Abstract
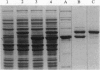
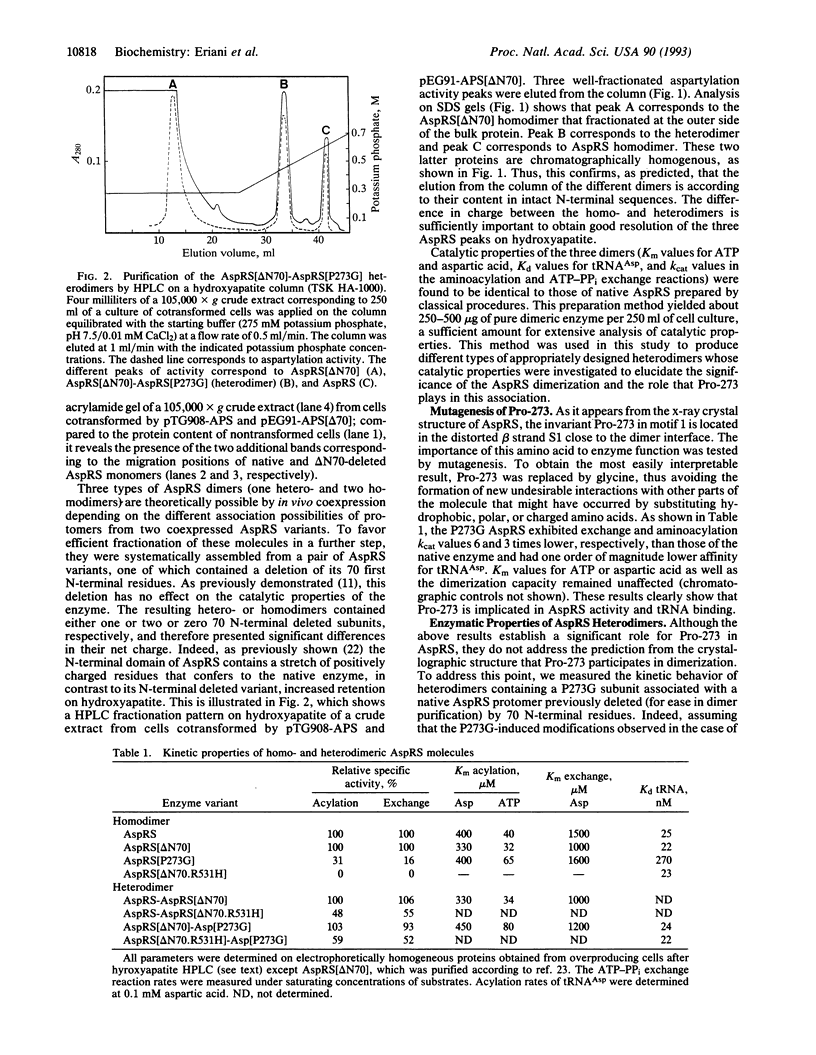
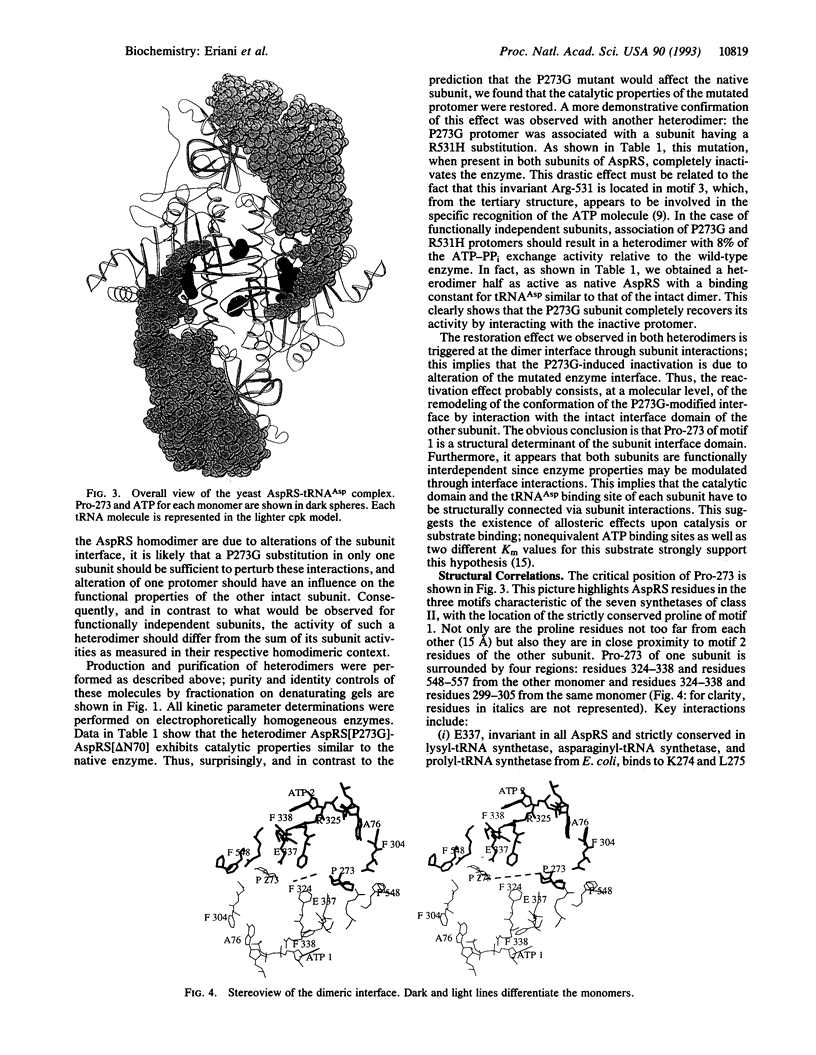
Cytoplasmic aspartyl-tRNA synthetase (AspRS; EC 6.1.1.12) from yeast is, as are most class II synthetases, an alpha 2 dimer. The only invariant amino acid in signature motif 1 of this class is Pro-273; this residue is located at the dimer interface. To understand the role of Pro-273 in the conserved dimeric configuration, we tested the effect of a Pro-273-->Gly (P273G) substitution on the catalytic properties of homo- and heterodimeric AspRS. Heterodimers of AspRS were produced in vivo by overexpression of their respective subunit variants from plasmid-encoded genes and purified to homogeneity in one HPLC step. The homodimer containing the P273G shows an 80% inactivation of the enzyme and an affinity decrease for its cognate tRNA(Asp) of one order of magnitude. The P273G-mutated subunit recovered wild-type enzymatic properties when associated with a native subunit or a monomer otherwise inactivated having an intact dimeric interface domain. These results, which can be explained by the crystal structure of the native enzyme complexed with its substrates, confirm the structural importance of Pro-273 for dimerization and clearly establish the functional interdependence of the AspRS subunits. More generally, the dimeric conformation may be a structural prerequisite for the activity of mononucleotide binding sites constructed from antiparallel beta strands.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amiri I., Mejdoub H., Hounwanou N., Boulanger Y., Reinbolt J. The complete amino acid sequence of cytoplasmic aspartyl-tRNA synthetase from Saccharomyces cerevisiae. Biochimie. 1985 Jun;67(6):607–613. doi: 10.1016/s0300-9084(85)80200-5. [DOI] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Construction of heterodimer tyrosyl-tRNA synthetase shows tRNATyr interacts with both subunits. