Abstract
Emerging gene-editing technologies are nearing a revolutionary phase in genetic medicine: precisely modifying or repairing causal genetic defects. This may include any number of DNA sequence manipulations, such as knocking out a deleterious gene, introducing a particular mutation, or directly repairing a defective sequence by site-specific recombination. All of these edits can currently be achieved via programmable rare-cutting endonucleases to create targeted DNA breaks that can engage and exploit endogenous DNA repair pathways to impart site-specific genetic changes. Over the past decade, several distinct technologies for introducing site-specific DNA breaks have been developed, yet the different biological origins of these gene-editing technologies bring along inherent differences in parameters that impact clinical implementation. This review aims to provide an accessible overview of the various endonuclease-based gene-editing platforms, highlighting the strengths and weakness of each with respect to therapeutic applications.
A Brief History of Gene Editing
Site-specific gene modifications were first achieved in the yeast Saccharomyces cerevisiae in the late 1970's. In these experiments, it was discovered that introduction of DNA sharing homology with an endogenous locus (into a cell's nucleus) could induce recombination using the host DNA repair machinery resulting in transfer of information from an exogenous DNA sequence to the endogenous chromosomal target.1,2 This technique became known as “gene targeting”, and depending on the design of the targeting construct, a variety of precise manipulations could be attained. In S. cerevisiae, using selection methods for the transferred DNA this process results in close to 100% correctly targeted clones.2
When gene-targeting was subsequently attempted in mouse embryonic stem cells to create model organisms, a different picture emerged. Targeting efficiencies dropped several orders magnitude, and only after extensive development of embryonic stem cell culture methods, plus several clever selection strategies to screen out random integrations, could precisely modified organisms could be recovered.3 The drop in targeting efficiency relative to S. cerevisiae was attributed to the low basal rate of homologous recombination in mammalian cells, the DNA repair pathway responsible for recombining the exogenous sequences.4 While these technologies revolutionized mouse genetics, the low targeting efficiencies and requirement for selection prohibited the use of conventional homologous recombination-based gene targeting in human therapeutics.
In 1983, Jack Szostak and Rodney Rothstein proposed the model that DNA double-strand breaks could induce homologous recombination.5 Subsequently, several instances of nature invoking this property were documented, such as yeast mating type switching using the HO endonuclease,6 and the super-Mendelian inheritance of the I-SceI homing endonuclease.7 The discovery of I-SceI homing endonuclease was particularly notable, as it was later determined to recognize and generate a double-strand break at an 18 bp DNA target sequence with little tolerance for mismatches across the target site.8 The length of the I-SceI target site implied it would rarely be found naturally (assuming a one in four chance of each DNA base occurring at each position of an 18 bp sequence suggests it will be found once in every 6.8E10 bp), and therefore was the first time a reagent was available that presumably had a sufficient level of specificity to introduce a single DNA break into a human-scale genome (~3.2E9 bp).
In a seminal experiment by the Jasin lab in 1994, a site-specific double-strand break induced by expression of I-SceI was shown to promote gene targeting with an exogenously provided donor template in mammalian cells through homology-directed repair pathways (HDR).9 In these experiments, gene targeting rates increased two to three orders of magnitude using the I-SceI homing endonuclease. Furthermore, insertion and deletion (“indel”) mutations attributed to an alternative DNA repair pathway called nonhomologous end joining (NHEJ) were also observed at the I-SceI break-point. These results formed the tenants of the current gene-editing paradigm, namely that a targeted DNA break could engage a variety of cell-intrinsic DNA repair mechanisms, each of which can be harnessed to achieve precise, yet diverse edits into large mammalian genomes.
