Abstract
Many applications of pluripotent stem cells (PSCs) require efficient editing of silent chromosomal genes. Here, we show that a major limitation in isolating edited clones is silencing of the selectable marker cassette after homologous recombination and that this can be overcome by using a ubiquitous chromatin opening element (UCOE) promoter-driven transgene. We use this strategy to edit the silent IL2RG locus in human PSCs with a recombinant adeno-associated virus (rAAV)-targeting vector in the absence of potentially genotoxic, site-specific nucleases and show that IL2RG is required for natural killer and T-cell differentiation of human PSCs. Insertion of an active UCOE promoter into a silent locus altered the histone modification and cytosine methylation pattern of surrounding chromatin, but these changes resolved when the UCOE promoter was removed. This same approach could be used to correct IL2RG mutations in X-linked severe combined immunodeficiency patient-derived induced PSCs (iPSCs), to prevent graft versus host disease in regenerative medicine applications, or to edit other silent genes.
Introduction
Many applications require that silent genes be edited. This is especially true for pluripotent stem cells (PSCs), which may not express the tissue-specific genes responsible for diseases. For example, in one common paradigm for regenerative medicine, PSCs reprogrammed from a patient's cells would be propagated as undifferentiated cells, the disease-causing mutations present in silent genes such as β-globin (HBB) or interleukin 2 receptor gamma (IL2RG) would be corrected by gene editing, and the corrected cells would then be differentiated into a therapeutic cell product. The creation of isogenic PSC-based cellular disease models requires the same types of genetic manipulations, as does the engineering of lineage specification genes.
Transcription has long been known to increase homologous recombination1 and gene targeting by transfection-based methods.2 One way to overcome this limitation is the introduction of sequence-specific double-strand breaks (DSBs) by engineered nucleases in order to enhance silent gene targeting. For example, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have been used to isolate human PSC clones edited at PITX3 (ref. 3), HBB,4,5 and IL2RG.6,7 However, it seems likely that these site-specific DSBs did not completely eliminate the bias against editing silent loci, since the targeting frequencies were lower than those of expressed genes in these same cell types.3,4 The use of engineered nucleases may also lead to a variety of genotoxic effects, including unwanted sequence changes at on- and off-target sites,6,8 which could be a disadvantage in some settings.
Alternatively, the lower targeting frequencies observed at silent loci could be due to inadequate expression of the selectable marker gene after it integrates, rather than a decrease in homologous recombination. This appears to have occurred when a hygromycin resistance cassette was inserted into the HBB gene in human PSCs, since the gene-edited cells lost hygromycin resistance over time.4 This example highlights the poorly understood epigenetic changes that presumably occur during silent gene editing, which include potential alterations induced by the recombination and repair enzymes acting on the locus, the effects of introducing an expressed selectable marker into silent chromatin, and in many cases, the subsequent removal of that same expressed marker after isolating an edited clone. In general, the epigenetic consequences of gene editing remain an important but largely unexplored area of research. Two notable exceptions are studies showing that gene expression and DNA methylation can be altered in mice derived from embryonic stem cells (ESCs) with gene-targeted, imprinted loci,9,10 and a recent report showing that DNA methylation can be rendered unstable at a gene-targeted locus in Arabidopsis.11 The epigenetic effects of gene editing in human cells have not yet been described.
In this study, we use recombinant adeno-associated virus (rAAV) vectors to edit silent genes in human PSCs. rAAV vectors deliver single-stranded linear DNA genomes that efficiently recombine with homologous chromosomal sequences in human cells,12 including PSCs.13,14,15 Under optimal conditions, between 0.1 and 1% of normal human cells exposed to rAAV targeting vectors undergo high fidelity gene editing at expressed target loci,12,16 without a requirement for site-specific nucleases. To date, rAAV vectors have not been used to edit silent genes in PSCs, although rAAV-mediated editing of silent genes has been demonstrated at lower frequencies in hepatocytes and fibroblasts.17,18,19 Here, we evaluate different selectable marker cassettes to develop a robust, silent gene-editing method for human PSCs that does not require a site-specific nuclease, we examine the epigenetic consequences of targeting silent loci, and we determine the developmental effects of IL2RG gene editing.
Results
Transgene promoter type determines targeted clone survival
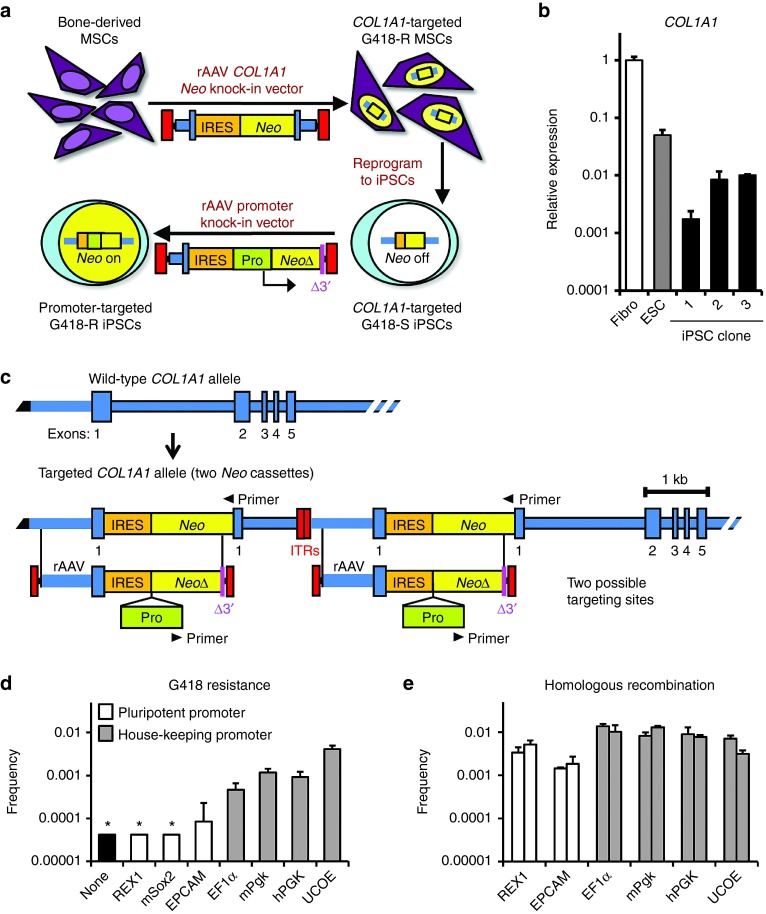
In order to optimize vector designs, we developed an assay to detect gene-editing events at a nontranscribed locus, in which only gene-targeted cells survive selection (Figure 1a). The assay uses induced pluripotent stem cells (iPSCs) containing a silenced Neo gene that can be activated by upstream promoter insertion. We first infected human mesenchymal stem/stromal cells (MSCs) with a rAAV knock-in vector designed to insert a Neo gene at the endogenous COL1A1 locus encoding type I collagen, which is highly expressed in MSCs. A polyclonal population of G418-resistant MSCs was then converted to iPSCs by expressing OCT4, SOX2, NANOG, and LIN28 transgenes.20 Three of these iPSC clones were analyzed further, and clone 1 had the lowest level of COL1A1 expression after reprogramming (Figure 1b). Southern blot analysis showed that this clone also had a duplication of the Neo transgene (Supplementary Figure S1c), which happens in a small percentage of targeted clones when vector genomes form dimers before recombination.16 Although this complicated our analysis, we confirmed that clone 1 was completely sensitive to G418 (Supplementary Figure S1a), so both Neo transgenes had been silenced and could therefore be activated by promoter insertion. The residual COL1A1 transcription detected in clone 1 cells may have been derived from the subpopulation of differentiating cells present in PSC cultures, which do not contribute to the PSC clones isolated by selection.
