R. (1997) Receptive properties of mouse sensory neurons innervating hairy skin. J. Neurophysiol. 78, 1841–1850 [DOI] [PubMed] [Google Scholar]
- 54. Watanabe H., Nagata E., Kosakai A., Nakamura M., Yokoyama M., Tanaka K., and Sasai H. (2000) Disruption of the epilepsy KCNQ2 gene results in neural hyperexcitability. J. Neurochem. 75, 28–33 [DOI] [PubMed] [Google Scholar]
- 55. Schroeder B. C., Kubisch C., Stein V., and Jentsch T. J. (1998) Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature 396, 687–690 [DOI] [PubMed] [Google Scholar]
- 56. Klein R., Lamballe F., Bryant S., and Barbacid M. (1992) The trkB tyrosine protein kinase is a receptor for neurotrophin-4. Neuron. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 57. Jones S., Brown D. A., Milligan G., Willer E., Buckley N. J., and Caulfield M. P. (1995) Bradykinin excites rat sympathetic neurons by inhibition of M current through a mechanism involving B2 receptors and Gαq/11. Neuron. 14, 399–405 [DOI] [PubMed] [Google Scholar]
- 58. Crozier R. A., Ajit S. K., Kaftan E. J., and Pausch M. H. (2007) MrgD activation inhibits KCNQ/M-currents and contributes to enhanced neuronal excitability. J. Neurosci. 27, 4492–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwake M., Pusch M., Kharkovets T., and Jentsch T. J. (2000) Surface expression and single channel properties of KCNQ2/KCNQ3, M-type K+ channels involved in epilepsy. J. Biol. Chem. 275, 13343–13348 [DOI] [PubMed] [Google Scholar]
- 60. Soh H., Pant R., LoTurco J. J., and Tzingounis A. V. (2014) Conditional deletions of epilepsy-associated KCNQ2 and KCNQ3 channels from cerebral cortex cause differential effects on neuronal excitability. J. Neurosci. 34, 5311–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. King C. H., Lancaster E., Salomon D., Peles E., and Scherer S. S. (2014) Kv7.2 regulates the function of peripheral sensory neurons. J. Comp. Neurol. 522, 3262–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rutlin M., Ho C.-Y., Abraira V. E., Cassidy C., Woodbury C. J., and Ginty D. D. (2014) The cellular and molecular basis of direction selectivity of Aδ-LTMRs. Cell 159, 1640–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]