Abstract
Bacteriophages and their hosts are continuously engaged in evolutionary competition. Here we isolated a lytic phage phiYe-F10 specific for Yersinia enterocolitica serotype O:3. We firstly described the phage receptor was regulated by DTDP-rhamnosyl transferase RfbF, encoded within the rfb cluster that was responsible for the biosynthesis of the O antigens. The deletion of DTDP-rhamnosyl transferase RfbF of wild type O:3 strain caused failure in phiYe-F10 adsorption; however, the mutation strain retained agglutination with O:3 antiserum; and complementation of its mutant converted its sensitivity to phiYe-F10. Therefore, DTDP-rhamnosyl transferase RfbF was responsible for the phage infection but did not affect recognition of Y. enterocolitica O:3 antiserum. Further, the deletions in the putative O-antigen biosynthesis protein precursor and outer membrane protein had no effect on sensitivity to phiYe-F10 infection. However, adsorption of phages onto mutant HNF10-ΔO-antigen took longer time than onto the WT, suggesting that deletion of the putative O-antigen biosynthesis protein precursor reduced the infection efficiency.
The primary determinant in the infection of a bacterial host by a bacteriophage is the adsorption of the phage to the host receptor. This receptor recognition is the most reported in phage- host interaction studies1,2. Yersinia enterocolitica is Gram-negative and a globally distributed foodborne human pathogen, belonging to the Enterobacteriaceae Yersinia species. There are about 60 different serotypes with variability in the O-antigen. Y. enterocolitica clinical isolates from humans predominantly belong to serotypes O:3, O:9, O:8 and O:5, 27 with variability on different continents3,4: serotypes O:3 and O:9 cause human infections and are most common in Europe, Canada, Japan, China and South Africa; while 1B/O:8 is the primary serotype infecting people in the Americas5,6. However, at present, serotype O:3 strains are becoming the most frequently detected pathogenic Y. enterocolitica all over the world7,8,9,10.
To date, the phage phiYeO3-12 and vB_YenP_AP5 display specificity for Y. enterocolitica O:3 and phage PY54 exhibits a host range restrict to Y. enterocolitica O:5 and O:5, 27 were previously described11,12,13,14,15. Phage viruses have stringent host specificities where the attachment of the virus particle requires specific recognition of phage receptor using a phage receptor binding protein (RBP)16,17. Several receptors are reported, including flagella/pilus related components18,19, lipopolysaccharides (LPS)20,21,22, capsular polysaccharides (CPS) and outer membrane proteins (OMP)23,24. Similar to other Gram-negative Enterobacteriaceae bacteria, the structure of the Yersinia enterocolitica LPS has three primary components: lipid A, core oligosaccharide and O-side-chain (O-antigen). LPS acts as an immune stimulatory agent (lipid A) or as a virulence factor (O-antigen)25,26,27. The lipid A portion is responsible for the endotoxin activity. The O-antigen functions as a barrier against complement-mediated lysis and resists killing of bacteria by microbicidal intracellular granules in polymorpho-nuclear leucocytes. Previously published assays showed that we can detect the serotypes of Y. enterocolitica strains using amplification of O-antigen-encoded genes28. The Yersinia enterocolitica O:3 lipopolysaccharide O-antigen is a homopolymer of 6-deoxy-L-altrose. The genes for the O:3 O-antigen translocation are located within the rfb gene cluster, including 10 open reading frames and eight of the genes, are organized into two operons, rfbABC and rfbDEFGH that are essential for O-antigen synthesis26. A specific detection of Y. enterocolitica serotype O:3 is obtained with fragment of the rfbC gene29. In this study, we evaluated the lysis ability of the phage phiYe-F10 on the Y. enterocolitica wild strain, a spontaneous rough mutant, three gene deletion strains and a rfbc compensation strain using phage adsorption tests and plaque formation tests. We are first to identify DTDP-rhamnosyl transferase RfbF as the receptor regulator protein, and another protein (putative O-antigen biosynthesis protein precursor) in a Y. enterocolitica O:3 strain is related to phiYe-F10 adsorption.
Results
The host range of phage phiYe-F10
The 188 Y. enterocolitica strains were (Supplemental data 1): 57 strains serotype O:3, 34 strains serotype O:9, 13 strains serotype O:8, 10 strains serotype O:5, 3 strains serotype O:5, 27, 5 strains of self-agglutinating Y. enterocolitica and 66 other serotypes biotypes 1A Y. enterocolitica (Among the six Y. enterocolitica biotypes, 1A is the most heterogeneous group, including more than 17 different serotypes, some of them are unable to serotyped30). At 25 °C and 37 °C, phiYe-F10 can formed plaques only on serotype O:3 strains, but not on Y. enterocolitica strains other O serogroups, or Y. pseudotuberculosis or Y. pestis strain (Table 1). All of the 57 sensitive strains were O:3 serotype Yersinia enterocolitica carrying the DTDP-rhamnosyl transferase RfbF encoding genes (rfbc), 49 of which were pathogenic Y. enterocolitica (including 39 strains biotype 3, 6 strains biotype 4, and 4 strains of biotype 5); 6 strains were biotype 1A nonpathogenic; and 2 strains were biotype 1A with ail genes (Table 2).
Table 1. Bacteriophage sensitivity test*.
| Y. enterocolitica |
Y. pseudotuberculosis |
||||
|---|---|---|---|---|---|
| Serotype | Resistant No. | Sensitive No. | Serotype | Resistant No. | Sensitive No. |
| O:10K | 1 | 0 | O:10 | 1 | 0 |
| O:19, 8 | 1 | 0 | O:5a | 1 | 0 |
| O:20 | 1 | 0 | O:9 | 1 | 0 |
| O:44 | 1 | 0 | O:2a | 2 | 0 |
| O:4, 32 | 2 | 0 | O:6 | 2 | 0 |
| O:5, 27 | 3 | 0 | O:8 | 2 | 0 |
| O:5 | 10 | 0 | O:15 | 3 | 0 |
| O:8 | 13 | 0 | O:1a | 3 | 0 |
| O:9 | 34 | 0 | O:14 | 4 | 0 |
| O:3 | 0 | 57 | O:1b | 4 | 0 |
| SA | 5 | 0 | O:3 | 5 | 0 |
| UD | 60 | 0 | O:4b | 5 | 0 |
| NT | 4 | 0 | |||
| Total | 131 | 57 | Total | 37 | 0 |
*Note: The one Yersinia pestis used in this study is not listed in this table.
UD: Undetermined; NT: Not typable; SA: self-agglutingating.
