Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most prevalent myopathies, affecting males and females of all ages. Both forms of the disease are linked by epigenetic derepression of the D4Z4 macrosatellite repeat array at chromosome 4q35, leading to aberrant expression of D4Z4-encoded RNAs in skeletal muscle. Production of full-length DUX4 (DUX4-fl) mRNA from the derepressed D4Z4 array results in misexpression of DUX4-FL protein and its transcriptional targets, and apoptosis, ultimately leading to accumulated muscle pathology. Returning the chromatin at the FSHD locus to its nonpathogenic, epigenetically repressed state would simultaneously affect all D4Z4 RNAs, inhibiting downstream pathogenic pathways, and is thus an attractive therapeutic strategy. Advances in CRISPR/Cas9-based genome editing make it possible to target epigenetic modifiers to an endogenous disease locus, although reports to date have focused on more typical genomic regions. Here, we demonstrate that a CRISPR/dCas9 transcriptional inhibitor can be specifically targeted to the highly repetitive FSHD macrosatellite array and alter the chromatin to repress expression of DUX4-fl in primary FSHD myocytes. These results implicate the promoter and exon 1 of DUX4 as potential therapeutic targets and demonstrate the utility of CRISPR technology for correction of the epigenetic dysregulation in FSHD.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is the most prevalent myopathy affecting males and females of all ages.1,2,3,4 Originally characterized as an autosomal dominant genetic myopathy,3,5 FSHD also displays striking features of an epigenetic disorder.6,7 Encompassing >95% of reported cases, FSHD1 (OMIM 158900) is linked to contractions of the D4Z4 macrosatellite repeat array at 4q35 (refs. 8,9,10). In healthy individuals, this array ranges from 11–100 D4Z4 repeats on both 4q chromosomes, whereas in FSHD1 patients, the array is contracted to 1–10 repeats on one 4q chromosome.10,11 In order to develop FSHD, this contraction must be in cis with a distal disease-permissive haplotype of 4q35 (refs. 12,13,14,15). While chromosome 10q26 contains a D4Z4 array that is highly homologous to the array at 4q35, and other polymorphic D4Z4 repeats are present throughout the genome, only D4Z4 contractions at 4q35 are pathogenic.14,16,17,18 FSHD2 patients, which represent <5% of reported cases, have no D4Z4 contraction at 4q35, but still carry at least one permissive 4q35 subtelomere.15,19,20,21
The extreme variability in FSHD onset, progression, and severity—ranging from asymptomatic to clinically severe3,22,23—suggests that multiple mechanisms acting together lead to disease, including genetic, epigenetic, developmental, and environmental factors. Indeed, both forms of FSHD are linked by common epigenetic alterations indicative of chromatin relaxation at the pathogenic locus.20,21,24,25,26,27,28,29,30,31 One consequence of the epigenetic disruption at 4q35 is the aberrant expression of the DUX4 retrogene in skeletal muscle.15,32,33 Although a copy of DUX4 resides in every D4Z4 repeat unit, only the full-length DUX4 mRNA (DUX4-fl) produced from the distal-most repeat is stably expressed, due to the presence of a polyadenylation signal in FSHD-permissive alleles.15,32 Production of DUX4-fl results in aberrant expression of the DUX4-FL protein and its transcriptional targets, which include germline genes, immune mediators, and retroelements,34,35 altered RNA and protein metabolism,36,37 and apoptosis,38,39,40,41,42 leading to muscle atrophy and accumulated pathology.34,35,40,43
While DUX4-FL and its downstream targets represent valid candidates for therapy, levels of DUX4-fl expression are highly variable among patients and do not necessarily correlate with disease severity.33,44 The epigenetic dysregulation at the FSHD locus, however, is strongly correlated with disease manifestation.44,45,46 In addition, the D4Z4 repeats encode multiple noncoding RNAs, which have the potential to play downstream pathogenic roles in FSHD.28,47 Thus, targeting the FSHD locus to return the chromatin to its nonpathogenic, repressed state might be more therapeutically beneficial than simply targeting the rare DUX4-fl mRNA or its downstream genes.
The nuclease-deficient component of the CRISPR/Cas9 genome engineering tool (dCas9) fused to transcriptional effectors has been instrumental in the targeted manipulation of gene expression.48 While previous studies have used CRISPR-based systems to modulate gene expression in more typical genomic regions, the pathogenic locus in FSHD is unusual in that only one of many D4Z4 repeat arrays in the human genome is pathogenic. Thus, it was unclear whether a CRISPR-based platform could effectively target the FSHD disease locus. Here, we demonstrate that CRISPR/dCas9 technology can successfully target transcriptional effectors to the pathogenic locus in primary FSHD skeletal myocytes, resulting in increased chromatin repression accompanied by decreased expression of DUX4-fl and its downstream targets. These results demonstrate the utility of a CRISPR effector platform for therapeutic targeting of the D4Z4 macrosatellite and correcting the epigenetic dysregulation in FSHD, and pave the way for mechanistic studies of endogenous DUX4 regulation.
