Abstract
Complete eradication of HIV-1 infection is impeded by the existence of cells that harbor chromosomally integrated but transcriptionally inactive provirus. These cells can persist for years without producing viral progeny, rendering them refractory to immune surveillance and antiretroviral therapy and providing a permanent reservoir for the stochastic reactivation and reseeding of HIV-1. Strategies for purging this latent reservoir are thus needed to eradicate infection. Here, we show that engineered transcriptional activation systems based on CRISPR/Cas9 can be harnessed to activate viral gene expression in cell line models of HIV-1 latency. We further demonstrate that complementing Cas9 activators with latency-reversing compounds can enhance latent HIV-1 transcription and that epigenome modulation using CRISPR-based acetyltransferases can also promote viral gene activation. Collectively, these results demonstrate that CRISPR systems are potentially effective tools for inducing latent HIV-1 expression and that their use, in combination with antiretroviral therapy, could lead to improved therapies for HIV-1 infection.
Introduction
Since its identification in 1983, HIV has remained a global pandemic. An estimated 35 million people worldwide are currently living with HIV type 1 (HIV-1), and an estimated 39 million people have died of AIDS-related illness since the onset of the epidemic (see UNAIDS report: http://unaids.org/globalreport/Global_report.htm). Strict adherence to combination antiretroviral therapy (cART) has significantly improved the quality of life and life expectancy of many infected individuals, transforming HIV-1 into a chronic, manageable illness in the industrialized world. However, despite its durable capacity to inhibit viral replication, cART does not cure HIV-1 infection since it cannot address the residual transcriptionally silent but replication-competent population of virus hidden within certain cells.1,2,3 This so-called latent reservoir is primarily established when infected CD4+ T cells revert to a resting memory state.4,5 The resulting cells are largely nonpermissive for viral gene expression and therefore do not generate new viral progeny.5,6 Because latent HIV-1 is refractory to both immunological surveillance and cART intervention, its existence poses a major obstacle to complete viral eradication.7,8,9
Though the mechanisms underlying latency are complex,10,11,12,13 several strategies have been developed to activate this HIV-1 reservoir,12 thereby rendering it susceptible to cART eradication. Because latent virus responds to T-cell activation signals, proinflammatory cytokines—such as interleukin 2 (IL-2)14 and IL-715—can be used to induce its emergence from resting CD4+ T cells. Protein kinase C agonists that stimulate viral gene expression via NF-κB signaling16—including bryostatin,17 prostratin,18,19 and other synthetic analogs20,21—are also candidates for this strategy. Additionally, because histone deacetylation can induce repressive changes in chromatin structure at the HIV-1 promoter, histone deacetylase (HDAC) inhibitors have emerged as a promising approach for reversing latency.22,23
Cytokine therapy, however, has been unable to completely purge latent virus24,25 and has even led to toxicity26 and/or long-term depletion of CD4+ T cells.27 Additionally, protein kinase C agonists may be unable to activate latent HIV-1 in repressive chromatin, and to date, several HDAC inhibitors have demonstrated suboptimal results in clinical trails.28,29 The use of high levels of HDAC inhibitors may also affect global gene expression,30 potentially leading to unwanted side effects.31 These limitations underscore the need for new strategies capable of stimulating latent HIV-1 expression in a robust and targeted manner.
The emergence of versatile genome modulation and editing technologies32,33—such as those based on zinc-finger domains,34 TAL effector proteins,35 and the RNA-guided CRISPR/Cas9 system36—can offer new means to combat HIV-1 infection. For example, the relative ease with which these tools can be configured, as well as their broad versatility, has endowed investigators with the ability to confer HIV-1 resistance upon primary cells37 and stem cells38,39,40 via modification of the CCR5 gene. Indeed, this approach recently showed evidence of efficacy in a phase 1 clinical trial.41 Other strategies facilitated by this technology include the disruption42 and excision43,44,45 of proviral DNA from HIV-infected cells, the disruption of endogenous host factors critical for HIV-1 integration,46,47 and the attenuation of HIV-1 replication via transcriptional repression.48,49 Among the three major DNA-targeting systems, CRISPR/Cas9 can readily be directed to nearly any genomic locus via RNA–DNA complementary base pairing using a chimeric single guide RNA (sgRNA).50 Though this system is typically used for inducing DNA cleavage,50,51,52 it can be co-opted for transcriptional modulation by fusing a catalytically inactivated variant of the Cas9 nuclease (referred to as dCas9)53 with a transcriptional activator54,55,56 or repressor57 domain. Because the only major restriction for CRISPR/Cas9 target site selection is the protospacer adjacent motif,50 which is recognized by the Cas9 protein and located immediately downstream of the sgRNA target site, CRISPR transcription activation systems54,55,58,59,60,61,62 have the potential for driving the expression of nearly any gene, including stimulating viral gene expression within latent HIV-1 reservoirs for shock-and-kill treatments. Importantly, unlike approaches that rely on the Cas9 nuclease to disrupt HIV-1 provirus, CRISPR-based activators pose no significant risk of introducing mutations within the host genome63 and have the added advantage of providing potentially reversible activity.
Here, we demonstrate that CRISPR-based transcription activators can be configured to induce latent HIV-1 expression and that combining these CRISPR tools with latency-reversing compounds can lead to robust activation of viral gene expression in cell-based models of HIV-1 latency.