Development of The Four Major Gene-Editing Platforms
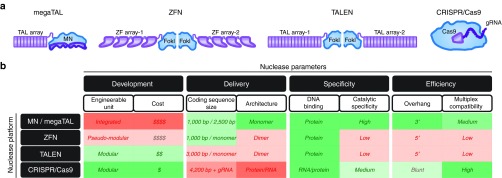
The demonstration that a site-specific double-strand break could be harnessed to achieve gene disruption and gene targeting in otherwise refractory cells inspired a concerted effort by several academic and industrial groups to develop programmable DNA recognition and cleavage technologies. Over the last 15 years, this effort has yielded several designer endonuclease platforms, including meganucleases,4,10 Zinc Finger Nucleases,11,12 TALENs,13,14 and CRISPR15 (Figure 1a, listed in chronological order of their demonstrated potential for gene editing). As each platform has a unique evolutionary origin and mechanism of DNA recognition, they are inherently endowed with distinct advantages and disadvantages for gene editing; such as relative ease of engineering toward a given DNA sequence, compatibility with delivery methods, target specificity, and editing efficiency to name a few (Figure 1b). Below is a brief synopsis of each system's attributes, with a particular emphasis on practical considerations when selecting a platform for a given application.
Figure 1.
Comparison of the four major gene-editing platforms. (a) Schematics of each endonuclease platform. Purple shaded regions represent areas mediating DNA binding; blue shaded areas represent areas mediating DNA catalysis. For megaTALs, the meganuclease (MN) domain represents a MN. (b) Table depicting relative comparison between each platform for various salient designer endonuclease parameters.
Meganucleases and megaTALs
Meganucleases (also referred to as homing endonucleases) were the first class of enzymes known to be capable of recognizing and cleaving a sufficiently long DNA sequence to allow for potential genome level specificity in human cells.4 They are naturally occurring selfish genetic elements present in a wide variety of microbes that propagate themselves by creating a double-strand break at a specific target site in a genome.16 The DNA break induces the host's homologous recombination machinery to copy the meganuclease (MN) coding sequence over to the recipient allele. This process is known as known as “homing” as nearly all progeny from MN+ and MN- microbes become MN+ upon mating.16
There are several distinct families of meganucleases, but the LAGLIDADG enzymes (named for the consensus amino acid sequences at the interface between the two halves of the protein) are the most tractable for genome editing. These enzymes function as either homodimers or monomeric pseudodimers, the latter of which can recognize fully asymmetric 18–22 bp target sites.10 Meganucleases are unique among the gene-editing platforms in that their DNA binding and cleavage activities are integrated. The DNA target can be viewed as two distinct half-sites that are directly contacted through antiparallel β-sheets belonging to each domain of the pseudodimer.10 In addition to direct readout of the DNA target, meganucleases also exhibit indirect specificity at the so-called “central 4” region of the target.17 In this region no direct contacts to the DNA bases are made, however there are strict sequence requirements to allow the correct DNA torsion that culminates in the hydrolysis of the DNA backbone.
While the integrated nature of meganuclease DNA recognition and cleavage makes them potentially highly specific, it also makes them complex to reprogram for the recognition of novel DNA targets. The β-sheets that make contact with the DNA on each half site of the target can be mutated to accommodate new specificities, but the enzymes are sufficiently compact that changing one contact at a given DNA base may have unanticipated impacts on specificity at neighboring bases,18,19 a phenomenon known as “context-dependence”. Furthermore, in addition to identifying DNA binding domains with the desired new recognition sequence, one must do so while maintaining the endonuclease activity, which is highly dependent on the topology of the whole target site when bound by the enzyme.19 However, there is a vast catalog of meganucleases with differing cognate target sequences that have been identified bioinformatically and subsequently validated experimentally,20,21 giving a variety of different starting points from which to engineer. Importantly, several protein engineering strategies, including yeast surface display and bacterial expression platforms, have been developed that address the specific challenges of reprogramming meganucleases. They each rely on a readout of endonuclease function (i.e., DNA binding and cleavage) as well as a modular selection approach based on overlapping modules of DNA targets, followed by reconstruction and optimization of the fully assembled enzyme.22,23 Custom meganucleases are not currently available through a commercial source, and therefore require a significant undertaking to obtain a reprogrammed enzyme. MegaTALs are a hybrid gene-editing platform comprised of a TAL effector DNA binding domain fused to a meganuclease (see below).