Results
Recruitment of dCas9 and VP64 to the DUX4 promoter or exon 1 activates DUX4-fl in FSHD myocytes
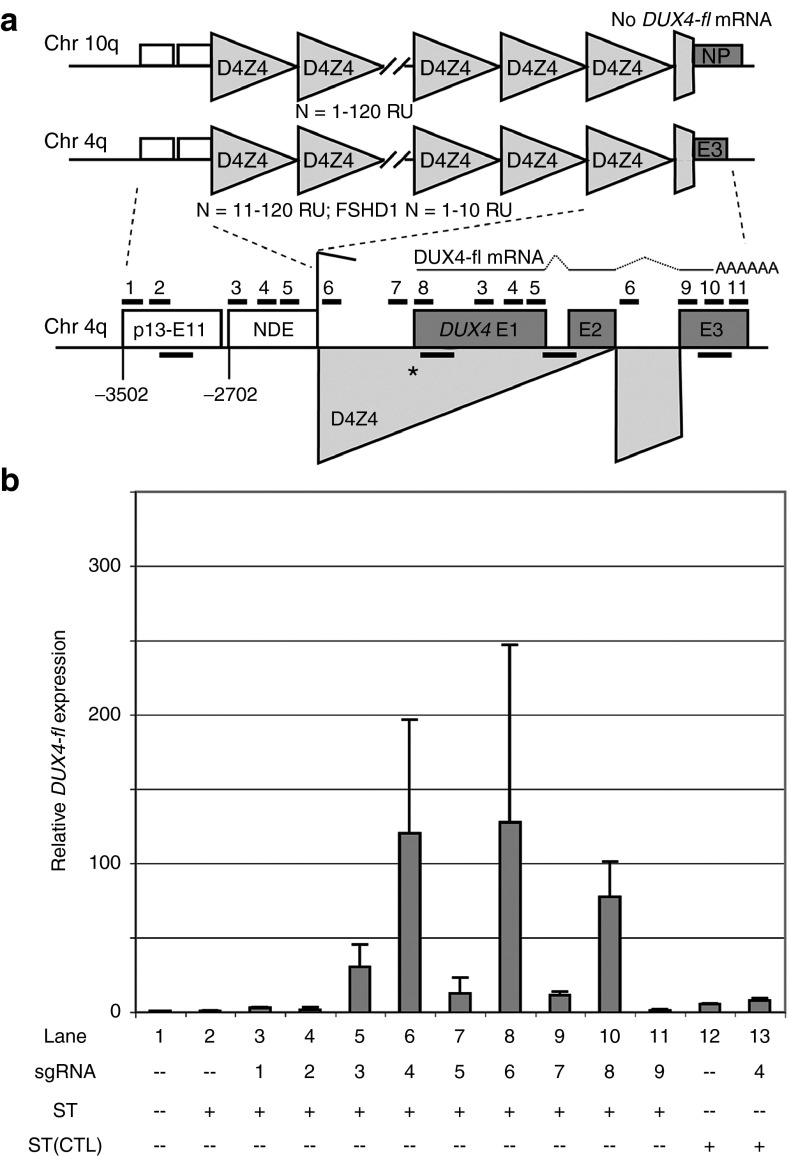
To search for potential FSHD therapeutic targets in vivo, we used the CRISPR/dCas9 system to test several candidate regions in or flanking the 4q35 D4Z4/DUX4 locus for the ability to modulate gene expression in the D4Z4 array. There are numerous D4Z4 repeat arrays in the genome49; however, we are interested in affecting expression from the array associated with FSHD. Therefore, we assayed polyadenylated DUX4-fl mRNA levels in FSHD1 myocytes as our read-out for gene expression which is specifically derived from the contracted 4q35 D4Z4 array. When targeted by small guide RNAs (sgRNAs), dCas9 transcriptional effector platforms are effective in modulating endogenous gene expression levels in mammalian cells.50,51,52,53,54,55,56,57,58,59,60,61 For our initial experiments, we used the SunTag system,62 which involves the dual activity of two constructs: (i) dCas9 fused to 10 copies of the GCN4 peptide and (ii) GCN4 antibody fused to the VP64 activator. The dCas9 fused directly to VP64 generally requires multiple, nonoverlapping sgRNAs to achieve strong activation of gene expression.50,54,58,59,60 In contrast, the SunTag system allows recruitment of multiple VP64 domains to a single dCas9, resulting in robust gene activation with only a single sgRNA.53,62 For these and the following experiments, we used myogenic cells from an FSHD1 patient (17Abic), which express consistent and relatively high levels of DUX4-fl when terminally differentiated.33,44,63
Sequence preferences for sgRNAs (e.g., requirement for protospacer adjacent motif (PAM), preference for purines, limited CpGs, no multiple Us in seed sequence, low secondary structure in spacer)64,65 preclude a comprehensive analysis of regions such as the DUX4 promoter, which contains long stretches of low-complexity sequence. Therefore, taking into account these constraints, we designed sgRNAs targeting two candidate regions upstream of the D4Z4 repeat (Figure 1a): the NDE (non-deleted element retained in FSHD patients) sequence28,66 and p13-E11, a region distinct in the genome that is used to identify D4Z4 arrays specific to chromosomes 4q35 and 10q26 (refs. 67,68). Within D4Z4, we designed sgRNAs targeting the promoter, exon 1, and exon 3 of DUX4. In addition to forming a macrosatellite repeat, each D4Z4 repeat unit also contains repetitive sequences, and part of the DUX4 exon 1 is duplicated in the NDE, which lies proximal to the array. Thus, three sgRNAs (#3–5) target both the NDE and DUX4 exon 1. In addition, sgRNA #6 targets DUX4 intron 2 as well as the DUX4 promoter. The rules governing sgRNA targeting are not yet fully understood, and poor targeting has been attributed to low stability, inefficient loading into dCas9, or low-affinity binding to DNA.52,64 Thus, for each target region, we tested four to five sgRNAs for the ability to recruit dCas9-VP64-HA, as assessed by chromatin immunoprecipitation (ChIP) using HA antibodies. We identified at least two sgRNAs for each region (p13-E11, DUX4 promoter, DUX4 exon 1/NDE, and DUX4 exon 3) that demonstrated correct targeting of dCas9-VP64-HA (Supplementary Table S1).
Figure 1.
Recruitment of dCas9 and VP64 to the DUX4 promoter or exon 1 activates DUX4-fl in facioscapulohumeral muscular dystrophy (FSHD) myocytes. (a) Schematic diagram of the FSHD locus at chromosome 4q35, with distances shown relative to the DUX4 MAL start codon (*). For simplicity, only the distal D4Z4 repeat unit of the macrosatellite array is depicted below. DUX4 exons 1 and 2 are located within the D4Z4 repeat, and exon 3 lies in the distal subtelomeric sequence. In FSHD skeletal myocytes, DUX4-fl mRNA from the distal repeat is stabilized by a polyadenylation signal in exon 3 that is present in disease-permissive haplotypes of 4qA. The p13-E11 diagnostic probe region67,68 and the NDE (non-deleted element)28,66 lie proximal to the D4Z4 array. The locations of sgRNA target sequences used in this study (#1–11) are indicated. Positions of chromatin immunoprecipitation amplicons are shown as unlabeled black bars (in order from 5' to 3': p13-E11, DUX4 exon 1, intron 1, and exon 3). Refer to text for more details. (b) Effects of targeting dCas9 and VP64 to the FSHD locus on DUX4-fl expression. FSHD myogenic cultures were subjected to four serial coinfections with lentiviral supernatants expressing either components of the SunTag system encoding dCas9 and VP64 (ST), a SunTag variant lacking VP64 (ST[CTL]) or individual sgRNAs (#1–9). After the final round of infection, cells were induced to differentiate and harvested ~48 hours later for analysis of DUX4-fl expression by quantitative reverse transcriptase polymerase chain reaction. Data are plotted as the mean + standard deviation (SD) value of three to five independent experiments, with relative mRNA expression for the mock-infected cells set to 1.
Primary myoblasts are notoriously difficult to transfect or infect; thus, we used the high-efficiency method of Springer and Blau69 in which ~100% infection efficiency is achieved by four serial rounds of viral exposure with centrifugation. After the final round of infection, the cells were induced to differentiate and harvested 48 hours later for analysis of DUX4-fl expression by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Expression of the SunTag system alone had no effect on DUX4-fl mRNA levels in FSHD myocytes (Figure 1b, lane 2). Likewise, recruitment of VP64 to the p13-E11 region or to exon 3 of DUX4 had little effect on DUX4-fl expression (Figure 1b, lanes 3, 4, and 11). In contrast to this, VP64 recruitment to the DUX4 promoter or exon 1/NDE yielded robust activation of DUX4-fl in FSHD myocytes (Figure 1b, lanes 5–10). Although we cannot rule out that VP64 recruited to the NDE has a positive effect on DUX4-fl, recruitment to p13-E11 (500 bp upstream) had no effect, whereas recruitment to the DUX4 promoter, directly upstream of exon 1, strongly activated DUX4-fl. Therefore, when guided by sgRNAs #3–5, the transcriptional effector is likely mediating its effects from DUX4 exon 1, and for simplicity, we will refer to these sgRNAs as targeting DUX4 exon 1. Although targeting by single sgRNAs proved sufficient for transcriptional activation, the functional capacity of sgRNAs targeting the same region was variable (e.g., ~120-fold activation with sgRNA #4 versus ~13-fold activation with sgRNA #5) (Figure 1b). This is consistent with the previous reports comparing sgRNA targeting and stability.51,52,64 As expected, when dCas9 lacking a transcriptional effector domain was recruited to DUX4 exon 1, it did not activate DUX4-fl expression (Figure 1b, lanes 12–13).