Table 2. The characteristics of the 57 phage sensitive Y. enterocolitica strains of O:3 serotype.
| Serotype | Genotype | Biotype |
Sensitive rate | |||
|---|---|---|---|---|---|---|
| 1A | 3 | 4 | 5 | (%) | ||
| O:3 | rfbc+, ail+, ystA+, ystB−, yadA+, virF+ | 0 | 36 | 3 | 4 | 75.44% |
| rfbc+, ail+, ystA+, ystB−, yadA−, virF− | 2 | 3 | 3 | 0 | 14.04% | |
| rfbc+, ail−, ystA−, ystB+, yadA−, virF− | 2 | 0 | 0 | 0 | 3.51% | |
| rfbc+, ail−, ystA−, ystB−, yadA−, virF− | 4 | 0 | 0 | 0 | 7.02% | |
Morphology
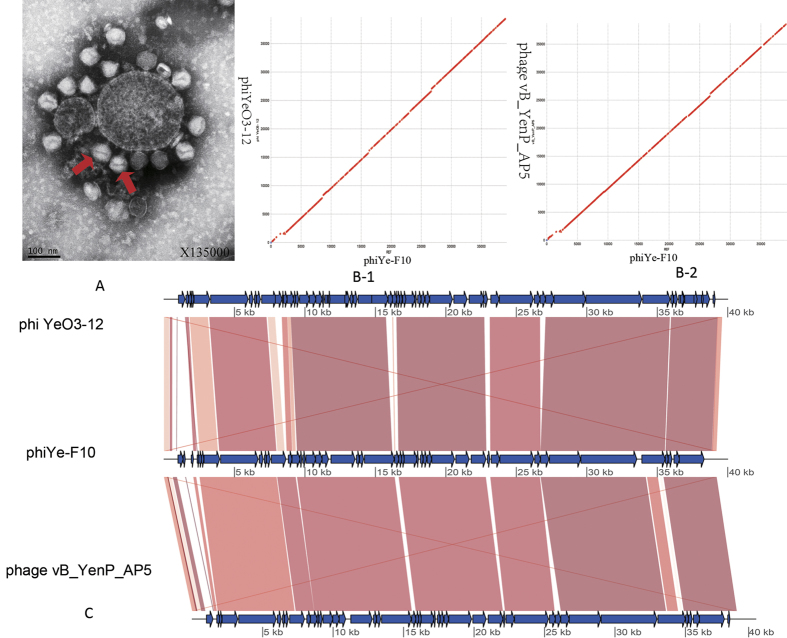
phiYe-F10 was negatively stained and examined using transmission electron microscopy (Fig. 1A showed with an arrow). The virions showed hexagonal outlines, indicating their icosahedral nature. The head connected with the tail through a short neck which exhibiting T7 symmetry in shape. The phage had a head at approximately 55.0 nm in diameter and a short non-contractile tail of ~11.0 nm in length. Extended tail fibers were not seen. Collectively, these morphological features indicated that this virus belongs to the family Podoviridae.
Figure 1.
(A) Electron micrograph of phiYe-F10. The phage is negatively stained with 2% potassium phosphotungstate. phiYe-F10 is shown at 135,000× magnification. Scale bar indicates size in nm. (B-1) Dot plot of genome sequences of phiYe-F10 and Yersinia phage phiYeO3-12. (B-2) Dot plot of genome sequences of phiYe-F10 and Yersinia phage vB_YenP_AP5. (C) Pairwise nucleotide sequence comparison of phiYe-F10, phiYeO3-12 and vB_YenP_AP5.
General features of the phiYe-F10 genome and comparative genomics
The phiYe-F10 genome was assembled as a circular molecule when the sequencing was completed. Using PCR, the results of terminal run-off sequencing confirmed that the phiYe-F10 has repeated sequences. The genomes of T7-like phages typically contain direct terminal repeats (DTRs) that are used during genome replication and packaging. For example, Yersinia phage phiYeO3-12 (Genbank No. NC_001271.1) and vB_YenP_AP5 (Genbank No. NC_025451.1) have DTRs of 232 bp and 235 bp, respectively. The lengths of the DTRs in phiYe-F10 (235 bp) were in agreement with the reported lengths of T7-like phage members. So we concluded the DNA sequence of the phiYe-F10 consists of a linear double stranded DNA of 39,210 bp, which correlates well with the size of other T7-like phage members. In total, 46 gene products were predicted in the phiYe-F10 genome; functions were assigned to 43 of them based on the similarities of the predicted products to known proteins.
Compared with the other two lytic phages for Y. enterocolitica serotype O:3, the genome of phiYe-F10 was highly similar to the 39,600 bp of Yersinia phage phiYeO3-12 (97%, 623/641) and the 38,646 bp of Yersinia phage vB_YenP_AP5 (89%, 583/648). Genomic comparisons indicated phages phiYe-F10, phi YeO3-12 and vB_YenP_AP5 were closely related. A dot plot comparison using the program Gepard was performed to illustrate the genomic similarities among phiYe-F10, phi YeO3-12 and vB_YenP_AP5. The red dots indicated the corresponding genome regions on the abscissa and the ordinate showed similarity (Fig. 1B-1,B-2). We also performed a graphical comparison of the three genome sequences. Highly related sequences were shown using red shadings. As shown in Fig. 1C, the genome sequence of phiYe-F10 had exactly similar genetic organization and large blocks of homologous synteny when compared to the other two phages. The differences were primarily for the 403 bp and 446 bp deletions (with the region from 26,675 bp to 27,078 bp of Yersinia phage phiYeO3-12 and the 25,710 bp to 26,156 bp of vB_YenP_AP5 deleted in the genome sequence of phiYe-F10). The missing genes encoded putative 13.5 protein for phage phiYeO3-12; encoded hypothetical protein and portion of internal virion protein A for phage vB_YenP_AP5. The Fig. 1C data was in concordance with the Fig. 1B-1,1 B-2 data, strongly indicated that phiYe-F10, phage phiYeO3-12 and vB_YenP_AP5 were closely associated.
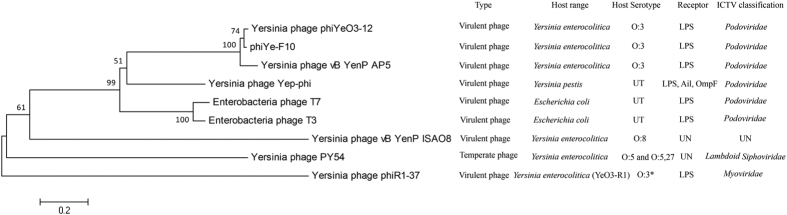
For bacteriophages, the primary determinant of the host receptor is the tail fiber protein, so we compared of the tail fiber protein of Yersinia phage which including the families Podoviridae, Myoviridae and Siphoviridae. The alignment showed high homology among phiYe-F10, phiYeO3-12 and vB_YenP_AP5. The six families Podoviridae members were classified into the same cluster, the other sequences of the families Myoviridae and Siphoviridae divided from that of the families Podoviridae, and located in different branches (Fig. 2).
Figure 2. The phylogenetic tree of tail fiber proteins.
