Abstract
The voltage-gated H+ channel (Hv) is a voltage sensor domain-like protein consisting of four transmembrane segments (S1–S4). The native Hv structure is a homodimer, with the two channel subunits functioning cooperatively. Here we show that the two voltage sensor S4 helices within the dimer directly cooperate via a π-stacking interaction between Trp residues at the middle of each segment. Scanning mutagenesis showed that Trp situated around the original position provides the slow gating kinetics characteristic of the dimer's cooperativity. Analyses of the Trp mutation on the dimeric and monomeric channel backgrounds and analyses with tandem channel constructs suggested that the two Trp residues within the dimer are functionally coupled during Hv deactivation but are less so during activation. Molecular dynamics simulation also showed direct π-stacking of the two Trp residues. These results provide new insight into the cooperative function of voltage-gated channels, where adjacent voltage sensor helices make direct physical contact and work as a single unit according to the gating process.
Keywords: ion channel, membrane biophysics, oocyte, proton transport, structure-function, channel gating, voltage-gated channel
Introduction
Voltage-gated ion channels sense the membrane potential and gate ion permeation to generate electronic signals in many cell types (1). Classical voltage-gated ion channels function as tetramers or pseudotetramers and feature four voltage sensors on the periphery and a pore in the center. Although the voltage sensors do not have direct contact with each other due to the spatially separated arrangement, the functional cooperativity arising from multimerization appears in the channel activation. Activation of all four voltage sensors together triggers the pore to open, whereas deactivating any one of the voltages sensors induces the pore to close (2). Full participation of the voltage sensors in the channel activation strengthens the voltage dependence and enhances its sensitivity to electrical signals in excitable cells (2).
The voltage-gated H+ channel (Hv)2 is a voltage sensor only protein consisting of four transmembrane domains (S1–S4) that correspond to the voltage sensor of voltage-gated channels (3, 4). The Hv passes H+ and forms a primary pathway for transmembrane H+ conduction in phagocytes and human sperm cells (5–7). The S4 helix contains positively charged residues and senses the voltage change across the membrane, as the voltage sensor does in the other voltage-gated channels (8). The functional unit of Hv is uniquely a dimer (9–11). The cytoplasmic coiled-coil domain just downstream of the S4 segment mediates the dimerization (12, 13), suggesting that the two voltage sensors are proximally positioned. It is known that, in the dimeric Hv, each channel protomer cooperates with each other during gating (13–16). This cooperativity enhances the voltage dependence of Hv activation and slows the gating kinetics, which works well for production of reactive oxygen species in phagocytes (7, 12, 13, 16). It has been proposed that within the dimer, the coiled-coil domain defines the configuration of the transmembrane helices (12, 13, 17) and modifies the functions of the two closely situated S4 helices during the channel activation (18). Besides this, there are various models for the relative positioning of the transmembrane segments of the two channel subunits (10, 15) and various scenarios for the cooperative function of the voltage-dependent gating (8, 14, 19). The structural mechanism of how the dimerization controls the gating, directly or indirectly involving the S4 helices, has been unknown.
Within voltage-gated channels, the S4 helix is considered to be the voltage sensor and to move up and down with rotation, following the membrane potential, thereby inducing a conformational change that opens or closes the channel (20–22). The amino acid sequence of S4 is well conserved in Hv, but unlike other voltage-gated channels, it contains a Trp residue at the middle of the S4 sequence that is 100% conserved among Hv species (Fig. 1A). Trp is a bulky aromatic residue that tends to be situated at membrane-water interfaces due to its amphipathic nature and the favorable interaction between its aromatic ring and lipid headgroups (23, 24). Thus, Trp situated at the middle of the S4 transmembrane segment in Hv is noteworthy. Furthermore, the bulky side chain of Trp should be less well tolerated at positions that are involved in tight protein-protein interactions (25, 26), which raises the question of why a Trp is situated in a region of close S4-S4 interaction within the dimeric channel. Moreover, the adjacency of the parallel S4 dipoles would seem to be energetically incompatible within a protein structure. Until now, however, these questions have not attracted much attention. Nonetheless, the crystal structure of the transmembrane region was, in fact, solved at low resolution and, unexpectedly, in a trimer (17), which provides less information about the function of Trp and the true structure than would the native dimeric channel. In the present study, therefore, to understand the functional and structural significance of the uniquely conserved Trp, we performed the electrophysiological analysis combined with the molecular dynamics simulation of WT and Trp mutants within Hv monomers and dimers. Here we show that the Trp residues form a direct π-stacking within a dimer only when the channel is closing and produce the characteristic slow deactivation phase observed in the native Hv.
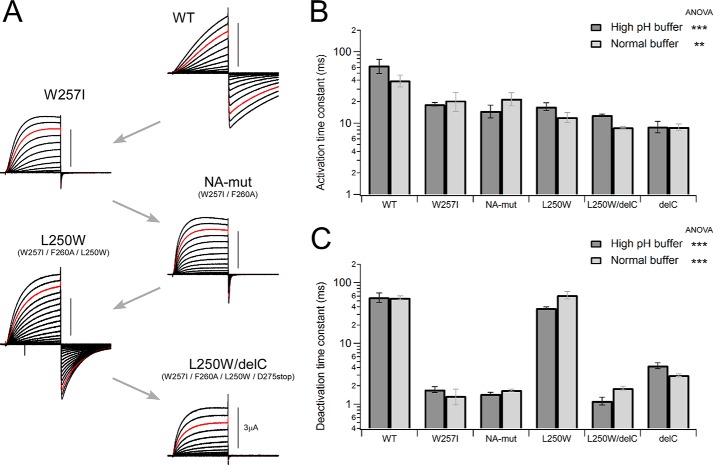
FIGURE 1.
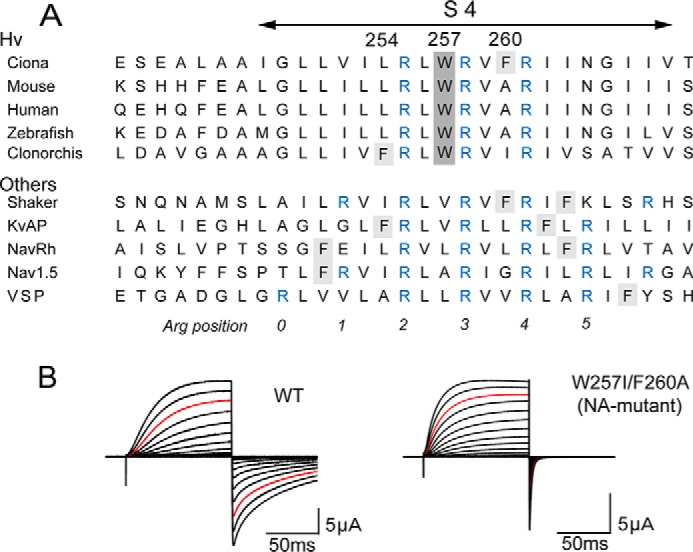
Conserved Trp residue within the S4 segment. A, alignment of the amino acid sequences of the S4 segment in the Hv family and other voltage-gated channels. The conserved Trp at the middle of S4 is highlighted in gray, and another aromatic residue, Phe, is highlighted in light gray. Conserved Arg, a voltage sensor, is highlighted in blue. B, representative current traces recorded from wild-type Hv (WT) and the NA-mutant (W257I/F260A). Currents were recorded with step pulses in the range of −60 to +120 mV for 100 ms with 10-mV increments. Red traces depict the currents recorded at +100 mV.
Experimental Procedures
In Vitro Mutagenesis and cRNA Synthesis
The original Ciona Hv1/VSOP channel cDNA was subcloned into pSD64TF vector. The original mouse Hv1/VSOP channel cDNA was subcloned into pIRES-EGFP vector. Single and double point mutants were made using a site-directed mutagenesis protocol and confirmed by DNA sequencing. To make the C terminus coiled-coil domain deletion constructs, a D275stop point mutation of Ciona Hv and a V216stop point mutation of mouse Hv were introduced, respectively; these make the channel monomeric (12, 13). To make the tandem channels, two subunits were linked with a flexible 19-amino acid linker, GGSGGSGGSGSGGSGGSGG, as reported previously (13). The coiled-coil assembly domain remains intact in a tandem channel, suggesting that the tandem channels form the same structure as the native dimeric channel. cRNAs encoding WT and mutant channels were prepared from linearized plasmid cDNA using an RNA transcription kit (Life Technologies Japan). In Fig. 1A, the accession numbers used for the alignments are as follows: Ciona Hv (NCBI Reference Sequence: NW_004190496.1), mouse Hv (NC000071), human Hv (NC000012), zebrafish Hv (NC007121), Clonorchis Hv (GAA49234.1), shaker (AAA28417.1), KvAP (BAA79939.1), NavRh (WP_009373403), Nav1.5 (NC-000003.12), and VSP (BAD98733.1).