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses DUX4-fl in FSHD myocytes
Reducing the aberrant expression of DUX4-fl in FSHD by returning the chromatin at the disease locus to a nonpathogenic, repressed state is a viable avenue of therapy. We first tested whether DUX4-fl expression could be reduced in FSHD myocytes using a dCas9-KRAB repressor. When guided by multiple sgRNAs, dCas9-KRAB has proven effective in reducing target gene expression in mammalian cells.51,52,53,55,56 Since dCas9-mediated recruitment of VP64 to the DUX4 promoter or exon 1 strongly activated DUX4-fl expression, we expected that these regions might be good candidates for therapeutic targeting. For these and the following experiments, we performed four serial coinfections of FSHD myogenic cultures. Cells were infected with various combinations of lentiviral supernatants expressing either dCas9-KRAB or individual sgRNAs targeting the candidate regions. After the final round of infection, the cells were induced to differentiate and harvested ~40 hours later for analysis of DUX4-fl expression by qRT-PCR.
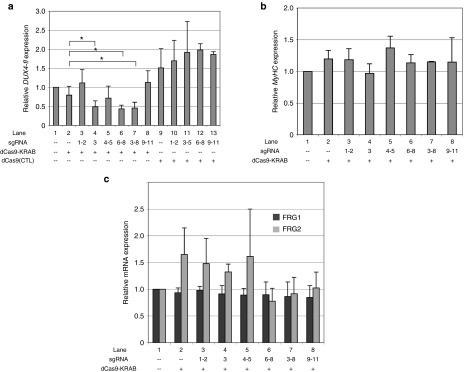
Expression of the dCas9-KRAB repressor alone had little effect on DUX4-fl levels (Fig. 2a, lane 2). Consistent with our results using the SunTag activator system, targeting dCas9-KRAB to either the p13-E11 region or DUX4 exon 3 had no effect on DUX4-fl expression (Figure 2a, lanes 3, 8). In contrast to this, targeting dCas9-KRAB to the DUX4 promoter or exon 1 reduced expression of DUX4-fl to ~45% of endogenous levels in FSHD myocytes (Figure 2a, lanes 4, 6–7). Although dCas9 effectors often require targeting by multiple, nonoverlapping sgRNAs to achieve significant transcriptional modulation,50,52,55,59,60 we found that in one case, a single sgRNA was effective in reducing DUX4-fl expression (Figure 2a, lane 4), and the combination of all six sgRNAs targeting these regions showed no enhanced effect (Figure 2a, lane 7).
Figure 2.
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses DUX4-fl in facioscapulohumeral muscular dystrophy (FSHD) myocytes. (a) Effects of targeting dCas9-KRAB to the FSHD locus on DUX4-fl expression. FSHD myogenic cultures were subjected to four serial co-infections with lentiviral supernatants expressing either dCas9-KRAB, a dCas9 variant lacking an effector domain (dCas9[CTL]), or individual sgRNAs (#1–11). After the final round of infection, cells were induced to differentiate and harvested ~40 hours later for analysis of DUX4-fl expression by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). (b) Recruitment of dCas9-KRAB to the FSHD locus does not impair skeletal myocyte differentiation. Expression of the terminal muscle differentiation marker Myosin heavy chain (MyHC) was assessed by qRT-PCR in the cultures described in Figure 2a. (c) Recruitment of dCas9-KRAB to the FSHD locus does not repress expression of FRG1 and FRG2. Levels of FSHD candidate genes FRG1 and FRG2 were measured by qRT-PCR in the cultures described in Figure 2a. For a–c, data are plotted as the mean + SD value of at least three independent experiments, with relative mRNA expression for the mock-infected cells set to 1. *P < 0.05 (Student's t-test).
Previous studies have demonstrated that in some contexts, dCas9 can inhibit transcription through steric hindrance of target regions.52,70,71 To determine whether the repressive effects we observed were due to an obstruction mechanism rather than KRAB-mediated repression, we tested the effect of a dCas9 variant lacking an effector domain. Recruitment of this protein to any of the target regions did not reduce levels of DUX4-fl (Figure 2a, lanes 9–13), demonstrating the importance of the KRAB domain for mediating DUX4-fl repression at the target regions.
In FSHD myogenic cultures, DUX4-FL expression is restricted to terminally differentiated myocytes. To rule out a nonspecific effect of dCas9-KRAB on muscle differentiation, we assessed levels of Myosin heavy chain (MyHC), a marker of terminal muscle differentiation, by qRT-PCR in the cells described above. Importantly, MyHC levels were equivalent in all cultures expressing dCas9-KRAB and sgRNAs (Figure 2b), indicating that lower levels of DUX4-fl are not due to impairment of muscle differentiation. We also measured expression of FRG1 and FRG2, two other FSHD candidate genes that lie proximal to the D4Z4 repeat. Although levels of FRG2 were variable, recruitment of the dCas9 repressor to any of the target regions did not reduce expression of either FRG1 or FRG2 mRNA (Figure 2c).