Figure 1.
Targeting a silent COL1A1-IRES-Neo cassette in human iPSCs. (a) Diagram of experimental design. (b) RT-qPCR of COL1A1 expression in undifferentiated iPSC clones containing COL1A1-IRES-Neo knockins. Fibro, human fibroblasts; ESC, undifferentiated H1 cells. (c) Structures of wild-type and IRES-Neo targeted COL1A1 alleles in iPSC clone 1 with rAAV promoter knock-in vector overlap indicated. The targeted COL1A1 locus contains two identical IRES-Neo cassettes, each of which can be targeted with rAAVs. Black triangles, primer-binding sites used for qPCR measurements of homologous recombination frequencies. (d) G418 resistance frequencies of iPSC clone 1 infected with promoter knock-in rAAVs. *less than 4 × 10−5. (e) Homologous recombination frequencies measured by qPCR with primers shown in c. Each infected cell population was analyzed with two primer pairs.
A series of gene editing vectors were designed to insert different promoters upstream of either silenced Neo transgene cassette, each of which contained a truncated Neo gene fragment in the right homology arm so that random integration could not confer G418-resistance, and only gene-edited clones would survive selection (Figure 1c). Two types of promoters were incorporated into the rAAV gene-editing vectors: developmentally regulated promoters that are expressed in human PSCs (REX1, murine Sox2, and EPCAM), and ubiquitously expressed promoters (EF1α, murine Pgk, human PGK, and ubiquitous chromatin opening element [UCOE]). When clone 1 PSCs with silenced Neo genes were infected with each of these vectors at the same multiplicity of infection, there were dramatic differences in the number of G418-resistant colonies obtained (Figure 1d), with the UCOE promoter producing the highest number. Southern blots showed that the G418-resistant colonies had been targeted at one of the two silent Neo gene targets (examples in Supplementary Figure S1c).
For a subset of vectors, gene-editing frequencies were also measured in unselected cells directly by qPCR, using one primer within the inserted promoter and one in chromosomal DNA outside of the homology arm (Figure 1e and Supplementary Figure S1b). This showed that despite a more than 100-fold difference in their ability to produce G418-resistant colonies, the REX1, EPCAM, EF1α, mPgk, hPGK, and UCOE promoter vectors all produced homologous recombinants at similar frequencies. Thus, rAAV vectors can edit a silent COL1A1 gene in human PSCs regardless of the transgene promoter they contain, but transgene selection requires a promoter that can drive expression at a silent locus. In the case of the UCOE promoter, the G418-resistance and homologous recombination frequencies were very similar, suggesting that almost every recombination event produced a G418-resistant cell. One potential drawback of the UCOE promoter is its relatively large size. Unfortunately, smaller promoter fragments did not produce as many G418-resistant colonies (Supplementary Figure S1d). These experiments demonstrate that the 1.2 kb UCOE promoter can be used to select for PSCs that undergo silent gene editing.
Editing of the silent IL2RG gene
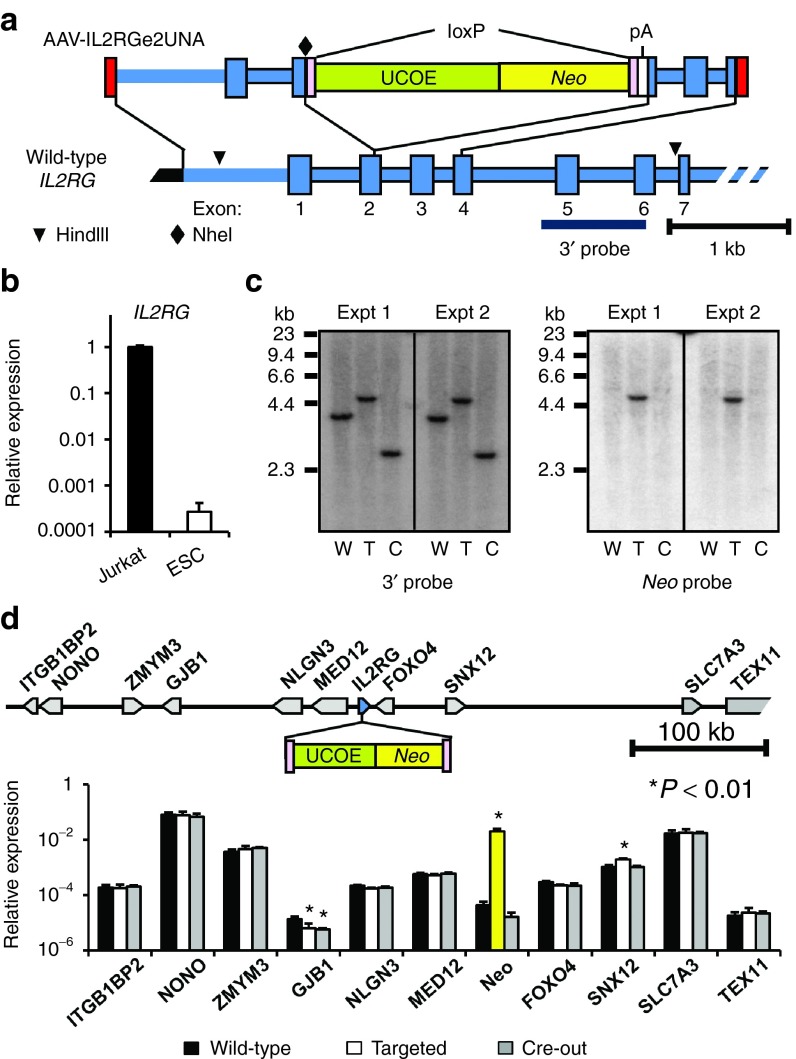
IL2RG encodes a common subunit for several cytokine receptors expressed in hematopoietic cells and is mutated in individuals with X-linked severe combined immunodeficiency (X-SCID). IL2RG is not expressed at a detectable level in human PSCs as measured by global expression arrays and RT-qPCR (data not shown and Figure 2b), is present as a single copy in male cells, and represents a promising target for developing PSC-based therapies. We therefore tested if a rAAV editing vector could be used to insert a UCOE-Neo-pA (UNA) cassette into exon 2 of IL2RG (Figure 2a). H1 human ESCs (a male cell line) were infected with vector AAV-IL2RGe2UNA and 3 of 18 G418-resistant colonies screened by PCR were targeted at the IL2RG gene (Supplementary Figure S2a). This represented 17% of G418-resistant colonies and 0.14% of the unselected cell population, which was similar to what we observed when targeting the COL1A1 locus, confirming that the UCOE promoter could be used to select for PSC clones with edited IL2RG genes.
