Abstract
Intelectins (X-type lectins), broadly distributed throughout chordates, have been implicated in innate immunity. Xenopus laevis embryonic epidermal lectin (XEEL), an intelectin secreted into environmental water by the X. laevis embryo, is postulated to function as a defense against microbes. XEEL is homologous (64% identical) to human intelectin-1 (hIntL-1), which is also implicated in innate immune defense. We showed previously that hIntL-1 binds microbial glycans bearing exocyclic vicinal diol groups. It is unknown whether XEEL has the same ligand specificity. Also unclear is whether XEEL and hIntL-1 have similar quaternary structures, as XEEL lacks the corresponding cysteine residues in hIntL-1 that stabilize the disulfide-linked trimer. These observations prompted us to further characterize XEEL. We found that hIntL-1 and XEEL have similar structural features. Even without the corresponding intermolecular disulfide bonds present in hIntL-1, the carbohydrate recognition domain of XEEL (XEELCRD) forms a stable trimer in solution. The structure of XEELCRD in complex with d-glycerol-1-phosphate, a residue present in microbe-specific glycans, indicated that the exocyclic vicinal diol coordinates to a protein-bound calcium ion. This ligand-binding mode is conserved between XEEL and hIntL-1. The domain architecture of full-length XEEL is reminiscent of a barbell, with two sets of three glycan-binding sites oriented in opposite directions. This orientation is consistent with our observation that XEEL can promote the agglutination of specific serotypes of Streptococcus pneumoniae. These data support a role for XEEL in innate immunity, and they highlight structural and functional conservation of X-type lectins among chordates.
Keywords: carbohydrate-binding protein, galactofuranose, innate immunity, lectin, protein cross-linking, X-ray crystallography, bacterial agglutination, glycerol phosphate, intelectin, omentin
Introduction
The carbohydrate coats of cells can be used by the immune system to distinguish self from non-self (1). Lectins can play a role in this discrimination because they have the ability to distinguish among carbohydrate residues. Several families of lectins, including the C-type lectins and ficolins, are classified as immune lectins. Their occurrence throughout the animal kingdom suggests that carbohydrate recognition is critical in immunity (2, 3). Recently, intelectins, also known as X-type lectins (4), have been proposed to function in immunity (5). This suggestion is intriguing, but little is known about the structure or function of this class of lectins.
Intelectins have been identified in diverse species from tunicates to humans (6). The limited functional studies conducted to date suggest a role in host defense. For example, tunicate intelectin can serve as an opsonin for phagocytosis by hemocytes (7). Amphioxus intelectin can agglutinate bacteria in vitro, suggesting that it could sequester them from the organism (8, 9). In some species, intelectin expression levels increase upon infection. Specifically, in rainbow trout and zebrafish, intelectins are up-regulated upon microbial infection (10–12). Sheep and mice increase intelectin production upon infection with intestinal parasitic nematodes (13–15). The up-regulation of intelectins upon exposure to microbes supports the hypothesis that these proteins function in animal immune defense.
The first intelectin was discovered in Xenopus laevis (16–18). At least five X. laevis intelectins have been described subsequently. X. laevis cortical granule lectins (XCGL-1 and XCGL-1) are found in oocytes and developing embryos (19), where they facilitate the formation of the fertilization envelope to block polyspermy (16, 17). X. laevis serum lectins (XSL-1 and XSL-2) are transcribed in response to lipopolysaccharide exposure (20, 21). The X. laevis embryonic epidermal lectin (XEEL)4 is secreted during various stages of development, but its level is highest around hatching time (22, 23). This lectin is produced by the goblet cells of the larval epidermis, which has properties akin to those of the human mucosal epithelia (24). Human goblet cells are critical for preserving the epithelial barrier and therefore for microbial defense. These parallels suggest that XEEL may function in innate immunity in Xenopus.
Humans encode two intelectins that have similarities to XEEL: hIntL-1 and hIntL-2 (25). These human lectins are expressed at mucosal barriers, including the lung and the intestine. The recognition properties of hIntL-1 are consistent with a role for this lectin in innate immunity. Specifically, it has been suggested that hIntL-1 recognizes furanose carbohydrate residues, including galactofuranose (Galf) and ribose (5). Our studies of hIntl-1 indicate that it does not bind ribose, but it does interact with β-Galf residues (26) and is thus not a general furanose-binding lectin. Analysis using glycan microarrays revealed that hIntL-1 does not bind any of the tested human glycans but rather binds diverse microbial glycan epitopes. Specifically, hIntL-1 can interact with β-Galf, d-glycerol-1-phosphate (GroP), heptoses, d-glycero-d-talo-oct-2-ulosonic acid, and 3-deoxy-d-manno-oct-2-ulosonic acid (26). None of these epitopes are present on human glycans. In accord with its glycan specificity, hIntL-1 binds microbial cell surfaces, supporting a role for hIntL-1 in the recognition of intact microbes (26). Given these recognition properties and the presence of intelectins in diverse species, an intriguing possibility is that the intelectins serve as microbial detectors.
XEEL and other intelectins share high sequence similarity, including a putative conserved fibrinogen-like domain (FBD). Although this domain is also found in ficolins (6), it comprises a mere 45 of the 300 amino acid residues in intelectins. Experiments suggest that carbohydrate binding is calcium ion-dependent. C-type lectins typically use calcium ions to coordinate their carbohydrate ligands, but the intelectins are not members of this family (16, 25, 27). We recently used x-ray crystallography to determine the structures of apo-hIntL-1 and hIntL-1 complexed to a β-Galf glycoside (26). Our results indicate that hIntL-1 binds the exocyclic vicinal diol present in β-Galf through direct calcium ion coordination. This exocyclic vicinal diol is the common feature of hIntL-1 ligands. Still, whether hIntL-1 ligands bind similarly and whether intelectins from different species use similar recognition modes was not known. In addition, whether different intelectins adopt different quaternary states is unclear. Because XEEL is secreted into environmental water, we hypothesized that this protein is stable and therefore would be conducive to crystallographic studies.
Here, we have described the biophysical properties and three-dimensional structure of the carbohydrate recognition domain (CRD) of XEEL (XEELCRD) with and without a bound GroP ligand. The structure of the complex of XEELCRD and GroP has striking similarities to that of hIntL-1 bound to β-Galf. The mechanism of ligand recognition, direct calcium ion coordination, is conserved. Although it is missing the intermolecular disulfide bonds present in hIntL-1, the XEELCRD is trimeric in solution. In light of the previously described hexameric XEEL (22), we propose that full-length XEEL is a dimer of trimers. This molecular arrangement suggests that XEEL could promote cell agglutination. We tested this hypothesis and found that XEEL can agglutinate serotypes of Streptococcus pneumoniae that display capsular polysaccharides with a terminal exocyclic 1,2-diol. In addition to highlighting that hIntL-1 and XEEL have a conserved structure and ligand-binding mechanism, the XEELCRD structure provides the means to identify and compare functional residues across species. We anticipate that the data will be useful as research into the ligand specificity and biological functions of intelectins expands.
Experimental Procedures
Expression and Purification of Xenopus laevis Embryonic Epidermal Lectin (XEEL)
The cDNA for XEEL (GenBankTM accession number BC087616) was purchased from Source BioScience and amplified with primer A (5′-TTTTTGCACTTGCATTTCCAGCAGGGCACGCTGGTTCATGGAGCCATCCGCAGTTTGAAAAGGGTTCATGTGAACAAGCTTCAATTTCTG-3′) and primer B (5′-GGTACCAAGCTCATTAACGGTAGAAGAGCATCACAGCTGCC-3′). The PCR product was then amplified with primers B and C (5′-GCGCGGATCCATGTTGTCATATAGCCTGTTGCTTTTTGCACTTGCATTTCCAGCAGGG-3′). The resulting PCR product was digested with BamHI and KpnI and ligated into a similarly digested pFastBac1 vector. The N terminus of the protein product is expected to be MLSYSLLLFALAFPAGHA GSWSHPQFEKGSCEQASIS. The expressed protein begins with residue 4, the second methionine in the annotated sequence. The underlined amino acids are the predicted secretion signal peptide, the bold amino acids denote the Strep-tag® II incorporated for purification, and the italicized amino acids correspond to the native, mature sequence. Residues 22 and 23 (GS) are repeated after the Strep-tag II. Following sequence verification, the vector was subjected to recombination into a baculovirus according to the manufacturer's protocol (Bac-to-Bac baculovirus expression system, Life Technologies). The resulting baculovirus genomic DNA was transfected (Insect GeneJuice, Novagen) into Sf21 insect cells to produce the first generation (P1) of recombinant baculovirus. Amplified baculovirus (P2) was produced using P1 to infect suspension Sf21 cells grown in SF900-II-SFM (Life Technologies). Virus-containing supernatant was harvested by centrifugation. Fetal bovine serum was added to 2% to stabilize the baculovirus.
XEEL was produced as a secreted protein using High Five cells (Life Technologies), a derivative of Trichopulsia ni. For expression of the selenomethionine (Se-Met)-labeled XEEL used for experimental phasing, High Five cells were suspension cultured in 921 Delta Series, methionine-deficient medium (Expression Systems, catalog No. 96-200, 200 ml) supplemented with 1× antibiotic-antimycotic (Life Technologies) and 10 μg/ml gentamicin (Life Technologies). Cells were infected when they reached a density of ≥2 × 106 cells/ml by the addition of 0.5 μl of baculovirus P2 conditioned medium/1 × 106 viable cells. After the first addition of l-selenomethionine (10 mg, Acros Organics) at 12 h post-infection, additional portions (10 mg) were added every 24 h until medium harvest. Cells producing XEEL were cultured for 5 days at 22 °C in a baffled flask with shaking at 90 rpm. The conditioned culture medium was harvested by centrifugation and passed through a 0.22-μm filter; the medium was stored at 4 °C for at least 1 week. This incubation period was crucial for the proteolytic cleavage that preceded successful crystallization of XEEL residues 54–342. Unlabeled XEEL was produced using the same conditions except Express Five SFM (Life Technologies) supplemented with 1× antibiotic-antimycotic, 10 μg/ml gentamicin, and 4 mm glutamine was used as a culture medium, and no Se-Met was added.