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1189–1192. doi: 10.1073/pnas.83.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Giege R., Comarmond M. B., Thierry J. C., Moras D. Crystallographic studies on the aspartyl-tRNA synthetase-tRNAAsp system from yeast. The crystalline aminoacyl-tRNA synthetase. J Mol Biol. 1980 Mar 25;138(1):129–135. doi: 10.1016/s0022-2836(80)80008-8. [DOI] [PubMed] [Google Scholar]
- Dock A. C., Lorber B., Moras D., Pixa G., Thierry J. C., Giégé R. Crystallization of transfer ribonucleic acids. Biochimie. 1984 Mar;66(3):179–201. doi: 10.1016/0300-9084(84)90063-4. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Prevost G., Kern D., Vincendon P., Dirheimer G., Gangloff J. Cytoplasmic aspartyl-tRNA synthetase from Saccharomyces cerevisiae. Study of its functional organisation by deletion analysis. Eur J Biochem. 1991 Sep 1;200(2):337–343. doi: 10.1111/j.1432-1033.1991.tb16190.x. [DOI] [PubMed] [Google Scholar]
- Giegé R., Lorber B., Ebel J. P., Moras D., Thierry J. C., Jacrot B., Zaccai G. Formation of a catalytically active complex between tRNAAsp and aspartyl-tRNA synthetase from yeast in high concentrations of ammonium sulphate. Biochimie. 1982 May;64(5):357–362. doi: 10.1016/s0300-9084(82)80440-9. [DOI] [PubMed] [Google Scholar]
- Hinchman S. K., Henikoff S., Schuster S. M. A relationship between asparagine synthetase A and aspartyl tRNA synthetase. J Biol Chem. 1992 Jan 5;267(1):144–149. [PubMed] [Google Scholar]
- Koch G. L., Bruton C. J. The subunit structure of methionyl-tRNA synthetase from Escherichia coli. FEBS Lett. 1974 Mar 15;40(1):180–182. doi: 10.1016/0014-5793(74)80922-1. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee M. L., Muench K. H. Prolyl transfer ribonucleic acid synthetase of Escherichia coli. I. Purification and evidence for subunits. J Biol Chem. 1969 Jan 25;244(2):223–230. [PubMed] [Google Scholar]