Figure 2.
Targeting a silent IL2RG gene in human ESCs. (a) Structure of the IL2RG locus and rAAV targeting vector, with the locations of restriction enzyme sites and probe used in c. (b) RT-qPCR of IL2RG expression in undifferentiated ESCs. (c) Southern blot performed on age-matched wild-type (W), targeted (T), and Cre-out (C) clones, digested with HindIII and NheI and probed with a 3′ chromosomal fragment outside the homology arm, which produces fragments of 3.8, 4.7, and 2.6 kb in wild-type, targeted, and Cre-out clones, respectively. The same blot was stripped and re-probed with a Neo fragment. (d) RT-qPCR of gene expression of IL2RG's genomic neighbors and Neo in age-matched wild-type, targeted, and Cre-out clones. Relative expression signals were normalized to GAPDH and plotted as 2−(CT of gene − CT of GAPDH). Statistical significance was calculated by comparing to wild-type cells. *P < 0.01.
Many gene-editing applications also require the removal of the selectable marker cassette, so that only a linked, subtle editing change remains. Vector AAV-IL2RGe2UNA was designed so that Cre-mediated recombination would remove the floxed Neo cassette and leave behind a polyadenylation signal and three stop codons to inactivate IL2RG. We infected two different IL2RG-targeted clones with a nonintegrating foamy virus vector that transiently expressed Cre,21 and efficiently removed the Neo transgene cassette from 6–28% of cells (Supplementary Figure S2b). Southern blots confirmed the structures of the targeted and Cre-out alleles, as well as the lack of random integrants in targeted clones (Figure 2c).
Ideally, the editing of a silent locus would not affect the expression of other genes, so we analyzed neighboring gene expression in 10 genes spanning a 700 kb window surrounding IL2RG (Figure 2d). One gene (SNX12) had slightly increased expression in targeted cells that still contained the UCOE-Neo cassette, reflecting a potential long distance effect of the UCOE promoter. A second gene (GJB1) had statistically significant changes in RT-qPCR measurements, but all values were below those of the Neo signal from control cells lacking a Neo gene, suggesting a lack of expression. The eight other genes had no significant changes in expression, including the three genes located closest to IL2RG (NLGN3, MED12, and FOXO4), indicating that IL2RG gene targeting and subsequent Cre-out had minimal impact on neighboring gene expression.
Epigenetic consequences of gene editing
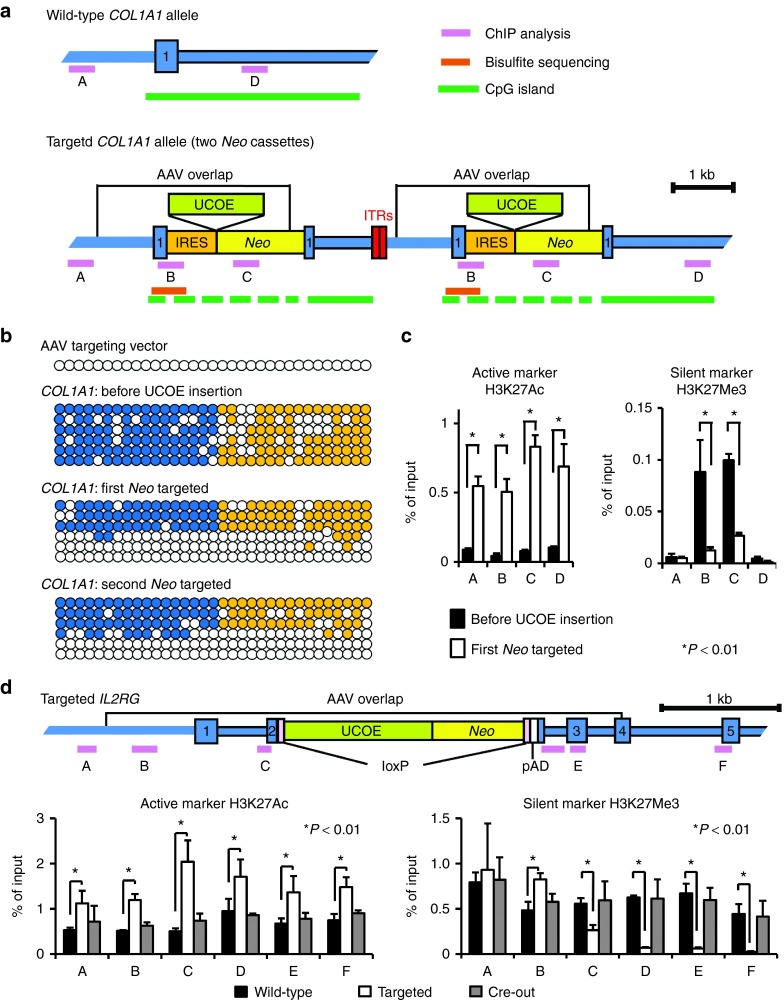
Insertion of an active promoter could change the epigenetic status of the surrounding chromatin, as could the recombination and repair proteins that carry out homologous recombination, yet the epigenetic consequences of gene editing remain a largely unexplored area of research. One possible effect is removal of 5-methylcytosine (5mC) residues in DNA, in particular at CpG islands, which are typically methylated in silent loci.22 The wild-type COL1A1 gene contains a CpG island that was duplicated in the PSC clone we used for promoter insertion studies (Figure 3a). When the locus is silent, these CpG islands are highly methylated, but after UCOE promoter insertion at either duplicated site, one of the islands became mostly unmethylated (Figure 3b). Although the bisulfite sequencing reaction does not distinguish between the two islands, these results are consistent with a localized region of hypomethylation at the CpG island nearest to the UCOE insertion site. Since this region is included within the 5′ homology arm of the UCOE insertion vector, incorporation of the unmethylated vector genome could have led directly to the loss of 5mC residues. In support of this hypothesis, we confirmed that the packaged rAAV vector genome was unmethylated (Figure 3b), and we showed previously that the entire homology arm sequence extending to the terminal repeats are typically incorporated into a targeted locus.23 Unfortunately, a similar analysis could not be performed at the IL2RG locus, which does not contain a CpG island within the homology region. We found no consistent difference in the methylation status of eight nonisland CpGs present in the IL2RG gene that were assayed in wild-type cells and knockout cells, suggesting that UCOE promoter insertion may not lead to demethylation of nonisland CpGs (Supplementary Figure S3).