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses DUX4-FL targets in FSHD myocytes
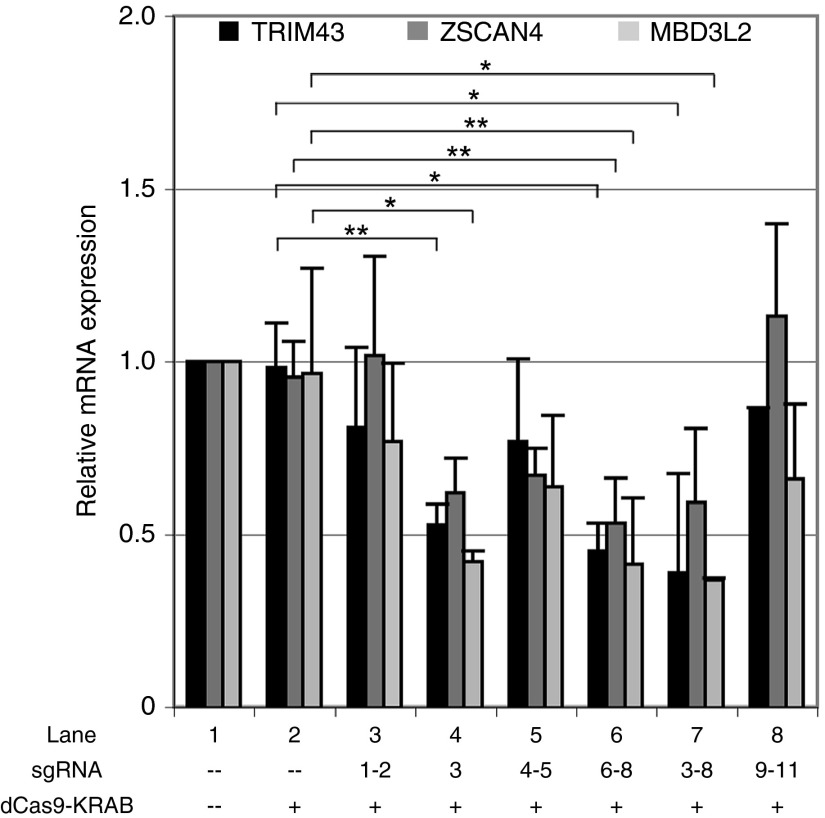
Expression of DUX4-FL in FSHD myocytes causes the aberrant upregulation of many downstream targets, including genes expressed in the germline and in early development.35 TRIM43, ZSCAN4, and MBD3L2 are downstream targets of DUX4-FL35 that were also found to be upregulated in the myogenic cultures used in this study (unpublished data). To determine whether dCas9-KRAB-mediated repression of DUX4-fl also results in repression of these DUX4-FL target genes, we measured levels of TRIM43, ZSCAN4, and MBD3L2 by qRT-PCR in the cells described above. As expected, expression of these genes was not significantly altered by expressing the dCas9-KRAB repressor alone or by targeting the repressor to p13-E11 or to DUX4 exon 3 (Figure 3, lanes 2, 3, and 8). However, as with DUX4-fl, targeting the KRAB repressor to the DUX4 promoter or exon 1 significantly reduced expression of all three DUX4-FL targets to ~35–60% of endogenous levels (Figure 3, lanes 4, 6–7). Thus, targeting dCas9-KRAB to the promoter or exon 1 of DUX4 results in efficient repression of both DUX4-fl and its target genes in FSHD myocytes.
Figure 3.
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses DUX4-FL target genes in facioscapulohumeral muscular dystrophy myocytes. Levels of the DUX4-FL target genes TRIM43, ZSCAN4, and MBD3L2 were assessed by quantitative reverse transcriptase polymerase chain reaction in the cultures described in Figure 2a. Data are plotted as the mean + SD value of at least three independent experiments, with relative mRNA expression for the mock-infected cells set to 1. *P < 0.05; **P < 0.01 (Student's t-test).
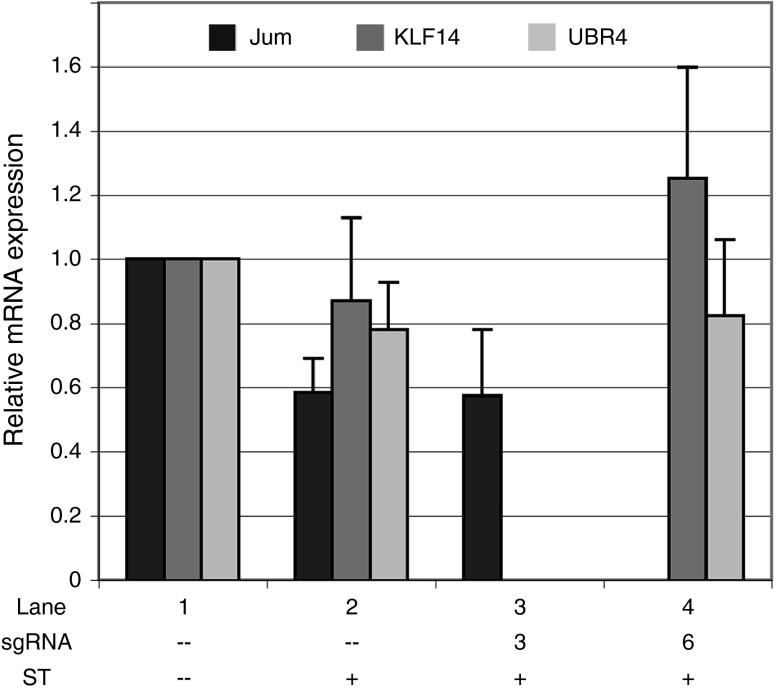
Next, we determined whether targeting a transcriptional effector to the DUX4 promoter or exon 1 has any effect on the expression of several predicted off-target genes. Of the sgRNAs used in this study to decrease expression of DUX4-fl and its downstream targets, #3 and #6 have the fewest off-target matches in the human genome (Supplementary Table S1). While an analysis of global effects on gene expression is beyond the scope of this study, we wanted to gain a preliminary assessment of the specificity of these sgRNAs. Therefore, we examined the expression of several genes at a range of distances from off-target matches to sgRNAs #3 and #6 in cells expressing the SunTag activator and either sgRNA. Since the binding specificity of an sgRNA is largely determined by the PAM-proximal sequence,72 we looked for genes in the vicinity of off-target matches to 9- or 12-bp seed sequences + NGG (PAM). For sgRNA #3, we assessed levels of the histone demethylase Jumonji, which contains an off-target match (12-bp seed + PAM) in intron 7. Importantly, while expression of the untargeted SunTag system alone had a slight repressive effect on levels of Jumonji (Figure 4, lane 2), targeting the activator with sgRNA #3 did not alter these levels (Figure 4, lane 3). For sgRNA #6, we assessed levels of the transcription factor KLF14 and the E3 ubiquitin ligase UBR4, which lie 28 and 76 kb downstream of off-target matches (9 bp seed + PAM). Similarly, neither gene showed altered expression in response to targeting the SunTag activator with this sgRNA (Figure 4, lane 4). These results are in stark contrast to the robust targeted activation of DUX4-fl (~30-fold and ~130-fold activation using sgRNAs #3 and #6, respectively; Figure 1b), and are consistent with the reports demonstrating limited off-target effects using dCas9 transcriptional effectors.52,53,56,57
Figure 4.
Targeting a transcriptional effector to the DUX4 promoter or exon 1 has no effect on expression of several off-target genes. Levels of Jumonji or KLF14 and UBR4 were assessed by quantitative reverse transcriptase polymerase chain reaction in mock-infected cultures or in cultures expressing the SunTag activator system alone or with sgRNA #3 or #6 (as in Figure 1b, lanes 1, 2, 5, and 8). Jumonji contains an off-target match (12-bp seed + PAM) to sgRNA #3 in intron 7. KLF14 and UBR4 lie 28 and 76 kb downstream of off-target matches (9 bp seed + PAM) to sgRNA #6. Refer to text for more details. Data are plotted as the mean + SD value of at least three independent experiments, with relative mRNA expression for the mock-infected cells set to 1.