Zinc finger nucleases
Zinc fingers are prototypical DNA binding domains found in many transcription factors in eukaryotic cells. Each finger is comprised of a ~30 amino acid ββα structure, which typically makes direct DNA contacts with 3 bps of DNA.24 Individual fingers can be tethered into an array with various linker domains to generate proteins that recognize increasingly long target sites.25 The modular nature of DNA recognition made them attractive for protein engineering, and several academic and industrial groups engineered libraries of individual zinc fingers with the goal of stringing several fingers together to generate an array long enough to recognize a unique target site. While this approach was somewhat successful, context-dependence also plays an important role in zinc finger protein (ZFP) DNA recognition and it was determined that some zinc fingers did not retain their expected triplet specificity once linked into a new zinc finger array.26,27 This necessitated building more complex library sets and selections to uncover solutions for 2-finger modules of zinc fingers.28
Zinc fingers do not possess endonuclease activity in and of themselves, and therefore must be endowed with an endonuclease domain to be employed as a DNA break-inducing gene-editing reagent. This is typically accomplished by linking a Zinc Finger array to the catalytic domain of the Type IIS restriction enzyme FokI to create zinc finger nucleases (ZFNs).29 As FokI requires dimerization to create a double strand break,30 a pair of zinc finger arrays with each being C terminally linked to FokI (one targeting the forward strand and one targeting the reverse strand) with a 5–7 bp spacer sequence between each zinc finger binding site is delivered in order to induce a DNA break with 4–5 bp, 5' overhangs.11 While there are several sources of ZFN DNA binding solutions from academic labs, success using these reagents to generate new enzymes is variable. Validated ZFNs generated from industrial libraries can currently be procured via Sigma-Aldrich.
TAL effector nucleases
Transcription-activator-like (TAL) effectors are a class of naturally occurring transcription factors that were originally identified as virulence factors in the bacterial plant pathogen Xanthomonas. They are expressed by plasmids and secreted from the bacterium via a Type III secretion system where they can subsequently be taken up by plant cells.31 The TALs then traffic to the nucleus by way of a nuclear localization signal where they alter the expression of host genes (hence the name “transcription activator-like” or TAL), some of which can lead to resistance and some of which cause virulence.31 DNA recognition by TAL effectors is mediated by a series of a 34 amino acid repeats that differ only at the 12th and 13th amino acids, known as the repeat variable di-residues (RVDs).32 A breakthrough occurred in 2009 when the DNA “binding code” for TAL effectors was determined to be governed by each RVD corresponding to a single DNA base (HD=C, NG or HG =T, NI=A, NN=G/A, NS=A/C/G, N*=C/T(*= a 33 amino acid repeat with a single N in the loop region)).32,33 This immediately suggested the potential for a highly modular DNA recognition domain, and it was quickly demonstrated that TAL effectors could be reprogrammed to new specificities simply by rearranging the RVDs.34,35,36 Each repeat forms a two-helix bundle that positions the RVD within a loop between the two coils.37,38 The first amino acid in the RVD typically makes a nonspecific DNA backbone contact, while the second makes a specific contact in the major groove of the DNA.37,38 Several groups have truncated the N and C termini of the parental TAL scaffold to generate backbone “architectures” with smaller sizes and more consistent performance.35,36 TAL effectors, like zinc fingers, lack endonuclease activity and therefore are also typically used as a pair fused to the FokI cleavage domain to enable them to create DNA breaks. These reagents are referred to as TAL effector-like nucleases (TALENs).
The simple DNA-binding code of TALENs made them highly attractive gene-editing reagents, as it eliminated the need for de novo protein engineering for each new DNA target. However, the repetitive nature of the scaffold creates a technical challenge for the quick assembly of novel RVD configurations. Several methods for constructing TALENs have been developed with varying degrees of throughput,39,40,41,42 but the process can still be a bit onerous for labs without experience. TALENs can be procured commercially through Thermo-Fisher.