Figure 3.
Epigenetic consequences of gene editing. (a) Structures of wild-type and IRES-Neo targeted COL1A1 loci shown with rAAV overlap, UCOE insertion sites, and CpG islands. DNA fragments (A to D) amplified in ChIP assays and bisulfite sequencing regions are marked. (b) Methylation status of the region spanning from exon 1 of COL1A1 (blue circles) to IRES (orange circles) in rAAV vector genomes, clone 1 genomic DNA, and clone 1 targeted at either the first or second Neo gene. Open and filled circles indicate unmethylated and methylated cytosines in CpGs, respectively. (c) The relative occupancies of H3K27Ac and H3K27Me3 in regions of the COL1A1 locus before and after UCOE insertion as measured by ChIP analysis. *P < 0.01. (d) Structure of the UCOE-Neo targeted IL2RG locus shown with rAAV overlap, loxP sites, and the locations of DNA fragments A to F amplified in ChIP assays. Histone occupancies were analyzed in age-matched wild-type, targeted, and Cre-out clones. *P < 0.01.
Histone modifications can also vary depending on the transcriptional activity of a locus and other factors. Although many such modifications have been described, here we studied acetylation and methylation at lysine 27 of histone H3 (H3K27Ac and H3K27Me3), which are associated with active and silent chromatin, respectively.24,25,26 In the case of COL1A1, the 1 kb region immediately surrounding the UCOE insertion site that includes the IRES element and Neo gene contained H3K27Me3 markers before UCOE insertion, indicative of silent chromatin (Figure 3c). After UCOE insertion, this pattern changed to one of active chromatin, with an increase in H3K27Ac levels that extended throughout the duplicated locus to sites 2.8 and 8.4 kb distal to the insertion site, as well as lower levels of H3K27Me3 in the more localized IRES and Neo regions. Interestingly, the change in H3K27Ac markers extended to regions outside of the homology arms, demonstrating that epigenetic changes had been propagated beyond the recombination site.
We studied the same histone markers at the IL2RG locus, only in this case, the analysis was more relevant because the target was single copy, the UCOE promoter was inserted at a wild-type gene that had not been previously targeted, and we could assay after both UCOE insertion and subsequent Cre-mediated UCOE removal. The wild-type locus had low levels of H3K27Ac and higher levels of H3K27Me3 throughout a 3.3 kb region surrounding the exon 2 insertion site, consistent with silent chromatin (Figure 3d). Insertion of the UCOE-Neo cassette activated this entire locus, including regions beyond the vector homology arms, as evidenced by increased H3K27Ac levels. However, unlike the COL1A1 locus, we did not observe a corresponding reduction in H3K27Me3 throughout this region. Instead, the decrease in H3K27Me3 was only observed downstream of the UCOE-Neo cassette. The basis for this asymmetry is unclear and was not shared by COL1A1, which had decreased H3K27Me3 levels on both sides of the UCOE insertion site. Importantly, once the UCOE-Neo cassette was removed by Cre-mediated recombination, the epigenetic status of the entire locus reverted back to that of the wild-type locus.
IL2RG-knockout human ESCs model X-SCID in vitro
While IL2RG mutation correction could be used to treat X-SCID, inactivation of IL2RG by gene editing could also have applications in regenerative medicine. For example, when differentiating pluripotent cells into hematopoietic progeny to be used for transplantation, it may be desirable to prevent the formation of natural killer (NK) or T cells that could react against host cells in a form of graft versus host disease. Based on the phenotype of X-SCID patients,27,28 and the known roles of the cytokine receptors that include IL2RG subunits,29 we predicted that IL2RG knockout stem cells should not produce NK or T cells in vitro.
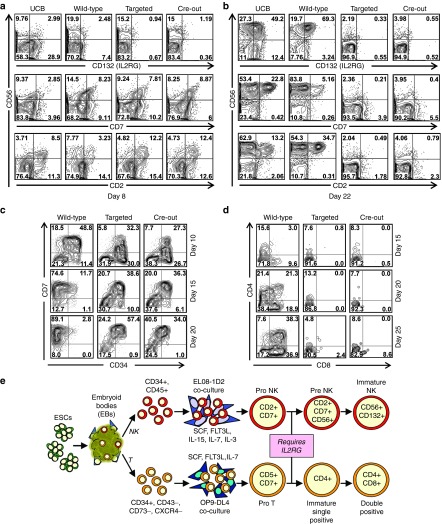
We first differentiated wild-type and IL2RG-edited ESC lines into embryoid bodies (EBs) containing hematopoietic progenitors, which were then selectively differentiated into NK or T cells as described.30,31 After 8 days of NK differentiation, all the ESC lines produced similar numbers of cells expressing the CD56 NK marker, including CD2+ and CD7+ early lymphoid subsets (Figure 4a). In comparison, NK differentiation of CD34+ cells isolated from umbilical cord blood cells produced fewer CD56+ cells and lower expression levels of CD7. However, after 22 days of differentiation, the umbilical cord blood and wild-type EB cultures both contained nearly 80% CD56+ NK cells with substantial CD2+ and CD7+ subpopulations that were largely absent from both IL2RG-edited cultures (targeted and Cre-out; Figure 4b). The wild-type and umbilical cord blood cultures also expressed IL2RG (CD132) and supported the expansion of CD14-, CD56+ NK cells that could lyse MHC class I-negative target cells as expected, while the gene-edited cells did not (Supplementary Figure S4a,b), confirming that both the targeted and Cre-out alleles were functional knockouts. Importantly, the IL2RG-edited lines were still able to produce monocytic and granulocytic CD14+ and CD15+ progeny (Supplementary Figure S4c). When EBs were cultured under T-cell differentiation conditions, the IL2RG-knockout cultures produced slightly fewer CD34+, CD7+ progenitors than wild-type cells at day 10, and these progenitors did not mature further and downregulate CD34 at later time points (Figure 4c). The IL2RG-knockout cells were capable of producing CD5+, CD7+ pro-T cell progeny (Supplementary Figure S4d) but failed to produce more mature CD4+, CD8+, and double-positive (CD4+CD8+) T cells (Figure 4d). Both IL2RG-edited lines generated normal numbers of primitive and definitive erythroid/myeloid progenitors (data not shown). These combined data show that in a human ESC in vitro differentiation model, IL2RG expression is required for progression from a CD7+ Pro-NK cell or Pro-T cell into more mature cell types (Figure 4e), confirming the well-known role of IL2RG in NK and T-cell development.