*Strain YeO3-R1, a virulence-plasmid-cured O antigen-negative derivative of Yersinia enterocolitica serotype O:3. The phage receptor of phi R1-37 is in the outer core hexasaccharide of Y. enterocolitica O:3 LPS. UT: untyped; UN: unknown.
The adsorption abilities and growth characteristics of the WT strain, its mutants, and compensation strain
Early studies showed the outer membrane protein A (OmpA) was not only a common phage receptor of Enterobacteriaceae23,31, but also a common immune protective antigen beyond the V antigen conserved within Yersinia species32,33. The phage Yep-phi, required not only LPS but also Ail and OmpF for its efficient infection, losing of the ail and ompF products may resulted in defective phage receptors34. In order to identified whether OmpA was involved in the phage infection of phiYe-F10, we constructed of HNF10-ΔompA knock-out mutants to test the growth characteristic and phage adsorption efficiency. The O-side-chain of Yersinia enterocolitica O:3 is a homopolymer of 6-deoxy-L-altrose35. Our research had suggested that the Yersinia enterocolitica serogroup O:3 were all carried with the fragment of the rfbC gene. The rfbc gene clustered in the rfb locus of the bacterial chromosome, and encoded the dTDP -rhamnosyl transferase RfbF. It directed the O-antigen biosynthesis through synthesize the O-antigen glycosyltransferases, which transferred the rhamnosyl to growing repeat units. As the phage phiYe-F10 specific for Y. enterocolitica serotype O:3, and the rfbc sequence was strictly found in sensitive strains, we wanted to identify whether dTDP -rhamnosyl transferase RfbF was involved in the phage infection of phiYe-F10.
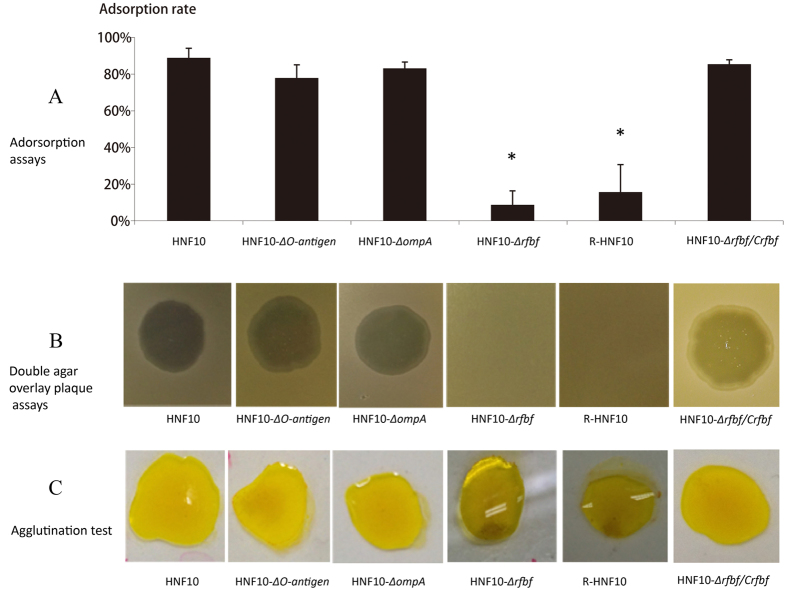
The mutants HNF10-Δrfbf, HNF10-ΔO-antigen and HNF10-ΔompA were verified by PCR (Supplemental data 2). The WT, knock-out mutant strains, the spontaneous rough mutant R-HNF10 (A spontaneous rough derivative of Y. enterocolitica serotype O:3 strain HNF10 carried with virulence-plasmid), and compensation strain HNF10-Δrfbf/Crfbf were all grown to log phase in LB broth at 25 °C with agitation reaching a similar value of OD600 for each group. After infecting with phage phiYe-F10 (at 4.5 h in total growth), every half hour the OD was graphed generating a growth curve for each group. The data showed WT, HNF10-ΔompA, HNF10-ΔO-antigen and HNF10-ΔrfbF/Crfbf were suppressed after infecting with phage phiYe-F10, and then began lysing (Fig. 3B). The OD600 value of the WT and HNF10-ΔompA showed a decrease after one hour (at 5.5 h in total growth) with phage phiYe-F10 infection; and then 2.5 hours later (at 7 h in total growth), both WT and HNF10-ΔompA were completely lysed at OD600 ≈0. phiYe-F10 adsorbed to the HNF10-ΔO-antigen and then began lysing (at 6 h in total growth), with half hour late than WT and HNF10-ΔompA (at 5.5 h in total growth). And 3 h (at 7.5 h in total growth) later, HNF10-ΔO-antigen reached complete lysis with an OD600 ≈0. The HNF10-Δrfbf and R-HNF10 resisted to phage infection with OD600 values of growth curves similar to their control groups (Fig. 3A,B). Consistent with these results, the adsorption assays showed a significant reduction in phage adsorption to R-HNF10 and HNF10-Δrfbf when compared to the adsorption rate of wild-type HNF10 strain, and the difference was statistically significant (Fig. 4A).
Figure 3. The growth curves of WT, its mutants, and compensation strain.
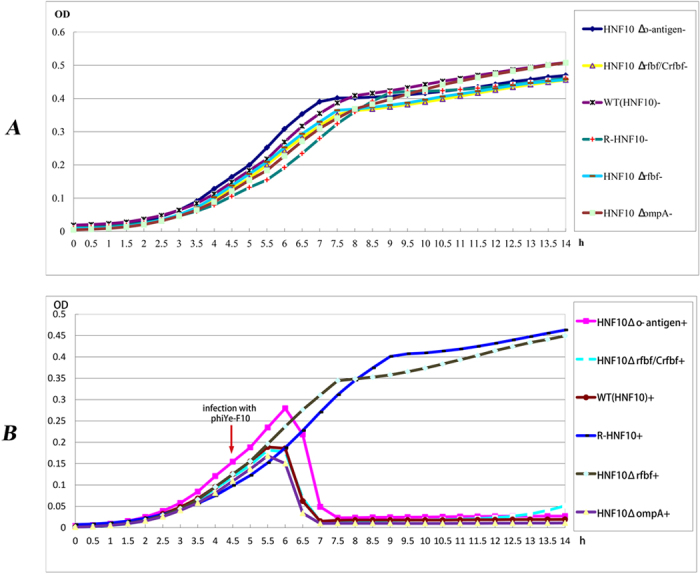
(A) The growth curves of strains without phage infection. (B) The growth curves of strains with phage infection. Approximately 6 × 107 PFU of phiYe-F10 in 1 ml was mixed with the 10ml bacterial culture (OD600 ≈0.1–0.2), and incubated at 25 °C for 9.5 h. Control cultures were grown without phage infection. The OD600 of each group was measured every half hour. At the time indicated by arrow, phage phiYe-F10 was added to the culture.