CRISPR/Cas9
The CRISPR/Cas9 system is a gene-editing platform derived from a recently elucidated adaptive immunity strategy employed by a diverse array of bacteria and archaea.43,44,45 While the DNA target recognition of the other nuclease platforms is governed by protein-DNA binding, CRISPR/Cas9 target recognition is primarily mediated via Watson-Crick base pairing. In nature, the CRISPR system works by a bacterium integrating foreign DNA sequences obtained from bacteriophages (known as “spacers”) into their genome in an expanding array known as a clustered regularly interspaced short palindromic repeats (CRISPR).46 The bacteria then express and process these spacers into short RNA molecules (crRNA), which together with an additional short RNA known as a tracrRNA form a complex with the CRISPR-associated endonuclease (Cas9) to target the enzyme to a 20 bp complementary DNA sequence.15 DNA breaks are induced at complementary target sites that contain the proper protospacer adjacent motif (PAMs) encoded by the base pairs immediately adjacent to the guide RNA (NGG for S. pyogenes Cas9), which are present only at the intended target site to prevent the host from self-cleavage.47,48 As Cas9 possesses inherent endonuclease activity, it does not require any additional domains to create a DNA break. The CRISPR/Cas9 system from S. pyogenes was the first to be adapted for gene editing when it was demonstrated in 2013 that expressing two RNA guides as they occur in bacteria or as a single chimeric fusion (sgRNA) with the Cas9 protein resulted in targeted DNA breaks in human cells.49,50,51,52 The facile reprogramming of CRISPR/Cas9 toward novel targets has resulted in a rapid adoption of this technology and enabled a variety of applications previously unachievable with prior gene-editing platforms. CRISPR tools and reagents including purified protein, expression vectors, and modified guide RNAs are available from many academic and commercial sources.
Delivery of Gene-Editing Technologies
Once an endonuclease is designed to cut a given target sequence, it must be delivered to the therapeutically relevant cell. While there are few limitations on delivery to cultured cell lines, delivery to primary cells ex vivo (such as hematopoietic stem cells and T-cells) and in vivo delivery (such as to the liver) have many of the same limitations as other classical “gene therapy” approaches, namely constraints on the immunogenicity and packaging capacity of the delivery modality. In addition, for many site-specific genome-editing applications it is desirable to have the endonuclease and donor template delivered to the cell transiently, such that a break can be made, the engineering can occur, and the enzyme and donor templates can dissipate from the cell. Several strategies have now been developed to accomplish this “hit and run” approach both ex vivo and in vivo, including nonviral gene transfer methods such as lipid nanoparticles, electroporation of in vitro transcribed mRNA, and protein transduction, as well as viral vector methods such as integrase-deficient lentivirus, adenovirus, baculovirus, and adeno-associated virus (AAV).53,54,55,56 HDR-based editing requires delivery of DNA donor templates which ultimately have to find their way to the nucleus. As plasmid DNA is typically toxic in primary cells in part through innate immune sensing,57 Adenovirus, Baculovirus, AAV, integrase-deficient lentivirus, and short oligonucleotides have become preferred strategies to provide a transient source of donor DNA.55,58,59 Each of these delivery modalities provides certain advantages and disadvantages such as packaging capacity, gene transfer rate in a particular cell type, safety, copy number, and toxicity, and thus must be chosen based on the downstream application and nuclease platform.
As mentioned above, meganucleases have evolved to be extremely compact to minimize the burden on their host, and thus are encoded by small ~1 kb ORFs.10 MegaTALs typically have an 8–12 repeat TAL array appended to the N terminus, making them ~2.5 kb per monomer.60 The small size and monomeric structure of meganuclease-based platforms makes them attractive from a delivery standpoint, and affords them the ability to be packaged into a variety of gene transfer vectors, including into a single AAV (which has a packaging capacity of ~4.4 kb before adversely affecting viral quality/titers61).
ZFNs have been successfully delivered using a variety of viral and non-viral methods, and been used in clinical trials since 2009 (ref. 62). In initial clinical trials targeting CCR5 for HIV therapy, ZFNs were delivered to T cells via adenoviral vectors,62 however current clinical protocols (NCT02388594 and NCT02500849) are now using electroporation of mRNA. As ZFNs are heterodimers, they require delivery of two distinct coding sequences or as bicistronic constructs. Depending on the number of fingers in the ZFN, they are approximately ~1 kb/monomer (~2 kb/pair), making it feasible to fit a bicistronic ZFN coding sequencing and promoter in a single AAV.