The conditioned medium was dialyzed extensively against 20 mm bis-Tris (pH 6.7), 150 mm NaCl, and 1 mm EDTA. The medium was slowly adjusted to pH 6.7, and a 1 m solution of CaCl2 was added to a final concentration of 10 mm. In addition, avidin (Calbiochem) was added to the conditioned medium (28 μg/ml) to absorb excess biotin, per the manufacturers protocol (IBA GmbH). The solution was then cleared by centrifugation. Strep-tag II XEEL was purified by binding to Strep-Tactin® Superflow resin (IBA GmbH, catalog No. 2-1206-002). The column was washed with 20 mm bis-Tris (pH 6.7), 150 mm NaCl, 10 mm CaCl2 and then 20 mm bis-Tris (pH 6.7), 150 mm NaCl, 0.5 mm EDTA. Protein was eluted with 5 mm d-desthiobiotin (Sigma Aldrich) in 20 mm bis-Tris (pH 6.7), 150 mm NaCl, and 0.5 mm EDTA and concentrated using a 10,000 MWCO Amicon Ultra centrifugal filter. During the concentration process, large sheet-like crystals began to form. Crystals were harvested by centrifugation at 2000 rpm and washed twice using 20 mm bis-Tris (pH 6.7), 150 mm NaCl, and 0.5 mm EDTA. The crystals were resuspended in 20 mm bis-Tris (pH 6.7), 150 mm NaCl, and 0.5 mm EDTA, and CaCl2 was added to 5 mm. Within 1 min, the crystals completely dissolved. The protein purity of the resulting solution was assessed by SDS-PAGE and Coomassie Blue staining to be >95%. The concentration of XEEL was determined using absorbance at 280 nm with an estimated ϵ = 75,455 cm−1m−1 for the monomer and a calculated molecular mass of 36,258 Da post-signal peptide removal. Typical yields were 0.5 mg/50 ml of conditioned medium.
Although the expected mass of the glycosylated protein produced is 36,258 Da, MALDI and ESI-TOF analysis returned masses of 32,884 and 32,802 Da, respectively. The observed mass difference indicated a truncation of at least 30 residues. N-terminal sequencing (ABI 494, Tufts University Core Facility) revealed the first five amino acids to be RSGGS. Therefore, the XEEL construct used in this study corresponds to residues 54–342. We refer to this as XEELCRD to denote the carbohydrate recognition domain.
Expression and Purification of Thioredoxin-fused XEEL (Residues 22–47)
The expression construct was synthesized as a gBlock (Integrated DNA Technologies) and then cloned into the MfeI and BamHI sites of pMAL-c5x (New England Biolabs). The expected protein sequence corresponds to full-length thioredoxin (underlined) followed by a linker, a histidine tag, and an enterokinase cleavage site (bold) fused to residues 22–47 of XEEL (italics): MSDKIIHLTDDSFDTDVLKADGAILVDFWAEWCGPCKMIAPILDEIADEYQGKLTVAKLNIDQNPGTAPKYGIRGIPTLLLFKNGEVAATKVGALSKGQLKEFLDANLAGSGSGHMHHHHHHSSGTDDDDKGSCEQASISEKKEKILNLLACWTEGN.
The expression plasmid was transformed into Escherichia coli SHuffle® Express (New England Biolabs). Cells were grown in Terrific broth at 30 °C until OD600 reached 0.6, at which time the temperature was lowered to 15 °C. Isopropyl 1-thio-β-d-galactopyranoside (0.1 mm) was added to induce expression, and fermentation was allowed to proceed for 16 h. The cells were lysed, and the protein was purified using nickel-nitrilotriacetic acid as described previously (28).
X-ray Crystallography
Selenomethionine-labeled XEELCRD was redissolved in 20 mm bis-Tris (pH 6.7), 150 mm NaCl, 0.5 mm EDTA, and 5 mm CaCl2 at a concentration of 2 mg/ml, and crystallization (hanging drop vapor-diffusion) was achieved by mixing 1 μl of the protein solution and 1 μl of well solution (100 mm Tris (pH 7.0) and 20–24% PEG 400). Crystals appeared in 2–3 h and matured to full size within 2–3 days. For cryoprotection, crystals were dipped briefly in well solution containing 30% PEG 400 and vitrified in liquid nitrogen.
Unlabeled XEELCRD protein was used to obtain the crystal structure with bound GroP (Sigma-Aldrich, G7886). Crystals were grown under similar conditions to Se-Met-labeled XEEL, with 50 mm GroP present during crystal growth and cryoprotection.
Single crystal x-ray diffraction experiments were performed on the Life Sciences Collaborative Access Team beamline 21-ID-D at the Advanced Photon Source, Argonne, IL. To determine the structure of Se-Met XEELCRD, diffraction data were collected at the Se K-edge (12,661.38 eV). Integration, scaling, and merging were performed with HKL2000 (29). The structure was solved using PHENIX by Se-SAD phasing with AutoSol (30). 42 selenium sites were expected; the substructure search yielded 39. The figures of merit were 0.28 and 0.66 before and after density modification, respectively. The GroP-bound XEELCRD structure was solved by molecular replacement using Phaser with monomeric Se-Met XEELCRD as a search model (31). Both structures were adjusted and refined using Coot and phenix.refine, respectively (32, 33). The coordinate restraint set for GroP was generated using PRODRG (34). MolProbity was used for validation (35). Data collection and refinement statistics are presented in Table 1. PyMOL was used to generate the figures depicting protein structures (Figs. 2, 4, 8B, 10, and 12) (36). Surface properties were calculated using PISA (37).
TABLE 1.
X-ray crystallographic data collection and refinement statistics
Statistics for the highest resolution shell are shown in parentheses. The coordinates and structure factors of Se-Met XEELCRD and XEELCRD-GroP complex were deposited in the Protein Data Bank under accession codes 4WMO and 4WN0, respectively.
| Statistics | Se-Met XEELCRD | XEELCRD-GroP complex |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97924 | 0.97856 |
| Resolution range (Å) | 30.10–2.30 (2.38–2.30) | 30.2–2.20 (2.28–2.20) |
| Space group | P 21 | P 6 |
| Unit cell (Å) | 123.6, 111.1, 123.6, | 124.6, 124.6, 55.6, |
| 90, 119.7, 90 | 90, 90, 120 | |
| Total reflections | 991,049 | 285,681 |
| Unique reflections | 129,043 (12,885) | 25,366 (2,526) |
| Multiplicity | 7.7 (7.7) | 11.3 (11.1) |
| Completeness (%) | 100 (100) | 100 (100) |
| Mean I/σ(I) | 13.7 (2.6) | 19.4 (3.2) |
| Wilson B-factor (Å2) | 21.0 | 29.6 |
| R-merge | 0.168 (0.625) | 0.127 (0.755) |
| R-meas | 0.180 (0.670) | 0.133 (0.792) |
| R-pim | 0.065 (0.241) | 0.040 (0.237) |
| Refinement | ||
| Resolution range (Å) | 30.10–2.30 (2.38–2.30) | 29.93–2.20 (2.25–2.20) |
| R-factor | 0.1597 (0.2152) | 0.1515 (0.1977) |
| R-free (5%) | 0.1881 (0.2673) | 0.1692 (0.2363) |
| Number of atoms | ||
| Protein | 12,927 | 2,151 |
| Calcium | 18 | 3 |
| GroP | 10 | |
| Water | 1,470 | 166 |
| Protein residues | 1,662 | 277 |
| r.m.s.d. (bonds, Å) | 0.011 | 0.010 |
| r.m.s.d. (angles, °) | 1.170 | 1.089 |
| Est. coordinate error (ML, Å) | 0.21 | 0.17 |
| Ramachandran favored (%) | 96.1 | 95.7 |
| Ramachandran outliers (%) | 0 | 0 |
| Average B-factor (Å2) | ||
| Protein | 13.0 | 24.2 |
| Calcium | 7.8 | 17.3 |
| GroP | 38.0 | |
| Solvent | 24.4 | 33.7 |
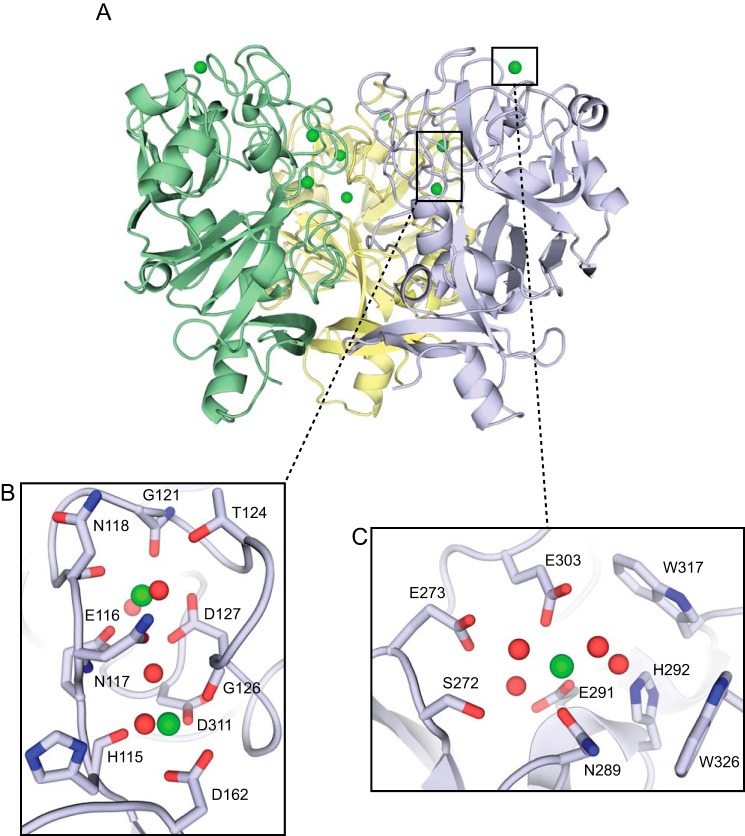
FIGURE 2.
XEEL is a trimer with multiple calcium ion binding sites. A, crystal structure of Se-Met-labeled XEELCRD trimer oriented with the intelectin-specific domains toward the top of the figure and the fibrinogen-like lobes below. The second trimer in the asymmetric unit is removed for clarity. B, structural calcium site with two calcium ions (green) and four ordered water molecules (red). C, ligand-binding site with one calcium ion (green) and four ordered water molecules (red).
FIGURE 4.