Figure 4. The results of phage adsorption assay, the double-layer plaque assay and Acriflavine agglutination test of WT, its mutants, and compensation strain.
(A) Adsorption assay of bacteriophage phiYe-F10 to the test strain. Approximately 6 × 106 PFU of phiYe-F10 in 100 μl was mixed with 500 μl samples of bacteria (OD600 ≈1.0). The adsorption rate of each strain was calculated as (Pt′-Pt)/Pt′. Error bars denote statistical variations. Significance was determined by Dunnett T3 test for comparison between the mutant group and the WT group. *P < 0.05. (B) The double-layer plaque assay. (C) Acriflavine Agglutination test.
dTDP-rhamnosyl transferase RfbF coding gene rfbc deletion in Y. enterocolitica resulted in resistance to phage phiYe-F10
The WT, knock-out mutants strains (HNF10-Δrfbf, HNF10-ΔO-antigen, HNF10-ΔompA), spontaneous rough mutant R-HNF10, and compensation strain HNF10-Δrfbf/Crfbf were tested the sensitivity to the phage with the double-layer plaque assay (Fig. 4B). The results showed the knock-out mutant strain HNF10-Δrfbf and spontaneous rough mutant R-HNF10 were resistant to the phage, producing no plaques and showing a weak positive in the Acriflavine agglutination test; while the WT, HNF10-ΔO-antigen, HNF10-ΔompA, and HNF10-Δrfbf/Crfbf strains produced plaques and showed negative in the Acriflavine agglutination test (Fig. 4B,C).
Discussion
In natural environments, bacteria and phage are competing and co-existing with each other. In this competition, the bacteria evolve resistance mechanisms against phage infection. These strategies include blocking receptors or altering receptor structures to prevent phage adsorption, preventing phage DNA entry, digesting phage nucleic acid, inhibiting replication of the phage genome, and cause abortive phage infection. Conversely, bacteriophages are capable of rapid adaptive responses to evolutionary changes in their hosts. As a counter-defensive measure, phages are able to modify their receptor binding proteins, e.g. the tail fiber, to achieve infection and kill the resistant bacterium. For example, Pseudomonas fluorescens SBW25 was found to coevolve with its lytic phage phi2 for more than 300 bacterial generations. This co-evolution is probably due to the continuous modification of the bacterial receptors and phage receptor binding protein36,37.
Yersinia enterocolitica is a common foodborne pathogen with O:3 and O:9 as the primary infectious serotypes in most countries8,38. It was reported that, there were two lytic phages which infected Yersinia enterocolitica O:3 had been fully sequenced: phiYeO3-12 (GenBank accession no. AJ251805.1) and vB_YenP_AP5 (GenBank accession no. KM253764.1). The genome sequences of YeO3-12 and vB_YenP_AP5 were submitted to the GenBank databases in 1999 by Pajunen, M.I. and colleagues and in 2014 by Leon-Velarde, C.G. and colleagues, respectively. The two phages were all belonged to the family Podoviridae12,14. In this study, the lytic Yersinia phage phiYe-F10, was isolated together with the pathogenic bioserotype 3/O:3 Y. enterocolitica HNF-10 from the same swine rectal swab sample. The whole genome sequence of phage phiYe-F10 showed great similarities to Yersinia phage PhiYeO3-12 and vB_YenP_AP5, excepted for minor insertions or deletions among the genome sequences in these phages (Fig. 1C). The genomic comparisons indicated the phiYe-F10, PhiYeO3-12 and vB_YenP_AP5 were closely genetically related, and most of their DNA sequences appeared to have descended from a single common ancestral phage. The Podoviridae family phage was common in Enterobacteriaceae, which showed the morphologic characteristics of an icosahedral head and a short non-retractable tail when observed under transmission electron microscopy, and the phages were known to have double strands linear genomes with direct terminal repeats39,40. Currently, the reported Yersinia enterocolitica phage were belonged to different families, phage YeO3-12 and vB_YenP_AP5, which specific to infect Y. enterocolitica of serotype O:3 belonged to the family Podoviridae12,14; however the temperate phage PY54 (GenBank accession no. AJ564013.1) belonged to the family Siphoviridae, which infected Y. enterocolitica of serotype O:5 and O:5, 2711,41; the phage PhiR1-37 (GenBank accession no. AJ972879.2) belonged to the family Myoviridae, which infected strain YeO3-R1 (a virulence-plasmid-cured O antigen-negative derivative of Yersinia enterocolitica serotype O:3)42. The tail fiber protein sequences alignment revealed that the phages belonged to the family Podoviridae showed a high similarity and were classified into the same cluster (Fig. 2), suggested that they follow a similar DNA ejection mechanism, with tail fibers adsorbed to the host surface LPS. Although the phage vB_YenP_ISAO8, PhiR1-37 and PY54 displayed lysis for Y. enterocolitica, the sequences of tail fiber proteins were clustered into different groups, and the hosts serotypes and phage receptors were different from that of phiYe-F10, phiYeO3-12 and vB_YenP_AP5 (Fig. 2).
Phage phiYe-F10 displayed strict specificity for Y. enterocolitica O:3 at 25 °C and 37 °C. Other serotypes (O:8, O:9, O:5, O:5, 27 et al.) of Y. enterocolitica as well as Y. pseudotuberculosis and Y. pestis were unaffected by the presence of phage phiYe-F10. Previous research indicated that Yersinia enterocolitica phage PhiYeO3-12 and PhiR1-37 used LPS as their receptors12,42. Here we first confirmed that the dTDP-rhamnosyl transferase RfbF, coded by the rfbc gene, played a critical role in synthesizing the phage receptor for phiYe-F10. The 57 phiYe-F10 sensitive Y. enterocolitica of O:3 serotype were all carrying the DTDP-rhamnosyl transferase RfbF encoding gene (rfbc). The rfbc gene, which responsible for the biosynthesis of the O side chain of Y. enterocolitica, was chosen for the specific detection target of O:3 serotype. Among the sensitive strains, four strains had different rfbc gene with multiple mutation sites compared with the rest of the strains. Although the protein sequence of dTDP-rhamnosyl transferase RfbF changes, the function of the phage receptors was remained and therefore these four strains were still phage sensitive. The rfbc gene located within the rfb gene cluster encoded the dTDP-rhamnosyl transferase RfbF, which was involved in the dTDP-L-rhamnose biosynthesis. L-Rhamnose is the receptor on the LPS core for the attachment of O polysaccharides43, it is an indispensable component of the lipopolysaccharide synthetic pathway35,44,45. Y. enterocolitica O:3 O-antigen is a homopolymer of 6-deoxy-L-altrose26; the rfbc gene regulates the LPS O antigen synthesis pathway. Deletion of dTDP-rhamnosyl transferase RfbF may resulted in the changes or losses of the O antigen, resulting in the failure of phages to bind and infect the strains. Complementation of dTDP-rhamnosyl transferase RfbF deletion mutant recovered its sensitivity to phiYe-F10. Mutation of dTDP-rhamnosyl transferase RfbF retained agglutination with O:3 antiserum, suggested some group modifications of O antigen were responsible for the phage infection but did not affect recognition of the O:3 antiserum.