TALENs are the largest of the gene-editing platforms at ~3 kb/monomer (~6 kb/pair) depending on the number of repeat domains used (typically 15–20). By their nature, they are highly repetitive sequences making delivery with commonly used retroviral vectors (gamma or lenti) difficult. However, this limitation can be overcome by codon diverging the RVDs.63 Due to complexity and size constraints, thus far adenoviral or mRNA-based gene transfer methods have been most widely used.14 TALENS entered the clinic for the first time in late 2015 when they were used to engineer a “universal” allogenic chimeric antigen receptor T-cell product that was administered to a single patient.64
The CRISPR/Cas9 system requires the co-delivery of the Cas9 nuclease and the guide RNA. While Cas9 from S. pyogenes is a relatively large protein (~4.2 kb coding sequence), it is adaptable to gene transfer using adenovirus, integrase-deficient lentivirus, mRNA or protein delivery.65 The ~100 bp sgRNA can be synthesized (enzymatically or synthetically) and codelivered separately or expressed in the cell via Pol II or Pol III promoters.65 While the originally adapted CRISPR/Cas9 from S. pyogenes is too large to fit into a single AAV with a guide RNA and promoter, Cas9 orthologs have been discovered which are smaller in size, such as the 3.2 kb Cas9 from S. Aureus.66
Specificity of Gene-Editing Technologies
Specificity of genome-editing reagents are paramount in therapeutics, as off-target mutations could lead to unintended side-effects. The inherent specificity of a given enzyme (independent of the target choice and its relative abundance of near-cognate matches in the genome) is dictated by both the DNA-binding specificity of enzyme and the catalytic mechanism employed to introduce the DNA break. While directly comparing specificity among the platforms can be challenging as it is difficult to target different enzymes to the exact same DNA sequence with similar efficiencies/delivery methods, some inferences can be drawn based on each platform's biochemistry and empirical performance independently.
Target DNA recognition is governed by base-specific contacts made at the interface of the enzyme and DNA. For MNs, ZFNs, and TALENs, these contacts are made by amino acid side chains (and some water mediated contacts for MNs67), whereas CRISPR DNA recognition is accomplished via RNA to DNA base pairing in the guide sequence as well as base-specific amino acid contacts in the PAM region.68,69 Meganucleases exhibit a high density of protein-DNA base contacts, as they can make upwards of ~52 base-specific amino acid side chain contacts over a 22 bp target.67 This compares to ~32 base-specific contacts made by a dimerized 2 × 4 finger ZFN at a 24 bp target,25 and ~30 base-specific contacts made by a dimerized 2 × 15 RVD containing TALEN at a 30 bp target.37,38 MN and TALEN binding can be effected by 5-methyl cytosine (5mC), making CpG motifs an important consideration when choosing a target with these platforms. However, both platforms can be engineered to accommodate the 5mC.70,71 S. pyogenes Cas9 makes four base-specific amino-acid contacts at the 3 bp PAM region while the remainder of the target specificity is provided via 20 Watson-crick base pairs, an inherently different mechanism of DNA recognition than that mediated by amino acid side chains.