Preparation of Xenopus Oocytes
Xenopus oocytes were collected from frogs anesthetized using tricaine. The isolated oocytes were treated with collagenase (0.5–1.0 mg/ml, type 1; Sigma) and then injected with 50 nl of cRNA solution. The injected oocytes were incubated for 3–5 days at 18 °C in ND96 frog Ringer solution. All experiments conformed to the guidelines of the Animal Experiments Committee of Osaka University.
Two-electrode Voltage Clamp Recordings
Macroscopic currents were recorded from Xenopus oocytes expressing Ciona Hv using the two-electrode voltage clamp technique with a bath clamp amplifier (OC-725C, Warner Co.), a digital/analog converter (Digidata 1440A, Molecular Devices), and recording software (pCalamp version 10, Molecular Devices). All recordings were made at room temperature (20 °C). Intracellular glass microelectrodes were filled with 3 m potassium acetate with 10 mm KCl (pH 7.2), and their electric resistances ranged from 0.1 to 0.4 megaohms. Two Ag-AgCl pellets (E205, Warner Co.) were used as bath electrodes for voltage sensing and current passing. The standard recording bath solution contained 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES (pH 7.15). In recordings under the high pH buffer condition (Fig. 2), we injected 50 nl of 1 m HEPES (pH 7.30) into each oocyte to prevent pHi change (8, 16) and recorded H+ currents with the bath solution containing 60 mm NaCl, 2 mm CaCl2, 1 mm MgCl2, and 120 mm HEPES (pH 7.30). Only data presenting small amplitudes of the H+ current (<5 μA) were used for analyses (Fig. 2).
FIGURE 2.
Effects of mutations on the gating kinetics. A, representative current traces of WT Hv and the mutant channels are shown. Recordings were performed under a high pH buffer condition (see “Experimental Procedures”). The gating kinetics was changed with respect to each mutation. B and C, comparison of the activation (B) and deactivation (C) time constants between two different recording buffer conditions for each channel, showing no condition dependence but phenotypes by mutation. Bars, means ± S.E. (error bars) (n = 3–5). Data in each solution were statistically analyzed using ANOVA (***, p ≤ 0.001; **, p ≤ 0.01).
All data were recorded by applying a set of step pulses using P/4 subtraction protocols. Cells were held at −60 mV, and voltage pulses were applied in the range of −60 to +120 mV for 100 ms with 10-mV increments. Actual clamped membrane potentials were monitored during current recordings, and data with an error of over 5 mV from the command potential were discarded. The activation time constant was obtained by fitting the activation phase of the outward currents upon depolarization to 100 mV, whereas the deactivation time constant was obtained by fitting the tail currents upon repolarization to −60 mV. Activation thresholds were evaluated by measuring the voltage at which a detectable tail current was elicited.
Patch Clamp Recordings
The cDNAs of mouse Hv for the WT and mutant channel constructs (Figs. 7 and 8) were transfected into HEK293T cells (human embryonic kidney cell line). The mouse Hv clone does not function well in oocytes but functions well in HEK293T cells. Electrophysiological recordings were carried out 18–30 h after transfection, and the transfected cells were identified by the fluorescent signal from GFP. Macroscopic currents were recorded in the whole-cell clamp configuration using an Axopatch-200B amplifier (Molecular Devices). The pipette resistance in the solution was 3–5 megaohms. 60–80% of the voltage error due to the series resistance was compensated for by a circuit in the amplifier. The external solution contained 75 mm N-methyl-d-glucamine, 1 mm CaCl2, 1 mm MgCl2, 10 mm glucose, and 180 mm HEPES (pH 7.0). The pipette solution contained 65 mm N-methyl-d-glucamine, 3 mm MgCl2, 1 mm EGTA, and 183 mm HEPES (pH 7.0). The recording temperature was controlled at 25 °C. Data were recorded by applying a set of step pulses without using subtraction protocols. Cells were held at −60 mV, and voltage pulses were applied in the range of −60 to +120 mV for 1 s with 10-mV increments. The activation time constant was obtained by fitting the activation phase of the outward currents upon depolarization to 100 mV, whereas the deactivation time constant was obtained by fitting the tail currents upon repolarization to −60 mV (Figs. 7 and 8). In the analysis of the limiting slope (Fig. 8, A and B), cells were held at −60 mV, and the slow ramp pulse (+1 mV/1 s) was applied in the range around the threshold, as reported previously (13). Valences of the effective charge movement (Zδ) of the gating were calculated by linear fitting of the chord conductance.
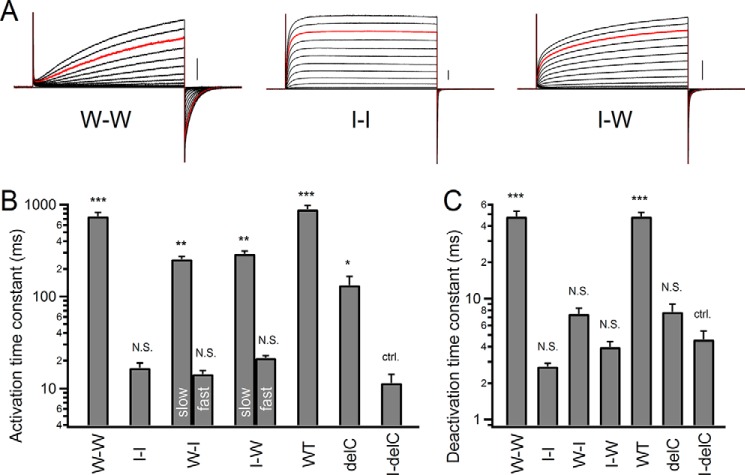
FIGURE 7.
Kinetic analysis for tandem dimers. A, representative current traces of tandem dimers. Currents were recorded with step pulses in the range of −60 to +120 mV for 1 s with 10-mV increments. Red traces, currents recorded at +100 mV. Scale bars, 100 pA in each mutant. B and C, comparison of the activation (B) and deactivation (C) time constants of the tandem constructs and the delC channel. Bars, means ± S.E. (error bars) (n = 5–6). Means in each mutant were compared statistically with the I-delC mutant using the Tukey-Kramer test (***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05; N.S., p > 0.05; ctrl., control). Two exponential components of the activation kinetics were analyzed in the I-W and W-I tandem dimers. We note that the deactivation kinetics in the I-I, I-W, W-I, delC, and I-delC (W203I delC) constructs were so fast that two components were inseparable.
FIGURE 8.
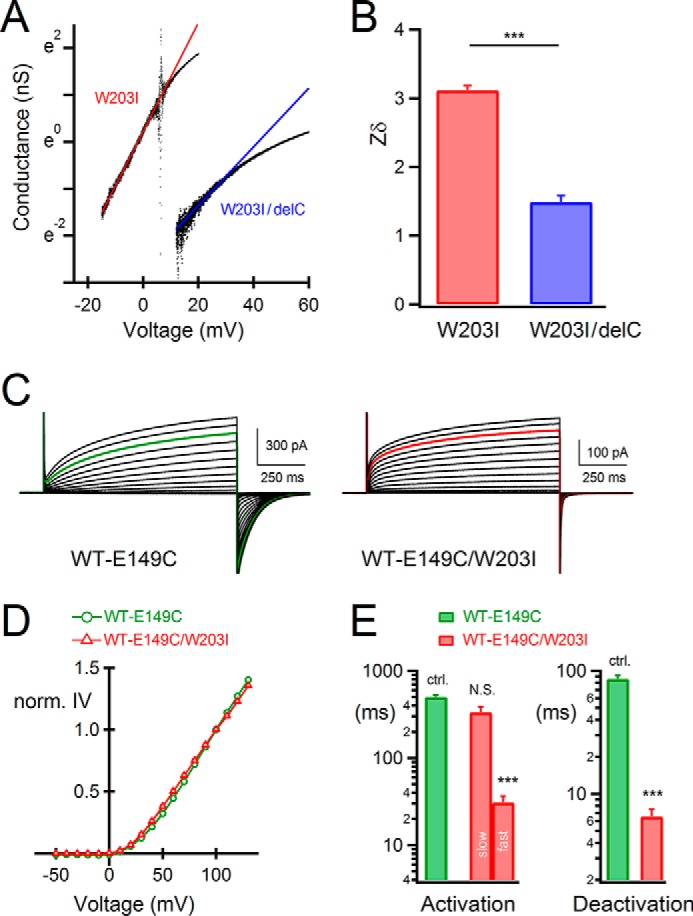
Analysis of the steady state cooperative gating in W203I. A, representative chord conductances and linear fitting lines of the W203I mutants. The small amplitudes of whole-cell currents in the range around threshold were recorded with slow ramp pulse protocols, and the chord conductances were obtained. B, the value of the effective gating charge (Zδ). Bars, means ± S.E. (error bars) (n = 5–6); means were compared statistically using Student's t test (***, p ≤ 0.001). C, representative current traces of tandem dimers. D, normalized current-voltage relationships of the tandem dimers. Current amplitudes were normalized at +100 mV. Bars, means ± S.E. (n = 7); errors are small and hidden behind the symbols. E, comparison of the activation (left) and deactivation (right) time constants of the tandem constructs. Bars, means ± S.E. (n = 7); means were compared statistically with data of the WT-E149C tandem dimer using the Tukey-Kramer test (***, p ≤ 0.001; NS, p > 0.05; ctrl., control). We analyzed the activation of the WT-E149C tandem dimer by fitting with a single-exponential function. Two-exponential components of the activation kinetics were analyzed in the WT-E149C/W203I tandem dimer.