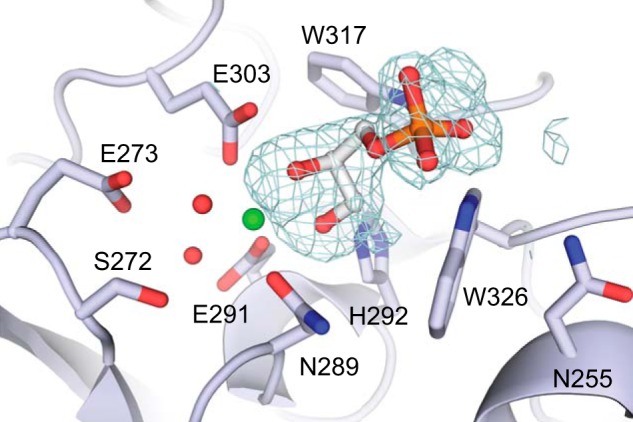
Crystal structure of XEELCRD-GroP complex. Calcium ion and ordered water molecules are shown in green and red, respectively. The mesh represents the 3σ contour in a difference electron density map (mFo − DFc, where m is figure of merit, and D is σA weight) after refinement without ligand.
FIGURE 8.
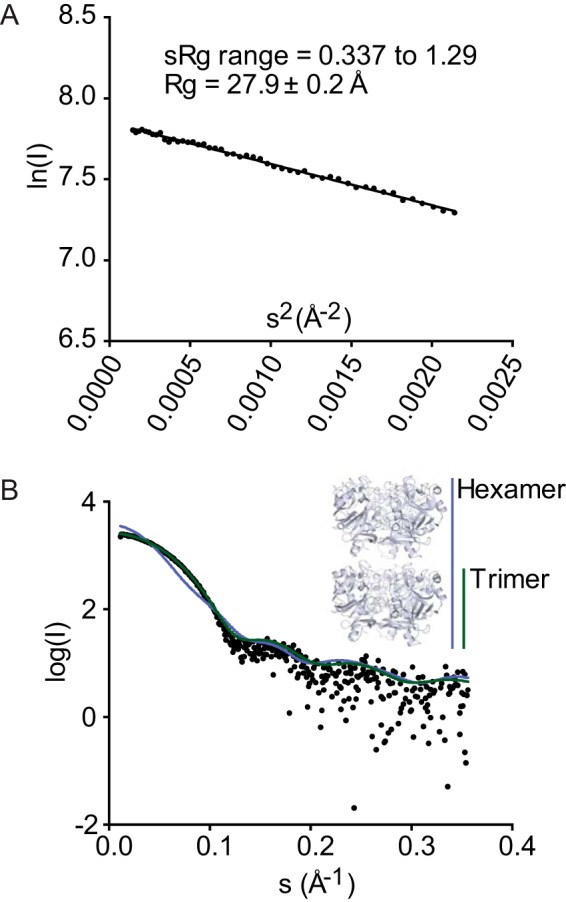
SAXS of XEELCRD. A, Guinier plot suggests Rg of 27.9 Å (consistent with a trimer) and no particle interactions. B, simulated scattering curves of trimer (green) and hexamer (blue) in the crystal structure compared with the experimental data (black).
FIGURE 10.
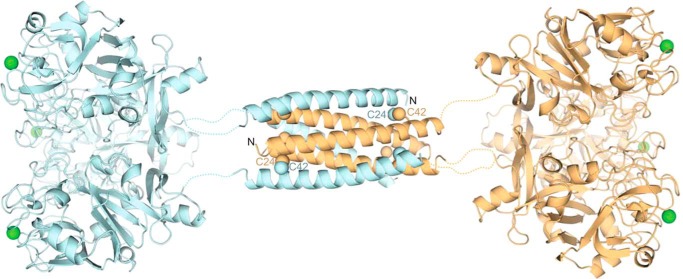
Predicted structure of full-length XEEL. The ligand-binding site on each carbohydrate-binding domain (derived from the crystal structure) is marked by its calcium ion (green). The predicted structure of the dimerization domain (from PDB ID: 2SIV (62)) is shown as a helical bundle, with two of the six Cys-24–Cys-42 disulfide bonds labeled.
FIGURE 12.
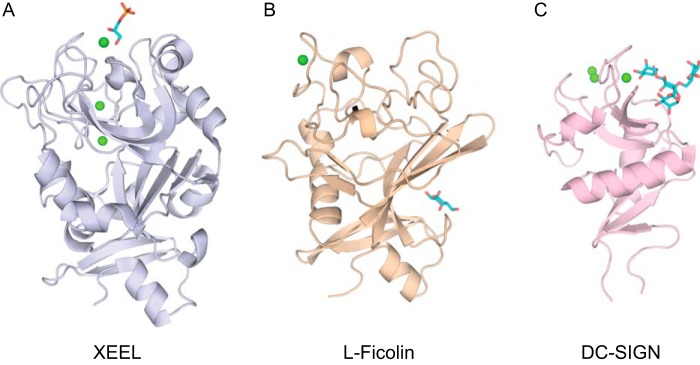
Comparison of lectin structures. A, XEEL from this work (PDB ID: 4WN0). B, L-ficolin CRD (PDB ID: 2J3U) (47). C, DC-SIGN (PDB ID: 1SL4) (50). Calcium ions are represented by green spheres, and the carbohydrate ligands are shown in cyan.
Surface Plasmon Resonance
Analysis of XEELCRD using surface plasmon resonance (SPR) was conducted on a ProteOn XPR36 (Bio-Rad) at the University of Wisconsin-Madison Department of Biochemistry Biophysics Instrumentation Facility. To measure XEELCRD binding to carbohydrate, ProteOn NLC sensor chips (NeutrAvidin-coated sensor chip) (Bio-Rad, catalog No. 176-5021) were used to capture biotinylated carbohydrate ligands. All experiments presented here were conducted at surface-saturated levels of ligand, ∼200 response units. In all experiments, captured biotin was used in flow cell 1 as a control. Samples containing purified XEELCRD were prepared by serial dilution into SPR running buffer (20 mm HEPES (pH 7.4), 150 mm NaCl, 1 mm CaCl2, and 0.005% Tween 20). Surfaces were regenerated with short injections of solutions of 10 mm HCl. Data were referenced using the biotin reference channel and processed using the Bio-Rad ProteOn software package. Competition binding experiments were conducted by adding glycerol to the SPR running buffer in which purified XEELCRD was diluted. Data were analyzed on Prism6 (GraphPad). For determination of the glycerol IC50, SPR data were fit to a one-site competition model in Prism6. The top of the fit was constrained to 110 response units based on the equilibrium response in the presence of 1 mm CaCl2.
Chemical Cross-linking
XEELCRD aliquots (1 mg/ml, 2 μl) were mixed with 1 μl of bis(sulfosuccinimidyl)suberate cross-linker (Pierce) stock solution to achieve final cross-linker concentrations ranging from of 0 to 5 mm. The buffer for all components was 20 mm bis-Tris (pH 6.7), 150 mm NaCl, 0.5 mm EDTA, and 5 mm CaCl2. Cross-linking was performed at room temperature for 30 min. Each reaction was diluted by adding 7 μl of the reaction buffer, and proteins were denatured by adding 2 μl of 6× SDS loading buffer (350 mm Tris (pH 6.8), 30% glycerol, 10% SDS, 9.3% DTT, and 0.06% bromphenol blue). Samples were heated at 95 °C for 3 min prior to analysis by SDS-PAGE stained with Coomassie Blue.
Small-angle X-ray Scattering
Scattering data for a solution of 2 mg/ml (63 μm monomer) XEELCRD were collected on a Bruker NANOSTAR small angle x-ray scattering (SAXS) system. Background scattering (20 mm bis-Tris (pH 6.7), 150 mm NaCl, 0.5 mm EDTA, and 5 mm CaCl2) was subtracted from the data. A preliminary analysis was performed using the ATSAS suite (PRIMUS and GNOM) (38–40). Predicted scattering curves from the crystal structure of XEELCRD were generated using CRYSOL (41).
Sedimentation Equilibrium Analytical Ultracentrifugation
Recrystallized XEELCRD was dissolved in 20 mm bis-Tris (pH 6.7), 150 mm NaCl, 0.5 mm EDTA, and 5 mm CaCl2, and the concentration was determined using UV-visible absorbance spectroscopy. Samples were prepared by dilution to concentrations of 9.0 μm (trimer), 5.2 μm, and 2.6 μm for analysis by sedimentation equilibrium analytical ultracentrifugation. Equilibrium data were collected at 20 °C in a Beckman Optima XLA Analytical Ultracentrifuge using 1.2-cm double sector charcoal-filled Epon centerpieces. Protein gradients were recorded at 276 nm every 2–3 h until two or more were superimposable. The equilibrium data were collected at speeds of 6,000, 8,000, 9,600, 11,500, 13,200, and 15,900 rpm. After the 15,900-rpm run, the contribution from non-sedimenting absorbance was determined by high speed depletion of the protein; this absorbance was ≤0.006 for the three samples. After depletion, subsequent re-equilibration at 11,500 rpm resulted in a gradient essentially superimposable on the original gradient at this speed, indicating no significant loss of protein to irreversible aggregation during the course of the experiment.
The solvent density (ρ) of 1.004 g/ml was computed from density increments (42), except that the bis-Tris buffer was not included, as it is not in the available table. The calculated partial specific volumes (υ) of intact XEEL and XEELCRD are the same and based on the amino acid sequences are 0.719 ml/g. The masses based on sequence (Ms) are 36,258 and 31,652, respectively.
Analysis of the equilibrium data followed an approach similar to that described previously (43) and was performed by Darrell R. McCaslin (University of Wisconsin-Madison, Department of Biochemistry, Biophysics Instrumentation Facility). The analysis utilized programs developed in Igor Pro (Wavemetrics Inc., Lake Oswega, OR) by Dr. D. R. McCaslin. The measured non-sedimenting absorbance for each sample was included as a fixed parameter. The data from three concentrations and five speeds were globally fit to models consisting of one or two macromolecular species. The single species model described the complete data set adequately and thus was employed. The reduced molecular weight (Mr) was used as the fitting parameter as it removes the impact of ambiguities in υ and ρ on the fit. The reduced molecular weight is defined as Mr = Mw(1-υρ), where Mw, in the single species case, is the molecular weight of the macromolecular complex. Mr must be an integral multiple of the sequence weight Ms; therefore, Mr = nMs(1-υρ), where n is equal to the oligomeric state of the protein.
S. pneumoniae Agglutination Assay
Formaldehyde-fixed S. pneumoniae serotypes were prepared as described previously (26). Full-length hexameric XEEL was expressed in High Five cells in a manner similar to the monomeric XEELCRD, except that 1% fetal bovine serum was included during expression and SIGMAFAST protease inhibitor (Sigma-Aldrich, S8820) was added after medium collection to suppress proteolysis. Each bacterial suspension was mixed with hexameric XEEL and imaged using a ×40 objective lens on a Nikon TE2000 inverted microscope equipped with an Andor IXON camera. Images were acquired with MetaMorph. A scale bar was added using ImageJ, with a scale of 0.162 μm/pixel.