Results
CRISPR/Cas9 transcription activation systems induce gene activation from the HIV-1 long terminal repeat
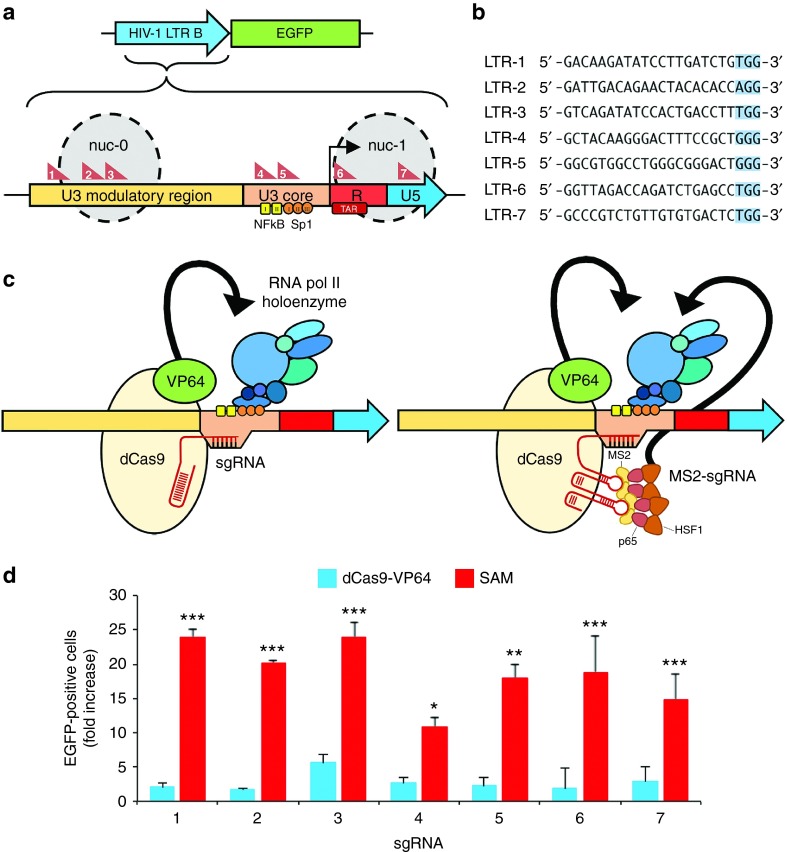
We have investigated whether CRISPR-based transcription factors can reverse latency by stimulating gene expression from the HIV-1 long terminal repeat (LTR) promoter.64 The HIV-1 subtype B LTR is 634 bp in length and can be subdivided into three regions (Figure 1a and Supplementary Figure S1): a 454-nucleotide (nt) segment upstream of the transcriptional start site termed U3, which includes binding sites for host transcription factors that drive HIV-1 gene expression as well as other cis-acting DNA elements; a 96-nt repeat region downstream of the transcriptional start site referred to as R, which encodes the trans-activation response element; and an 84-nt segment downstream of the R domain designated U5.
Figure 1.
CRISPR transcriptional activators induce gene expression from the HIV-1 LTR. (a) Schematic of the EGFP reporter system used to evaluate Cas9-mediated gene activation and the architecture of the HIV-1 subtype B long terminal repeat (LTR) promoter. Gray circles indicate approximate locations of nucleosome-0 (nuc-0) and nuc-1 in the LTR. Red triangles indicate location of the sgRNA target site. Black arrow denotes the transcriptional start site (TSS). Yellow squares and orange circles indicate NF-κB and Sp-1 binding sites, respectively. (b) sgRNA target sites from the LTR. PAM is highlighted in blue. (c) Cartoon illustrating the potential modes of transcriptional activation by dCas9-VP64 and SAM. Black arrows indicate VP64- and MS2-p65-HSF1-mediated recruitment of cellular transcription factors and the chromatin-remodeling proteins. (d) Fold increase in the percentage of EGFP-positive HEK293T cells after transfection with EGFP reporter plasmid and expression vectors encoding dCas9-VP64 with sgRNA (blue) or SAM with sgRNA (red). EGFP expression was normalized to cells transfected with reporter plasmid, dCas9-VP64, and an empty sgRNA expression cassette. EGFP expression was measured 48 hours after transfection and was observed in 0.5 ± 0.6% of negative control cells. Error bars indicate SD (n = 3; *P < 0.005; **P < 0.001; ***P < 0.0001; Student's t-test).
We designed seven sgRNA to overlap with key features of the LTR (Figure 1a,b). The U3 region was targeted by sgRNAs 1–5. sgRNAs 4 and 5, in particular, were designed to overlap with the binding sites for NF-κB and Sp-1, respectively, since these transcription factors contribute directly to viral gene expression and latency.65,66,67,68,69 The R region, which encodes a repressive mRNA hairpin structure (trans-activation response) that inhibits RNA pol II processivity70,71 in the absence of the viral Tat protein, was targeted by sgRNA 6. Finally, the U5 region was targeted by sgRNA 7.
We initially evaluated the ability of two distinct CRISPR complexes to induce gene activation: (i) dCas9-VP64 (refs. 54,55) and (ii) the synergistic activation mediator (SAM) complex.61 dCas9-VP64 comprises dCas9 fused with a tetrameric repeat of the herpes simplex virus VP16 transactivation domain (VP64; ref. 72; Figure 1c). The SAM system similarly contains dCas9-VP64 but also: (i) a modified sgRNA harboring an aptamer that binds to the MS2 bacteriophage coat protein and (ii) a tripartite fusion protein consisting of the MS2 protein, the NF-κB trans-activating subunit p65, and the activation domain from the human heat-shock factor 1 protein (HSF1) (MS2–p65–HSF1) (Figure 1c). Both complexes function by recruiting cellular transcription factors73 and chromatin-remodeling proteins73,74 to targeted genomic loci, leading to transcriptional activation.
To assess the ability of each Cas9-based complex to stimulate gene expression, we co-transfected human embryonic kidney (HEK) 293T cells with either dCas9-VP64 or SAM, as well as a reporter plasmid containing the full-length HIV-1 subtype B promoter upstream of an EGFP reporter gene (Figure 1a). Both dCas9 complexes were tested with each of the seven individual sgRNAs, and the level of Cas9-mediated gene activation was directly correlated with EGFP fluorescence. We observed a minimal increase in EGFP expression in cells transfected with dCas9-VP64, with only sgRNA 3 yielding a significant increase (P < 0.05) in the number of EGFP-positive cells (Figure 1d). Surprisingly, “tiling” the LTR promoter with multiple sgRNAs, which has been shown to lead to increased activation of endogenous genes in HEK293T cells,54,55 had minimal effect on EGFP expression in this transient reporter system (Supplementary Figure S2).
In contrast to the dCas9-VP64 complex, HEK293T cells co-transfected with reporter plasmid and the SAM system showed a 12- to 24-fold increase in the number or percentage of EGFP-positive cells, with the most robust levels of activation observed using sgRNAs 1, 2, and 3 (Figure 1d). As previously reported,61 no substantial increase in SAM-mediated activation was observed by tiling the promoter with multiple sgRNAs (Supplementary Figure S3). As a control for target specificity, we evaluated EGFP expression in cells co-transfected with reporter plasmid and SAM encoding a random sgRNA library. No significant increase in EGFP was observed in transfected cells (Supplementary Figure S4), indicating that SAM induced specific activation from the HIV-1 LTR promoter.