Data Analysis
Activation/deactivation phases of the recorded currents were fitted with a single exponential function,
where τ is the time constant of activation/deactivation, and A and C are constants. With the I257W mutation of the non-aromatic mutant (NA-mutant), deactivation appeared to follow a biexponential time course, and the combined kinetics (τcomb) were calculated using a double exponential function.
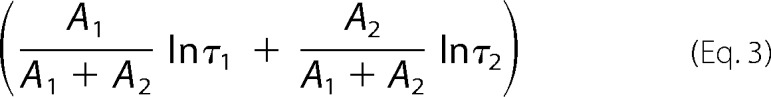 |
To evaluate the recovery of deactivation kinetics in the Trp scanning mutagenesis (Fig. 4B), we calculated the relative recovery ratio in each mutant using the logarithm of mean time constant on the basis of WT (as 100%) and the NA-mutant (0%).
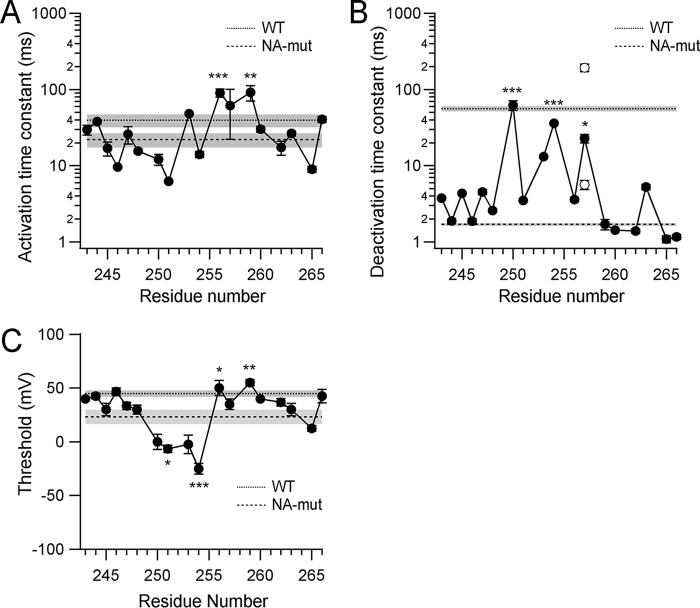
FIGURE 4.
Analysis of data from the Trp scanning mutagenesis. A and B, the activation phases (A) and the deactivation tail currents (B) were fitted with a single exponential function, and the time constants of the fittings are plotted. Two open circles for the I257W mutant depict the fast and the slow components of the deactivation kinetics fitted using double exponential functions. Bars, means ± S.E. (error bars) (n = 3–8). A dotted line represents the mean time constant (n = 4) obtained from WT with the S.E. (gray shadow), and a dashed line with a gray shadow represents data from the NA-mutant (n = 3). C and D, weighted perturbation energy (ΔΔGw) of activation (C) and deactivation (D). To quantitatively analyze the effect of Trp introduction on the kinetics, we analyzed the perturbation energy (ΔΔG) between the Trp mutants and the NA-mutant at each position, and to determine whether the effects of Trp introduction were biased by the change in side chain volume, we calculated ΔΔGw (see “Experimental Procedures”). E, thresholds were measured and plotted, and bars depict means ± S.E. (n = 3–4). Dotted (WT) and dashed (NA-mutant) lines with gray shadows depict the data as in A. Data were significantly different with ANOVA, p ≤ 0.001. Mean values in each mutant were compared statistically with the NA-mutant using the Tukey-Kramer test (***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05).
To quantify the effect of each mutant on the rate of activation/deactivation, we calculated a perturbation energy (27),
where R is the gas constant and T is the absolute temperature. τmut and τcont are kinetic parameters obtained from the specific mutant of interest and the NA-mutant, respectively.
To evaluate the structural coupling between two Trp in each channel protomer, we applied the method of mutant cycle analysis. The apparent coupling free energy (ΔΔGint) was calculated (28, 29),
where ΔΔG2mer is the apparent free energy difference in the perturbation energy between the monomer and dimer, and ΔΔGTrp is the apparent energy difference caused by the introduction of Trp. If the two Trp are independent within the dimeric channel, ΔΔGint = 0, but if they are coupled/interacting, then ΔΔGint ≠ 0. Absolute coupling energy greater than ∼0.89 kcal/mol represents a significant interaction between the two residues (29).
Data were analyzed using Clampfit (Molecular Devices), Igor Pro (WaveMetrics Inc.), and Excel (Microsoft Corp.) software. Experimental data are shown as means ± S.E., whereas processed data, such as ΔΔG, are shown as difference values between two averaged data sets without deviations. In all data sets, the means of two groups were statistically compared using Student's t test, whereas pairs of means among three or more groups were compared using the Tukey-Kramer test, and the difference among the means of multiple groups was tested using one-way ANOVA (supplemental Table 1).
Structure Modeling
A dimeric Hv structure model was built based on two crystal structures. One is the crystal structure of the cytoplasmic coiled-coil domain of mouse Hv (Protein Data Bank entry 3VMX), showing a natural dimeric form that mediates dimeric assembly of the channel (13). The other is the crystal structure of the nominal full-length Hv (Protein Data Bank entry 3WKV) that consists of the transmembrane region of a mouse-Ciona chimeric Hv and the cytoplasmic region of a yeast coiled-coil (17). This full-length structure forms an unnatural trimer in crystal (17), but it shows that the coiled-coil domain and the transmembrane S4 segment form a continuous helix and that S4 helices constitute an interaction interface between channel protomers, which is consistent with a recent cross-linking analysis of the native mouse Hv dimer (18). Because multimerization of Hv is reportedly determined by the oligomerization state of the coiled-coil domain (12), we constructed a dimer model by connecting the transmembrane structure of the full-length Hv (Protein Data Bank entry 3WKV) with the cytoplasmic assembly foundation of the dimeric coiled-coil of mouse Hv (Protein Data Bank entry 3VMX) (13, 17). Superimposition of the coiled-coil regions of the full-length chimeric Hv (Ile224–Leu241) and mouse Hv (Ile224–Leu241) was performed using the superpose program within COOT. The resultant dimer model was used for the molecular dynamics simulation.
Molecular Dynamics Simulation
We have complemented invisible regions in the dimeric Hv model using the MODELLER program (30). The symmetric axis of the dimer was set to the z axis. Adequate hydrogen atoms were added under a neutral pH condition. The coiled-coil region was discarded in consideration of the calculation cost. The dimeric channel was set to be in the center of a pre-equilibrated lipid bilayer consisting of 320 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine lipid molecules under periodic boundary conditions in the x-y plane, and overlapping lipid molecules were removed. Chemical bonds associated with hydrogen atoms were constrained using the LINCS algorithm (31). The channel and lipid bilayer energy was minimized to remove hard contacts among atoms. Equilibrated waters were added to the upper (+z) and lower (−z) sides of the lipid bilayer. To electrostatically neutralize the system, sodium and chloride ions were replaced with water to achieve an ionic strength of 150 mm. The resulting simulation system included about 80,000 atoms, and the size of the periodic box (x × y × z) = (96 × 96 × 89 Å). After energy minimization, the system was equilibrated using NVT with a Nose-Hoover thermostat at 310 K (32). The system was further equilibrated using NPT simulations with the Parrinello-Rahman method (33). In the equilibration, restraining forces were imposed on the heavy atoms of the proteins. The simulations were integrated using the leapfrog method with a time step of 2 fs. Electrostatic interactions were calculated using the particle mesh Ewald method (34) with a real-part cut-off of 12 Å, and the cut-off radius for van der Waals interactions was set to 12 Å. After the equilibrations, a long production run (NPT) was performed with no restraints, saving the trajectory every 2 ps. All of the simulations were conducted using Gromacs version 4.5.5 (35). We also prepared and calculated the structure of a W257I mutant in the same manner. For the simulations, 200-ns trajectories were obtained and used for analysis. The residue numbers are based on the sequence of Ciona Hv.