Results
An Oligomerization Domain at the XEEL N Terminus
We used an insect cell expression system for XEEL, both because of its ability to yield the large quantities of protein needed for biophysical characterization and because it can be used to incorporate Se-Met into the protein for crystallographic structure determination. The expression construct encodes full-length XEEL with an N-terminal Strep-tag II purification tag sequence linked to the predicted signal peptide (44). Cleavage of the signal peptide should yield XEEL with a two-amino acid linker followed by an eight-amino acid Strep-tag II at the N terminus. Purification of secreted XEEL from conditioned culture supernatant using Strep-Tactin affinity chromatography (45) afforded a protein product, but mass spectrometry of the purified protein indicated a truncation of at least 30 N-terminal residues had occurred. Analysis by N-terminal sequencing revealed that secondary proteolysis had yielded a protein product corresponding to residues 54–342 (XEELCRD). The XEELCRD lacks the Strep-tag II purification tag, yet the protein could be purified using a Strep-Tactin resin. We postulate that the lectin has modest affinity for agarose resin. The production of this truncated protein provided the opportunity to examine the role of the N-terminal sequence in oligomerization.
The N-terminal sequences of the intelectins are not conserved, and previous studies suggest that this region controls the intelectin oligomeric state (6). The recombinant XEELCRD lacks the N-terminal intermolecular disulfide bonds responsible for covalent trimerization of hIntL-1. As expected, XEELCRD migrates as a monomer on reducing and non-reducing SDS-PAGE (Fig. 1A). When the aforementioned construct was expressed in HEK293T cells, full-length XEEL was obtained. The full-length protein migrated as a monomer under reducing conditions but, as observed previously, it ran as a disulfide-linked hexamer under non-reducing conditions (22). These results are consistent with predictions that the XEEL N terminus engages in intermolecular disulfide formation. The disulfide bonds are likely mediated by cysteines Cys-24 and Cys-42. We tested this possibility by generating a fusion protein in which residues 22–47 of XEEL were appended to the C terminus of thioredoxin. The resulting recombinant protein forms covalent dimers and hexamers (Fig. 1B). The data indicate that this 26-amino acid peptide is a disulfide-mediated oligomerization domain.
FIGURE 1.
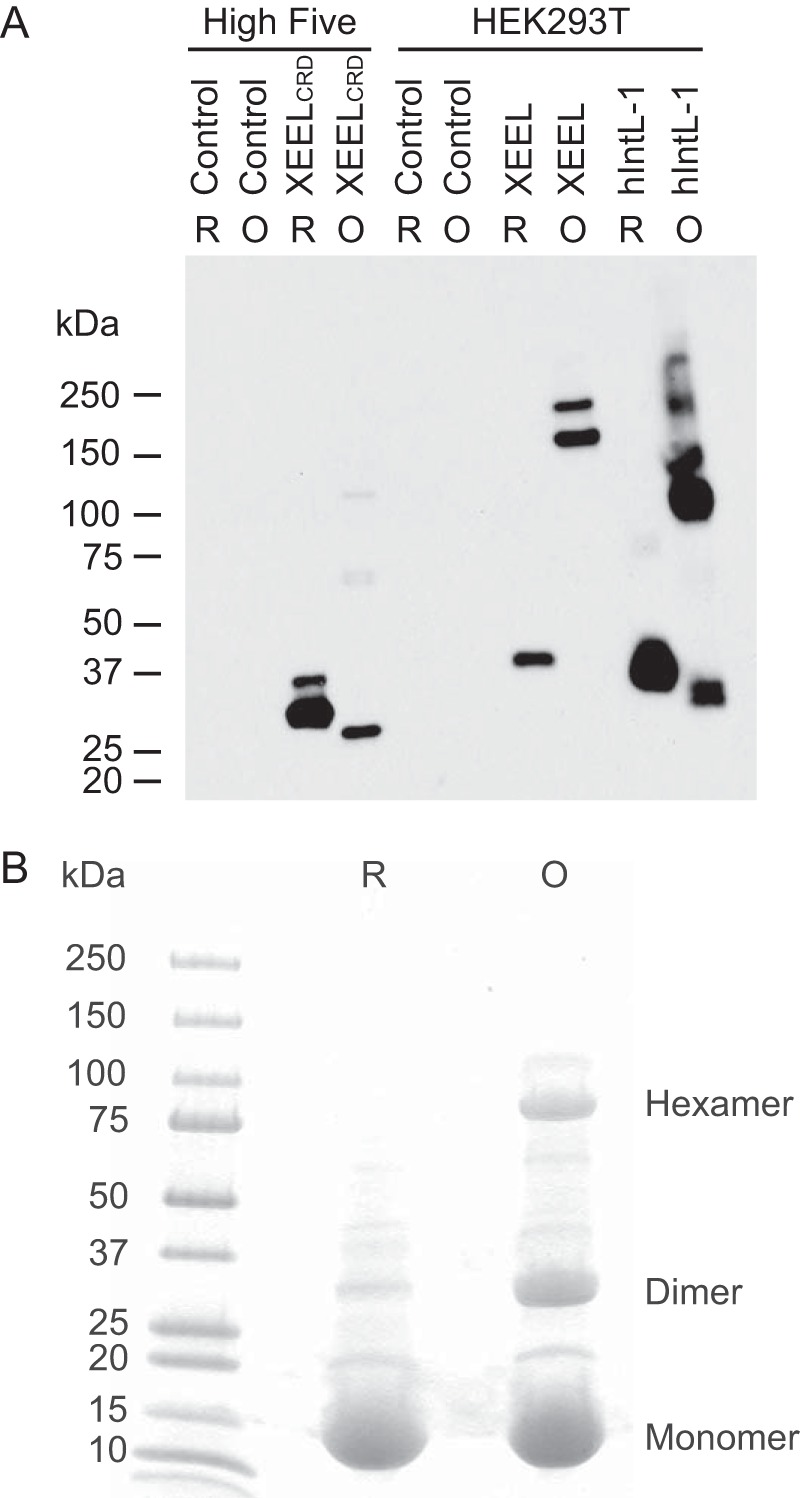
Analysis of recombinant XEEL. A, Western blotting of supernatant from High FiveTM (BTI-TN-5B1-4) cells expressing the construct encoding XEEL fused to a Strep-tag II or HEK293T cells expressing the identical construct in either reducing (R) or non-reducing (oxidized (O)) conditions. A sample of hIntL-1 is included for comparison. The hIntL-1 antibody used (Proteintech, 11770-1-AP) cross-reacts with XEEL. Spent culture media from non-infected or non-transfected cells were used as negative controls. Intelectins are glycoproteins, and thus the higher molecular weight bands likely represent different glycoforms. Under non-reducing conditions, disulfide-containing proteins are nonlinear polypeptides and thus may have different mobility than their reduced, linear polypeptide counterparts. B, SDS-PAGE of thioredoxin fused to N-terminal residues 22–47 of XEEL under reducing and oxidizing conditions. The expected masses are 17, 34, and 102 kDa for monomer, dimer, and hexamer, respectively.
Three-dimensional Structure of XEELCRD
To analyze the ligand-binding site of XEEL and to compare the oligomeric state of XEELCRD to that of hIntL-1, we used x-ray crystallography to determine the structure of XEELCRD. For phasing purposes, we produced Se-Met-labeled XEELCRD. Mass spectrometry analysis indicated that Se-Met was substituted at all seven methionine residues. Native and Se-Met-labeled XEELCRD crystallized under similar conditions. The structure of Se-Met XEEL was solved by Se-SAD (with 67.5% solvent content) and refined to 2.3 Å resolution. The six molecules in each asymmetric unit form two trimers (Fig. 2A) related to one another by a pseudotranslation vector of 55.6 Å approximately parallel to the b axis. Although the C terminus is clearly resolved in the electron density, the first resolvable residue on the N terminus is Gly-66.
The trimeric carbohydrate recognition domains of XEEL and hIntL-1 are highly related, each possessing a globular subunit that does not fall into a well known structural family. Both have a fibrinogen-like lobe that contains a split and twisted seven-stranded β-sheet. The intelectin-specific region is characterized by a large fraction of random coil and a three-stranded curved β-sheet. The domains are encircled by 12 short solvent-exposed α-helical stretches (Fig. 2A).
Three well ordered calcium ions are a defining feature of the intelectin-specific region of XEELCRD. Two calcium ions are buried deep in the protein core and are presumed to play a structural role (Fig. 2B). The structural calcium ion that is closer to the protein surface is heptacoordinated. Calcium ion ligands include two distal waters and five protein atoms: the carboxylates of Glu-116 and Asp-127 (both oxygen atoms) and the backbone carbonyls of Asn-118 and Gly-121. Asn-117 forms additional hydrogen bonds to calcium-bound water molecules through both its side chain and backbone amide nitrogen. The structural calcium ion located farther inside the trimeric protein is coordinated directly by the carboxylates of Asp-162 (both oxygen atoms) and Asp-311, backbone amide carbonyls of His-115 and Gly-126, and two water molecules. Finally, the His-115 side chain also forms a hydrogen bond with a calcium-bound water molecule. No single amino acid or water molecule coordinates both calcium ions simultaneously.
The remaining calcium ion is positioned analogously to that in the β-Galf-bound hIntL-1 structure (PDB ID: 4WMY) (26) (Fig. 2C). This congruence suggests that this ion also marks the carbohydrate recognition pocket in XEELCRD. This calcium ion is directly coordinated by Asn-289, Glu-291, Glu-303, and four ordered water molecules. The pocket geometry is determined by a network of 16 hydrogen bonds distributed among the four calcium-bound water molecules and the aforementioned residues, Ser-272, Glu-273, and His-292. Side chains from Trp-317 and Trp-326 form the back walls of this conserved binding site. The structural similarity of this region of XEEL and hIntL-1 suggest that these lectins may recognize similar glycans.