- Lorber B., Kern D., Dietrich A., Gangloff J., Ebel J. P., Giegé R. Large scale purification and structural properties of yeast aspartyl-tRNA synthetase. Biochem Biophys Res Commun. 1983 Nov 30;117(1):259–267. doi: 10.1016/0006-291x(83)91569-3. [DOI] [PubMed] [Google Scholar]
- Lorber B., Mejdoub H., Reinbolt J., Boulanger Y., Giegé R. Properties of N-terminal truncated yeast aspartyl-tRNA synthetase and structural characteristics of the cleaved domain. Eur J Biochem. 1988 May 16;174(1):155–161. doi: 10.1111/j.1432-1033.1988.tb14076.x. [DOI] [PubMed] [Google Scholar]
- Prevost G., Eriani G., Kern D., Dirheimer G., Gangloff J. Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase. Eur J Biochem. 1989 Mar 15;180(2):351–358. doi: 10.1111/j.1432-1033.1989.tb14655.x. [DOI] [PubMed] [Google Scholar]
- Ruff M., Cavarelli J., Mikol V., Lorber B., Mitschler A., Giege R., Thierry J. C., Moras D. A high resolution diffracting crystal form of the complex between yeast tRNAAsp and aspartyl-tRNA synthetase. J Mol Biol. 1988 May 5;201(1):235–236. doi: 10.1016/0022-2836(88)90450-0. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellami M., Fasiolo F., Dirheimer G., Ebel J. P., Gangloff J. Nucleotide sequence of the gene coding for yeast cytoplasmic aspartyl-tRNA synthetase (APS); mapping of the 5' and 3' termini of AspRS mRNA. Nucleic Acids Res. 1986 Feb 25;14(4):1657–1666. doi: 10.1093/nar/14.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellami M., Prévost G., Bonnet J., Dirheimer G., Gangloff J. Isolation and characterization of the yeast aspartyl-tRNA synthetase gene. Gene. 1985;40(2-3):349–352. doi: 10.1016/0378-1119(85)90060-5. [DOI] [PubMed] [Google Scholar]
- Van Heeke G., Schuster S. M. Expression of human asparagine synthetase in Escherichia coli. J Biol Chem. 1989 Apr 5;264(10):5503–5509. [PubMed] [Google Scholar]
- Ward W. H., Jones D. H., Fersht A. R. Protein engineering of homodimeric tyrosyl-tRNA synthetase to produce active heterodimers. J Biol Chem. 1986 Jul 25;261(21):9576–9578. [PubMed] [Google Scholar]