Results
Unique Trp at the Middle of the S4 Helix
Hv is a voltage sensor domain-like protein that was cloned on the basis of a homology search for the voltage sensor sequence of other voltage-gated channels (3, 4). The amino acid sequence of Hv is well conserved among all Hv species and the voltage-gated channel family. Sequence alignments for S4, the voltage sensor helix, are shown in Fig. 1A. Within the sequence, the positively charged Arg residues at 3-residue intervals are known to promote voltage sensing and are well conserved among all voltage-gated channels (Fig. 1A, blue). We also noticed that a Trp at the middle of S4 is 100% conserved among Hv species but is absent from other voltage-gated channels (Fig. 1A). Another aromatic residue, Phe, also appears in Hv from Ciona and Clonorchis. We constructed a W257I/F260A mutant channel (NA-mutant) from the Ciona Hv and analyzed its electrophysiological properties. Representative current traces from the wild-type (WT) and mutant channels evoked by step pulses are shown in Fig. 1B. The activation kinetics were 1.8-fold accelerated in time constant by removal of the aromatic residues, and the deactivation kinetics were also accelerated to an even greater extent (33-fold decrease in time constant). Even a W257I single mutation also showed the same effects as those of the NA-mutant (Fig. 2). Given this obvious impact of the aromatic residues in S4 on Hv gating, we designed a set of experiments focusing on the function of the unique Trp residue within the S4 helix. In addition, we also recorded H+ currents of WT and the representative mutants in this study under conditions in which pH was more strictly controlled (19) (see “Experimental Procedures”), and the changes of kinetics by the mutations were reproducibly observed (Fig. 2). One-way ANOVA showed significant differences, suggesting that kinetics changes were caused by the mutations (Fig. 2).
Trp Scanning Mutagenesis
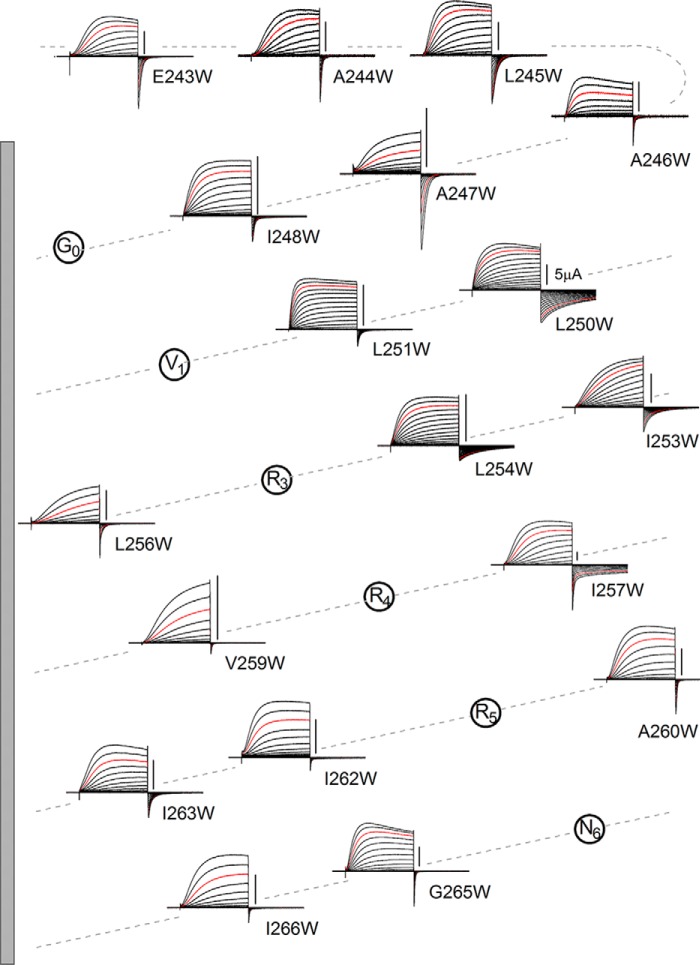
To determine whether the slow gating kinetics could be restored to the NA-mutant by introduction of Trp and the importance of its position in the sequence, we used a Trp scanning mutagenesis approach (27, 36–38) in which a Trp residue was introduced at different positions within and around the S4 segment in the NA-mutant. Two-electrode voltage clamp recordings with oocytes were used for the following high throughput analyses (Figs. 2–6). We made 18 Trp mutants, substituting residues, one-by-one, from the position of Glu243 in the S3-S4 linker region to Ile-266 at the end of S4. We did not substitute the Arg positions because mutation of the voltage sensor Arg reportedly changes the gating properties so significantly that they would probably mask the effects of Trp introduction (8, 39–42). Representative current traces recorded from the mutants are shown in a helical net diagram in Fig. 3, and analyzed data are plotted against the residue number in Fig. 4. The most noticeable feature of some of the mutants is their slow deactivation kinetics. We found that the introduction of Trp at position 257 (I257W) restored the slow deactivation kinetics of the native channel (Fig. 3). We also observed that L250W, I253W, and L254W mutations, all in the N-terminal half of the S4 helix, slowed the deactivation kinetics clearly (Figs. 3 and 4B). 74.2% (in I257W), 103.0% (L250W), and 87.3% (L254W) of recoveries in the logarithmic scale were counted (Fig. 4B), and they were statistically significant (supplemental Table 1). In addition, the deactivation kinetics of I257W were biphasic, and the relative weights of the fast and slow components were not voltage-dependent but constant (about 40% for the fast component; data not shown). We analyzed the total time constant for comparison using a double-exponential fitting (see “Experimental Procedures”). Given the time constants of I257W, the slow component would correspond to the slow deactivation in WT, whereas the fast one would correspond to the fast deactivation of the NA-mutant, which is discussed under “Direct Interaction within the Hv Dimer.”
FIGURE 3.
Trp scanning mutagenesis. Representative current traces for each mutant channel are arranged on a helical net diagram of the S4 region. Trp was introduced into the NA-mutant. Currents were recorded as in Fig. 1. Red traces depict the currents recorded at +100 mV. Scale bars, 5 μA in each mutant. The gray bar on the left depicts the S4 transmembrane region (residues 249–267).
FIGURE 5.
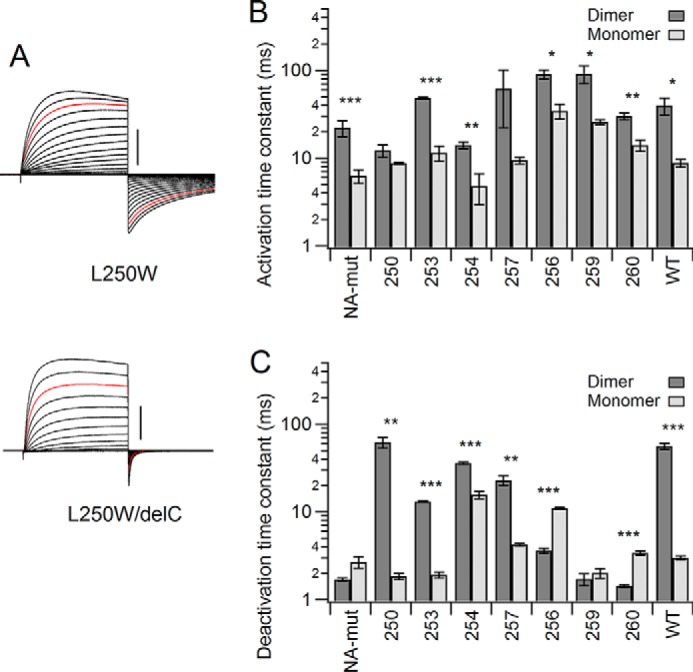
Comparison of the effects of introducing Trp into dimeric and monomeric Hv. A, representative changes in the activation/deactivation phases by deletion of the coiled-coil assembly domain (delC). Currents were recorded as in Fig. 1. Red traces, currents at +100 mV. Scale bars, 5 μA in each mutant. B and C, comparison of the activation (B) and deactivation (C) time constants between the monomer and dimer for each mutant. Bars, means ± S.E. (error bars) (n = 3–4). Means between dimer and monomer were compared statistically using Student's t test (***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05).
FIGURE 6.
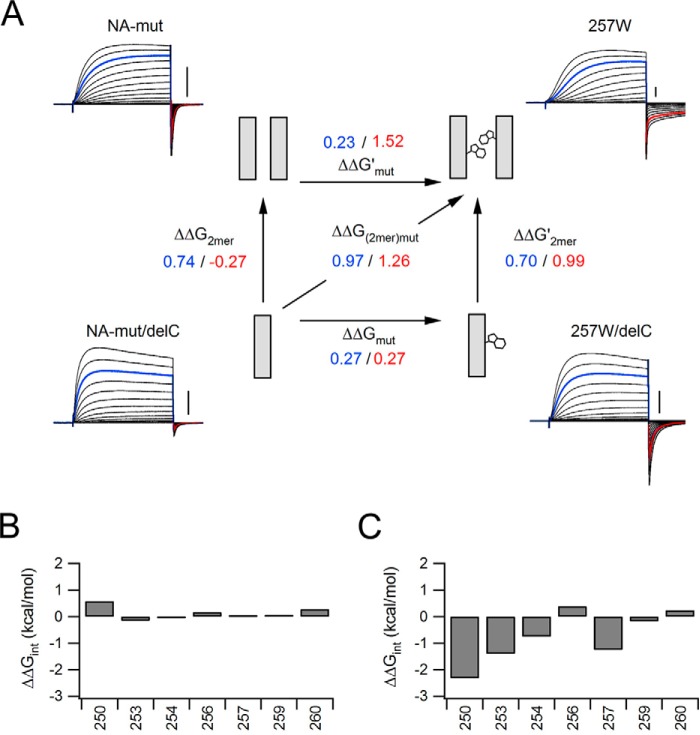
Schematic discussion of the Hv gating. A, Schematic illustration of the Trp257 introduction. The scheme shows the coupling energy between the monomer and dimer. The numbers indicate the ΔΔG (blue, activation; red, deactivation). B and C, interaction energy profiles for channel mutation. The criterion for two well correlated residues is a value greater than 0.89. Interaction energies for positions 250, 253, and 257 show strong interaction of the two Trp residues during the deactivation phase.