The XEELCRD Binds Ligands Containing a Terminal 1,2-Diol
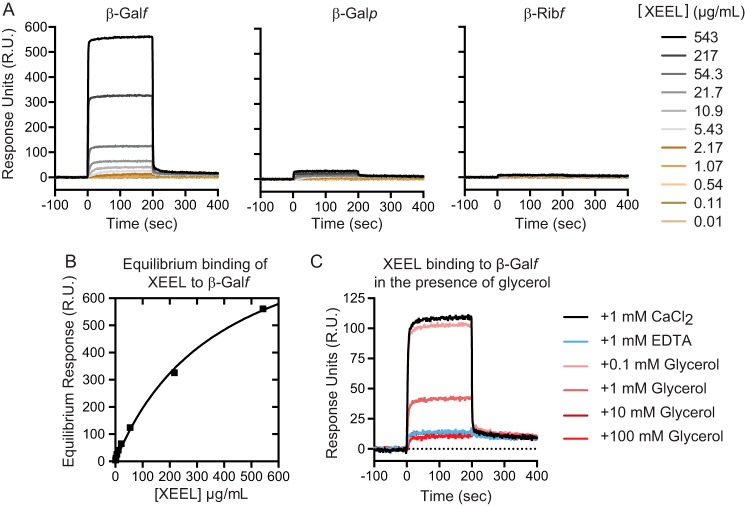
The human homolog of XEEL, hIntL-1, interacts with a range of glycan epitopes, each of which possesses a terminal 1,2-diol (i.e. a diol of the form RCH(OH)-CH2OH) (26). To address whether this ligand specificity is conserved between XEEL and hIntL-1, we used SPR to monitor XEELCRD ligand binding properties (Fig. 3A). XEELCRD displays concentration-dependent binding to immobilized β-Galf. The on- and off-rates are rapid, suggesting that minimal structural reorganization of XEELCRD occurs upon ligand binding. As a measure of the affinity of XEELCRD for immobilized β-Galf, we used an equilibrium binding model to analyze the SPR data (Fig. 3B). The apparent affinity of the XEELCRD trimer for β-Galf is 4.1 ± 0.5 μm. No specific binding to β-galactopyranose (β-Galp) or β-ribofuranose was observed. These results suggest that, like hIntL-1, XEEL does not bind generally to furanoside ligands but instead recognizes the exocyclic 1,2-diol epitope of β-Galf.
FIGURE 3.
Binding of XEEL to carbohydrates, characterized using SPR. A, SPR sensorgrams of XEELCRD binding to immobilized carbohydrates. B, equilibrium binding analysis of XEELCRD to immobilized β-Galf. These data were used to determine the apparent affinity of XEELCRD for β-Galf. C, glycerol was assayed for its ability to inhibit XEELCRD binding to β-Galf. These data were used to determine the IC50 of glycerol. This experiment was reproduced with an independently purified protein sample.
To test whether the exocyclic vicinal diol on β-Galf is the epitope recognized by XEELCRD, we conducted competition binding studies with the simplest terminal 1,2-diol, glycerol. Glycerol inhibits XEELCRD binding to immobilized β-Galf, indicating that it is a competitive inhibitor (Fig. 3C). These experiments suggest that XEEL binds both β-Galf and GroP through their common exocyclic vicinal diol epitope. From the inhibition data, we determined the IC50 of glycerol to be 0.6 ± 0.6 mm. The glycerol inhibition data fit well to a one-site competition model. Thus, the determined IC50 value serves as an estimate of the Kd of an exocyclic vicinal diol-containing compound for a single XEEL-binding site.
Human and Xenopus Intelectins Use Similar Ligand Recognition Modes
To compare ligand recognition of XEEL and hIntL-1, we co-crystallized the XEEL with GroP. Alignment of Se-Met XEELCRD and GroP-bound XEELCRD yields an r.m.s.d. value of 0.17 Å over 276 Cα atoms, suggesting that no drastic structural changes occur upon either Se-Met labeling or ligand binding. Specifically, no significant movement of residues in the vicinity of the ligand-binding site is observed upon XEEL complexation to GroP.
The structure reveals that two hydroxyl groups from GroP coordinate directly to a calcium ion (Fig. 4). Upon binding, these hydroxyls displace two ordered water molecules in the ligand-binding site. This mode of ligand binding is nearly identical to what was observed in the structure of the hIntL-1-β-Galf complex. In XEEL, the diol moiety of the GroP binds in a pocket formed by the side chain of Trp-317 and Trp-326. Trp-326 serves as an aromatic wall; it is positioned such that it allows binding of a diol terminating with a hydroxymethylene (-CH2OH) group but not a more substituted diol. A similar binding mode is found for hIntL-1, but the human protein possesses a tyrosine residue in place of Trp-326 in XEEL.
The indole NH of Trp-326 forms a hydrogen bond with the phosphate group of GroP. Electron density around the phosphate group reveals elongation toward the indole NH of Trp-317, suggesting dynamic equilibration of hydrogen bonding between Trp-317 and Trp-326 and the phosphate group. The primary hydroxyl of GroP forms a hydrogen bond with His-292. In addition to calcium coordination, Glu-303 forms a hydrogen bond with the secondary hydroxyl group of GroP. These additional hydrogen-bonding interactions enhance the ability of the diol hydroxyl groups to serve as ligands for the calcium ion.
Intelectin Structures Provide Insight into Residue Conservation and Function
The availability of the XEEL and hIntL-1 structures allows the assignment of residue function to the broader class of intelectins. We aligned the sequences of intelectins from humans and a range of model organisms, using ClustalW2 (Fig. 5) (46). The structural calcium sites are highly conserved. The only exception is the amino acid corresponding to Asn-118 of XEEL, which coordinates the calcium ion through its backbone amide carbonyl. The majority of the structural calcium site residues cluster between amino acids 115 and 127, with the consensus sequence of HENXXXGXCTXGD. Cys-123 is conserved in the intelectin fold but has no role in calcium ion binding. Although not present in the consensus sequence, aspartates 162 and 311 are also conserved residues that ligate the structural calcium ion. The high conservation of amino acids in this region suggests that most, if not all, intelectins contain two structural calcium ions.
FIGURE 5.
Sequence alignment of XEEL and intelectins from human (hIntL-1 and hIntL-2) and common model organisms (XCGL-1, XCGL-2, XSL-1, and XSL-2 from X. laevis; mIntL-1 and mIntL-2 from mouse; DrIntL-1 and DrIntL-2 from zebrafish). Corresponding GenBankTM accession numbers are included in the second column. Residues in the structural calcium site and ligand-binding site are highlighted in blue and green, respectively.
Compared with the structural calcium ion site, more amino acid variation is found in the ligand-binding site (Fig. 5). Among the five X. laevis intelectins, residues that directly coordinate the ligand-binding site calcium ion are completely conserved. Trp-317 and Trp-326, which form a box around the vicinal diol ligand, are conserved amongst XEEL, XSL-1, and XSL-2, all of which are proposed to participate in innate immunity. These observations suggest that XSL-1 and XSL-2 also can recognize the terminal 1,2-diols prevalent in microbial glycans. By contrast, the corresponding residues in XCGL-1 and XCGL-2 are phenylalanine and asparagine, respectively. XCGL-1 and XCGL-2 are involved in fertilization membrane formation (16, 17), and they therefore likely recognize self-carbohydrate epitopes. Indeed, glycan array screening data suggest that XCGL-1 binds Galα(1–3)GalNAc (Consortium for Functional Glycomics, primscreen_758). This dissacharide is not specific to microbes.
Intelectins and Ficolins Are Structurally Divergent
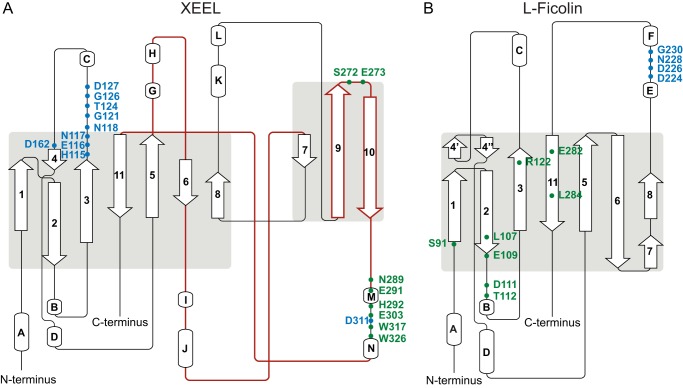
Prior to our determinations of the hIntL-1 and XEELCBD structures, intelectins were proposed to be part of the ficolin structural family. Although both lectin classes share an FBD, our results indicate that the intelectins and ficolins are structurally divergent. Indeed, our attempts at molecular replacement using ficolin structures did not yield interpretable maps. Topology diagrams of XEELCRD and L-ficolin (PDB ID: 2J3U) rationalize this observation and highlight the differences in these two lectin families (Fig. 6) (47). Compared with XEELCRD, L-ficolin has three deletions: one between strands 5 and 6 of XEEL, one between strands 6 and 7 of XEEL, and one encompassing the XEEL ligand-binding site (corresponding to XEEL β-strands 9 and 10 as well as a large loop). In L-ficolin, a single calcium-binding site lies between helices E and F, which differ in location and structure to helices K and L in XEELCRD. The structural calcium sites in XEELCRD are clustered between strands 3 and 4, with the exception of Asp-311, which is located within the ligand-binding site. Strand 4′ in L-ficolin has no equivalent in the XEELCRD structure. Specifically, Asp-162 in XEEL is a part of the structural calcium site, and this interaction would disrupt the secondary structure of L-ficolin. The ligand-binding residues of XEELCRD are located between strands 9 and 11, whereas in L-ficolin the residues that interact with carbohydrates are dispersed (Fig. 6). Finally, the ligand-binding site of XEEL contains a calcium ion that directly interacts with the ligand, a feature absent in l-ficolin. Therefore, while XEEL contains an FBD, the ligand binding and structural calcium ion sites are distinct from other lectins. Thus, XEEL, and intelectins in general, represent a novel class of lectins with a unique structural fold.
FIGURE 6.
Topology diagram of XEEL (A) and l-ficolin (PDB ID: 2J3U) (B), highlighting residues in the structural calcium sites (blue) and ligand-binding sites (green). Regions found in XEEL but not in L-ficolin correspond roughly to the brown traces in XEEL. Images were generated using Pro-origami (61).
XEELCRD Is Trimeric in Solution
Despite lacking the N-terminal region that engages in disulfide bond formation, the structure of XEELCRD indicates it forms a non-covalent trimer. Two head-to-tail trimers are observed in the asymmetric unit of the XEELCRD structure, yet there are few contacts between them. This observation is consistent with the fragility of XEELCRDcrystals and their tendency to separate into thin sheets. These properties led us to suspect that this particular crystallographic packing arrangement is not biologically relevant. To examine the oligomerization state of XEEL in solution, we performed chemical cross-linking using bis(sulfosuccinimidyl)suberate (Fig. 7). Trimers predominated at high cross-linker concentration, consistent with the intimate crystallographic trimer. No species larger than trimeric XEELCRD were observed.