Figure 4.
NK and T-cell differentiation of IL2RG-targeted human ESCs. (a) Flow cytometry analysis of CD2, CD7, CD132, and CD56 expression in day 8 NK cell differentiation cultures derived from UCB or wild-type, IL2RG-targeted or Cre-out ESC-derived EBs. (b) Same as a but assayed at 22 days. (c) Flow cytometry analysis of CD34 and CD7 expression T-cell differentiation cultures of wild-type, IL2RG-targeted or Cre-out ESCs harvested at the indicated day. (d) Same as c but analyzing CD4 and CD8 expression. (e) Working model of the differentiation block of IL2RG-knockout ESCs. EB, embryoid bodies; NK, natural killer; UCB, umbilical cord blood.
Discussion
Here, we have described a robust method for editing silent loci in human PSCs without employing a nuclease. Our approach requires the use of a UCOE promoter-driven selectable marker gene that resists silencing, rAAV vectors that efficiently deliver the targeting construct to PSCs, and subsequent selectable marker removal if desired. The UCOE promoter was superior to the other ubiquitously expressed promoters we tested, consistent with its known ability to maintain active chromatin.32,33,34 With this approach, 0.1–1% of the entire cell population undergoes gene editing, and these clones can be isolated by antibiotic selection. The Neo selection cassette used for IL2RG targeting could also function after random integration, and 1 out of 6 of G418-selected clones were accurately edited in those experiments, which is comparable to results obtained when using rAAV vectors to edit active human genes.35 Cre-mediated recombination can be used to efficiently excise the transgene and produce a minimally altered locus that reverts to silent chromatin.
The raw (unselected) editing frequencies we obtained were similar to what has been reported for nuclease-based editing of silent genes in PSCs. For example, both our approach (Supplementary Figure S2a) and TALEN-based targeting of IL2RG6 led to gene editing in 0.14% of unselected PSCs. After antibiotic selection, 0.3–60% of PSC clones were edited by ZFN- or TALEN-based targeting of silent PITX3 or HBB genes,3,5 which demonstrates the variability observed in these types of experiments, but still encompasses the 17% editing frequency we observed in G418-selected clones. An important advantage of our approach is that the rAAV vector does not include any nuclease or integrase proteins that might lead to unwanted on- or off-target mutations.6,8 And while rAAV can integrate randomly at spontaneously occurring chromosomal DSBs, infection with rAAV does not increase background mutation rates in cellular genes.36 Random rAAV integrants are rarely found in edited PSC clones and can be easily ruled out by PCR or Southern blots for vector sequences.13 In contrast, the small in-del mutations produced by nonhomologous end joining at off-target, nuclease-induced DSBs can only be identified in an unbiased manner by full genome sequencing. This reduced genotoxicity of rAAV-mediated gene editing may be an advantage when preparing cells for clinical applications.
In settings where nuclease-induced genotoxicity can be tolerated or lowered to an acceptable level, our findings may also lead to further improvements in these gene-editing methods. Prior studies of silent gene editing in PSCs with ZFNs or TALENs used a PGK promoter to express the selectable marker,3,4,5 and our results suggest that using the UCOE promoter instead would have increased the number of edited clones that survived selection approximately fourfold (Figure 1d). Sequence-specific nucleases could also be combined with rAAV vectors for efficient delivery of both nuclease genes and UCOE-based targeting constructs to PSCs. Target-site DSBs can increase rAAV-mediated gene editing significantly,37,38 and rAAV-encoded ZFNs have been combined with rAAV targeting vectors for efficient in vivo gene editing,39 demonstrating the potential of this approach. Finally, the recently developed Clustered, Regularly Interspaced Short Palindromic Repeat systems that employ guide RNAs to induce sequence-specific DSBs could also be combined with rAAV and provide further enhancements in silent gene editing, given the promising results obtained so far in CRIPSR-based editing of expressed genes in human PSCs.40,41,42
Our study begins to describe the epigenetic changes that can occur at an edited locus. We found that insertion of a UCOE promoter into silent chromatin can lead to a loss of CpG island 5mC residues and convert histone modifications to a more active signature, but the details of these changes can be complex. For example, UCOE insertion increased H3K27Ac levels throughout the transcribed Neo cassette and into both upstream and downstream regions, but only reduced H3K27Me3 levels over a more localized region in the case of COL1A1, and only in downstream sequences in the case of IL2RG. The gene-editing process itself could have played a role in some of these changes, for example, by incorporating unmethylated vector DNA into the chromosome. Or alternatively, UCOE-dependent transcription could have indirectly altered the epigenetic signature, which may explain why some changes in histone modifications extended beyond the region of vector homology. Importantly, removal of the UCOE-Neo cassette caused the edited locus to return to an inactive epigenetic signature indistinguishable from the unedited, parental locus, based on the limited analysis we performed. However, a more detailed examination of the many other histone modifications that have been described,26 as well as the identification of DNA-binding proteins,43,44 DNase hypersensitive sites,45 and long-range chromatin interactions46,47 found at the locus, would presumably reveal additional epigenetic changes associated with gene editing, and it remains to be seen if all these changes convert back to a wild-type signature after removing the UCOE-Neo cassette.