Meganucleases are unique among gene-editing platforms in that the DNA catalysis active site is directly integrated into the DNA binding interface.67 As such, MNs do not need to be appended to a separate DNA cleavage domain to be used for targeted DNA-break based genome editing, unlike ZFN and TALEN platforms. MNs have extremely precise requirements for the correct DNA binding-induced topology at the active site in order to introduce a break (a mechanism of indirect DNA specificity), and therefore it is virtually impossible for it to cleave DNA without first binding a target site.67 The megaTAL variant of the MN take advantage of these monomeric properties by adding on a TAL array to further drive affinity at specific targets, while using the catalytic discrimination of the MN to act as a failsafe at off-target sites.60
ZFNs and TALENs primarily rely on the nonspecific FokI endonuclease domain to create DNA breaks.55 While both ZFNs and TALENs are designed such that two distinct DNA binding domains in the correct orientation must be present in order for the FokI enzyme to dimerize and become active, homo-dimers can also form which may enable cleavage at off-target sites. This possibility has been minimized by the development of engineered variants of FokI which have had the dimer interface mutated to create exclusive pairs. These “obligate heterodimer” variants exhibit increased activity and specificity as a result of reduced homo-dimer formation.72,73,74,75
Cas9 is a multi-domain, “bi-lobed” enzyme which contains a target recognition lobe and an endonuclease lobe comprised of the HNH and RuvC domains which cleave the complementary and noncomplementary DNA sequences respectively.69 The specificity of Cas9 is implemented primarily at two levels, the PAM and guide RNA. First, the PAM DNA duplex is contacted via amino acids in the Cas9 PAM interacting domain, including base-specific contacts that readout the PAM sequence.68 This recognition induces local unwinding of the target duplex and allows for R loop formation with the “seed” sequence at the PAM proximal part of the guide RNA (provided the guide seed and target are complementary).68 The R loop is then further extended toward the 5' PAM distal end of the 20 bp guide. While guide RNA mismatches at the 5' end still allow for Cas9 DNA binding, it has been demonstrated that more stringent complementarity is required to induce the allosteric conformational change in the endonuclease lobe required for cleavage,76 suggesting some level of catalytic specificity in addition to DNA binding. There were several early reports that the off-target rate of the CRISPR/Cas9 system could be significant,77,78,79,80 raising concerns about the clinical application of this technology. Subsequent improvements of the CRISPR/Cas9 system, including truncated guides,81 better target-site optimization algorithms,82 dimeric FokI-based versions,83,84 and protein engineering85 suggest that specificity can be improved. Furthermore, recent whole-genome sequencing of individual edited clones86,87 and genetically engineered mouse models88 suggest that CRISPR can be quite specific when using the right guide RNAs and delivery conditions.
An additional strategy to improve the specificity of endonuclease platforms has been the use of single-strand DNA breaks as opposed to double-strand breaks. Single-strand breaks, or “nicks”, are typically seamlessly religated, but can also be repaired through HDR-like pathways. Each of the platforms are able to accommodate specific mutations within their catalytic sites to render them “nickases” as opposed the “cleavases”. Nickases have been shown to be favorably repaired by the HDR pathway as opposed to NHEJ-based pathways89,90,91 (through mechanisms which are still being defined92), and therefore may be particularly useful for gene repair approaches and limiting genotoxicity at off-target sites. “Paired nickases” (two nickases that have target sites in close proximity) are now being explored as an approach to reduce off-target mutagenesis by effectively only introducing a double strand break where two nicks are made next to each other at the same time.81,93
While inherent attributes of the platforms such as number of amino acid contacts and catalytic mechanisms may provide some a priori hypothesis about relative specificity among them, it is most important to understand the actual specificity of a given reagent entering the clinic. Traditionally, endonuclease specificity had been inferred via in vitro profiling and subsequently rational/in silico predictions based on the empirical data were used to identify putative off-target sites.94 These potential off-targets could then be evaluated individually to determine whether or not they were bona fide (within the limit of detection of available assays). Of course, this approach is flawed as it relies on assumptions made about the enzyme in vitro and our ability to use that information to infer where the enzyme might cut. To address this short-coming, several new tools have been developed to perform “unbiased” off-target identification and detect larger genomic rearrangements such as translocations.95,96,97,98,99 These types of analyses may become particularly important as the field moves towards multiplexing where multiple breaks are made simultaneously.