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses the D4Z4 locus in FSHD myocytes
We performed the current study in FSHD1 muscle cells that display relatively high levels of DUX4-fl, which serves both as a measure of pathogenic gene expression and as a read-out of chromatin relaxation at the contracted allele. Since targeting a dCas9 repressor to the DUX4 promoter or exon 1 reduced DUX4-fl, we wanted to assess changes in the chromatin at the pathogenic locus. However, although DUX4 is present in every D4Z4 repeat unit at both 4q and 10q alleles, the chromatin at three of these alleles is already in a compacted, heterochromatic state. Thus, any attempt to assess repression at the contracted allele will be dampened by the presence of the other three alleles, and we expected that any observable changes in chromatin proteins or histone modifications would be small.
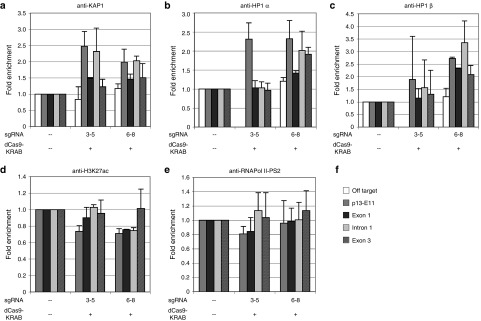
To determine whether changes in the D4Z4 chromatin structure could be detected, we infected FSHD myogenic cultures with combinations of lentiviral supernatants expressing dCas9-KRAB and sgRNAs targeting the DUX4 promoter or exon 1, induced the cells to differentiate, then fixed and harvested ~40 hours later for analysis by ChIP. Recruitment of the dCas9 repressor to the DUX4 promoter resulted in a trend toward increased levels of the KAP1/TRIM28 corepressor, which is recruited by the KRAB domain, as well as HP1α and HP1β, which are recruited by KAP1 to heterochromatin (Figure 5a–c, sgRNAs #6–8). These repressive changes were detectable across DUX4 as well as in the proximal p13-E11 region. Although levels of enrichment were slight (~2–3-fold), this was not surprising considering that the heterochromatic D4Z4 repeats at the uncontracted 4q allele and both 10q alleles were included in the assay. Changes in overall levels of the repressive histone marks H3K9me3 and H3K27me3 were undetectable across the D4Z4 repeats (data not shown), and targeting dCas9-KRAB to the DUX4 promoter resulted in only a slight decrease in the activating H3K27ac mark across DUX4 exon 1, intron 1, and p13-E11 (Figure 5d, sgRNAs #6–8).
Figure 5.
Recruitment of dCas9-KRAB to the DUX4 promoter or exon 1 represses the D4Z4 locus in facioscapulohumeral muscular dystrophy (FSHD) myocytes. Chromatin immunoprecipitation (ChIP) assays were performed using FSHD myogenic cultures infected with combinations of lentiviral supernatants expressing either dCas9-KRAB or individual sgRNAs targeting the DUX4 promoter (#6–8) or exon 1 (#3–5). Following infection, cells were induced to differentiate for ~40 hours, as in Figures 2 and 3. Chromatin was immunoprecipitated using antibodies specific for (a) KAP1, (b) HP1α, (c) HP1β, (d) H3K27ac, or (e) the elongating form of RNA Pol II (Pol II-PS2), and analyzed by qPCR using primers to the (f) p13-E11 region of 4q35 or exon 1, intron 1, or exon 3 of DUX4. Location of primers is shown in Figure 1a. In cases where enrichment of the specific factor was observed across the DUX4 locus, an off-target region was also assessed. Data are presented as fold enrichment of the target region by each specific antibody normalized to α-histone H3, with enrichment for the mock-infected cells set to 1. For all panels, each bar represents the average of at least three independent ChIP experiments.
Recruitment of the dCas9 repressor to DUX4 exon 1 increased levels of KAP1 at DUX4 intron 1, but had little observable effect on levels of HP1 or H3K27 acetylation across the gene (Figure 5a–d sgRNAs #3–5). By contrast, repressive changes (enrichment of KAP1 and HP1α, and slightly reduced levels of H3K27 acetylation) were more readily detected at p13-E11, likely as a result of recruitment to the NDE (Figure 5a–d, sgRNAs #3–5). There was also a trend toward slightly lower levels of elongating RNA Pol II at both exon 1 of DUX4 and p13-E11 (Figure 5e, sgRNAs #3–5). Considering that repressive effects are only expected at the 5 D4Z4 repeat units on the contracted 4qA allele, and these effects must be assessed amongst a background of >100 other heterochromatic D4Z4 repeats, these results are consistent with a model in which recruitment of dCas9-KRAB to the DUX4 promoter and exon 1 increases chromatin repression at the contracted 4q locus, resulting in decreased expression of the pathogenic DUX4-fl transcript.
Discussion
While CRISPR technology has been used successfully in early studies of genome editing, this is the first report in which a CRISPR/dCas9 system has been used to ameliorate pathogenic gene expression in FSHD. This is also, to our knowledge, the first time the technique has been used successfully in primary muscle cells. We overcame the technical hurdle of infecting primary myoblasts using serial infections with centrifugation,69 critical for achieving the high infection efficiency required to decrease DUX4-fl mRNA, which is only expressed in rare FSHD myocytes at any given time.32,33,44 Recently, exogenous siRNAs targeting the DUX4 promoter and coding sequence were successfully used to enhance silencing of D4Z4 by the DICER/Argonaute system in FSHD myocytes.73 Our study demonstrates a complementary approach, using a CRISPR/dCas9 effector to repress DUX4-fl and its misexpressed target genes, and supporting the usefulness of the DUX4 promoter and exon 1 as potential therapeutic targets.