Deletion of the putative O-antigen biosynthesis protein precursor encoding gene may reduced the infection efficiencies; however, the mutant strain was still phiYe-F10 sensitive. When infected with phage phiYe-F10 at a similar OD in logarithmic phase, the WT strain began to lyse after 1 h and reached complete lysis 2.5 hour later; whereas HNF10-ΔO-antigen started lysis at 1.5 h and reached complete lysis 3 hour later. This showed phiYe-F10 adsorbed to HNF10-ΔO-antigen slower than WT; but the time needed from the beginning of lysis to complete lysis was the same for the two strains (Fig. 3B). Consequently, we speculated that the putative O-antigen biosynthesis protein precursor played a role in the first steps of virus-host interaction, the deletion of this protein may affected phage adsorption and phage DNA entry, e.g., blocking the phage tail fiber recognition and binding the receptor; or altering the host receptor structure on the bacterial cell surface to prevent phage adsorption, or preventing phage DNA entry. This needs further investigation.
Many studies showed that OmpA was a common phage receptor of Enterobacteriaceae23,31, but also a common immune protective antigen beyond the V antigen conserved within Yersinia species32,33. Unlike Y. pseudotuberculosis and Y. enterocolitica, Y. pestis had a rough LPS without O antigen where Ail and OmpF were identified to act as the receptors of Yep-phi in addition to the rough lipopolysaccharide of Y. pestis34. In our study, the OmpA deletion mutant of Y. enterocolitica did not affect the binding and lysis of phage phiYe-F10, showing OmpA was not the receptor of phiYe-F10.
A gene altered by artificial or natural mutation will lead to the lack or change of the phage receptor; then alter the phage resistance. In Y. pestis, some spontaneous mutations in the core polysaccharide brought about the loss of the LPS core, which resulted in phage resistance46. A sequence comparison between the spontaneous resistant strain (R-HNF10) and the WT strain showed the coding genes of dTDP-rhamnosyl transferase RfbF were completely the same. However, R-HNF10 formed obviously rough colonies on agar; and the Acriflavine agglutination test also showed O-antigen deficiency, which further suggested that the phiYe-F10 resistance mechanism resulted from an O-antigen deficiency. Some of the R-HNF10 resistant strains did not agglutinate with O:3 antiserum and monoclonal antibodies, but the others with serotype not changed. This finding suggested small modifications or other changes of O antigen were responsible for the phage infection (binding) but did not affect recognition of the O:3 antiserum. We inferred the epitopes determining serogroup had more and larger distribution than that of phage receptor within the LPS O side-chains.
This study firstly showed the phage receptor of phiYe-F10 was regulated by dTDP-rhamnosyl transferase RfbF which was encoded by the LPS O side-chain synthesis related gene, rfbc. Presently, O:3 serotype Y. enterocolitica infections occur widely in the world and bring heavy burden to the social community and families8,47. The phage is proposed as a promising alternative to conventional antibiotics for treating bacterial infections. The emergence and high occurrence of this serotype strain requires in-depth research of the phage specific for Y. enterocolitica serotype O:3. The finding of the phage specific for Yersinia enterocolitica serotype O:3 and the successful detection of its receptor regulatory protein may provide a foundation for the prevention of infection and transmission of Y. enterocolitica.
Methods
Bacteriophage isolation and sensitivity test
A lytic phage, named phiYe-F10, specific for Yersinia enterocolitica was isolated from a swine rectal swab sample. The phage concentration in the swine rectal swab, was estimated using the plaque assay on Y. enterocolitica HNF-10 (Bioserotype 3/O:3, virulence plasmid positive). The phage was isolated together with strain HNF-10 from the same sample in a routine prevalence surveillance for Yersinia in China.
From these specimens, phiYe-F10 was chosen for detailed study because of its ability to infect Y. enterocolitica strains of serotype O:3. The host range of phiYe-F10 was determined using a double-layer plaque assay at 25 °C and at 37 °C, with one Y. pestis, 188 Y. enterocolitica and 37 Y. pseudotuberculosis belonging to different serotypes. Strains used in the bacteriophage sensitivity test were listed in Table 1. Among the 188 different Y. enterocolitica, 183 were widely distributed within 17 provinces of China from 1982 to 2014; and 5 reference strains were provided by H. Fukushima at the Shimane Prefectural Institute of Public Health, Matsue, Japan. The strains were collected from routine monitoring of animals, food and patients with diarrhea (Supplemental data 1).
Electron microscopy
Filtered phage lysates (about 2 × 1010 PFU/mL) were pelleted at 25,000 × g for 1 h at 4 °C, using a Beckman high-speed centrifuge with a JA-18.1 fixed-angle rotor (Beckman, Palo Alto, CA, USA). The phage pellet was washed twice in neutral 0.1 M ammonium acetate. The final phage sediment was re-suspended in 150 μL of SM-buffer supplemented with 5 mM CaCl2. Samples were then deposited onto a carbon-coated Formvar film on copper grids, and stained with 20 μl of 2% potassium phosphotungstate (PT, pH 7.2). Get rid of the dye with filter paper, air dried, and examined under a TECNAI 12 transmission electron microscope (FEI, Hillsboro, OR, USA) at 120 KEv. Images were collected and analyzed using Digital Micrograph™ Software (Gatan, Pleasanton, CA, USA).
The Genome sequencing of phage DNA, assembly and bioinformatics analysis
A random “shotgun” library was constructed using a fast nebulization method48 and the fragments were ligated to the vector pUC118 (TaKaRa Code: 3322, pUC118 HincII/BAP). Cycle sequencing reactions from the plasmid inserts were performed using the ABI PRIS Mw Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems) with a Gene Ampw PCR System 9700 (Applied Biosystems). Sequence reactions were analyzed using an ABI PRISMTM 3730XL DNA Analyzer (Applied Biosystems). Sequencing continued until an eight-fold coverage of the sequenced plasmid was attained. Assembly of the sequences was performed using the SeqMan module of the Lasergene software (DNASTAR Inc.). Persisting gaps were closed using primer-walking sequencing of the genomic DNA.
A dot plot comparison and graphical comparison were conducted using BLAST 2.25 and displayed using ACT49. The minimum score cutoff was 100 and the minimum identity cutoff was 50%.
Sequences of tail fiber proteins were collected from NCBI (6 phages for Y. enterocolitica, 1 phage for Y. pestis, and the Escherichia coli phages T7 and T3), the phylogenetic tree was generated using the neighbor-joining method implemented in MEGA6. Bootstrap values representing the percent in 1,000 replicates were shown in the tree.