Reactivation of latent HIV expression by CRISPR activator complexes
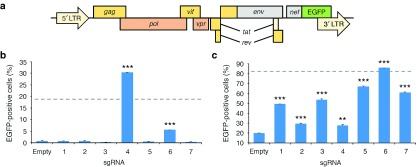
We next evaluated whether SAM could reactivate viral gene expression in established cell line models of HIV-1 latency. Specifically, we sequentially infected the Jurkat-derived lymphocytic cell lines J-Lat 9.2 and J-Lat 10.6 (ref. 75) with lentiviruses encoding each SAM element (Figure 2a). J-Lat cells harbor an integrated but transcriptionally silent HIV-1 provirus that expresses GFP in lieu of the nef and env genes but still recapitulates the natural transcriptional complexity of the HIV promoter, thereby mimicking HIV latency. We specifically chose the J-Lat 9.2 and 10.6 cell lines since they display distinct gene activation thresholds arising from differences in chromatin accessibility at the HIV-1 LTR.76 Specifically, J-Lat 9.2 is a strongly repressed clone that requires high transcriptional induction to overcome its chromatin environment, whereas J-Lat 10.6 cells possess a lower activation threshold.
Figure 2.
SAM-mediated activation of latent HIV-1 expression in cell line models of latency. (a) Diagram of the HIV-1 proviral genome in J-Lat cells. Gray boxes indicate genes whose expression has been disabled. (b and c) Percentage of GFP-positive (b) J-Lat 9.2 and (c) J-Lat 10.6 cells stably expressing SAM with sgRNA. “Empty” indicates J-Lat cells expressing SAM with an empty sgRNA expression cassette. Dotted line indicates the percentage of naive GFP positive (b) J-Lat 9.2 and (c) J-Lat 10.6 cells after treatment with 20 ng/µl tumor necrosis factor α (TNF-α). GFP fluorescence was measured 14 days after final infection. Error bars indicate SD (n = 3; **P < 0.001; ***P < 0.0001; Student's t-test).
We observed GFP expression in ~30% and ~5% of J-Lat 9.2 cells stably expressing SAM with sgRNAs 4 and 6, respectively (Figure 2b). Additionally, we observed reactivation in up to 85% of J-Lat 10.6 cells expressing SAM with sgRNA 6 (Figure 2c). As an efficient positive induction control, we used tumor necrosis factor alpha (TNF-α), a proinflammatory cytokine that stimulates HIV expression through activation of NF-κB, but whose in vivo toxicity77 precludes its use as a therapeutic. TNF-α treatment yielded ~18% and ~82% GFP-positive J-Lat 9.2 and 10.6 cells, respectively (Supplementary Figure S5). Compared to an empty sgRNA cassette, each sgRNA we tested induced an increase (P < 0.05) in the number of GFP-positive J-Lat 10.6 cells (Figure 2c). However, only two sgRNA (4 and 6) induced a significant increase (P < 0.05 for both) in the number of GFP-positive cells in the more repressed J-Lat 9.2 cell line (Figure 2b). Taken together, these data indicate that CRISPR/Cas9-based transcription activation systems delivered via lentivirus can stimulate latent HIV gene expression but that differences in chromatin accessibility at the HIV-1 LTR can affect the ability of Cas9 to stimulate transcription.
Combining CRISPR activators with an HDAC inhibitor and prostratin synergistically increases latent HIV activation
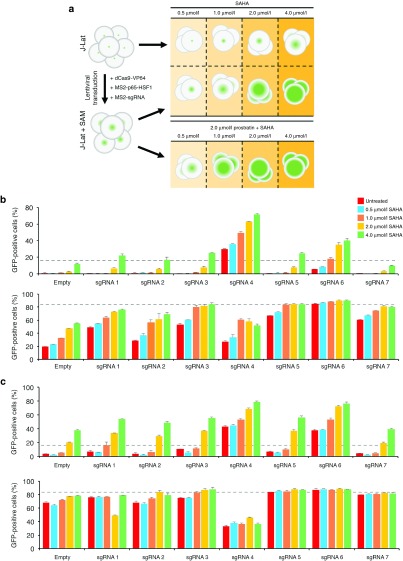
HDAC inhibitors, such as valproic acid78 or suberoylanilide hydroxamic acid (i.e., SAHA or Vorinostat),29 are capable of partially reversing the effects of chromatin silencing and alleviating HIV latency. We hypothesized that such compounds could also enhance the ability of Cas9 to induce HIV-1 expression, particularly in repressive chromatin contexts. To this end, we treated J-Lat 9.2 and 10.6 cells stably expressing SAM with increasing doses of SAHA (Figure 3a), a compound that inhibits the Class I HDAC isotypes 1, 2, 3, and 8. SAHA was recently shown to be safe and well tolerated in a phase 1 clinical trial, though it was unable to provide a long-term increase in HIV-1 expression in patients.28
Figure 3.
SAHA and prostratin enhance SAM-mediated activation of latent HIV-1 expression. (a) Schematic illustrating experimental setup. Naive or SAM-expressing J-Lat cells were treated with SAHA alone or in the presence of prostratin. (b and c) Percentage of GFP-positive J-Lat 9.2 (upper) and J-Lat 10.6 (lower) cells stably expressing SAM and treated with (b) increasing amounts of SAHA only or (c) increasing amounts of SAHA with 2 µmol/l prostratin. “Empty” indicates J-Lat cells expressing SAM with an empty sgRNA expression cassette. Dotted line indicates the percentage of naive GFP-positive J-Lat cells after treatment with 20 ng/µl of TNF-α. Cells were treated 17 days after final infection, and GFP expression was measured 24 hours after treatment. Error bars indicate SD (n = 3).
Incubation of SAM-expressing J-Lat cells with SAHA led to a robust and dose-dependent increase in the number of GFP-positive cells (Figure 3b). Specifically, reactivation was evident in ~70% of J-Lat 9.2 cells with sgRNA 4, and ~80% of J-Lat 10.6 cells with sgRNAs 3, 5, 6, and 7 after treatment with 4 μmol/l SAHA, substantially higher than control cells expressing an empty sgRNA expression cassette (Figure 3b). Depending on the sgRNA used, high-doses of SAHA led to a twofold to fivefold increase in activation in J-Lat 9.2 cells (Figure 3b), indicating that CRISPR-based transcriptional modulators and HDAC inhibitors can act synergistically to reactivate HIV-1.
We next investigated whether the use of multiple latency-reversing compounds in combination with Cas9 could further increase HIV-1 expression. We treated J-Lat 9.2 and 10.6 cells expressing SAM with escalating doses of SAHA and 2 μmol/l prostratin (Figure 3a). The latter small molecule stimulates IKK-dependent phosphorylation and degradation of Iκ-Bα,79 leading to the rapid nuclear translocation of NF-κB and activation of latent provirus.18,19 Depending on the sgRNA used, SAM-expressing J-Lat 9.2 cells co-treated with SAHA plus prostratin showed a twofold to threefold increase in the number of GFP-positive cells compared to those treated with SAHA only (Figure 3c). Notably, sgRNAs 4 and 6 stimulated GFP expression in up to 80% of J-Lat 9.2 cells treated with SAHA and prostratin (Figure 3c). J-Lat cells expressing SAM and treated with prostratin in the absence of SAHA, however, showed less activation than those treated with both compounds (Supplementary Figure S6).