FIGURE 7.
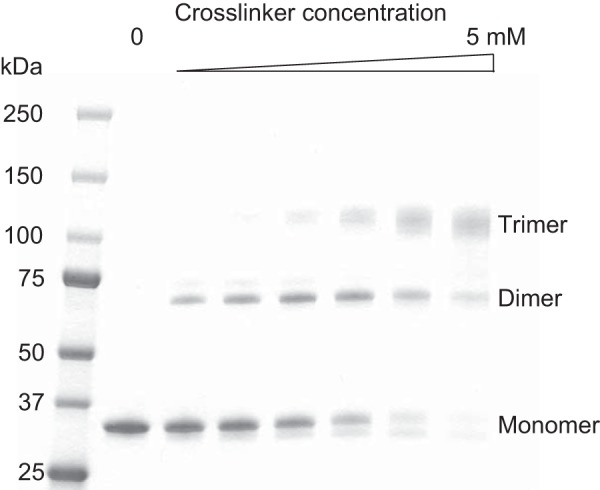
Cross-linking of XEELCRD with bis(sulfosuccinimidyl)suberate. The final concentrations employed of this cross-linker are 0, 0.1, 0.25, 0.5, 1, 2.5, and 5 mm. The expected masses are 31.6, 63.2, and 94.8 kDa for monomer, dimer, and trimer, respectively.
To measure more directly the oligomeric status in solution, we employed SAXS. A Guinier plot of the data from analysis of XEELCRD in solution indicates a radius of gyration (Rg) of 27.9 ± 0.2 Å (Fig. 8A). This Rg value is consistent with the calculated Rg of 27.2 Å from our trimeric XEELCRD crystal structure. The calculated Rg values for monomeric XEELCRD and hexameric XEELCRD (18.7 and 39.1, respectively) do not agree with the experimental data. In addition, a comparison of simulated SAXS curves for monomeric (not shown), trimeric, or crystallographic head-to-tail hexameric XEEL with the experimental data indicated a superior fit for trimeric XEELCRD (Fig. 8B). Therefore, our SAXS findings indicate that XEELCRD is trimeric in solution.
As a third independent assessment of the oligomeric state, we characterized XEELCRD by equilibrium analytical ultracentrifugation. The absence of curvature in the plots of ln(absorbance) versus radial position squared indicated the presence of a single molecular species (Fig. 9). The reduced mass (Mr) obtained from a global fit was 27,120 daltons over all of the XEELCRD concentrations tested. The calculated molecular mass was 96,811 Da, which returns an aggregation number of 3.08, based on the molecular mass of the N-terminally truncated XEELCRD. This result is consistent with our SAXS data indicating that XEELCRD is trimeric. The analytical ultracentrifugation data show that the XEELCRD trimer is the preferred species because no free monomeric XEELCRD was observed at any point during the experiment. Thus, the XEELCRD forms a stable trimer in solution, despite lacking intermolecular disulfide bonds.
FIGURE 9.
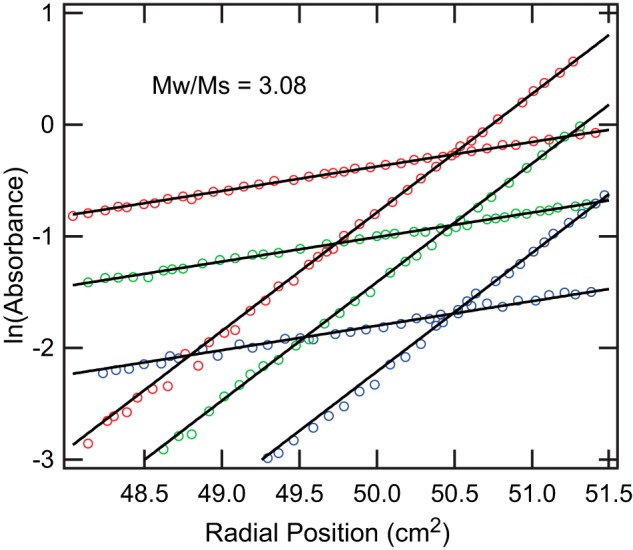
Sedimentation equilibrium analytical ultracentrifugation of XEELCRD. ln(absorbance) versus radial position squared (cm2) plot of XEELCRD at 6,000 rpm (lower slope) or 13,200 rpm (higher slope) for each XEELCRD concentration (2.6 μm (blue), 5.2 μm (green), and 9.0 μm (red)). Only every third raw data point is shown for clarity. The solid lines represent the single species fit of the data used to calculate the reduced mass.
Analysis of the interfaces in the XEELCRD structure indicated that each monomer has an average buried surface area of 1571 Å2 resulting in a total of 4715 Å2 buried upon trimerization. The interface surfaces are polar but not highly charged. Interactions between each monomer include 18–19 hydrogen bonds and 1–3 salt bridges. Superposition of trimeric hIntL-1 (PDB ID: 4WMY) (26) and GroP-bound trimeric XEELCRD using all Cα atoms yielded an r.m.s.d. value of 0.63 Å, suggesting a similar trimeric arrangement. The ligand-binding sites of both trimers occupy the same space. Therefore, the trimeric architecture of XEELCRD and hIntL-1 are conserved in three-dimensional space. Amino acid sequences at the interface are not highly conserved, as determined by ConSurf (48) using both automatic sequence alignment of 150 homologs or intelectins from model organisms (Fig. 5). Given the large buried surface area and the only two unique intelectin structures available to date, the relationship between sequence variation in the monomer interface and the oligomeric state of the intelectins remains to be explored.
XEEL Dimer of Trimers Structure Predicts Agglutination Activity
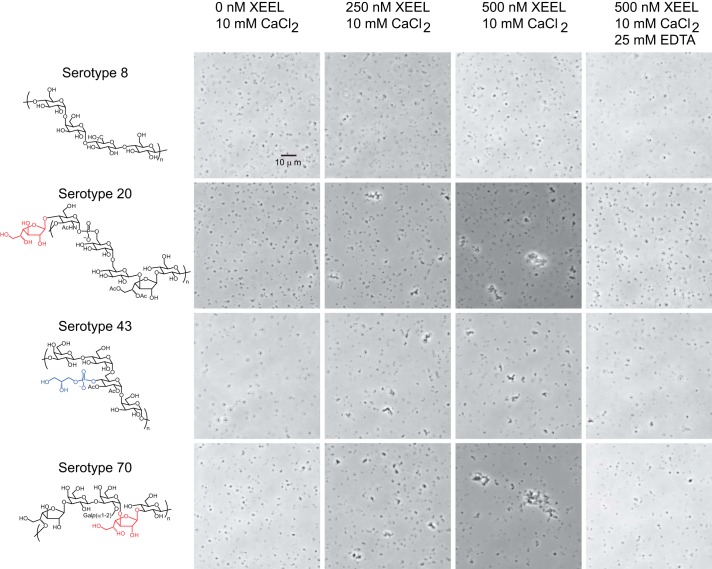
Our crystal structure and solution measurements of the XEELCRD all corroborate that it is a trimer, but full-length XEEL has been reported to be hexameric (22). As fusion of amino acids 22–47 of XEEL to thioredoxin could promote oligomerization (Fig. 1B), we hypothesized that this sequence mediates the assembly of two XEEL trimers into the observed hexamer. A secondary structure prediction strongly suggests that this region is helical (49). We postulated that XEEL could form an anti-parallel six-helix bundle held together by six disulfide bonds, with the consequence being that the protein would adopt a barbell-like arrangement, with each trimer displaying three ligand-binding sites oriented in opposite directions (Fig. 10). This model suggests that XEEL could bind two microbes simultaneously and thereby mediate agglutination. To test this hypothesis, we investigated whether full-length XEEL can agglutinate specific serotypes of S. pneumoniae (Fig. 11). No agglutination was observed with serotype 8, of which the capsule lacks an exocyclic 1,2-diol-containing carbohydrate residue. In contrast, S. pneumoniae serotypes 43 (which contains GroP) and serotypes 20 and 70 (which contain β-Galf residues in their capsular polysaccharide) were agglutinated in the presence of XEEL. Furthermore, this agglutination activity was calcium ion-dependent, as it was abolished upon the addition of the calcium ion chelator EDTA.
FIGURE 11.
Agglutination of S. pneumoniae serotypes by wild-type, full-length XEEL. Known surface glycan epitopes from each strain (63) are depicted, with the moieties containing a vicinal diol highlighted in blue (GroP) or red (β-Galf). The data shown are representative of those obtained in three replicate experiments (different protein and S. pneumoniae preparations).
Discussion
Intelectins are produced by organisms ranging from tunicates to humans, and these soluble lectins have been suggested to participate in innate immunity. Despite this proposal and the high sequence similarity among intelectins, little is known regarding intelectin function. An understanding of structure and carbohydrate-binding specificity can illuminate intelectin functional roles. The human intelectin hIntL-1 binds a range of epitopes found only in microbial glycans (26). To examine whether other homologs share similar binding modes, we determined the structure of the carbohydrate recognition domain of the Xenopus homolog XEEL, alone and complexed to GroP.
The x-ray crystal structures of XEELCRD and hIntL-1 indicate that the intelectins are a discrete lectin class. Although the intelectins and ficolins share an FBD, our x-ray structures show that they are structurally divergent. Intelectins, ficolins, and many C-type lectins possess structural calcium ions. Still, the structural calcium site of XEEL bears no resemblance to the single structural calcium site present in ficolins nor to the two structural calcium ions in the C-type lectin DC-SIGN (Fig. 12). The structural calcium ions of intelectins are buried, which contrasts with the solvent-exposed site in L-ficolin. In addition, intelectins use protein residues from three loops (Fig. 6) to coordinate their structural calcium ions, whereas ficolins employ side chains within a short single loop (47). The intelectin site is also distinct from the two non-conserved structural calcium ions in the C-type lectin DC-SIGN, which are solvent-exposed and are coordinated by two protein loops (50, 51).
The ligand-binding site of the intelectins is also different from that of other lectins. In the intelectin structures solved to date, a calcium ion directly coordinates an exocyclic vicinal diol present in the carbohydrate epitopes. Ficolin structures show no direct interactions between the calcium ion and the ligand. Alternatively, C-type lectins, such as DC-SIGN, recognize their carbohydrate ligands through calcium coordination to adjacent secondary hydroxyl groups within a pyranose ring (Fig. 12). Additionally, the ligand-binding site of DC-SIGN lacks the two electron-rich aromatic residues that surround the calcium-coordinated ligand in hIntL-1 and the XEELCRD. These observations underscore that intelectins are a distinct lectin class.