Our choice of the IL2RG gene illustrates some of the potential applications of silent gene editing in PSCs. Edited PSCs can be used as cellular disease models to study the function of lineage-restricted genes. IL2RG-knockout PSCs were unable to differentiate into NK or T cells, confirming a central role for IL2RG-dependent signaling in the developing immune system29 and demonstrating that the lack of NK and T cells observed in X-SCID patients27,28 is due to a differentiation block at the Pro T and Pro NK stage of lymphopoiesis. A similar rAAV editing strategy could be used to correct the IL2RG point mutations that typically cause X-SCID48 so that patient-derived, gene-edited iPSCs could in principle be differentiated ex vivo into hematopoietic cells and transplanted into autologous recipients. In vivo selection should enrich for edited cells, and only a few cells would be required to correct the disease based on the mild phenotype of patients with spontaneous reversion mutations49,50 and the success of IL2RG gene therapy.51 TALEN-mediated gene editing was recently used to correct an IL2RG mutation in X-SCID iPSCs.7
IL2RG knockouts could also be used to prevent graft versus host disease when transplanting PSCs for therapeutic purposes, by eliminating PSC-derived T cells that may react against HLA-mismatched host cells. This would be especially valuable when using allogeneic PSC-derived cells to produce nonlymphoid hematopoietic cell types, such as macrophages and neutrophils to treat chronic granulomatous disease, or erythrocytes to treat hemoglobinopathies. In these settings, transplanted, PSC-derived hematopoietic stem/progenitor cells capable of long-term engraftment would continuously produce terminally differentiated therapeutic cell types in the absence of host-reactive allogeneic lymphoid cells. IL2RG-knockout cells may even be an advantage when transplanting autologous cells, because PSC-derived T and NK cells do not develop in a normal embryo and may not be educated appropriately to tolerate autologous host cells. Other scenarios can also be envisioned where preventing the expression of a lineage-specification gene could produce a therapeutic advantage, such as PSCs with edited glucagon or somatostatin genes that can differentiate into insulin-secreting beta cells for the treatment of diabetes without producing the alpha or delta cells that frequently contaminate PSC-derived pancreatic islet cell preparations.52,53
Materials and Methods
Cell culture. H1 human ESCs54 and human iPSC lines were cultured on mouse embryo fibroblasts as described.20,55 COL1A1-targeted G418-sensitive iPSCs were derived by reprogramming of (COL1A1-IRES-Neo)-targeted MSCs with lentiviral vectors as described.55 50 μg/ml active G418 was used for selection. Jurkat cells were cultured in RPMI-1640 with10% FBS and 1% Pen/Strep (Life Technologies, Carlsbad, CA).
Viral vectors. AAV vector plasmids were assembled from PCR products by standard methods and confirmed by DNA sequencing. Homology arm fragments were amplified from the target cell type, and promoter fragments were amplified from H1 human ESCs and CF1 mice, respectively. The 1.2 kb UCOE fragment has been described and consists of nucleotides 26240199 to 26241411 of chromosome 7 (GRCh37/hg19).56 Plasmid sequences are available upon request. AAV vectors were packaged in serotype 3b capsids by co-transfection of vector plasmids and packaging plasmid pDGM3B into 293T cells, purified by iodixanol step gradients, and their titers were determined by Southern blots as described.57
Gene targeting. In promoter comparison experiments, 4 × 105 iPSCs were seeded in triplicate in 35-mm wells and transduced with rAAVs at a multiplicity of infection of 104 genome-containing particles/cell the next day. G418 selection was initiated 2 days after infection, and the surviving colonies were counted, picked, and expanded for further analysis. The total number of colony-forming units (CFU) was calculated by culturing 4 × 103 cells in a 6-cm dish without selection. In IL2RG targeting experiments, 5 × 105 wild-type H1 ESCs were seeded in a 10-cm dish and transduced with AAV at a multiplicity of infection of 3 × 103 genome-containing particles/cell the next day. Four days later, transduced ESCs were disaggregated into single cells using Accutase (Stemgent, Lexington, MA) and plated in serial dilutions in 10-cm dishes for G418 selection. 5 × 103 transduced H1 ESCs were also plated in a 10-cm dish without selection to determine the total number of CFUs. G418-resistant colonies were counted, picked, and screened initially by PCR to identify targeted clones.
Cre-mediated transgene removal. A polyclonal population of wild-type and IL2RG-targeted H1 ESCs was transduced as described with the nonintegrating foamy vector, NIFV-EokCreW, that expresses Cre recombinase.55 Four days later, infected ESCs were disaggregated into single cells with Accutase, and serial dilutions were plated in 10-cm dishes. The surviving colonies were randomly picked and screened by PCR to identify age-matched clones with wild-type, UCOE-Neo-containing, or Cre-out IL2RG alleles for subsequent experiments.
DNA and RNA isolation. Genomic DNA was prepared from PSCs as described.57,58 Total cellular RNA was extracted by the Trizol method (Life Technologies) and used to generate cDNA with M-MLV reverse transcriptase and oligo-dT primers according to the manufacturer's protocol (Life Technologies).
Quantitative PCR and RT-PCR. cDNA qRT-PCR reactions were performed in triplicate with SYBR Select Master Mix (Life Technologies) on a StepOnePlus Real-Time PCR System (Life Technologies), and the relative gene expression levels were calculated by the ΔΔCT method. Homologous recombination frequencies were measured by infecting iPSCs with rAAV vectors, culturing for 5 days without selection, and determining the number of promoter-targeted alleles in 1 μg of genomic DNA by Taqman qPCR (Life Technologies). Plasmids containing promoter-targeted COL1A1 sequences were constructed by conventional cloning methods and used in qPCR reactions containing 0 to 104 plasmid molecules and 1 μg of wild-type genomic DNA (1.5 × 105 diploid genome equivalents) to generate standard curves.
Bisulfite sequencing. Genomic DNA was treated as described in the EZ DNA Methylation-Gold Kit (ZYMO Research, Irvine, CA) and used to PCR amplify COL1A1 CpG island fragments. These PCR products were cloned into the pGEM-Teasy vector (Promega, Madison, WI), and the recombinant plasmids were sequenced.
Chromatin immunoprecipitation (ChIP). The ChIP protocol was adapted from Abcam's “X-ChIP protocol” (http://www.abcam.com) as follows. Three weeks after transduction with an AAV or Cre vector, PSCs with verified genotypes were dissociated with Accutase and fixed with PBS containing 1% formaldehyde at room temperature for 10 minutes with constant mixing. Fixation was stopped by adding glycine to a final concentration of 125 mmol/l. Samples were sonicated until chromatin was sheared to 500 to 1,000 bp. For binding, chromatin from 107 cells, 10 μg antibodies, and 30 μl solid protein-A/G beads (Santa Cruz Biotechnology, Dallas, TX) were combined and incubated at 4 °C overnight. After washing the beads, chromatin was eluted and incubated at 65 °C overnight to reverse cross-linking. The eluted chromatin was purified using a PCR purification kit (Qiagen, Valencia, CA) and used in qPCR reactions performed in triplicate with SYBR Select Master Mix (Life Technologies) on a StepOnePlus Real-Time PCR System (Life Technologies). Relative occupancy was calculated by the ΔΔCT method. Antibodies used were against H3K27me3 (Millipore, Billerica, MA), H3K27Ac (Abcam, Cambridge, MA), and normal rabbit IgG (Santa Cruz Biotechnology) as a control.