Interestingly, J-Lat 10.6 cells expressing SAM with sgRNA 4—whose target site overlaps with the binding sites for NF-κB—showed a marked decrease in GFP-positive cells after treatment with prostratin (Figure 3c), indicating the possibility that SAM and NF-κB may be competing for LTR-binding sites and that this interplay led to suboptimal viral gene expression. Collectively, these results indicate that HIV latency-reversing compounds can enhance the efficacy of Cas9-mediated activation of HIV-1, particularly in repressive chromatin contexts.
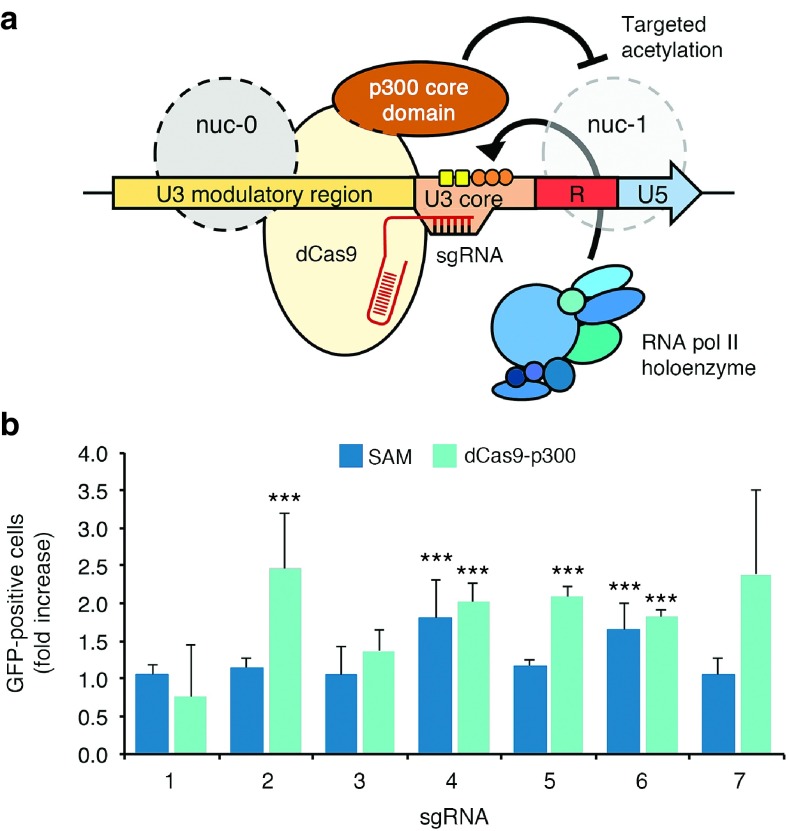
Reactivation of latent HIV by epigenome editing using CRISPR/Cas9-based acetyltransferases
Multiple mechanisms contribute to HIV latency, including repressive local chromatin effects stemming from histone deacetylation of the nucleosomes (nuc-0 and nuc-1) that form within the LTR.80 While HDAC inhibitors such as SAHA can stimulate latent HIV expression by promoting an open chromatin environment around the provirus promoter, their activity can also influence the expression of host genes. In contrast, emerging epigenome-modifying technologies81 have the potential to directly alter the chromatin structure of the LTR and thereby affect target gene expression in a site-specific manner. For instance, fusion of dCas9 with the catalytic histone acetyltransferase core domain of the human E1A-associated protein p300 (dCas9-p300) was recently shown to facilitate transcriptional activation via targeted acetylation of histone H3 in the promoter region of targeted genes (Figure 4a).82 We thus hypothesized that dCas9-p300 could also reactivate latent HIV-1 expression by acetylation of nuc-0 or nuc-1. To examine this possibility, we evaluated GFP expression in J-Lat 10.6 cells nucleofected with expression vectors encoding dCas9-p300 and the aforementioned LTR-targeted sgRNAs. In particular, sgRNAs 1, 2, and 3 mediate binding near nuc-0, whereas the target sites for sgRNAs 6 and 7 overlap with nuc-1 (Figure 1a). dCas9-p300 introduced into cells by nucleofection-induced significant gene expression when targeted by sgRNAs 2, 4, 5, and 6 (P < 0.05 for each; Figure 4b), achieving levels of activation comparable to each corresponding SAM complex (which were ~2-fold lower than that by lentiviral-mediated expression; Figure 2). In the case of sgRNA 2, whose target site overlaps with nuc-0, dCas9-p300 induced a significant increase (P < 0.05) in HIV-1 expression over SAM (Figure 4b). Likely due to their more repressive transcriptional environment, no significant increase in activation was observed when J-Lat 9.2 cells were nucleofected with dCas9-p300 (data not shown). However, these findings collectively indicate that dCas9-p300 offers strong potential to stimulate transcription from the HIV-1 LTR.
Figure 4.
Reactivation of latent HIV-1 expression using CRISPR-based acetyltransferases. (a) Cartoon illustrating the potential mode of action by a CRISPR-based acetyltransferase (dCas9-p300) on the HIV LTR. Gray circles indicate approximate locations of nuc-0 and nuc-1 in the LTR. (b) Fold increase in the percentage of GFP-positive J-Lat 10.6 cells after nucleofection with SAM and sgRNA (dark blue) or dCas9-p300 with sgRNA (teal). Data were normalized to J-Lat 10.6 cells nucleofected with SAM or dCas9-p300 and an empty sgRNA expression cassette. GFP fluorescence was measured 48 hours after nucleofection. Error bars indicate SD (n = 3; ***P < 0.0001; Student's t-test).