Bacteria, plasmids, and growth media
The Y. enterocolitica wild type strain HNF10 was isolated from a swine rectal swab sample from Henan province. R-HNF10 was a spontaneous rough mutant of HNF10. The knock-out mutants HNF10-ΔO-antigen, HNF10-ΔompA, and HNF10-Δrfbf were constructed in this work. The strains and plasmids used for gene cloning and mutation were listed in Table 3. The serotypes, biotypes, and pathogenicity of these strains were determined as previously describe8,50. The E. coli and Y. enterocolitica strains were incubated at 37 °C and 28 °C, respectively. Solid and soft agar media contained 2.0% and 0.5% (w/v) agar, respectively. Antibiotics were used at the following concentrations: kanamycin: 50 μg/ml for agar plates and 100 μg/ml for broth; chloramphenicol, 34 μg/ml; cefsulodin, 15 μg/ml; and novobiocin, 2.5 μg/ml.
Table 3. Strains and plasmids used for gene cloning and mutation in this study.
| Strains and plasmids | Description or comments | Source orreference |
|---|---|---|
| Strains | ||
| Y. entercolitica HNF10 | BioSerotype 3/O:3 with pYV plasmid, isolated in Henan, China from a swine rectal swab | This study |
| HNF10-ΔO-antigen | Strain HNF10 with a deletion of gene for putative O-antigen biosynthesis protein precursor | This study |
| HNF10-ΔompA | Strain HNF10 with a deletion of ompA gene | This study |
| HNF10-Δrfbf | Strain HNF10 with a deletion of rfbc gene | This study |
| HNF10-Δrfbf/Crfbf | rfbc complementation of HNF10-Δrfbf | |
| R-HNF10 | Spontaneous rough mutant of HNF10 | This study |
| S17 λpir | λ-pir R6K (thi thr leu ton lacY supE recA::RP4-2Tc::Mu) | 52 |
| Plasmids | ||
| pDS132 | Conditionally replicating vector; R6K origin, mobRK4 transfer origin, sucrose-inducible sacB, CmR | 51 |
| pDS132-O-antigen | containing a fusion fragment of upstream and downstream of the O-antigen gene | This study |
| pDS132-ompA | containing a fusion fragment of upstream and downstream of the ompA gene | This study |
| pDS132-rfbf | containing a fusion fragment of upstream and downstream of the rfbc gene | This study |
| pSRKKm | Expression vector containing lac promoter and lacIq, KmR | 53 |
| pSRKKm -rfbf | rfbc orf cloned into pSRKKm, KmR | This study |
Construction of HNF10-ΔrfbF, HNF10-ΔO-antigen and HNF10-ΔompA knock-out mutants
The deletion mutants were constructed using homologous recombination with the suicide plasmid pDS132 51. To obtain the knock-out mutants, the corresponding two sets of primers were used to amplify two different fragments respectively. The p1 and p2 primers amplified the upstream region of genes, the P3 and P4 primers amplified the downstream region of genes (Table 4). pDS132-O-antigen, pDS132-ompA and pDS132-rfbf were generated using the In-Fusion HD Cloning Kit (Clontech) following the manufacturer’s instructions. The recombinant plasmids were induced into the competent cell S17λpir52, and then mobilized into Y. enterocolitica HNF10 through biparental conjugation. Transconjugants were selected after growth on LB plates containing Yersinia complement (cefsulodin and novobiocin) and chloramphenicol. Bacteria from individual colonies were pooled and allowed to grow in LB without antibiotic overnight at 25 °C. Bacterial cultures were serially diluted in LB without NaCl containing 10% sucrose; the plates were incubated at 25 °C. The recombinants that survived in the 10% sucrose were examined for their antibiotic resistance. The appropriate replacement of the wild-type alleles by the mutants was confirmed using PCR and sequencing.
Table 4. Primers used in this study.
| Primer | Sequence (5′–3′) |
|---|---|
| Construction of mutants | |
| rfbf p1 | AAAGGATCGATCCTCTAGAGGCGATCCTTATGGGTAC |
| rfbfp2 | GCTAGAAACTCACTAGAAAGATTTAAGAGACCGAAAA |
| rfbfp3 | TTTTCGGTCTCTTAAATCTTTCTAGTGAGTTTCTAGC |
| rfbfp4 | CCCGGGAGAGCTCGATATCGCAACATCACGGAATCTT |
| ompA p1 | AAAGGATCGATCCTCTAGAGACAGAAATTGAGTCGTTTAT |
| ompA p2 | GTTTTTTACGGAAGCAGTAAACTTTACGCCTCGTTATC |
| ompA p3 | GATAACGAGGCGTAAAGTTTACTGCTTCCGTAAAAAAC |
| ompA p4 | CCCGGGAGAGCTCGATATCTAGTAGGGTGAAATTCT |
| Oag P1 | AAAGGATCGATCCTCTAGAGGAGGTTGTTACTGTTAC |
| Oag P2 | AAGAATTTTTCTCTTCATATAATTTCCCTCCAAGAGA |
| Oag P3 | TCTCTTGGAGGGAAATTATATGAAGAGAAAAATTCTT |
| Oag P4 | CCCGGGAGAGCTCGATATCGTTGGTGCGTCACTAAGA |
| Complementation of mutation HNF10-Δrfbf | |
| rfbfCF | AATCCATATGAATGCATCGCAATATTC |
| rfbfCR | AATCGAGCTCTTAATTTAACTTACCTG |
Note: Restriction endonuclease sites are underlined.
Complementation of mutations
Primers rfbfCF and rfbfCR, incorporating NdeI and SacI restriction sites (Table 4), were used to amplify the ORF of rfbc, including the 904-bp region in the HNF10 genome. The amplicon was digested with NdeI and SacI (New England BioLabs) and ligated into the NdeI- and SacI-digested plasmid pSRKKm (D3050; TaKaRa, Japan)53, producing plasmid pSRKKm–rfbf (the rfbc ORF cloned into pSRKKm, KmR); introduced into the competent cell S17λpir; and then mobilized into Y. enterocolitica HNF10-Δrfbf using biparental conjugation. The recombinant clones HNF10-Δrfbf/Crfbf were confirmed using PCR and sequenced to ensure they contained the correct insert sequence.
Phage adsorption assays
The WT, R-HNF10, HNF10-Δrfbf, HNF10-ΔO-antigen HNF10-ΔompA knock-out mutant strains and compensation strain HNF10-Δrfbf/Crfbf were cultured overnight on BHI at 25 °C. Approximately 6 × 106 PFU of phiYe-F10 in 100 μl was mixed with 500 μl samples of bacteria (OD600 ≈1.0). The suspension was incubated at room temperature for 5 min and centrifuged at 16,000 × g for 3 min, after which the phage titer remaining in the supernatant was determined as Pt. BHI was used as a non-adsorbing control in each assay, and the phage titer in the control supernatant was set to Pt′. The adsorption rate of each strain was calculated as (Pt′-Pt)/Pt′× 100%, each stain was repeated three times.