One of the difficulties inherent in studying FSHD is the presence of large genomic duplications and chromosomal rearrangements in the 4q35 region. Assessing increased repression at the FSHD locus (the contracted, permissive 4qA allele in an FSHD1 patient) is complicated by: (i) the presence of the noncontracted, heterochromatic 4q allele and both heterochromatic 10q alleles, and (ii) the inability of primers to distinguish between these and other homologous, repetitive sequences. In spite of these caveats, recruitment of a dCas9 repressor to the DUX4 promoter resulted in a detectable increase in repressive chromatin regulators and a decrease in an activating histone mark across the region. Together with the decrease in DUX4-fl transcription, these results are consistent with enhanced repression of chromatin at the pathogenic locus. Analyzing effects on global gene expression via transcriptome profiling is beyond the scope of this proof-of-principle study; however, our examination of several genes in the vicinity of off-target matches for sgRNAs that target DUX4 revealed no changes in gene expression. In addition, virtually no off-target matches for the sgRNAs used here occur outside of D4Z4 homologues (Supplementary Table S1).49,74
While off-target binding of Cas9 and its derivatives is a serious concern for CRISPR-based therapeutics, the catalytically inactive dCas9 has the advantage of not generating double-stranded breaks in DNA, which are hotspots for chromosomal translocations. It is encouraging that studies of dCas9 effector platforms have also reported no significant or very low-level off-target effects on genome-wide transcription.52,53,56,57 This can be attributed in part to the narrow genomic window in which dCas9 effectors can mediate effects on gene expression (mainly enhancers and near the TSS of genes).53,56 Consistent with this, we found that dCas9 targeting of VP64 or KRAB to regions near D4Z4 (the proximal p13-E11 region and the distal DUX4 exon 3) had no effect on DUX4-fl expression. The off-target effects of a CRISPR repressor targeted to D4Z4 should be minimally toxic, as virtually all sequence matches for the sgRNAs used in our study occur in repressed, heterochromatic regions. In addition, the repressive activity of dCas9-KRAB is highly sensitive to mismatches in sgRNA target sequence; even single bp mismatches substantially reduce the level of repression observed.53 A recent study using CRISPR/Cas9 editing to correct the genetic lesion in a mouse model of Duchenne muscular dystrophy reported no difference in the frequency of indel mutations in 32 off-target regions among gene-edited and control mice, suggesting that Cas9 function is less promiscuous in vivo than in vitro.75
The development of increasingly sophisticated CRISPR-based systems is actively underway,57,71 as are methods for delivering Cas9 and its derivatives in vivo (e.g., via AAV vectors). From a therapeutic standpoint, the identification of sgRNAs that successfully target DUX4-fl in FSHD is likely to prove useful even as effector platforms and delivery methods evolve. By demonstrating feasibility, we have laid the groundwork for testing other dCas9 platforms and effectors in both cultured cells and in more therapeutically amenable in vivo models. Safe, efficient delivery of a dCas9-based platform that mediates the combinatorial recruitment of specific regulators—both protein and RNA—should pave the way for more effective and stable correction of FSHD and other epigenetic diseases.
With increasing evidence that the repeat genome (comprising nearly half the human genome) plays important roles in gene regulation, additional diseases will likely be found associated with aberrant repetitive genomic sequences.76,77,78,79 We have provided the first evidence that the repeat genome can be targeted via the CRISPR system, which is likely to prove useful as this hitherto overlooked portion of the genome is decoded.
Materials and Methods
Plasmids and antibodies. pHAGE EF1-dCas9-VP64 was a gift from Rene Maehr & Scot Wolfe (Addgene plasmid #50918).55 pHAGE EF1-dCas9-KRAB was a gift from Rene Maehr & Scot Wolfe (Addgene plasmid #50919).55 pLKO.1-puro U6 sgRNA BfuAI stuffer was a gift from Rene Maehr & Scot Wolfe (Addgene plasmid #50920).55 pHRdSV40-dCas9-10xGCN4-v4-P2A-BFP was a gift from Ron Vale (Addgene plasmid #60903).62 pHRdSV40-scFv-GCN4-sfGFP-VP64-GB1-NLS was a gift from Ron Vale (Addgene plasmid #60904).62 pHR-scFv-GCN4-sfGFP-GB1-NLS-dWPRE was a gift from Ron Vale (Addgene plasmid #60906).62 ChIP-grade antibodies used in this study were: α-KAP1 (ab3831), α-HP1α (ab77256), α-HP1β (ab10811), α-histone H3 (ab1791), α-histone H3K27acetyl (ab4729), and α-RNA Polymerase II CTD phospho S2 (ab5095) from Abcam (Cambridge, MA). Other antibodies used for ChIP were α-HA high affinity (clone 3F10, Roche, Indianapolis, IN) and normal mouse IgG (sc-2025, Santa Cruz Biotechnology, Dallas, TX).
sgRNA design and plasmid construction. We used the publically available sgRNA design tool from the Broad Institute (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) to identify high-scoring candidate sgRNAs to four target regions within and flanking the D4Z4 repeat array (Figure 1a; Supplementary Table S1). Predicted off-target matches were determined by BLASTing each sequence against the human genomic database (https://blast.ncbi.nlm.nih.gov) (Supplementary Table S1). High-scoring, nonoverlapping candidates with the fewest CpGs and off-target matches (four to five sgRNAs for each target region) were cloned individually into BfuAI sites in the pLKO.1-puro U6 sgRNA BfuAI stuffer plasmid and sequence-verified.
Cell culture, transient transfections, and lentiviral infections. Myogenic cultures derived from biceps muscle of an FSHD1 patient (17Abic) were used in this study. Patient 17A has two permissive 4qA alleles (~5 repeat units on a contracted 4A161 allele; ~26 repeat units on the non-contracted 4A-L161 allele; each 10q allele has ~37 repeat units). 17Abic myoblasts were grown in Ham's F-10 medium supplemented with 20% FBS (Hyclone), 0.5% chick embryo extract, 1% antibiotics and antimycotics, and 1.2 mmol/l CaCl2. 293T packaging cells were grown in DMEM + 10% FBS + 0.1% penicillin-streptavidin. At ~80% confluency, 293T cells were transfected with lentiviral packaging plasmid (pCMV-dR8.91), envelope plasmid (VSV-G), and sgRNA expression plasmid using the TransIT-LT1 transfection reagent (Mirus), according to the manufacturer's protocol. Lentiviral supernatants were harvested at 11-hour intervals from 72–108 hours post-transfection. At ~70–80% confluency, 17Abic myoblasts were subjected to four serial infections essentially as described.69 Briefly, lentiviral supernatants + 8 μg/ml polybrene were added to myoblasts and the plates were incubated for 15 minutes at 37 °C, then wrapped well with parafilm before centrifuging for 30 minutes at 1,100 g (32 °C). Following centrifugation, the viral supernatants were replaced with growth medium and cells were allowed to recover for ~8 hours prior to the next round of infection. Following the last round of infection, cells were switched to differentiation medium (DM) (DMEM/F-12 medium (1:1, Hyclone) plus 2% horse serum (Lonza)) for ~40–48 hours prior to harvesting.
qRT-PCR. Total RNAs were extracted using TRIzol (Invitrogen) and purified using the RNeasy Mini kit (Qiagen) after on-column DNase I digestion. Total RNA (2 µg) was used for cDNA synthesis using Superscript III Reverse Transcriptase (Invitrogen), and 200 ng of cDNA were used for qPCR analysis as previously described.33 Oligonucleotide primer sequences are provided in Supplementary Table S2.
ChIP. ChIP assays were performed with lentiviral-infected 17Abic differentiated myocytes using the Fast ChIP method80 with some modifications. Cells were fixed in 1% formaldehyde in DMEM for 10 minutes and dounced 10× prior to sonication. Cells were sonicated for 12 rounds of 15-second pulses at 65% power output on a Branson Sonifier 450 (VWR Scientific) to shear the DNA to a ladder of ~200–800 bp, and efficiency of shearing was verified by agarose gel electrophoresis. Chromatin was immunoprecipitated using 2 μg of specific antibodies or normal IgG. SYBR green quantitative PCR assays were performed for 40 cycles of: 94 °C for 15 seconds, 55 °C for 30 seconds, and 72 °C for 30 seconds. PCR products were analyzed on a 1.5% agarose gel to verify correct size of products and specificity of primer annealing. Oligonucleotide primer sequences are provided in Supplementary Table S2.