Discussion
The presence of residual latent but replication-competent HIV-1 reservoirs is a major hurdle impeding viral eradication. Recent work has indicated that HIV expression can be induced using zinc-finger83 and TAL effector-based84 transcription factors engineered to bind the LTR promoter. Here, we show that CRISPR/Cas9-based transcriptional effectors—which can be retargeted to nearly any DNA sequence without protein engineering—can reactivate viral gene expression in cell line models of HIV-1 latency. Using LTR-targeted sgRNAs, we tested the ability of two distinct Cas9 complexes to stimulate transcription: dCas9-VP64 (refs. 54,55) and the SAM complex,61 which consists of dCas9-VP64 and an accessory transactivation domain designed to recruit a complementary suite of transcription and chromatin remodeling factors. We found that SAM activated gene expression from the LTR in transient reporter assays at levels that exceeded dCas9-VP64. This result is consistent with recent data indicating that, for some genes, first-generation dCas9-based transcription factors induce only modest levels of transcription.53,54,55,56,57
While all seven LTR-targeted sgRNAs induced robust expression from the full-length LTR promoter in the context of transient reporter assays, they demonstrated variable activity when challenged with proviral DNA. Only sgRNAs 4 and 6, whose target sites overlap with the binding sites for NF-κB and the trans-activation response element, induced significant changes in expression in J-Lat 9.2, with the latter possibly through a mechanism that enhances transcriptional elongation. Despite the use of identical sgRNAs, different levels of Cas9-mediated activation were also observed between these cell lines (especially with sgRNA 4), suggesting that epigenetic and/or genetic85,86 factors can influence the ability of Cas9 to stimulate transcription. However, when combined with latency-reversing compounds, such as SAHA and prostratin, SAM-induced HIV-1 transcription at levels that exceeded those observed using TNF-α simulation, particularly in J-Lat 9.2 cells. Interestingly, we observed reduced transcription in J-Lat 10.6 cells expressing SAM with sgRNA 4 and treated with prostratin. Possible explanations for this include competition for LTR-binding sites between Cas9 and prostratin-activated NF-κB, or an unforeseen inhibitory effect between endogenous NF-κB protein and the transactivation domains present in the SAM complex.
The SAM system used here61 contains both the activation domain of the HSF1 protein and the NF-κB trans-activating subunit p65, which can stimulate transcriptional elongation during proviral activation.87 This notwithstanding, we observed no increase in HIV activation in J-Lat cells expressing dCas9-VP64, MS2-p65-HSF1, and an empty sgRNA cassette, indicating that HIV-1 expression was due to sgRNA targeting to the LTR. The modularity of the tripartite MS2–p65–HSF1 fusion protein could be exploited in the future to create a Cas9 activator complex specifically tailored for CD4+ T cells or other cell types relevant to HIV latency.
We also show that epigenome editing using a Cas9 acetyltransferase (dCas9-p300)82 can lead to a significant increase in GFP expression in J-Lat 10.6 cells. dCas9-p300-induced changes in transcription similar to SAM for multiple sgRNAs including sgRNA 2, which overlaps with a nucleosome (nuc-0) that contributes to proviral silencing. Because SAM and dCas9-p300 function through complementary mechanisms, emerging Cas9 orthologs88 could be co-opted and used in tandem for multiplexed induction of viral gene expression via epigenome editing and promoter-directed transcriptional activation. Future studies are necessary to determine the effectiveness of these technologies in primary cell models of HIV-1 latency, as well as the DNA-binding specificity of each Cas9 activator89 and the exact epigenetic modifications induced by dCas9-p300. In addition, successful clinical translation of this approach necessitates the safe and effective delivery of CRISPR transcriptional activators into cells that harbor latent HIV-1. Neither SAM nor dCas9-p300 (~7.5 kb and ~6.1 kb in length, respectively) can be packaged into a single adeno-associated virus vector. As an alternative, nonintegrated lentiviruses could be used to facilitate their delivery ex vivo, as these vectors have enabled zinc-finger nuclease-mediated modification of the CCR5 gene in patient-derived resting CD4+ T cells.90 In vivo delivery of Cas9-sgRNA could prove challenging, however. The use of dCas9-VP64 ribonucleoprotein91,92 conjugates engineered to specifically recognize,93 and subsequently be internalized by CD4+ T cells, or emerging lentiviral vectors capable of infecting human lymphocytes upon systemic delivery94 could help to overcome this limitation.
Finally, as with RNA interference-based gene therapies for HIV-1,95,96 CRISPR-mediated transcriptional activation of HIV-1 could be vulnerable to viral escape, as recent work has indicated that even the latent reservoir can become populated with escape mutants.97 However, due to the ease with which sgRNA can be designed, CRISPR-resistant strains of HIV-1 could be addressed through the use of new sgRNA tailored for specific escape mutants. These sgRNA could conceivably be introduced into cells already expressing a dCas9 variant via an aptamer-based bridge design similar to those previously described for combating HIV-1 resistance using RNAi.98,99 The use of a CRISPR-based cocktail consisting of multiple sgRNA from the onset of treatment could also be used to help block HIV-1 escape.
In summary, we show that CRISPR activation systems have the potential to induce HIV-1 expression in cell-based models of latency. CRISPR systems, in combination with cART, may lead to new treatments for HIV-1 infection.
Materials and Methods
Plasmid construction. pLV-dCas9-VP64-Blast (Addgene plasmid #61425), pLV-MS2-p65-HSF1-Hygro (Addgene plasmid #61426), and pLV-sgRNA(MS2)-Zeo (Addgene plasmid #61427)61 were gifts from Feng Zhang. pcDNA-dCas9-VP64 (Addgene plasmid #47107), pSP-sgRNA (Addgene plasmid #47108),54 and pcDNA-dCas9-p300 (Addgene plasmid #61357)82 were gifts from Charles A. Gersbach.
Reporter plasmid encoding the HIV-1 subtype B LTR upstream of the EGFP gene (LTR-EGFP) was previously described.13 Oligonucleotides encoding sgRNA target sites were custom ordered (Elim Biopharm, Hayward, CA), phosphorylated by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA), hybridized, and ligated into the BbsI and BsmBI restriction sites of pSP-gRNA and pLV-sgRNA(MS2)-Zeo, respectively. Correct construction of each sgRNA was verified by sequence analysis. Oligonucleotides used in this study are shown in Supplementary Figure S7.
Cell culture. HEK293T and Jurkat cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). J-Lat 9.2 and J-Lat 10.6 cells were obtained from the National Institutes of Health (NIH) AIDS Reagent Program (Germantown, MD). HEK293T cells were maintained in Iscove's modified Dulbecco's medium supplemented with 10% (vol/vol) fetal bovine serum (Life Technologies, Carlsbad, CA) and 1% (vol/vol) antibiotic-antimycotic (Anti-Anti; Life Technologies) in a humidified 5% CO2 atmosphere at 37 °C. Jurkat and J-Lat cells were maintained in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum and 1% Anti-Anti in a humidified 5% CO2 atmosphere at 37 °C.
Lentivirus production and infections. HEK293T cells were seeded onto 10-cm plates at a density of 2 × 106 cells per plate in serum-containing medium. After 24 hours of seeding, cells were transfected with 10 µg of pLV-dCas9-VP64-Blast, pLV-MS2-p65-HSF1-Hygro, or pLV-sgRNA(MS2)-Zeo, as well as 5 µg of pMDL g/p RRE, 3.5 µg of pMD2.G, and 1.5 µg of pRSV-Rev using polyethylenimine, as described.100 After 48 and 72 hours of transfection, cell culture medium was harvested, concentrated by ultracentrifugation (L8-55M Ultracentrifuge; Beckman Coulter, Pasadena, CA), and resuspended in 200 µl of phosphate-buffered saline with 20% sucrose. Lentivirus was stored in single-use aliquots at −80 °C.