Agglutination test
The WT, R-HNF10, knock-out mutant strains and compensation strain were agglutinated using concentrations of Acriflavine. This procedure offered a simple way to distinguish LPS defective bacteria. Bacteria were suspended in 0.2% Acriflavine solution, the LPS defective strains agglutinated, whereas the LPS positive strains showed no agglutination54,55.
The growth characteristic of WT, mutant strains and compensation strain
The WT, R-HNF10, HNF10-Δrfbf, HNF10-ΔO-antigen HNF10-ΔompA and HNF10-Δrfbf/Crfbf were grown overnight and 1:100 diluted in fresh LB medium at the zero time point. After 4.5 h culture, 1 ml of phiYe-F10 (6 × 107 PFU/ml) was mixed with the bacterial culture (10 ml), and incubated at 25 °C for 9.5 h. Control cultures were grown without phage infection. The OD600 of each group was measured every half hour. Each group was repeated three times.
Ethics statement
The sample collection and detection protocols were carried out in accordance with relevant guidelines and regulations. All experimental procedures were approved by the Ethics Review Committee [Institutional Review Board (IRB)] of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. Signed informed consent was obtained from all study participants.
Additional Information
Accession codes: The annotated genome sequence for the phage phiYe-F10 was deposited in the NCBI nucleotide database under the accession number KT008108.
How to cite this article: Liang, J. et al. DTDP-rhamnosyl transferase RfbF, is a newfound receptor-related regulatory protein for phage phiYe-F10 specific for Yersinia enterocolitica serotype O:3. Sci. Rep. 6, 22905; doi: 10.1038/srep22905 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (General Project, no. 31500117, 81470092) and the National Sci-Tech Key Project (2012ZX10004201, 2013ZX10004203-002). We thank Liuying Tang and Jim Nelson for critical reading and helpful comments on our manuscript.
Footnotes
Author Contributions X.W. and H.J. conceived and designed the experiments; J.L., X.L., T.Z., Y.C. and Y.X. performed the experiments; M.S. participated in the sequence alignment; H.H., C.L. and R.D. performed the statistical analysis, J.L., X.L., R.D. and Y.C. wrote the paper. All authors read and approved the final manuscript.
References
- Heilpern A. J. & Waldor M. K. CTXphi infection of Vibrio cholerae requires the tolQRA gene products. Journal of bacteriology 182, 1739–1747 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M. et al. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS microbiology letters 211, 77–83 (2002). [DOI] [PubMed] [Google Scholar]
- Fredriksson-Ahomaa M. & Korkeala H. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: a methodological problem. Clinical microbiology reviews 16, 220–229 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice M. B., Cope D. & Swann R. A. The epidemiology of Yersinia enterocolitica infection in the British Isles 1983–1988. Contributions to microbiology and immunology 12, 17–25 (1991). [PubMed] [Google Scholar]
- Bottone E. J. Yersinia enterocolitica: the charisma continues. Clinical microbiology reviews 10, 257–276 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P. O. et al. Variation in the prevalence of enteropathogenic Yersinia in slaughter pigs from Belgium, Italy, and Spain. Foodborne pathogens and disease 8, 445–450, 10.1089/fpd.2009.0461 (2011). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Distribution of pathogenic Yersinia enterocolitica in China. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 28, 1237–1244, 10.1007/s10096-009-0773-x (2009). [DOI] [PubMed] [Google Scholar]
- Liang J. et al. Prevalence of Yersinia enterocolitica in pigs slaughtered in Chinese abattoirs. Applied and environmental microbiology 78, 2949–2956, 10.1128/AEM.07893-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. A. et al. Yersinia enterocolitica O:3: an emerging cause of pediatric gastroenteritis in the United States. The Yersinia enterocolitica Collaborative Study Group. The Journal of infectious diseases 163, 660–663 (1991). [DOI] [PubMed] [Google Scholar]
- Poljak Z. et al. Prevalence of Yersinia enterocolitica shedding and bioserotype distribution in Ontario finisher pig herds in 2001, 2002, and 2004. Preventive veterinary medicine 93, 110–120, 10.1016/j.prevetmed.2009.10.003 (2010). [DOI] [PubMed] [Google Scholar]
- Hertwig S. et al. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. Journal of molecular biology 331, 605–622 (2003). [DOI] [PubMed] [Google Scholar]
- Pajunen M., Kiljunen S. & Skurnik M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. Journal of bacteriology 182, 5114–5120 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajunen M. I., Kiljunen S. J., Soderholm M. E. & Skurnik M. Complete genomic sequence of the lytic bacteriophage phiYeO3-12 of Yersinia enterocolitica serotype O:3. Journal of bacteriology 183, 1928–1937, 10.1128/JB.183.6.1928-1937.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Velarde C. G. et al. Complete genome sequence of bacteriophage vB_YenP_AP5 which infects Yersinia enterocolitica of serotype O:3. Virology journal 11, 188, 10.1186/1743-422X-11-188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig S., Klein I. & Appel B. Properties of the temperate Yersinia enterocolitica bacteriophage PY54. Advances in experimental medicine and biology 529, 241–243, 10.1007/0-306-48416-1_46 (2003). [DOI] [PubMed] [Google Scholar]
- Garcia-Doval C. & van Raaij M. J. Bacteriophage receptor recognition and nucleic acid transfer. Subcell Biochem 68, 489–518, 10.1007/978-94-007-6552-8_17 (2013). [DOI] [PubMed] [Google Scholar]
- Bielmann R. et al. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 477, 110–118, 10.1016/j.virol.2014.12.035 (2015). [DOI] [PubMed] [Google Scholar]
- Raimondo L. M., Lundh N. P. & Martinez R. J. Primary adsorption site of phage PBS1: the flagellum of Bacillus subtilis. Journal of virology 2, 256–264 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldvinsson S. B., Sorensen M. C., Vegge C. S., Clokie M. R. & Brondsted L. Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Applied and environmental microbiology 80, 7096–7106, 10.1128/AEM.02057-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camprubi S., Merino S., Benedi V. J. & Tomas J. M. Isolation and characterization of bacteriophage FC3-10 from Klebsiella spp. FEMS microbiology letters 67, 291–297 (1991). [DOI] [PubMed] [Google Scholar]
- al-Hendy A., Toivanen P. & Skurnik M. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infection and immunity 60, 870–875 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiljunen S. et al. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. Journal of bacteriology 193, 4963–4972, 10.1128/JB.00339-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Chen-Schmeisser U., Hindennach I. & Henning U. Apparent bacteriophage-binding region of an Escherichia coli K-12 outer membrane protein. Journal of bacteriology 153, 581–587 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Matsuzaki S. & Tanaka S. A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS microbiology letters 125, 101–105 (1995). [DOI] [PubMed] [Google Scholar]