The mutations also affected the activation kinetics (p ≤ 0.001, ANOVA). Several of the mutations slowed the activation kinetics to some degree (e.g. L253W, L256W, V259W, and I266W), whereas some slightly accelerated the activation kinetics (A246W, L251W, and G265W) (Fig. 3 and supplemental Table 1). Overall, the data indicate that whereas the activation kinetics vary to a small degree throughout the scanning mutagenesis (Fig. 4A), the deactivation kinetics show obvious changes in the N-terminal half of S4 (Figs. 3 and 4B). It was difficult to accurately analyze the conductance-voltage (G-V) relationship for Hv due to the weak voltage dependence and high threshold potential, as compared with other voltage-gated channels. The local H+ accumulation also makes it impossible to accurately analyze steady state currents and the inverse tail current of large current amplitudes evoked by strong depolarization (7), because the pH change significantly shifts the G-V relationship and the reversal potential of H+ (7), which is one of the major characteristics of the Hv gating. Hence, we did not analyze the G-V relationship in this study, and we instead analyzed the activation threshold for each mutant to approximately estimate the effect of mutation on the gating transition. Introduction of Trp into the N-terminal half of S4 reduced the threshold, just as it affected the deactivation kinetics (Fig. 4E). Thus, Trp introduction in a scanning mutagenesis protocol showed the presence of a hot spot affecting channel gating at positions 250, 253, 254, and 257 in the N-terminal half of S4.
Effects of the Trp Introduction on the Monomeric Channel
Native Hv is known to be a dimer with subunits linked via the coiled-coil assembly domain in the cytoplasmic C terminus, deletion of which makes the channel monomeric (delC). Deletion of the C terminus also reportedly accelerates channel activation and deactivation, reflecting the lack of gating cooperativity between channel protomers (12, 13). We next assessed the impact of Trp introduction on gating of the monomeric channel. We analyzed L250W, I253W, L254W, and I257W as slowly deactivating mutants, L256W and V259W as slowly activating mutants, and A260W as an ineffective mutant (Fig. 5). The slow deactivation tail currents of L250W and I257W observed with the dimeric channel were not seen in the delC channel (Figs. 5A and 6). The accumulated data show that, overall, C terminus deletion slightly (but not significantly in some mutants) increases the activation rate in all mutants (Fig. 5B and supplemental Table 1), reflecting a lack of gating cooperativity during activation. C terminus deletion significantly (2.3–33-fold) accelerated deactivation of the slowly deactivating mutants, L250W, I253W, L254W, and I257W, whereas the other mutants were not affected or showed slight deceleration in some mutants (Fig. 5C and supplemental Table 1). Thus, the Trp-induced slowing of the deactivation kinetics in the L250W, I253W, L254W, and I257W mutants (Figs. 3 and 4) was only observed when Trp was introduced in the dimeric channel.
We also note that the effects of C terminus deletion on the kinetics in WT showed a common tendency, when compared with the slowly deactivating mutants, in which the activation kinetics showed mild acceleration (4.5-fold decrease in time constant) and the deactivation showed striking acceleration in WT (18.7-fold decrease in time constant) (Fig. 5, B and C). This suggests that the effects of C terminus deletion on kinetics reflect the interaction effect in a dimer rather than the bulk effect of the mutations into the S4 helix. We also observed that the degree of change varied by mutation, where L250W showed the most prominent change (35-fold acceleration; similar to WT) in the deactivation kinetics, and L254W showed the smallest change (2.3-fold acceleration) (Fig. 5C). I253W accelerated deactivation (6.9-fold decrease in time constant), but the speeding effect was weaker than in WT (Fig. 5C). Thus, although the common tendency was observed among the slowly deactivating mutants, including WT, the degree of change varied by the mutation position and the additional background mutation. We analyze this point more quantitatively below.
Schematic Discussion of the Effects of Trp for Monomers Versus Dimers
Until now, our focus has been on the relationship between the effects of Trp introduction and channel dimerization. The Trp residues within the two close-set S4 helices of the dimeric channel could be spatially close to one another and could affect the channel gating. The Trp introduction, in fact, showed some effects on the gating kinetics in the monomer, although it seemed stronger in the dimer (Fig. 5). To evaluate the effects of the various mutations, we next calculated the perturbation energy (ΔΔG) for the activation and deactivation kinetics, which will reflect the relative change of height of barrier potential for the closed-open state transition by mutation. On the assumption that the effects of Trp introduction on the activation and deactivation kinetics reflect almost exclusively the changes of height of the barrier, we schematically discussed the structure-based mechanism of the Hv gating by application of the mutant cycle analysis (see “Experimental Procedures”). Although mutations may change not only the height of the barrier but also the energy difference between the closed-open states, we simplified the model and estimated interactions between the two Trp residues in the dimeric channel (Fig. 6A). If the effect of Trp introduction on the gating kinetics is independent of the effect of channel dimerization, the ΔΔG changes caused by the two mutations will be additive (i.e. the sum of the apparent free energy changes caused by the single mutations will be equal to the apparent free energy change of the double mutation). As an example, Fig. 6A shows the results obtained for Trp at position 257 within the monomeric and dimeric channels. In the analysis of the activation kinetics, the effects of the mutations are apparently independent (Fig. 6A, blue values). Introduction of Trp slowed activation slightly (ΔΔGTrp = 0.27 kcal/mol), and the dimerization also slowed activation (ΔΔG2mer = 0.74 kcal/mol) (Fig. 6A, blue values). Introduction of Trp additively slowed activation when applied to the dimeric channel (ΔΔG2mer,Trp = 0.97 kcal/mol) (Fig. 6A, blue values), suggesting independent effects on activation. By contrast, if the effect of Trp introduction interacted with that of channel dimerization, there would be a difference between the sum of the apparent free energy changes of the two single mutations and the apparent free energy change of the double mutation. In the analysis of the deactivation kinetics, the introduction of Trp slowed deactivation slightly (ΔΔGTrp = 0.27 kcal/mol), whereas dimerization accelerated the deactivation slightly (ΔΔG2mer = −0.27 kcal/mol) (Fig. 6A, red values). Introduction of Trp slowed the deactivation more than expected when applied to the dimeric channel (ΔΔG2mer,Trp = 1.26 kcal/mol) (Fig. 6A, red values), suggesting that the two effects are apparently energetically coupled during channel deactivation.
The difference in the apparent free energy is here defined as the apparent interaction (coupling) energy (ΔΔGint) (see “Experimental Procedures”). An absolute value of the interaction energy (|ΔΔGint|) of >0.89 kcal/mol reportedly represents a significant interaction between two mutations (29). We calculated the apparent interaction energy for several mutation sites during activation and deactivation of the channel (Fig. 6, B and C). For activation, all |ΔΔGint| values were less than 0.89 (Fig. 6B), suggesting that Trp slowed channel activation independently of the channel dimerization. For deactivation, |ΔΔGint| values were >0.89 when Trp was introduced at position 250, 253, or 257, and it was slightly lower than the borderline when Trp was introduced at position 254 (ΔΔGint = −0.75 kcal/mol). The effects of Trp at the other sites tested were apparently independent of channel dimerization (Fig. 6C). Thus, the slow deactivation observed in L250W, I253W, and I257W is assumed to be caused by the Trp residues within the two close-set S4 helices in the dimeric channel.
Tandem Dimer Experiments
Two-electrode voltage clamp recordings with oocytes expressing Ciona Hv highlighted the effects of Trp on the channel gating (Figs. 1–6). We next analyzed the mouse Hv constructs by patch clamp recording, to obtain more detailed information about the channel gating and also to confirm whether the effects of Trp on the channel gating were commonly observed in other species of Hv. We analyzed the mouse Hv WT (a native dimer), the mouse Hv delC mutant (a monomer), and the tandem dimer constructs using the patch clamp recordings with HEK293T cells. The W203I of mouse Hv, corresponding to the Ciona W257I mutant, was linked to the mouse Hv WT (the I-W tandem or W-I tandem), and we examined what would happen in the tandem heterodimers. Representative current traces of the tandem constructs are shown (Fig. 7A), and the data are summarized (Fig. 7, B and C). The WT tandem constructs did not show significant change in kinetics in comparison with the native dimer (Fig. 7, B and C, W-W tandem versus WT, and supplemental Table 1). The acceleration effects of the Trp mutation on the gating kinetic were expectedly observed despite species (Fig. 7A, I-I tandem). It is noteworthy that biphasic activation kinetics, the fast and slow components, were observed in the I-W and W-I tandems (Fig. 7, A and B). Relative weights of the fast component were 41 ± 4% in the I-W tandem and 44 ± 2% in the W-I tandem, suggesting that the biphasic kinetics would be derived from each different protomer. The fast components were considered to be derived from the W203I protomer that corresponds to the I-delC mutant here, whereas the slow components were considered to be derived from the WT protomer, on the basis of the time constants (Fig. 7B and supplemental Table 1). Time constants of the slow components in the tandem heterodimers were a bit faster than that in the W-W tandem, which would reflect the monomerization effect of the WT protomer in activation kinetics, as seen in the delC mutant (Fig. 7B and supplemental Table 1). In contrast to the activation phase, only rapid deactivations, as seen in the I-I dimer, were observed in the heterodimers (Fig. 7, A and C). The delC mutant, a monomeric channel with an intact Trp in S4, also showed rapid deactivation (Fig. 7C). The deactivation kinetics were assigned to two major classifications, rapid and slow, and the native slow deactivation was only seen in the W-W dimer (Fig. 7, A and C, and supplemental Table 1), suggesting that the two Trp residues in each S4 helix are required to produce the slow deactivation. Taken together, these results support the idea that, as proposed in the schematic discussion (Fig. 6), the deceleration effect by each Trp in S4 is additive within the dimer during activation but coupled within the dimer during deactivation.