For infections, J-Lat 9.2 and 10.6 cells were seeded onto 24-well plates at a density of 2 × 105 cells per well in serum-containing medium. Lentiviral infections were performed sequentially. After 24 hours of seeding, cells were infected with LV-dCas9-VP64-Blast, LV-MS2-p65-Hygro, or LV-sgRNA(MS2)-Zeo using a multiplicity of infection of 0.1. After 1 week, infected cells were selected with either 10 µg/ml of blasticidin (Sigma, St. Louis, MO) for LV-dCas9-VP64-Blast, 1 mg/ml of hygromycin (Sigma) for LV-MS2-p65-Hygro, or 100 µg/ml of zeocin (Sigma) for LV-sgRNA(MS2)-Zeo. Each selection was performed for 1 week. Seven days after the final selection, cells were washed once with phosphate-buffered saline, and GFP expression was evaluated by flow cytometry (BD LSR Fortessa X-20; BD Biosciences, San Jose, CA). For each sample, 10,000 live events were collected, and data were analyzed using FlowJo (Tree Star, Ashland, OR).
For small molecule treatments, 10 days after the final selection, J-Lat cells expressing the SAM complex were seeded into a 96-well plate at a density of 4 × 104 cells per well with 0.5, 1.0, 2.0, or 4.0 µmol/l SAHA (Sigma) in the presence or absence of 2.0 µmol/l prostratin (Sigma). After 24 hours of treatment, cells were harvested, and EGFP expression was measured by flow cytometry. For each sample, 10,000 live events were collected, and data were analyzed using FlowJo.
Transient transfections. HEK293T cells were seeded into 96-well plates at a density of 4 × 104 cells per well in serum-containing medium. For SAM, after 16–24 hours of seeding, cells were transfected with 20 ng of LTR-EGFP, 60 ng of pLV-dCas9-VP64-Blast, 60 ng of pLV-MS2-p65-HSF1-Hygro, and 60 ng of pLV-sgRNA(MS2)-Zeo using polyethylenimine. For dCas9-VP64 transfections, after16–24 h of seeding, cells were transfected with 20 ng of LTR-EGFP, 90 ng of pcDNA-dCas9-VP64, and 90 ng of pSP-sgRNA using polyethylenimine. Transfection efficiency was measured to be >90%.
For nucleofections, J-Lat cells were seeded into a T-25 cell culture flask at a density of 1 × 105 cells per ml. For SAM, after 24 hours of seeding, 2 × 105 cells per nucleofection were centrifuged at 90xg for 10 minutes at room temperature and resuspended in Nucleofector Solution SE (Lonza, Basel, Switzerland) with 650 ng of pLV-dCas9-VP64-Blast, 650 ng of pLV-MS2-p65-HSF1-Hygro, 650 ng of pLV-sgRNA(MS2)-Zeo, and 40 ng of pEntry-CMV-puro-mTagBFP, which was used as a transfection control. For dCas9-p300, 2 × 105 cells per nucleofection were resuspended in Nucleofector Solution SE with 1 μg of pcDNA-dCas9-p300 Core, 1 μg of pSP-sgRNA, and 40 ng of pEntry-CMV-puro-mTagBF. Cells were transferred to a 16-well Nucleocuvette Strip (Lonza) and electroporated using the 96-well Shuttle Device (Lonza) with the program CL-120, according to the manufacturer's instructions. Transfection efficiency was measured to be ~20%.
After 48 hours of nucleofection, cells were washed once with phosphate-buffered saline, and GFP expression was evaluated by flow cytometry. For each sample, 10,000 live events were collected, and data were analyzed using FlowJo.
Statistical analysis. Data represents the mean (data points) and SD (error bars) of three independent replicates. Statistical significance was calculated using a one-tailed independent two-sample Student's t-test (Microsoft Excel 2013, Microsoft, Redmond, WA).
SUPPLEMENTARY MATERIAL Figure S1. Long-terminal repeat (LTR) sequence from the HIV-1 subtype B strain HXB2 (NCBI accession number: K03455.1). Figure S2. “Tiling” multiple sgRNAs across the HIV-1 LTR promoter does not dramatically enhance the efficiency of dCas9-VP64-mediated gene activation. Figure S3. Tiling multiple sgRNAs across the HIV-1 LTR promoter does not significantly increase the efficiency of SAM-mediated gene activation. Figure S4. Co-transfection of SAM with a library of non-specific sgRNA does not lead to a significant increase in gene activation from the HIV-1 LTR. Figure S5. Tumor necrosis factor alpha (TNF-α) stimulation of J-Lat 9.2 and J-Lat 10.6 cells. Figure S6. Prostratin treatment of J-Lat cells expressing SAM is not as effective at stimulating viral gene expression as combination treatment with SAHA. Figure S7. Primer sequences used to construct the sgRNA used in this study.
Acknowledgments
We thank J.A. Doudna for use of the Nucleofector equipment. This work was supported by the National Institutes of Health (NIH) grant R01 GM073058. T.G. was supported by a Ruth L. Kirschstein National Research Service Award (F32GM113446).
The authors have no competing financial interests.