- Reeves P. R. et al. Bacterial polysaccharide synthesis and gene nomenclature. Trends in microbiology 4, 495–503 (1996). [DOI] [PubMed] [Google Scholar]
- Zhang L., al-Hendy A., Toivanen P. & Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-L-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Molecular microbiology 9, 309–321 (1993). [DOI] [PubMed] [Google Scholar]
- Zhao G., Wu B., Li L. & Wang P. G. O-antigen polymerase adopts a distributive mechanism for lipopolysaccharide biosynthesis. Applied microbiology and biotechnology 98, 4075–4081, 10.1007/s00253-014-5552-7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzetti D. et al. A molecular scheme for Yersinia enterocolitica patho-serotyping derived from genome-wide analysis. International journal of medical microbiology: IJMM 304, 275–283, 10.1016/j.ijmm.2013.10.007 (2014). [DOI] [PubMed] [Google Scholar]
- Weynants V., Jadot V., Denoel P. A., Tibor A. & Letesson J. J. Detection of Yersinia enterocolitica serogroup O:3 by a PCR method. Journal of clinical microbiology 34, 1224–1227 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant S. M., Grant T. H. & Robins-Browne R. M. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS immunology and medical microbiology 38, 127–137 (2003). [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L. Role of the receptor for bacteriophage lambda in the functioning of the maltose chemoreceptor of Escherichia coli. Journal of bacteriology 124, 119–126 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. et al. Gene polymorphism analysis of Yersinia enterocolitica outer membrane protein A and putative outer membrane protein A family protein. BMC genomics 15, 201, 10.1186/1471-2164-15-201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. Homology analysis and cross-immunogenicity of OmpA from pathogenic Yersinia enterocolitica, Yersinia pseudotuberculosis and Yersinia pestis. Molecular immunology, 10.1016/j.molimm.2015.09.016 (2015). [DOI] [PubMed] [Google Scholar]
- Zhao X. et al. Outer membrane proteins ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yep-phi. Journal of virology 87, 12260–12269, 10.1128/JVI.01948-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J., Lindberg B. & Brubaker R. R. Structural studies of the O-specific side-chains of the lipopolysaccharide from Yersinia enterocolitica Ye 128. Carbohydrate research 78, 212–214 (1980). [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Samson J. E. & Moineau S. Bacteriophage resistance mechanisms. Nature reviews. Microbiology 8, 317–327, 10.1038/nrmicro2315 (2010). [DOI] [PubMed] [Google Scholar]
- Tock M. R. & Dryden D. T. The biology of restriction and anti-restriction. Current opinion in microbiology 8, 466–472, 10.1016/j.mib.2005.06.003 (2005). [DOI] [PubMed] [Google Scholar]
- Bonardi S. et al. Detection, enumeration and characterization of Yersinia enterocolitica 4/O:3 in pig tonsils at slaughter in Northern Italy. International journal of food microbiology 177, 9–15, 10.1016/j.ijfoodmicro.2014.02.005 (2014). [DOI] [PubMed] [Google Scholar]
- Zhao X. et al. Characterization of phiCFP-1, a virulent bacteriophage specific for Citrobacter freundii. Journal of medical virology, 10.1002/jmv.24401 (2015). [DOI] [PubMed] [Google Scholar]
- Hardies S. C. et al. Identification of structural and morphogenesis genes of Pseudoalteromonas phage phiRIO-1 and placement within the evolutionary history of Podoviridae. Virology 489, 116–127, 10.1016/j.virol.2015.12.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl J. A., Klein I., Appel B. & Hertwig S. Interplay between the temperate phages PY54 and N15, linear plasmid prophages with covalently closed ends. Journal of bacteriology 189, 8366–8370, 10.1128/JB.01066-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiljunen S. et al. Yersiniophage phiR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology 151, 4093–4102, 10.1099/mic.0.28265-0 (2005). [DOI] [PubMed] [Google Scholar]
- Rahim R., Burrows L. L., Monteiro M. A., Perry M. B. & Lam J. S. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146(Pt 11), 2803–2814, 10.1099/00221287-146-11-2803 (2000). [DOI] [PubMed] [Google Scholar]
- Balsanelli E. et al. Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environmental microbiology 12, 2233–2244, 10.1111/j.1462-2920.2010.02187.x (2010). [DOI] [PubMed] [Google Scholar]
- Wartenberg K., Knapp W., Ahamed N. M., Widemann C. & Mayer H. Temperature-dependent changes in the sugar and fatty acid composition of lipopolysaccharides from Yersinia enterocolitica strains. Zentralblatt fur Bakteriologie, Mikrobiologie und Hygiene. 1. Abt. Originale A, Medizinische Mikrobiologie, Infektionskrankheiten und Parasitologie = International journal of microbiology and hygiene. A, Medical micro 253, 523–530 (1983). [PubMed] [Google Scholar]
- Filippov A. A. et al. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PloS one 6, e25486, 10.1371/journal.pone.0025486 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone E. J. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes and infection/Institut Pasteur 1, 323–333 (1999). [DOI] [PubMed] [Google Scholar]
- Pohl T. M. & Maier E. Sequencing 500 kb of yeast DNA using a GATC 1500 Direct Blotting Electrophoresis System. BioTechniques 19, 482–486 (1995). [PubMed] [Google Scholar]
- Carver T. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676, 10.1093/bioinformatics/btn529 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Pathogenic strains of Yersinia enterocolitica isolated from domestic dogs (Canis familiaris) belonging to farmers are of the same subtype as pathogenic Y. enterocolitica strains isolated from humans and may be a source of human infection in Jiangsu Province, China. Journal of clinical microbiology 48, 1604–1610, 10.1128/JCM.01789-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N., Alcaraz J. P., Coursange E., Geiselmann J. & Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255, 10.1016/j.plasmid.2004.02.003 (2004). [DOI] [PubMed] [Google Scholar]
- Thoma S. & Schobert M. An improved Escherichia coli donor strain for diparental mating. FEMS microbiology letters 294, 127–132 (2009). [DOI] [PubMed] [Google Scholar]
- Khan S. R., Gaines J., Roop R. M. 2nd & Farrand S. K. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Applied and environmental microbiology 74, 5053–5062, 10.1128/AEM.01098-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alton G. G., Jones L. M. & Pietz D. E. Laboratory techniques in brucellosis. Monogr Ser World Health Organ 55, 1–163 (1975). [PubMed] [Google Scholar]
- Corbel M. J. & Thomas E. L. The in vivo activity of a smooth phage-resistant variant of Brucella abortus strain 19. The British veterinary journal 132, 121–123 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.