Cooperative Gating in Kinetics Versus Steady State
In this study, we showed that, in the dimeric Hv, Trp in S4 decelerated the kinetics in deactivation. It is also reported that the cooperative gating between each channel protomer in a dimer affects the steady state activation, enhancing the voltage dependence of gating (13–16). We next examined whether the effects of Trp introduction on kinetics coexist with the cooperative gating of the steady state activation. We used the limiting slope analysis to estimate the effective gating charge in the W203I mutants. Limiting slopes were measured by linear fitting of the chord conductances (Fig. 8A), keeping the H+ current to a minimum in the range around threshold. The value of the effective gating charge (Zδ) in W203I was 3.11 ± 0.07 (n = 5), and it was decreased to 1.49 ± 0.09 (n = 6) upon deletion of the coiled-coil domain (Fig. 8B). This 2-fold Zδ value of W203I compared with the monomeric W203I/delC suggests that cooperative gating remains intact in W203I. Thus, the W203I mutation that changes the “kinetics” did not break the cooperativity in a “steady state.”
We also analyzed the E149C mutant tandem dimers. Tombola et al. reported that the E149C mutation intensely shifted the G-V relationship to the hyperpolarized direction, but the G-V was brought back to a similar range to that of WT when concatenated with WT in a tandem dimer (WT-E149C) (14). This influence on G-V by the partner in a dimer is illustrative of the cooperative gating in a steady state. We analyzed the W203I mutation on the background of the WT-E149C tandem dimer which is reported to show the steady state cooperative gating (14). Representative current traces recorded by whole-cell patch clamp are shown (Fig. 8C). As shown in Fig. 7, the biphasic activation kinetics and the fast deactivation kinetics were observed by the introduction of W203I in WT-E149C (WT-E149C/W203I) (Fig. 8C). The W203I mutation had little influence on I-V of WT-E149C (Fig. 8D). These results suggest that W203I primarily influenced the time constants of gating but did not significantly affect the steady state gating. Each channel protomer showed its own kinetics in activation, whereas the deactivation kinetics was influenced by one another (Fig. 8, C and E); the existence of two Trp decelerated the deactivation as in Fig. 7. We also note that the biphasic activation kinetics of WT-E149C was not clear, although the two different channel protomers, WT and E149C, might have different kinetics. Because the W203I mutation changes the kinetics much more clearly than E149C, the differences in the activation kinetics between the two protomers were able to be observed. Thus, the cooperative slowing effect of Trp on the deactivation “kinetics” in a dimer showed a qualitatively different relationship with the cooperative gating of the “steady state” activation.
Direct Interaction within the Hv Dimer
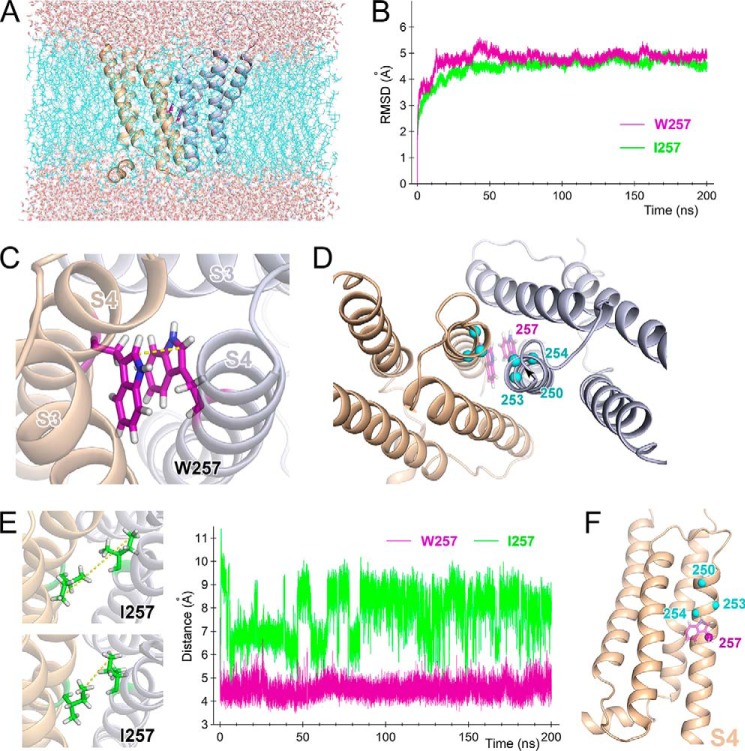
Given the significant energetic coupling between Trp introduction and channel dimerization (Fig. 6), we hypothesized that the two Trp residues within the close-set S4 helices made direct contact to produce the slow gating kinetics during deactivation. We tested that idea in a molecular dynamics simulation (Fig. 9). The dimeric model of the transmembrane region of Hv was built by superposing the crystal structure of chimeric mouse Hv (17) onto the dimeric structure of the cytoplasmic coiled-coil (13). Molecular dynamics simulations for the WT and W257I channels were run for 200 ns after the structure achieved equilibrium (Fig. 9, A and B). In the simulation with the WT channel, Trp was situated at the interaction interface between the dimeric subunits and interacted with its partner Trp via π-stacking (Fig. 9, C and D). The parallel displaced formation of the π-stacking was continuously observed throughout the simulation (Fig. 9C), where the CH/π interactions between the benzene ring of indole and Cβ-H/Cϵ-H were formed. The distance between the Cδ1 atoms of the two Trp257 residues within the dimer remained constant throughout the simulation (Fig. 9, C and E, right), suggesting a stable interaction. On the other hand, the distance between the Cδ atoms of the two Ile257 residues was wider within the dimer and showed stepwise fluctuation during the simulation, suggesting that the side chains of Ile257 take various conformations with rotation (Fig. 9E). Collectively, these results suggest that the W257I mutation destabilizes the interaction between the two S4 helices at the 257-position. The overall structures of WT and W257I did not significantly differ (Fig. 9B).
FIGURE 9.
Molecular dynamics simulation for the dimeric channel. A, snapshot of the transmembrane domain from a viewpoint parallel to the membrane 197 ns into the molecular dynamics simulation. Two channel protomers are shown as ribbon models (light orange and light blue). Trp257 is shown as purple sticks between the two protomers. Cyan wires represent bulk lipid molecules, and CPK sticks outside the bilayer represent water molecules. B, time series of root mean square deviations for Cα along the entire protein for WT (purple) and the W257I (green) mutant in the molecular dynamics simulation. C, representative conformation of Trp257 within a dimer. Two channel protomers are shown as ribbon models (light orange and light blue). Trp257 is shown as purple sticks between the two protomers. D, Cα positions in the Trp residues introduced at positions 250, 253, and 254 are plotted on the resultant model structure after the molecular dynamics simulation (cyan spheres). Original Trp residues at position 257 are shown as purple sticks. E, comparison of the side chain motions of WT and W257I. Traces (on the right) depict the distances between two Cδ1s in Trp257 in the dimeric WT channel (purple) and between two Cδs in Ile257 in the dimeric W257I mutant (green). Two representative conformations of the Ile257 side chain are also shown (left). Yellow dotted lines, distance between the two Cδs (cf. Fig. 9C). F, positions 250, 253, 254, and 257 are plotted on the resultant model structure after the molecular dynamics simulation (spheres). Only one subunit is shown here.
The positions of the Trp introduced at 250, 253, 254, and 257, which showed energetic coupling within the dimer (Fig. 6), are plotted on the resulting structure model in Fig. 9D. Within each subunit, all four positions are situated at the periphery of the S4 helix (Fig. 9F), and the model shows that Trp in one subunit is able to contact its partner in the neighboring subunit when the rotamers are favorable (Fig. 9D). We noted that Trp residues at position 254 could not fully interact with one another due to rotametric restriction (Fig. 9D), resulting in low energetic coupling (Fig. 6). We also note that the deactivation kinetics of I257W were biphasic (Figs. 3 and 4), and, given the time constant, two populations of channels are considered to exist on the membrane. The loss of interaction of Trp may arise in some channels of I257W, and Trp at the 250-position may interact more tightly than Trp at 257 on the background of the NA-mutant (W257I/F260A). Thus, the molecular dynamics simulation demonstrates that within the dimeric channel, the Trp residues in the parallel S4 helices are able to directly interact with each other, which produces the channel's characteristically slow deactivation kinetics.