Supplementary Material
References
- Strain, MC, Little, SJ, Daar, ES, Havlir, DV, Gunthard, HF, Lam, RY et al. (2005). Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis 191: 1410–1418. [DOI] [PubMed] [Google Scholar]
- Archin, NM, Vaidya, NK, Kuruc, JD, Liberty, AL, Wiegand, A, Kearney, MF et al. (2012). Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci USA 109: 9523–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlama, C, Deeks, SG, Autran, B, Martinez-Picado, J, van Lunzen, J, Rouzioux, C et al. (2013). Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 381: 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, TW, Finzi, D, Margolick, J, Chadwick, K, Schwartz, D and Siliciano, RF (1995). In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Chun, TW, Engel, D, Berrey, MM, Shea, T, Corey, L and Fauci, AS (1998). Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA 95: 8869–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermankova, M, Siliciano, JD, Zhou, Y, Monie, D, Chadwick, K, Margolick, JB et al. (2003). Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J Virol 77: 7383–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi, D, Hermankova, M, Pierson, T, Carruth, LM, Buck, C, Chaisson, RE et al. (1997). Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278: 1295–1300. [DOI] [PubMed] [Google Scholar]
- Chun, TW, Stuyver, L, Mizell, SB, Ehler, LA, Mican, JA, Baseler, M et al. (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA 94: 13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, JD, Kajdas, J, Finzi, D, Quinn, TC, Chadwick, K, Margolick, JB et al. (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9: 727–728. [DOI] [PubMed] [Google Scholar]
- Battistini, A and Sgarbanti, M (2014). HIV-1 latency: an update of molecular mechanisms and therapeutic strategies. Viruses 6: 1715–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelas, DS and Greene, WC (2013). An integrated overview of HIV-1 latency. Cell 155: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin, NM, Sung, JM, Garrido, C, Soriano-Sarabia, N and Margolis, DM (2014). Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol 12: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger, LS, Burnett, JC, Toettcher, JE, Arkin, AP and Schaffer, DV (2005). Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122: 169–182. [DOI] [PubMed] [Google Scholar]
- Chun, TW, Engel, D, Mizell, SB, Hallahan, CW, Fischette, M, Park, S et al. (1999). Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med 5: 651–655. [DOI] [PubMed] [Google Scholar]
- Wang, FX, Xu, Y, Sullivan, J, Souder, E, Argyris, EG, Acheampong, EA et al. (2005). IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest 115: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Duffhues, G, Vo, MQ, Pérez, M, Calzado, MA, Moreno, S, Appendino, G et al. (2011). Activation of latent HIV-1 expression by protein kinase C agonists. A novel therapeutic approach to eradicate HIV-1 reservoirs. Curr Drug Targets 12: 348–356. [DOI] [PubMed] [Google Scholar]
- Kortmansky, J and Schwartz, GK (2003). Bryostatin-1: a novel PKC inhibitor in clinical development. Cancer Invest 21: 924–936. [DOI] [PubMed] [Google Scholar]
- Gulakowski, RJ, McMahon, JB, Buckheit, RW Jr, Gustafson, KR and Boyd, MR (1997). Antireplicative and anticytopathic activities of prostratin, a non-tumor-promoting phorbol ester, against human immunodeficiency virus (HIV). Antiviral Res 33: 87–97. [DOI] [PubMed] [Google Scholar]
- Kulkosky, J, Culnan, DM, Roman, J, Dornadula, G, Schnell, M, Boyd, MR et al. (2001). Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98: 3006–3015. [DOI] [PubMed] [Google Scholar]
- Beans, EJ, Fournogerakis, D, Gauntlett, C, Heumann, LV, Kramer, R, Marsden, MD et al. (2013). Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc Natl Acad Sci USA 110: 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChristopher, BA, Loy, BA, Marsden, MD, Schrier, AJ, Zack, JA and Wender, PA (2012). Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat Chem 4: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa, K, Chavez, L, Hakre, S, Calvanese, V and Verdin, E (2013). Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol 21: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint, C, Emiliani, S, Ott, M and Verdin, E (1996). Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 15: 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Dybul, M, Hidalgo, B, Chun, TW, Belson, M, Migueles, SA, Justement, JS et al. (2002). Pilot study of the effects of intermittent interleukin-2 on human immunodeficiency virus (HIV)-specific immune responses in patients treated during recently acquired HIV infection. J Infect Dis 185: 61–68. [DOI] [PubMed] [Google Scholar]
- Stellbrink, HJ, van Lunzen, J, Westby, M, O'Sullivan, E, Schneider, C, Adam, A et al. (2002). Effects of interleukin-2 plus highly active antiretroviral therapy on HIV-1 replication and proviral DNA (COSMIC trial). AIDS 16: 1479–1487. [DOI] [PubMed] [Google Scholar]
- Coiras, M, López-Huertas, MR, Pérez-Olmeda, M and Alcamí, J (2009). Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7: 798–812. [DOI] [PubMed] [Google Scholar]
- van Praag, RM, Prins, JM, Roos, MT, Schellekens, PT, Ten Berge, IJ, Yong, SL et al. (2001). OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion. J Clin Immunol 21: 218–226. [DOI] [PubMed] [Google Scholar]
- Archin, NM, Liberty, AL, Kashuba, AD, Choudhary, SK, Kuruc, JD, Crooks, AM et al. (2012). Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin, NM, Bateson, R, Tripathy, MK, Crooks, AM, Yang, KH, Dahl, NP et al. (2014). HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 210: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, KB, Staver, MJ, Waring, JF, Stender, J, Ulrich, RG and Davidsen, SK (2003). Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2: 151–163. [PubMed] [Google Scholar]
- Subramanian, S, Bates, SE, Wright, JJ, Espinoza-Delgado, I, and Piekarz, RL (2010). Clinical Toxicities of Histone Deacetylase Inhibitors. Pharmaceuticals 3: 2751–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, T, Gersbach, CA and Barbas, CF 3rd (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, D (2014). Genome engineering with targetable nucleases. Annu Rev Biochem 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Gersbach, CA, Gaj, T and Barbas, CF 3rd (2014). Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Acc Chem Res 47: 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, JK and Sander, JD (2013). TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, JA and Charpentier, E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Perez, EE, Wang, J, Miller, JC, Jouvenot, Y, Kim, KA, Liu, O et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, PK, Ferreira, LM, Collins, R, Meissner, TB, Boutwell, CL, Friesen, M et al. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L, Wang, J, Beyer, AI, Teque, F, Cradick, TJ, Qi, Z et al. (2014). Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA 111: 9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas, P, Stein, D, Tang, WW, Frank, I, Wang, SQ, Lee, G et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, HK, Gu, Y, Diaz, A, Marlett, J, Takahashi, Y, Li, M et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6: 6413. [DOI] [PubMed] [Google Scholar]
- Qu, X, Wang, P, Ding, D, Li, L, Wang, H, Ma, L et al. (2013). Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res 41: 7771–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina, H, Misawa, N, Kanemura, Y and Koyanagi, Y (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3: 2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W, Kaminski, R, Yang, F, Zhang, Y, Cosentino, L, Li, F et al. (2014). RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA 111: 11461–11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel, HJ, Morrison, JH, Saenz, DT, Fuchs, JR, Kvaratskhelia, M, Ekker, SC et al. (2014). TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol 88: 9704–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia, R, Pauls, E, Riveira-Munoz, E, Clotet, B, Esté, JA and Ballana, E (2014). Zinc finger endonuclease targeting PSIP1 inhibits HIV-1 integration. Antimicrob Agents Chemother 58: 4318–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, DJ, Gonçalves, J, Eberhardy, S, Swan, CH, Torbett, BE, Li, X et al. (2004). Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J Biol Chem 279: 14509–14519. [DOI] [PubMed] [Google Scholar]