SUPPLEMENTARY MATERIAL Table S1. sgRNAs targeting the FSHD Locus. Table S2. Sequences of oligonucleotide primers (5' to 3'). References
Acknowledgments
This work was financially supported by the National Institute of Arthritis, Musculoskeletal, and Skin Diseases grant #1R01AR062587 and the Association Française contre les Myopathies grant #AFM15700. The authors thank Kathryn R. Wagner and the UMMS Wellstone Center for providing cells, and the Chris Carrino Foundation for FSHD for their support of our FSHD research projects. The authors declare no conflicts of interest.
Supplementary Material
References
- Deenen, JC, Arnts, H, van der Maarel, SM, Padberg, GW, Verschuuren, JJ, Bakker, E et al. (2014). Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 83: 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanet. 2011. Prevalence of rare diseases: bibliographic data in Orphanet report series: rare diseases collection. http://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence_of_rare_diseases_by_alphabetical_list.pdf.
- Padberg, GW. Facioscapulohumeral Disease [Thesis]. Leiden University, Leiden, the Netherlands, 1982. pp. 243. [Google Scholar]
- van der Maarel, SM, Miller, DG, Tawil, R, Filippova, GN and Tapscott, SJ (2012). Facioscapulohumeral muscular dystrophy: consequences of chromatin relaxation. Curr Opin Neurol 25: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil, R (2008). Facioscapulohumeral muscular dystrophy. Neurotherapeutics 5: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda, CL, Jones, TI and Jones, PL (2015). Facioscapulohumeral muscular dystrophy as a model for epigenetic regulation and disease. Antioxid Redox Signal 22: 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger, L, Tapscott, SJ and van der Maarel, SM (2015). Genetic and epigenetic contributors to FSHD. Curr Opin Genet Dev 33: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom, JC, Wijmenga, C, van Tienhoven, EA, Gruter, AM, Hewitt, JE, Padberg, GW et al. (1993). FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2: 2037–2042. [DOI] [PubMed] [Google Scholar]
- Wijmenga, C, Frants, RR, Brouwer, OF, Moerer, P, Weber, JL and Padberg, GW (1990). Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet 336: 651–653. [DOI] [PubMed] [Google Scholar]
- Wijmenga, C, Hewitt, JE, Sandkuijl, LA, Clark, LN, Wright, TJ, Dauwerse, HG et al. (1992). Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2: 26–30. [DOI] [PubMed] [Google Scholar]
- van Deutekom, JC, Bakker, E, Lemmers, RJ, van der Wielen, MJ, Bik, E, Hofker, MH et al. (1996). Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet 5: 1997–2003. [DOI] [PubMed] [Google Scholar]
- Lemmers, RJ, Wohlgemuth, M, Frants, RR, Padberg, GW, Morava, E and van der Maarel, SM (2004). Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet 75: 1124–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, RJ, de Kievit, P, Sandkuijl, L, Padberg, GW, van Ommen, GJ, Frants, RR et al. (2002). Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32: 235–236. [DOI] [PubMed] [Google Scholar]
- Lemmers, RJ, Wohlgemuth, M, van der Gaag, KJ, van der Vliet, PJ, van Teijlingen, CM, de Knijff, P et al. (2007). Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet 81: 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, RJ, van der Vliet, PJ, Klooster, R, Sacconi, S, Camaño, P, Dauwerse, JG et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329: 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y, Forner, J, Fournet, S and Jeanpierre, M (2001). Improved characterization of FSHD mutations. Ann Genet 44: 105–110. [DOI] [PubMed] [Google Scholar]
- Rossi, M, Ricci, E, Colantoni, L, Galluzzi, G, Frusciante, R, Tonali, PA et al. (2007). The Facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med Genet 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJL, de Kievit, P, van Geel, M, van der Wielen, MJ, Bakker, E, Padberg, GW et al. (2001). Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol 50: 816–819. [DOI] [PubMed] [Google Scholar]
- Gilbert, JR, Stajich, JM, Wall, S, Carter, SC, Qiu, H, Vance, JM et al. (1993). Evidence for heterogeneity in facioscapulohumeral muscular dystrophy (FSHD). Am J Hum Genet 53: 401–408. [PMC free article] [PubMed] [Google Scholar]
- de Greef, JC, Lemmers, RJ, van Engelen, BG, Sacconi, S, Venance, SL, Frants, RR et al. (2009). Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat 30: 1449–1459. [DOI] [PubMed] [Google Scholar]
- Lemmers, RJ, Tawil, R, Petek, LM, Balog, J, Block, GJ, Santen, GW et al. (2012). Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 44: 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil, R, Forrester, J, Griggs, RC, Mendell, J, Kissel, J, McDermott, M et al. (1996). Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. The FSH-DY Group. Ann Neurol 39: 744–748. [DOI] [PubMed] [Google Scholar]
- Tawil, R and Van Der Maarel, SM (2006). Facioscapulohumeral muscular dystrophy. Muscle Nerve 34: 1–15. [DOI] [PubMed] [Google Scholar]
- van Overveld, PG, Lemmers, RJ, Sandkuijl, LA, Enthoven, L, Winokur, ST, Bakels, F et al. (2003). Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35: 315–317. [DOI] [PubMed] [Google Scholar]
- Yang, F, Shao, C, Vedanarayanan, V and Ehrlich, M (2004). Cytogenetic and immuno-FISH analysis of the 4q subtelomeric region, which is associated with facioscapulohumeral muscular dystrophy. Chromosoma 112: 350–359. [DOI] [PubMed] [Google Scholar]
- Jiang, G, Yang, F, van Overveld, PG, Vedanarayanan, V, van der Maarel, S and Ehrlich, M (2003). Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet 12: 2909–2921. [DOI] [PubMed] [Google Scholar]
- Hartweck, LM, Anderson, LJ, Lemmers, RJ, Dandapat, A, Toso, EA, Dalton, JC et al. (2013). A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology 80: 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca, DS, Casa, V, Bodega, B, Xynos, A, Ginelli, E, Tanaka, Y et al. (2012). A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W, de Greef, JC, Chen, YY, Chien, R, Kong, X, Gregson, HC et al. (2009). Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 5: e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi, S, Boyden, SE, Estrella, EA, Jones, TI, Rahimov, F, Yu, TW et al. (2013). Exome sequencing identifies a novel SMCHD1 mutation in facioscapulohumeral muscular dystrophy 2. Neuromuscul Disord 23: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacconi, S, Lemmers, RJ, Balog, J, van der Vliet, PJ, Lahaut, P, van Nieuwenhuizen, MP et al. (2013). The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet 93: 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, L, Geng, LN, Lemmers, RJ, Kyba, M, Ware, CB, Nelson, AM et al. (2010). Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 6: e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, TI, Chen, JC, Rahimov, F, Homma, S, Arashiro, P, Beermann, ML et al. (2012). Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum Mol Genet 21: 4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, JM, Whiddon, JL, Yao, Z, Kasinathan, B, Snider, L, Geng, LN et al. (2013). DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet 9: e1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, LN, Yao, Z, Snider, L, Fong, AP, Cech, JN, Young, JM et al. (2012). DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma, S, Beermann, ML, Boyce, FM and Miller, JB (2015). Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann Clin Transl Neurol 2: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q, Snider, L, Jagannathan, S, Tawil, R, van der Maarel, SM, Tapscott, SJ et al. (2015). A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife 4:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow, V, Marcowycz, A, Ansseau, E, Conde, CB, Sauvage, S, Mattéotti, C et al. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 17: 611–623. [DOI] [PubMed] [Google Scholar]