The structures of XEELCRD and hIntL-1 reveal conserved features: the intelectin fold, the trimeric CRD core, and the ligand-binding mode. We attribute the ligand-binding specificity of XEEL to Trp-317 and Trp-326, which form a tight box around the vicinal diol (Fig. 4). The structure of GroP-bound XEELCRD confirms that GroP does indeed bind in the same site as β-Galf binds to hIntL-1. Both hIntL-1 and XEEL contain an aromatic residue (Trp-326 in XEEL and Tyr-297 in hIntL-1) in the binding site that serves as an aromatic wall, a steric barrier that prevents the binding of more substituted diols. Moreover, the placement of the aromatic residue suggests that it can engage in a CH-π interaction with the methylene group of the 1,2-diol-containing ligands (52). Coordination of the diol to a calcium ion should increase the strength of the CH-π interaction (53). We anticipate that intelectins with this dual aromatic residue signature will show similar ligand-binding modes and specificities.
The structures of XEEL and hIntL-1 provide the means to further interpret intelectin sequence alignment data. We compared human intelectins to those from common laboratory model organisms. The amino acid sequence of the structural calcium site is exquisitely conserved. Moreover, the overall tertiary structure observed for hIntL-1 and XEELCRD is conserved throughout chordate species. At least one intelectin in each model organism shows a high degree of conservation in the ligand-binding site with XEEL and hIntL-1, suggesting that a microbe-binding intelectin likely exists in most animals.
The conservation of the ligand-binding site in some, but not all, members of the intelectin family across the evolutionary tree may distinguish those intelectins that recognize microbial surface glycan epitopes from those involved in other functions. Interestingly, intelectin paralogs within the same species (e.g. XEEL and XCGL-2) show appreciable variation in ligand-binding site residues (Fig. 5). The corresponding gene sequences of XCGL-1 and XCGL-2 encode binding site residue substitutions (W317F and W326N), and these lectins appear to have altered ligand specificity, as mentioned earlier. Thus, one intriguing hypothesis is that these two residues control carbohydrate specificity and can be varied to discriminate between different carbohydrate epitopes. Indeed, a further examination of sequence alignment predicts intelectins with novel ligand binding properties. For example, mouse intelectin-2 (mIntL-2) does not have a predicted aromatic box, as it has a W317A substitution. In addition, there are marked variations in the ligand-binding site of the zebrafish intelectins (Fig. 5). The differences between XEEL and hIntL-2 are especially intriguing. For example, the equivalent of XEEL Trp-326 (or hIntL-1 Tyr-297) in hIntL-2 is a serine residue. As a result, hIntL-2 is unlikely to recognize terminal 1,2-diols, but rather it binds some other motif. Furthermore, Glu-303, which directly coordinates the ligand-binding site calcium ion in XEEL and hIntL-1, is a glutamine in hIntL-2. This change may alter the ability of hIntL-2 to chelate a calcium ion. The consequences of these changes on intelectin structure and ligand specificity warrant further study.
The oligomeric state of intelectins appears to vary among family members; hIntL-1 is a disulfide-linked trimer, whereas XEEL is reported to be a disulfide-linked hexamer. Here we show that the CRD of XEEL is trimeric in solution, despite lacking intermolecular disulfide bonds. The intermolecular disulfide bonds that exist in full-length XEEL can be mapped to Cys-24 and Cys-42, the only two cysteine residues missing from XEELCRD. Thus in XEEL, the intermolecular disulfide bond may be required for the formation of stable high-order hexamers but is not required for trimerization of the carbohydrate binding core. The two intelectin x-ray structures suggest that this twice observed trimer arrangement, which orients three ligand-binding sites on one face, is a basic unit. Other domains can be used to mediate its further assembly into higher order oligomers. Previous functional studies have explored the significance of intelectin oligomeric states by altering these N-terminal cysteine residues. It was concluded that the resulting protein was monomeric by denaturing SDS-PAGE (54, 55). This experimental design cannot reveal whether the protein forms a non-covalent oligomer. Our findings that XEELCRD exists as a non-covalent trimer highlight the utility of using additional techniques such as cross-linking, SAXS, equilibrium analytical ultracentrifugation, and crystal packing analysis to establish the oligomeric state of an intelectin.
Intermolecular disulfide bonds are found in several important proteins such as antibodies, hormones, and growth factors (56–58). Their presence can provide stability and is often crucial for biological function (59). For example, TGF-β3, in which the intermolecular disulfide bond was mutated, is monomeric and has diminished biological activity (58). It has been suggested that disulfide bonds are conserved once acquired through evolution and that the acquisition of disulfide bond is associated with organismal complexity (60). Whether this trend is observed for all intermolecular disulfide bonds is not clear, as we did not find a systematic evolutionary study relating the acquisition or loss of an intermolecular disulfide bond to protein oligomeric state. With regard to intelectins, mouse intelectins do not contain intermolecular disulfide bonds, but their oligomeric state is not yet known. Given our work on XEELCRD and the large sequence variation in the N terminus of intelectins, we suggest that biophysical characterization of the intelectin oligomeric state in solution, in combination with sequence analysis, could provide insight into intermolecular disulfide bond evolution.
The structure of XEELCRD presented here provided a basis to predict the structure of full-length XEEL. We hypothesize that the structure is a barbell-shaped dimer of trimers held together by the N-terminal helical bundle that was removed by proteolysis in our experimental system. Because the bundle would align the two carbohydrate binding faces in opposite orientations, we hypothesized that XEEL can agglutinate bacteria. This hypothesis is supported by the findings that XEEL agglutinates S. pneumoniae in a ligand- and calcium ion-dependent manner. We postulate that this agglutinating activity could be further optimized and utilized for pathogen detection.
In conclusion, the structure and ligand recognition mechanism of intelectins are unique among lectins. With structures of the XEELCRD and hIntL-1, features conserved between intelectins can be identified. Our results also lay a foundation for further structural analysis of the intelectin family, which will provide insight into the biological function of individual intelectins. Because both XEELCRD and hIntL-1 bind microbe-specific carbohydrate residues, these intelectin structures provide blueprints for engineering intelectin variants for microbe recognition and targeting.
Author Contributions
K. W., K. T. F., and L. L. K. conceived the study and wrote the paper. K. W. generated the expression constructs and recombinant baculoviruses, and K. W. and D. A. W. purified and crystallized XEEL. K. W. and K. T. F. determined the structure. D. A. W. conducted SPR analysis of XEEL ligands. K. W. analyzed SAXS data and performed the cross-linking experiments and the agglutination assays. J. A. V. conducted the Western blot analysis depicted in Fig. 1. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
For access to equipment and technical assistance, we gratefully acknowledge E. Kondrashkina (LS-CAT), M. M. Benning (Bruker AXS, Madison, WI), D. R. McCaslin (University of Wisconsin (UW)-Madison, Biophysics Instrumentation Facility (BIF)), and G. Sabat, M. Rajendarm, G. Auer, L. Zarling, and D. B. Weibel (UW-Madison). We thank R. A. Brown and A. W. Baker (UW-Madison) for critical feedback. This research used the resources of the Advanced Photon Source, a United States Dept. of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of the Stanford Synchrotron Radiation Lightsource (SSRL) (SLAC National Accelerator Laboratory) is supported by the DOE Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by National Institutes of Health Grant P41GM103393 from NIGMS. SPR and analytical ultracentrifugation experiments were conducted at the UW-Madison Biophysics Instrumentation Facility supported by UW-Madison, National Science Foundation Grant BIR-9512577, and National Institutes of Health Grant S10 RR13790.
This work was supported by National Institutes of Health Grants R01GM55984 and R01AI063596 (to L. L. K.) and Chemistry-Biology Interface Training Grant T32 GM008505 (to D. A. W.) and by National Science Foundation Grants IOS1353674 (to K. T. F.) and DGE-1256259 (to D. A. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 4WMO and 4WN0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- XEEL
- Xenopus laevis embryonic epidermal lectin
- hIntL
- human intelectin
- Galf
- galactofuranose
- GroP
- d-glycerol-1-phosphate
- FBD
- fibrinogen-like domain
- CRD
- carbohydrate recognition domain
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SPR
- surface plasmon resonance
- r.m.s.d.
- root mean square deviation
- SAXS
- small angle x-ray scattering
- PBD
- Protein Data Bank.