- Eberhardy, SR, Goncalves, J, Coelho, S, Segal, DJ, Berkhout, B and Barbas, CF 3rd (2006). Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol 80: 2873–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA and Charpentier, E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, LS, Larson, MH, Gilbert, LA, Doudna, JA, Weissman, JS, Arkin, AP et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera, P, Kocak, DD, Vockley, CM, Adler, AF, Kabadi, AM, Polstein, LR et al. (2013). RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, ML, Linder, SJ, Cascio, VM, Fu, Y, Ho, QH and Joung, JK (2013). CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard, F, Perli, SD and Lu, TK (2013). Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Larson, MH, Morsut, L, Liu, Z, Brar, GA, Torres, SE et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Aach, J, Stranges, PB, Esvelt, KM, Moosburner, M, Kosuri, S et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Horlbeck, MA, Adamson, B, Villalta, JE, Chen, Y, Whitehead, EH et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S, Ji, H, Kabadi, AM, Gersbach, CA, Christoforou, N and Leong, KW (2014). A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports 3: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S, Brigham, MD, Trevino, AE, Joung, J, Abudayyeh, OO, Barcena, C et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan, JG, Lee, ME, Almeida, R, Gilbert, LA, Whitehead, EH, La Russa, M et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X, Scott, DA, Kriz, AJ, Chiu, AC, Hsu, PD, Dadon, DB et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, KA and Peterlin, BM (1994). Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem 63: 717–743. [DOI] [PubMed] [Google Scholar]
- Williams, SA, Chen, LF, Kwon, H, Ruiz-Jarabo, CM, Verdin, E and Greene, WC (2006). NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, SA, Kwon, H, Chen, LF and Greene, WC (2007). Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J Virol 81: 6043–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, YK, Bourgeois, CF, Pearson, R, Tyagi, M, West, MJ, Wong, J et al. (2006). Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 25: 3596–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic, M, Marcello, A, Cereseto, A and Giacca, M (2003). Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett, JC, Miller-Jensen, K, Shah, PS, Arkin, AP and Schaffer, DV (2009). Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog 5: e1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout, B, Silverman, RH and Jeang, KT (1989). Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59: 273–282. [DOI] [PubMed] [Google Scholar]
- Dingwall, C, Ernberg, I, Gait, MJ, Green, SM, Heaphy, S, Karn, J et al. (1989). Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA 86: 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli, RR, Segal, DJ, Dreier, B and Barbas, CF 3rd (1998). Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA 95: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K, Stuehler, T and Meisterernst, M (2002). The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 7: 49–58. [DOI] [PubMed] [Google Scholar]
- Neely, KE, Hassan, AH, Wallberg, AE, Steger, DJ, Cairns, BR, Wright, AP et al. (1999). Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4: 649–655. [DOI] [PubMed] [Google Scholar]
- Jordan, A, Bisgrove, D and Verdin, E (2003). HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Jensen, K, Dey, SS, Pham, N, Foley, JE, Arkin, AP and Schaffer, DV (2012). Chromatin accessibility at the HIV LTR promoter sets a threshold for NF-κB mediated viral gene expression. Integr Biol (Camb) 4: 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauss, F, Dröge, W and Männel, DN (1987). Tumor necrosis factor mediates endotoxic effects in mice. Infect Immun 55: 1622–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman, G, Hogue, IB, Palmer, S, Jennings, C, Spina, CA, Wiegand, A et al. (2005). Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366: 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, SA, Chen, LF, Kwon, H, Fenard, D, Bisgrove, D, Verdin, E et al. (2004). Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem 279: 42008–42017. [DOI] [PubMed] [Google Scholar]
- Verdin, E, Paras, P Jr and Van Lint, C (1993). Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keung, AJ, Joung, JK, Khalil, AS and Collins, JJ (2015). Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet 16: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, IB, D'Ippolito, AM, Vockley, CM, Thakore, PI, Crawford, GE, Reddy, TE et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P, Qu, X, Wang, X, Zhu, X, Zeng, H, Chen, H et al. (2014). Specific reactivation of latent HIV-1 with designer zinc-finger transcription factors targeting the HIV-1 5'-LTR promoter. Gene Ther 21: 490–495. [DOI] [PubMed] [Google Scholar]
- Wang, X, Wang, P, Fu, Z, Ji, H, Qu, X, Zeng, H et al. (2015). Designed transcription activator-like effector proteins efficiently induced the expression of latent HIV-1 in latently infected cells. AIDS Res Hum Retroviruses 31: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y, Lin, YB, An, W, Xu, J, Yang, HC, O'Connell, K et al. (2008). Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed, A, Laydon, DJ, Gillet, NA, Tanaka, Y, Taylor, GP and Bangham, CR (2013). Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog 9: e1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, MJ, Lowe, AD and Karn, J (2001). Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol 75: 8524–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, KM, Mali, P, Braff, JL, Moosburner, M, Yaung, SJ and Church, GM (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein, LR, Perez-Pinera, P, Kocak, DD, Vockley, CM, Bledsoe, P, Song, L et al. (2015). Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res 25: 1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G, Choi, JG, Bharaj, P, Abraham, S, Dang, Y, Kafri, T et al. (2014). CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids 3: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, S, Kwaku Dad, AB, Beloor, J, Gopalappa, R, Lee, SK and Kim, H (2014). Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, K, Lin, S, Boyer, E, Simeonov, DR, Subramaniam, M, Gate, RE et al. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA 112: 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P, Ban, HS, Kim, SS, Wu, H, Pearson, T, Greiner, DL et al. (2008). T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 134: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q, Uhlig, KM, Muth, A, Kimpel, J, Lévy, C, Münch, RC et al. (2015). Exclusive Transduction of human CD4+ T cells upon systemic delivery of CD4-targeted lentiviral vectors. J Immunol 195: 2493–2501. [DOI] [PubMed] [Google Scholar]
- Boden, D, Pusch, O, Lee, F, Tucker, L and Ramratnam, B (2003). Human immunodeficiency virus type 1 escape from RNA interference. J Virol 77: 11531–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, JN, Shah, PS, Burnett, JC and Schaffer, DV (2008). HIV evades RNA interference directed at TAR by an indirect compensatory mechanism. Cell Host Microbe 4: 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K, Pertea, M, Rongvaux, A, Wang, L, Durand, CM, Ghiaur, G et al. (2015). Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, CP, Zhou, J, Remling, L, Kuruvilla, J, Zhang, J, Li, H et al. (2011). An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med 3: 66ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J, Neff, CP, Swiderski, P, Li, H, Smith, DD, Aboellail, T et al. (2013). Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol Ther 21: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo, JR, Prieto, Y, Oramas, N and Sánchez, O (2009). Polyethylenimine-based transfection method as a simple and effective way to produce recombinant lentiviral vectors. Appl Biochem Biotechnol 157: 538–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.