- Bosnakovski, D, Xu, Z, Gang, EJ, Galindo, CL, Liu, M, Simsek, T et al. (2008). An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 27: 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski, D, Daughters, RS, Xu, Z, Slack, JM and Kyba, M (2009). Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One 4: e7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuebbles, RD, Long, SW, Hanel, ML and Jones, PL (2010). Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int J Clin Exp Pathol 3: 386–400. [PMC free article] [PubMed] [Google Scholar]
- Wallace, LM, Garwick, SE, Mei, W, Belayew, A, Coppee, F, Ladner, KJ et al. (2011). DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol 69: 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplanck, C, Ansseau, E, Charron, S, Stricwant, N, Tassin, A, Laoudj-Chenivesse, D et al. (2011). The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One 6: e26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, TI, King, OD, Himeda, CL, Homma, S, Chen, JC, Beermann, ML et al. (2015). Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin Epigenetics 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, RJ, Goeman, JJ, van der Vliet, PJ, van Nieuwenhuizen, MP, Balog, J, Vos-Versteeg, M et al. (2015). Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum Mol Genet 24: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, TI, Yan, C, Sapp, PC, McKenna-Yasek, D, Kang, PB, Quinn, C et al. (2014). Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin Epigenetics 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, L, Asawachaicharn, A, Tyler, AE, Geng, LN, Petek, LM, Maves, L et al. (2009). RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18: 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, JA and Charpentier, E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Zeng, W, Chen, YY, Newkirk, DA, Wu, B, Balog, J, Kong, X et al. (2014). Genetic and epigenetic characteristics of FSHD-associated 4q and 10q D4Z4 that are distinct from non-4q/10q D4Z4 homologs. Hum Mutat 35: 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, AW, Wang, H, Yang, H, Shi, L, Katz, Y, Theunissen, TW et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X, Tsang, JC, Gaba, F, Wu, D, Lu, L and Liu, P (2014). Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res 42: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Larson, MH, Morsut, L, Liu, Z, Brar, GA, Torres, SE et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Horlbeck, MA, Adamson, B, Villalta, JE, Chen, Y, Whitehead, EH et al. (2014). Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Lei, Y, Wong, WK, Liu, S, Lee, KC, He, X et al. (2014). Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res 42: 4375–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, NA, Genga, RM, Enuameh, MS, Garber, M, Wolfe, SA and Maehr, R (2014). Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, NA, Pham, H, Tabak, B, Genga, RM, Silverstein, NJ, Garber, M et al. (2015). Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12: 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S, Brigham, MD, Trevino, AE, Joung, J, Abudayyeh, OO, Barcena, C et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, ML, Linder, SJ, Cascio, VM, Fu, Y, Ho, QH and Joung, JK (2013). CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Aach, J, Stranges, PB, Esvelt, KM, Moosburner, M, Kosuri, S et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera, P, Kocak, DD, Vockley, CM, Adler, AF, Kabadi, AM, Polstein, LR et al. (2013). RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, IB, D'Ippolito, AM, Vockley, CM, Thakore, PI, Crawford, GE, Reddy, TE et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum, ME, Gilbert, LA, Qi, LS, Weissman, JS and Vale, RD (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda, CL, Debarnot, C, Homma, S, Beermann, ML, Miller, JB, Jones, PL et al. (2014). Myogenic enhancers regulate expression of the facioscapulohumeral muscular dystrophy-associated DUX4 gene. Mol Cell Biol 34: 1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X, Scott, DA, Kriz, AJ, Chiu, AC, Hsu, PD, Dadon, DB et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H, Xiao, T, Chen, CH, Li, W, Meyer, CA, Wu, Q et al. (2015). Sequence determinants of improved CRISPR sgRNA design. Genome Res 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, RJ, Osborn, M, Haaf, T, Rogers, M, Frants, RR, Padberg, GW et al. (2003). D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology 61: 178–183. [DOI] [PubMed] [Google Scholar]
- Wijmenga, C, Sandkuijl, LA, Moerer, P, van der Boorn, N, Bodrug, SE, Ray, PN et al. (1992). Genetic linkage map of facioscapulohumeral muscular dystrophy and five polymorphic loci on chromosome 4q35-qter. Am J Hum Genet 51: 411–415. [PMC free article] [PubMed] [Google Scholar]
- Bakker, E, Wijmenga, C, Vossen, RH, Padberg, GW, Hewitt, J, van der Wielen, M et al. (1995). The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve Suppl 2: S39–44. [PubMed] [Google Scholar]
- Springer, ML and Blau, HM (1997). High-efficiency retroviral infection of primary myoblasts. Somat Cell Mol Genet 23: 203–209. [DOI] [PubMed] [Google Scholar]
- Qi, LS, Larson, MH, Gilbert, LA, Doudna, JA, Weissman, JS, Arkin, AP et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan, JG, Lee, ME, Almeida, R, Gilbert, LA, Whitehead, EH, La Russa, M et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu, C, Arslan, S, Singh, R, Thorpe, J and Adli, M (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32: 677–683. [DOI] [PubMed] [Google Scholar]
- Lim, JW, Snider, L, Yao, Z, Tawil, R, Van Der Maarel, SM, Rigo, F et al. (2015). DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD. Hum Mol Genet 24: 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altherr, MR, Bengtsson, U, Markovich, RP and Winokur, ST (1995). Efforts toward understanding the molecular basis of facioscapulohumeral muscular dystrophy. Muscle Nerve Suppl 2: S32–38. [PubMed] [Google Scholar]
- Long, C, McAnally, JR, Shelton, JM, Mireault, AA, Bassel-Duby, R and Olson, EN (2014). Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 345: 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, LL and Lawrence, JB (2010). XIST RNA and architecture of the inactive X chromosome: implications for the repeat genome. Cold Spring Harb Symp Quant Biol 75: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa, V and Gabellini, D (2012). A repetitive elements perspective in Polycomb epigenetics. Front Genet 3: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone, DM and Lawrence, JB (2013). Heterochromatin instability in cancer: from the Barr body to satellites and the nuclear periphery. Semin Cancer Biol 23: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken, J, Zeller, P and Gasser, SM (2015). Repeat DNA in genome organization and stability. Curr Opin Genet Dev 31: 12–19. [DOI] [PubMed] [Google Scholar]
- Nelson, JD, Denisenko, O and Bomsztyk, K (2006). Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.