References
- 1. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., and Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 2. Holmskov U., Thiel S., and Jensenius J. C. (2003) Collections and ficolins: humoral lectins of the innate immune defense. Annu. Rev. Immunol. 21, 547–578 [DOI] [PubMed] [Google Scholar]
- 3. Fujita T., Matsushita M., and Endo Y. (2004) The lectin-complement pathway–its role in innate immunity and evolution. Immunol. Rev. 198, 185–202 [DOI] [PubMed] [Google Scholar]
- 4. Lee J. K., Baum L. G., Moremen K., and Pierce M. (2004) The X-lectins: a new family with homology to the Xenopus laevis oocyte lectin XL-35. Glycoconj. J. 21, 443–450 [DOI] [PubMed] [Google Scholar]
- 5. Tsuji S., Uehori J., Matsumoto M., Suzuki Y., Matsuhisa A., Toyoshima K., and Seya T. (2001) Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J. Biol. Chem. 276, 23456–23463 [DOI] [PubMed] [Google Scholar]
- 6. Yan J., Xu L., Zhang Y., Zhang C., Zhang C., Zhao F., and Feng L. (2013) Comparative genomic and phylogenetic analyses of the intelectin gene family: implications for their origin and evolution. Dev. Comp. Immunol. 41, 189–199 [DOI] [PubMed] [Google Scholar]
- 7. Abe Y., Tokuda M., Ishimoto R., Azumi K., and Yokosawa H. (1999) A unique primary structure, cDNA cloning and function of a galactose-specific lectin from ascidian plasma. Eur. J. Biochem. 261, 33–39 [DOI] [PubMed] [Google Scholar]
- 8. Yan J., Wang J., Zhao Y., Zhang J., Bai C., Zhang C., Zhang C., Li K., Zhang H., Du X., and Feng L. (2012) Identification of an amphioxus intelectin homolog that preferably agglutinates Gram-positive over Gram-negative bacteria likely due to different binding capacity to LPS and PGN. Fish Shellfish Immunol. 33, 11–20 [DOI] [PubMed] [Google Scholar]
- 9. Yan J., Zhang C., Zhang Y., Li K., Xu L., Guo L., Kong Y., and Feng L. (2013) Characterization and comparative analyses of two amphioxus intelectins involved in the innate immune response. Fish Shellfish Immunol. 34, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 10. Lin B., Cao Z., Su P., Zhang H., Li M., Lin Y., Zhao D., Shen Y., Jing C., Chen S., and Xu A. (2009) Characterization and comparative analyses of zebrafish intelectins: highly conserved sequences, diversified structures and functions. Fish Shellfish Immunol. 26, 396–405 [DOI] [PubMed] [Google Scholar]
- 11. Russell S., Hayes M. A., and Lumsden J. S. (2009) Immunohistochemical localization of rainbow trout ladderlectin and intelectin in healthy and infected rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 26, 154–163 [DOI] [PubMed] [Google Scholar]
- 12. Russell S., Young K. M., Smith M., Hayes M. A., and Lumsden J. S. (2008) Identification, cloning and tissue localization of a rainbow trout (Oncorhynchus mykiss) intelectin-like protein that binds bacteria and chitin. Fish Shellfish Immunol. 25, 91–105 [DOI] [PubMed] [Google Scholar]
- 13. Datta R., deSchoolmeester M. L., Hedeler C., Paton N. W., Brass A. M., and Else K. J. (2005) Identification of novel genes in intestinal tissue that are regulated after infection with an intestinal nematode parasite. Infect. Immun. 73, 4025–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. French A. T., Knight P. A., Smith W. D., Brown J. K., Craig N. M., Pate J. A., Miller H. R., and Pemberton A. D. (2008) Up-regulation of intelectin in sheep after infection with Teladorsagia circumcincta. Int. J. Parasitol. 38, 467–475 [DOI] [PubMed] [Google Scholar]
- 15. Pemberton A. D., Knight P. A., Gamble J., Colledge W. H., Lee J. K., Pierce M., and Miller H. R. (2004) Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J. Immunol. 173, 1894–1901 [DOI] [PubMed] [Google Scholar]
- 16. Roberson M. M., and Barondes S. H. (1982) Lectin from embryos and oocytes of Xenopus laevis: purification and properties. J. Biol. Chem. 257, 7520–7524 [PubMed] [Google Scholar]
- 17. Nishihara T., Wyrick R. E., Working P. K., Chen Y. H., and Hedrick J. L. (1986) Isolation and characterization of a lectin from the cortical granules of Xenopus laevis eggs. Biochemistry 25, 6013–6020 [DOI] [PubMed] [Google Scholar]
- 18. Lee J. K., Buckhaults P., Wilkes C., Teilhet M., King M. L., Moremen K. W., and Pierce M. (1997) Cloning and expression of a Xenopus laevis oocyte lectin and characterization of its mRNA levels during early development. Glycobiology 7, 367–372 [DOI] [PubMed] [Google Scholar]
- 19. Shoji H., Ikenaka K., Nakakita S., Hayama K., Hirabayashi J., Arata Y., Kasai K., Nishi N., and Nakamura T. (2005) Xenopus galectin-VIIa binds N-glycans of members of the cortical granule lectin family (xCGL and xCGL2). Glycobiology 15, 709–720 [DOI] [PubMed] [Google Scholar]
- 20. Ishino T., Kunieda T., Natori S., Sekimizu K., and Kubo T. (2007) Identification of novel members of the Xenopus Ca2+ -dependent lectin family and analysis of their gene expression during tail regeneration and development. J. Biochem. 141, 479–488 [DOI] [PubMed] [Google Scholar]
- 21. Nagata S., Nishiyama S., and Ikazaki Y. (2013) Bacterial lipopolysaccharides stimulate production of XCL1, a calcium-dependent lipopolysaccharide-binding serum lectin, in Xenopus laevis. Dev. Comp. Immunol. 40, 94–102 [DOI] [PubMed] [Google Scholar]
- 22. Nagata S. (2005) Isolation, characterization, and extra-embryonic secretion of the Xenopus laevis embryonic epidermal lectin, XEEL. Glycobiology 15, 281–290 [DOI] [PubMed] [Google Scholar]
- 23. Nagata S., Nakanishi M., Nanba R., and Fujita N. (2003) Developmental expression of XEEL, a novel molecule of the Xenopus oocyte cortical granule lectin family. Dev. Genes Evol. 213, 368–370 [DOI] [PubMed] [Google Scholar]
- 24. Dubaissi E., Rousseau K., Lea R., Soto X., Nardeosingh S., Schweickert A., Amaya E., Thornton D. J., and Papalopulu N. (2014) A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development 141, 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J. K., Schnee J., Pang M., Wolfert M., Baum L. G., Moremen K. W., and Pierce M. (2001) Human homologs of the Xenopus oocyte cortical granule lectin XL35. Glycobiology 11, 65–73 [DOI] [PubMed] [Google Scholar]
- 26. Wesener D. A., Wangkanont K., McBride R., Song X., Kraft M. B., Hodges H. L., Zarling L. C., Splain R. A., Smith D. F., Cummings R. D., Paulson J. C., Forest K. T., and Kiessling L. L. (2015) Recognition of microbial glycan epitopes by human intelectin. Nat. Struct. Mol. Biol. 22, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weis W. I., Taylor M. E., and Drickamer K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev. 163, 19–34 [DOI] [PubMed] [Google Scholar]
- 28. Fullerton S. W., Daff S., Sanders D. A., Ingledew W. J., Whitfield C., Chapman S. K., and Naismith J. H. (2003) Potentiometric analysis of UDP-galactopyranose mutase: stabilization of the flavosemiquinone by substrate. Biochemistry 42, 2104–2109 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode, in Macromolecular Crystallography, Part A (Carter C. W. Jr., and Sweet R. M., eds) pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 30. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Afonine P. V., Grosse-Kunstleve R. W., and Adams P. D. (2005) The Phenix refinement framework. CCP4 Newsletter 42, 8 [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schüttelkopf A. W., and van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 35. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeLano W. L. (year?) The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC, New York [Google Scholar]
- 37. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 38. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 40. Svergun D. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 41. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 42. Laue T. M., Shah B. D., Ridgeway T. M., and Pelletier S. L. (1992) Computer-aided interpretation of analytical sedimentation data for proteins, in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., and Horton J. C., eds) pp. 90–125, Royal Society of Chemistry, Cambridge [Google Scholar]
- 43. Laue T. M. (1995) Sedimentation equilibrium as thermodynamic tool. Methods Enzymol. 259, 427–452 [DOI] [PubMed] [Google Scholar]
- 44. Käll L., Krogh A., and Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction: the Phobius Web server. Nucleic Acids Res. 35, W429–W432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmidt T. G., and Skerra A. (2007) The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535 [DOI] [PubMed] [Google Scholar]
- 46. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 47. Garlatti V., Belloy N., Martin L., Lacroix M., Matsushita M., Endo Y., Fujita T., Fontecilla-Camps J. C., Arlaud G. J., Thielens N. M., and Gaboriaud C. (2007) Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 26, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mooij W. T., Mitsiki E., and Perrakis A. (2009) ProteinCCD: enabling the design of protein truncation constructs for expression and crystallization experiments. Nucleic Acids Res. 37, W402–W405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., and Drickamer K. (2004) Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11, 591–598 [DOI] [PubMed] [Google Scholar]
- 51. Feinberg H., Powlesland A. S., Taylor M. E., and Weis W. I. (2010) Trimeric structure of langerin. J. Biol. Chem. 285, 13285–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Asensio J. L., Ardá A., Cañada F. J., and Jiménez-Barbero J. (2013) Carbohydrate-aromatic interactions. Acc. Chem. Res. 46, 946–954 [DOI] [PubMed] [Google Scholar]
- 53. Hudson K. L., Bartlett G. J., Diehl R. C., Agirre J., Gallagher T., Kiessling L. L., and Woolfson D. N. (2015) Carbohydrate-aromatic interactions in proteins. J. Am. Chem. Soc. 137, 15152–15160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsuji S., Yamashita M., Hoffman D. R., Nishiyama A., Shinohara T., Ohtsu T., and Shibata Y. (2009) Capture of heat-killed Mycobacterium bovis bacillus Calmette-Guerin by intelectin-1 deposited on cell surfaces. Glycobiology 19, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsuji S., Yamashita M., Nishiyama A., Shinohara T., Li Z., Myrvik Q. N., Hoffman D. R., Henriksen R. A., and Shibata Y. (2007) Differential structure and activity between human and mouse intelectin-1: human intelectin-1 is a disulfide-linked trimer, whereas mouse homologue is a monomer. Glycobiology 17, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 56. Röthlisberger D., Honegger A., and Plückthun A. (2005) Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J. Mol. Biol. 347, 773–789 [DOI] [PubMed] [Google Scholar]
- 57. Chang S. G., Choi K. D., Jang S. H., and Shin H. C. (2003) Role of disulfide bonds in the structure and activity of human insulin. Mol. Cells 16, 323–330 [PubMed] [Google Scholar]
- 58. Zúñiga J. E., Groppe J. C., Cui Y., Hinck C. S., Contreras-Shannon V., Pakhomova O. N., Yang J., Tang Y., Mendoza V., López-Casillas F., Sun L., and Hinck A. P. (2005) Assembly of TbetaRI:TβRII:TGFβ ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J. Mol. Biol. 354, 1052–1068 [DOI] [PubMed] [Google Scholar]
- 59. Fass D. (2012) Disulfide bonding in protein biophysics. Annu. Rev. Biophys. 41, 63–79 [DOI] [PubMed] [Google Scholar]
- 60. Wong J. W., Ho S. Y., and Hogg P. J. (2011) Disulfide bond acquisition through eukaryotic protein evolution. Mol. Biol. Evol. 28, 327–334 [DOI] [PubMed] [Google Scholar]
- 61. Stivala A., Wybrow M., Wirth A., Whisstock J. C., and Stuckey P. J. (2011) Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics 27, 3315–3316 [DOI] [PubMed] [Google Scholar]
- 62. Malashkevich V. N., Chan D. C., Chutkowski C. T., and Kim P. S. (1998) Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. U.S.A. 95, 9134–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bentley S. D., Aanensen D. M., Mavroidi A., Saunders D., Rabbinowitsch E., Collins M., Donohoe K., Harris D., Murphy L., Quail M. A., Samuel G., Skovsted I. C., Kaltoft M. S., Barrell B., Reeves P. R., Parkhill J., and Spratt B. G. (2006) Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]