NK cell differentiation. NK differentiation was performed as described.30,59 Briefly, day 13 EBs were co-cultured with EL08-1D2 stromal cells in media supplemented with IL-3, IL7, IL-15, SCF, and FLT3L (Peprotech, Rocky Hill, NJ), and cells were harvested at appropriate time points for analysis. In chromium release assays, derived NK cells were stimulated by Clone 9.mbIL-21 aAPCs as described60 and incubated with radioactive 51Cr-labeled K562 target cells, and lysis was measured by scintillation counter using the equation: % specific lysis = 100 × (test release − spontaneous release)/(maximal release − spontaneous release) as described.61 Flow cytometry was performed with a BD LSRII (BD Biosciences, Franklin Lakes, NJ) flow cytometer, and the data were analyzed by FlowJo software version 10.0 (Tree Star, Ashland, OR). Antibodies, which were used according to the manufacturers' recommendations, were from BD Biosciences unless otherwise indicated. Live cells were distinguished from dead cells by CYTOX blue dead cell stain (Life Technologies). Antibodies used for NK cell phenotype analysis were: CD56 (PE-Cy7-clone B159); CD7 (Alexa Fluor 700-clone M-T701); CD2 (PE-CF594-clone RPA-2.10); and CD132 (PE-clone TUGh4, eBioscience, San Diego, CA).
T-cell differentiation. T-cell differentiation and analysis were performed as described previously.31 Briefly, at day 8 of EB differentiation, 2 × 104 CD34+ CD43- CD73- CXCR4- cells isolated by fluorescence activated cell sorting were plated onto individual wells of a six-well plate containing OP9-DL4 stromal cells in the presence of rhFLT3L and rhIL-7. rhSCF was added for the first 7 days only (R&D Systems, Minneapolis, MN). Every 5 days, co-cultures were passaged onto fresh OP9-DL4 stromal cells. Cells were harvested and assayed at various time points. Cell suspensions were stained and analyzed on a BD LSR II flow cytometer. Data analysis was performed using FlowJo Software by gating on live cells followed by lack of DAPI uptake. Fluorophore-conjugated antibodies against CD4, CD5, CD7, CD8, CD34, and CD45 were purchased from BD Biosciences and eBioscience.
PCR Primers. All primer sequences are listed in Supplementary Table S1.
Statistical analysis. Statistical significance was assessed using the two-tailed Student's t-test. P values less than 0.01 were considered statistically significant. Data represent mean ± SEM of three.
SUPPLEMENTARY MATERIAL Figure S1. Targeting a silent COL1A1-IRES-Neo cassette. Figure S2. IL2RG targeting and UCOE-Neo removal. Figure S3. CpG methylation at the IL2RG locus. Figure S4. Characterization of T and NK cells derived from ESCs. Table S1. Primer sequence.
Acknowledgments
The authors thank Kristi Pilat, Roli Hirata, and Raisa Stolitenko for technical assistance and Els Henckaerts for helpful discussions. This work was supported by grants from the US NIH to D.W.R. (HL53750, DK55759), D.S.K. (Minnesota Partnership for Biotechnology), and G.K. (NIH U01HL100395 and CIHR MOP126117).
D.W.R. holds equity in Horizon Discovery and Universal Cells Inc. The other authors declare no competing financial interests.
Supplementary Material
References
- Nickoloff, JA and Reynolds, RJ (1990). Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol 10: 4837–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan, B, Johnson, BL and Campbell, C (1995). The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res 23: 2784–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer, D, Soldner, F, Beard, C, Gao, Q, Mitalipova, M, DeKelver, RC et al. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J, Mali, P, Huang, X, Dowey, SN and Cheng, L (2011). Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 118: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, N and Zhao, H (2014). Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng 111: 1048–1053. [DOI] [PubMed] [Google Scholar]
- Hendel, A, Kildebeck, EJ, Fine, EJ, Clark, JT, Punjya, N, Sebastiano, V et al. (2014). Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep 7: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, T, Firth, AL, Scripture-Adams, DD, Galic, Z, Qualls, SJ, Gilmore, WB et al. (2015). Lymphoid regeneration from gene-corrected SCID-X1 subject-derived iPSCs. Cell Stem Cell 16: 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y, Foden, JA, Khayter, C, Maeder, ML, Reyon, D, Joung, JK et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche, MA, Kress, C, Poirier, F and Dandolo, L (1997). Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Tsai, TF, Bressler, J, Jiang, YH and Beaudet, AL (2003). Disruption of the genomic imprint in trans with homologous recombination at Snrpn in ES cells. Genesis 37: 151–161. [DOI] [PubMed] [Google Scholar]
- Lieberman-Lazarovich, M, Melamed-Bessudo, C, de Pater, S and Levy, AA (2013). Epigenetic alterations at genomic loci modified by gene targeting in Arabidopsis thaliana. PLoS One 8: e85383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, DW and Hirata, RK (1998). Human gene targeting by viral vectors. Nat Genet 18: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, IF, Hirata, RK, Wang, PR, Li, Y, Kho, J, Nelson, A et al. (2010). Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther 18: 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, LB, Chang, KH, Wang, PR, Hirata, RK, Papayannopoulou, T and Russell, DW (2012). Trisomy correction in Down syndrome induced pluripotent stem cells. Cell Stem Cell 11: 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, K, Suzuki, K, Aizawa, E, Kawase, E, Suemori, H, Nakatsuji, N et al. (2009). Gene targeting in human pluripotent stem cells with adeno-associated virus vectors. Biochem Biophys Res Commun 388: 711–717. [DOI] [PubMed] [Google Scholar]
- Chamberlain, JR, Schwarze, U, Wang, PR, Hirata, RK, Hankenson, KD, Pace, JM et al. (2004). Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science 303: 1198–1201. [DOI] [PubMed] [Google Scholar]
- Wang, PR, Xu, M, Toffanin, S, Li, Y, Llovet, JM and Russell, DW (2012). Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci USA 109: 11264–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, CS, Hao, Y, Rokhlina, T, Samuel, M, Stoltz, DA, Li, Y et al. (2008). Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest 118: 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X, Yan, Z, Yi, Y, Li, Z, Lei, D, Rogers, CS et al. (2008). Adeno-associated virus-targeted disruption of the CFTR gene in cloned ferrets. J Clin Invest 118: 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J, Vodyanik, MA, Smuga-Otto, K, Antosiewicz-Bourget, J, Frane, JL, Tian, S et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- Deyle, DR, Li, Y, Olson, EM and Russell, DW (2010). Nonintegrating foamy virus vectors. J Virol 84: 9341–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton, AM and Bird, A (2011). CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyle, DR, Li, LB, Ren, G and Russell, DW (2014). The effects of polymorphisms on human gene targeting. Nucleic Acids Res 42: 3119–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton, MP, Cheng, AW, Welstead, GG, Kooistra, T, Carey, BW, Steine, EJ et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias, A, Bajpai, R, Swigut, T, Brugmann, SA, Flynn, RA and Wysocka, J (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, J, Kheradpour, P, Mikkelsen, TS, Shoresh, N, Ward, LD, Epstein, CB et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, M, Yi, H, Rosenblatt, HM, Filipovich, AH, Adelstein, S, Modi, WS et al. (1993). Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73: 147–157. [DOI] [PubMed] [Google Scholar]