Discussion
Recognition that dimerization is the key framework underlying the physiological functionality of Hv in phagocytes (12, 13, 16) has prompted an effort to resolve the dimeric structure of Hv using several approaches. Cross-linking analysis showed two types of contacts between transmembrane segments: an S1-S1 interaction at the extracellular end of the protein and an S4-S4 interaction along the entirety of the two helices, which seemingly conflict with one another (10, 18). Voltage clamp fluorometry showed that mutation of the S1-S1 contact site changed the cooperative movement of S4 during gating (19). On the other hand, systematic mutagenesis focusing on the linker region between S4 and the coiled-coil demonstrated that the two S4 segments form continuous helices with the dimer coiled-coil and function as a single unit for channel gating, which supports the idea of an S4-S4 interaction interface within the dimeric channel (18). In the present study, we obtained additional evidence of direct S4-S4 interaction: apparent energetic coupling (Fig. 6) and π-stacking (Fig. 9) of the two Trp residues in the S4 segments within the dimer. Trp residues introduced at position 250, 253, or 257 showed apparent energetic coupling in the dimeric channel (Fig. 6), and the distances between the Trp pairs are consistent with direct interactions in the N-terminal half of S4 (Fig. 9E), further supporting the idea of direct S4-S4 interaction. It is also noteworthy that the energetic coupling between Trp residues introduced into S4 in the dimeric channel did not affect the activation phase, only deactivation (Fig. 6), suggesting that the transmembrane arrangement of the two channel subunits changes during the gating process (i.e. activation and deactivation go through different conformational steps, and the Trp residues in the S4 segments make contact only during the deactivation phase). This interpretation may provide a clue to resolving the seemingly incompatible features of the interaction interface within the dimeric protein (10, 18).
The observation that Trp mutations significantly alter the deactivation kinetics suggests that the π-stacking conformation (Fig. 9) elevates the intermediate activation energy when the channel transits from the open state to the closed state. We propose a gating model for the dimeric WT channel based on the results of the Trp interactions between the two subunits (Fig. 10A). In this model, the two Trp residues interact only during the deactivation phase, and the structural stabilization of the two S4 helices mediated by the π-stacking of Trp prohibits the movement of S4 when the channel transits from the open state to the closed state (Fig. 10A, right). On the other hand, when the channel transits from the closed state to the open state, the two Trp residues do not interact; nor do they prevent S4 movement (Fig. 10A, left). High activation energy when the channel transits from the open state to the closed state results in the stabilization of the open state, which was observed as the shift of threshold (Fig. 4E). Several studies of voltage-gated K+ channels have shown that the S4 helix moves up and down with a certain degree of rotation during the channel gating (41, 43, 44). This means that in the case of the dimeric Hv, the angles of the Trp position relative to the central z axis of the dimer may change during gating. If so, the two Trp residues in the dimer can potentially interact only if movement of the two S4 helices is precisely timed. If the timing of the two S4 helices is out of sync, there will be no interaction between the Trp residues, and the two S4 helices will be able to move freely (Fig. 10A, left). This separate movement of S4 helices is also suggested in an earlier study focusing on the gating cooperativity during activation (19), and our tandem experiments further support the idea (Fig. 7). On the other hand, if the timing of the two S4 movements is synchronized within the dimer, the interaction of the Trp residues will influence the movement of S4 (Fig. 10A, right). From our findings, it appears that this occurs only during the deactivation phase. Our observations thus highlight the existence of gating cooperativity within the dimeric channel, which slows the kinetics of channel deactivation and provides structural insight into the process.
FIGURE 10.
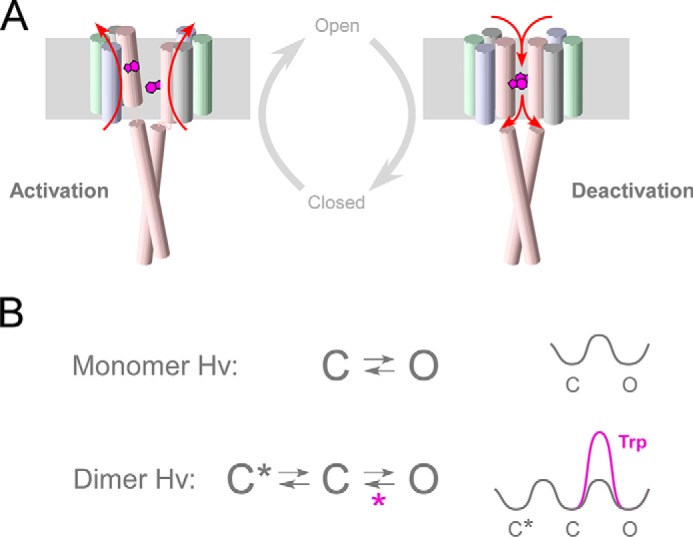
Hv gating model, focusing on the Trp interaction and S4 motion. A, cylinders depict transmembrane segments, and Trp257 in S4 is shown as purple plates. The motion of S4 is represented by red arrows. The activation model depicts the unsynchronized motion of the S4 helices. The π-stacking interaction between the two Trp residues appears only when the channel is closing. B, simple state models for a monomer and dimer. In the monomer Hv, channels transit from the closed state to the open state voltage-dependently. In the dimer Hv, a new onset closed state (C*) induced by dimerization produces an extra voltage-dependent step (C*-C) that underlies the strengthened effective charge of gating and the sigmoidal activation. Trp increases the activation energy in the deactivation phase (right), and one more cooperativity (purple asterisk) is proposed during the C-O transition during deactivation in the WT dimer.
We present kinetic schemes for the voltage-driven transition (Fig. 10B), where the simplest models are proposed in each case of monomer and dimer. Several earlier studies reported that Hv dimerization mediated by the coiled-coil domain generates cooperative gating that results in slow sigmoidal activation kinetics and enhances the voltage dependence of the activation (12–14, 16, 18). From those findings, it was proposed that a new onset closed state (C*) induced by dimerization underlies the cooperative slow sigmoidal activation (Fig. 10B). Based on our observation that the coupling of Trp slowed the deactivation kinetics, another cooperativity should exist during the C-O transition during deactivation in the WT dimer (Fig. 10B). Because the cooperative deactivation showed a qualitatively different relationship from the cooperative activation (Figs. 6–8), the dimer cooperativity affecting Hv gating has at least two mechanisms: the extra closed state before the channel opening and the friction caused by π-stacking during deactivation, both of which induce a slowing of the gating kinetics. The simplest model may actually need further modification, including the idea of single-line transition in Fig. 10A, to illustrate the split activation kinetics observed in the tandem heterodimer (Fig. 7), and further approaches focusing on the structural-functional dynamics of the Hv gating will be required for proposing a comprehensive model.
Our structural model based on the molecular dynamics simulation also provides a clue to the functioning state at the atomic level. The recently resolved crystal structure of the transmembrane region of Hv is thought to reflect a closed state of the channel primarily because the structure includes a Zn2+ ion, which is a physiological blocker (5, 6), and its permeation pathway is split by hydrophobic residues (17). In an alternative interpretation, however, the S4 helix in the crystal structure is situated at a position one helix turn higher than the previously predicted position for the resting (closed) form of the voltage sensor (reflected by the salt bridge formation between the Arg of S4 and the countercharge in S1), and the S4 helix appears to have the space to move up and down (28, 45, 46). In addition, the crystal structure was not a natural dimeric channel but a crystallization trimer, and the resolution was not high (17). Consequently, the functional states of the dimeric structure remain a matter of debate. Considering that in the present study, the Trp mutation affected chiefly the gating kinetics, we may have detected one of the intermediate states of the gating, namely the parallel conformation of the two S4 helices stabilized by the π-stacking between the Trp residues. In the molecular dynamics simulation, we did not observe any signs of H+ permeation, such as the water passing and the transmembrane hydrogen bond formation (47); hence, the π-stacking structure would reflect one of the non-conducting intermediate states rather than an open state, which also supports the idea of the gating model (Fig. 10). If the S4 helix moves upward or downward with rotation from the position in the crystal structure, the π-stacking between the Trp residues will be absent, which may indicate that the crystal structure represents an intermediate state of gating.