- Buckley, RH, Schiff, RI, Schiff, SE, Markert, ML, Williams, LW, Harville, TO et al. (1997). Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr 130: 378–387. [DOI] [PubMed] [Google Scholar]
- Leonard, WJ (2001). Cytokines and immunodeficiency diseases. Nat Rev Immunol 1: 200–208. [DOI] [PubMed] [Google Scholar]
- Knorr, DA, Ni, Z, Hermanson, D, Hexum, MK, Bendzick, L, Cooper, LJ et al. (2013). Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl Med 2: 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M, Awong, G, Sturgeon, CM, Ditadi, A, LaMotte-Mohs, R, Zúñiga-Pflücker, JC et al. (2012). T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2: 1722–1735. [DOI] [PubMed] [Google Scholar]
- Lindahl Allen, M and Antoniou, M (2007). Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE). Epigenetics 2: 227–236. [DOI] [PubMed] [Google Scholar]
- Müller-Kuller, U, Ackermann, M, Kolodziej, S, Brendel, C, Fritsch, J, Lachmann, N et al. (2015). A minimal ubiquitous chromatin opening element (UCOE) effectively prevents silencing of juxtaposed heterologous promoters by epigenetic remodeling in multipotent and pluripotent stem cells. Nucleic Acids Res 43: 1577–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff, N, Lachmann, N, Ackermann, M, Kohlscheen, S, Brendel, C, Maetzig, T et al. (2013). A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 31: 488–499. [DOI] [PubMed] [Google Scholar]
- Hirata, R, Chamberlain, J, Dong, R and Russell, DW (2002). Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol 20: 735–738. [DOI] [PubMed] [Google Scholar]
- Miller, DG, Petek, LM and Russell, DW (2004). Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet 36: 767–773. [DOI] [PubMed] [Google Scholar]
- Porteus, MH, Cathomen, T, Weitzman, MD and Baltimore, D (2003). Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol 23: 3558–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, DG, Petek, LM and Russell, DW (2003). Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol Cell Biol 23: 3550–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H, Haurigot, V, Doyon, Y, Li, T, Wong, SY, Bhagwat, AS et al. (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z, Zhang, Y, Propson, NE, Howden, SE, Chu, LF, Sontheimer, EJ et al. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA 110: 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q, Regan, SN, Xia, Y, Oostrom, LA, Cowan, CA and Musunuru, K (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko, PV, Tolstorukov, MY and Park, PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman, RE, Rynes, E, Humbert, R, Vierstra, J, Maurano, MT, Haugen, E et al. (2012). The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, A, Lajoie, BR, Jain, G and Dekker, J (2012). The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E, Bouwman, BA, Zhu, Y, Klous, P, Splinter, E, Verstegen, MJ et al. (2013). The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature 501: 227–231. [DOI] [PubMed] [Google Scholar]
- Puck, JM, Pepper, AE, Henthorn, PS, Candotti, F, Isakov, J, Whitwam, T et al. (1997). Mutation analysis of IL2RG in human X-linked severe combined immunodeficiency. Blood 89: 1968–1977. [PubMed] [Google Scholar]
- Kuijpers, TW, van Leeuwen, EM, Barendregt, BH, Klarenbeek, P, aan de Kerk, DJ, Baars, PA et al. (2013). A reversion of an IL2RG mutation in combined immunodeficiency providing competitive advantage to the majority of CD8+ T cells. Haematologica 98: 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann, C, Pannicke, U, Wiech, E, Schwarz, K, Fisch, P, Friedrich, W et al. (2008). Clinical and immunologic consequences of a somatic reversion in a patient with X-linked severe combined immunodeficiency. Blood 112: 4090–4097. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo, M, Hacein-Bey, S, de Saint Basile, G, Gross, F, Yvon, E, Nusbaum, P et al. (2000). Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288: 669–672. [DOI] [PubMed] [Google Scholar]
- D'Amour, KA, Bang, AG, Eliazer, S, Kelly, OG, Agulnick, AD, Smart, NG et al. (2006). Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24: 1392–1401. [DOI] [PubMed] [Google Scholar]
- Jiang, J, Au, M, Lu, K, Eshpeter, A, Korbutt, G, Fisk, G et al. (2007). Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells 25: 1940–1953. [DOI] [PubMed] [Google Scholar]
- Thomson, JA, Itskovitz-Eldor, J, Shapiro, SS, Waknitz, MA, Swiergiel, JJ, Marshall, VS et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Deyle, DR, Khan, IF, Ren, G, Wang, PR, Kho, J, Schwarze, U et al. (2012). Normal collagen and bone production by gene-targeted human osteogenesis imperfecta iPSCs. Mol Ther 20: 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F, Frost, AR, Blundell, MP, Bales, O, Antoniou, MN and Thrasher, AJ (2010). A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol Ther 18: 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, IF, Hirata, RK and Russell, DW (2011). AAV-mediated gene targeting methods for human cells. Nat Protoc 6: 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou, EP and Sadelain, M (2011). Derivation of genetically modified human pluripotent stem cells with integrated transgenes at unique mapped genomic sites. Nat Protoc 6: 1274–1289. [DOI] [PubMed] [Google Scholar]
- McCullar, V, Oostendorp, R, Panoskaltsis-Mortari, A, Yun, G, Lutz, CT, Wagner, JE et al. (2008). Mouse fetal and embryonic liver cells differentiate human umbilical cord blood progenitors into CD56-negative natural killer cell precursors in the absence of interleukin-15. Exp Hematol 36: 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman, CJ, Senyukov, VV, Somanchi, SS, Phatarpekar, PV, Kopp, LM, Johnson, JL et al. (2012). Membrane-bound IL-21 promotes sustained ex vivo proliferation of human natural killer cells. PLoS One 7: e30264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, JS, Oelkers, S, Verfaillie, C and McGlave, P (1992). Role of monocytes in the expansion of human activated natural killer cells. Blood 80: 2221–2229. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.