The mutagenesis approach with Trp introduction into the transmembrane region has been applied to several membrane proteins to assess interhelix contact. In most cases, the bulkiness of Trp caused the helix to become disordered and, in some instances, disrupted the protein function (25–27, 37, 38, 42). In the case of Hv, it is unique and noteworthy that within the native amino acid sequence, a pair of Trp residues are situated face-to-face in their transmembrane helices, and they affect channel function through their interaction. Hv channels are expressed in phagocytes and sperm cells, which are not typical excitable cells, such as neurons and cardiomyocytes (5–7). In these cells, fast channel kinetics are not required and may even be a disadvantage to achieving pH homeostasis through H+ conductance. Considering that the slow deactivation is imposed by a pair of Trp residues in the dimeric channel and that all Hv species contain a Trp residue in the middle of the S4 voltage sensor segment, Hv appears to be optimally designed as a dimeric voltage-gated channel with a solid foundation derived from its primary structure.
Author Contributions
Y. F. designed the experiments; H. O. performed the two-electrode voltage clamp recordings; Y. F. performed the patch clamp recordings; Y. Y. and Y. T. performed the molecular dynamics analysis; Y. O. assisted with the electrophysiology experiments. Y. F., H. O., Y. Y., and Y. T. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank M. Kobayashi for technical assistance and Drs. S. Sakata, A. Nakagawa, K. Takeshita, K. Kinoshita, M. Shirota, and H. Kondo for discussion.
This work was supported by a grant-in-aid for scientific research, (26291030), the Uehara Memorial Foundation, the Takeda Science Foundation, and the Naito Foundation. This work was supported by CREST, Japan Science and Technology Agency. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table 1.
- Hv
- voltage-gated H+ channel
- NA-mutant
- non-aromatic mutant
- ANOVA
- analysis of variance.
References
- 1. Armstrong C. M., and Hille B. (1998) Voltage-gated ion channels and electrical excitability. Neuron 20, 371–380 [DOI] [PubMed] [Google Scholar]
- 2. Bezanilla F. (2000) The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80, 555–592 [DOI] [PubMed] [Google Scholar]
- 3. Sasaki M., Takagi M., and Okamura Y. (2006) A voltage sensor-domain protein is a voltage-gated proton channel. Science 312, 589–592 [DOI] [PubMed] [Google Scholar]
- 4. Ramsey I. S., Moran M. M., Chong J. A., and Clapham D. E. (2006) A voltage-gated proton-selective channel lacking the pore domain. Nature 440, 1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lishko P. V., Botchkina I. L., Fedorenko A., and Kirichok Y. (2010) Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell 140, 327–337 [DOI] [PubMed] [Google Scholar]
- 6. Morgan D., Capasso M., Musset B., Cherny V. V., Ríos E., Dyer M. J., and DeCoursey T. E. (2009) Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 106, 18022–18027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Decoursey T. E. (2003) Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83, 475–579 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalez C., Rebolledo S., Perez M. E., and Larsson H. P. (2013) Molecular mechanism of voltage sensing in voltage-gated proton channels. J. Gen. Physiol. 141, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tombola F., Ulbrich M. H., and Isacoff E. Y. (2008) The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron 58, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee S. Y., Letts J. A., and Mackinnon R. (2008) Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc. Natl. Acad. Sci. U.S.A. 105, 7692–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koch H. P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., and Larsson H. P. (2008) Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. U.S.A. 105, 9111–9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujiwara Y., Kurokawa T., Takeshita K., Nakagawa A., Larsson H. P., and Okamura Y. (2013) Gating of the designed trimeric/tetrameric voltage-gated H+ channel. J. Physiol. 591, 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujiwara Y., Kurokawa T., Takeshita K., Kobayashi M., Okochi Y., Nakagawa A., and Okamura Y. (2012) The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H+ channel Hv1. Nat. Commun. 3, 816. [DOI] [PubMed] [Google Scholar]
- 14. Tombola F., Ulbrich M. H., Kohout S. C., and Isacoff E. Y. (2010) The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat. Struct. Mol. Biol. 17, 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Musset B., Smith S. M., Rajan S., Cherny V. V., Sujai S., Morgan D., and DeCoursey T. E. (2010) Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J. Physiol. 588, 1435–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez C., Koch H. P., Drum B. M., and Larsson H. P. (2010) Strong cooperativity between subunits in voltage-gated proton channels. Nat. Struct. Mol. Biol. 17, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeshita K., Sakata S., Yamashita E., Fujiwara Y., Kawanabe A., Kurokawa T., Okochi Y., Matsuda M., Narita H., Okamura Y., and Nakagawa A. (2014) X-ray crystal structure of voltage-gated proton channel. Nat. Struct. Mol. Biol. 21, 352–357 [DOI] [PubMed] [Google Scholar]
- 18. Fujiwara Y., Kurokawa T., and Okamura Y. (2014) Long α helices projecting from the membrane as the dimer interface in the voltage-gated H+ channel. J. Gen. Physiol. 143, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu F., Rebolledo S., Gonzalez C., and Larsson H. P. (2013) Subunit interactions during cooperative opening of voltage-gated proton channels. Neuron 77, 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Q., Wanderling S., Paduch M., Medovoy D., Singharoy A., McGreevy R., Villalba-Galea C. A., Hulse R. E., Roux B., Schulten K., Kossiakoff A., and Perozo E. (2014) Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 21, 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tombola F., Pathak M. M., and Isacoff E. Y. (2005) How far will you go to sense voltage? Neuron 48, 719–725 [DOI] [PubMed] [Google Scholar]
- 22. Bezanilla F. (2002) Voltage sensor movements. J. Gen. Physiol. 120, 465–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deol S. S., Bond P. J., Domene C., and Sansom M. S. (2004) Lipid-protein interactions of integral membrane proteins: a comparative simulation study. Biophys. J. 87, 3737–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Killian J. A., and von Heijne G. (2000) How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25, 429–434 [DOI] [PubMed] [Google Scholar]
- 25. Collins A., Chuang H., Jan Y. N., and Jan L. Y. (1997) Scanning mutagenesis of the putative transmembrane segments of Kir2.1, an inward rectifier potassium channel. Proc. Natl. Acad. Sci. U.S.A. 94, 5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choe S., Stevens C. F., and Sullivan J. M. (1995) Three distinct structural environments of a transmembrane domain in the inwardly rectifying potassium channel ROMK1 defined by perturbation. Proc. Natl. Acad. Sci. U.S.A. 92, 12046–12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subbiah R. N., Kondo M., Campbell T. J., and Vandenberg J. I. (2005) Tryptophan scanning mutagenesis of the HERG K+ channel: the S4 domain is loosely packed and likely to be lipid exposed. J. Physiol. 569, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chamberlin A., Qiu F., Rebolledo S., Wang Y., Noskov S. Y., and Larsson H. P. (2014) Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc. Natl. Acad. Sci. U.S.A. 111, E273–E282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranganathan R., Lewis J. H., and MacKinnon R. (1996) Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron 16, 131–139 [DOI] [PubMed] [Google Scholar]
- 30. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., and Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Protein Sci. 10.1002/0471140864.ps0209s50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edberg R., Evans D. J., and Morriss G. P. (1986) Constrained molecular dynamics: simulations of liquid alkanes with a new algorithm. J. Chem. Phys. 84, 6933 [Google Scholar]
- 32. Nose S. (1984) A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511 [Google Scholar]
- 33. Parrinello M., and Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Physics 52, 7182 [Google Scholar]
- 34. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577 [Google Scholar]
- 35. Hess B., Kutzner C., van der Spoel D., and Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 36. Ishii T. M., Nakashima N., and Ohmori H. (2007) Tryptophan-scanning mutagenesis in the S1 domain of mammalian HCN channel reveals residues critical for voltage-gated activation. J. Physiol. 579, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hackos D. H., Chang T. H., and Swartz K. J. (2002) Scanning the intracellular S6 activation gate in the shaker K+ channel. J. Gen. Physiol. 119, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monks S. A., Needleman D. J., and Miller C. (1999) Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J. Gen. Physiol. 113, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Musset B., Smith S. M., Rajan S., Morgan D., Cherny V. V., and Decoursey T. E. (2011) Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature 480, 273–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berger T. K., and Isacoff E. Y. (2011) The pore of the voltage-gated proton channel. Neuron 72, 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakrapani S., Cuello L. G., Cortes D. M., and Perozo E. (2008) Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure 16, 398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li-Smerin Y., Hackos D. H., and Swartz K. J. (2000) α-helical structural elements within the voltage-sensing domains of a K+ channel. J. Gen. Physiol. 115, 33–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glauner K. S., Mannuzzu L. M., Gandhi C. S., and Isacoff E. Y. (1999) Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature 402, 813–817 [DOI] [PubMed] [Google Scholar]
- 44. Cha A., Snyder G. E., Selvin P. R., and Bezanilla F. (1999) Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813 [DOI] [PubMed] [Google Scholar]
- 45. Pathak M. M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P., Kohout S., Tombola F., and Isacoff E. Y. (2007) Closing in on the resting state of the Shaker K+ channel. Neuron 56, 124–140 [DOI] [PubMed] [Google Scholar]
- 46. Tombola F., Pathak M. M., and Isacoff E. Y. (2005) Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron 45, 379–388 [DOI] [PubMed] [Google Scholar]
- 47. Ramsey I. S., Mokrab Y., Carvacho I., Sands Z. A., Sansom M. S., and Clapham D. E. (2010) An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat. Struct. Mol. Biol. 17, 869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.