An important consideration for the field moving forward is what to do with this information. While uncovering an off-target in a known tumor-suppressor gene would be grounds for refining the enzyme or choosing a new one, the function of many genes in a given cell type will be unknown, and evaluating cumulative effect in combination would be particularly complex. While work is ongoing to establish functional assays for off-target effects, determining the detection limit and proper controls for these assays will be essential.94 Therefore, although the genome-wide specificity of a reagent is important to understand relative risk, the preclinical safety of the edited cell product may also need to be assessed by more classical in vitro and in vivo safety/toxicology assays and ultimately in clinical trials. Acceptable off-target rates/profiles will be influenced by the target cell and disease that is being treated by gene editing. For example, the acceptable off-target risk for editing in terminally differentiated cells (such as a mature T cell) for an adult with metastatic refractory cancer will be different than for editing a stem cell in a patient with a non–life-threating genetic disease. In this sense, preclinical off-target rates of a few percent (at a limited set of defined sites)100 was considered acceptable to the FDA in combination with more traditional toxicology studies for a phase 1 clinical trial using ZFNs to target the CCR5 locus in autologous lymphocytes for HIV disease.
Efficiency of Gene Editing
A number of factors contribute to the absolute efficiency of a given gene-editing procedure, but most paramount is the quality of the nuclease. Below we discuss considerations and observations for the overall editing rate, the number of edits that can be made simultaneously, and how editing outcome can potentially be influenced by the unique biochemistry of the different platforms. In addition to the quality and attributes of the nuclease, efficiency of editing can depend on several platform independent variables including the cell type,4 cell cycle,101 epigenetics at the target site,70,102 and delivery kinetics.
Owing to reasons not yet fully understood, the overall editing rate each of the different platforms is capable of achieving is still dependent on empirically testing a number of different enzymes for a given target. Within a platform, there is a wide variation in editing efficiency nuclease to nuclease, and therefore one platform may be able to achieve a higher editing rate at one target, while another may be better for a different target. As a result of these idiosyncrasies, the individual researcher is best advised to build several enzymes/guides within their chosen platform and test in parallel to uncover an active nuclease.
MegaTALs, ZFNs, TALENs, and CRISPR/Cas9 have each been able to achieve efficient gene editing with optimal reagents in cell culture lines where high-level delivery and toxicity are not a concern. In primary human T cells, editing rates have been reported >70% for megaTALs,60 ZFNs,103 and TALENs104 when electroporating mRNA. While coelectroporation of mRNA for both Cas9 and guide RNAs has not produced significant editing in T cells,105 direct delivery of precomplexed Cas9 ribonucleoproteins (RNPs)106,107,108 has shown ~30% editing in combination with HDR-driven oligo knockins,109 and recent efforts utilizing chemical modifications to protect the guide RNA ends have resulted in very high editing rates (~80%) at some primary T-cell loci.110 Additional approaches to improve the level and persistence of guide RNAs in hematopoietic cells, such as expression via AAV, is ongoing. Both NHEJ- and HDR-based gene editing has been demonstrated in human hematopoietic stem cells with ZFNs111,112,113 and gene knockout has recently been demonstrated in CRISPR/Cas9.105 However, editing rates pre- and postengraftment into immunocompromised mice have been variable, suggesting that further optimization in human long-term hematopoietic stem cells may be required.
While ex vivo editing in hematopoietic cells is currently in the clinic, several groups have recently reported the feasibility of in vivo editing for ZFNs and CRISPR in mice.114,115,116,117 Currently, the most tractable in vivo clinical approaches would be delivery of nucleases and/or donor templates to the liver using either AAV or mRNA-shuttling lipid nanoparticles. This paradigm may provide a means to treat liver-based diseases or enable a platform for stable production of protein replacement therapies, the latter of which is currently advancing to the clinic.118
For certain gene therapy applications, it may be desirable to target several genes for modification. MegaTALs and similar hybrid enzymes119 are the only monomeric gene-editing system, which readily lends itself to multiplex delivery of potentially several nucleases in a single gene transfer procedure. One drawback to using FokI-based dimerization for the catalytic activity of ZFNs and TALENs is the requirement for two proteins per target site. This could limit the ability to multiplex several edits simultaneously, as expressing multiple monomers in parallel can result in mis-pairing between them and thus potential new off-target sites. One strategy to address this is the use of different pairs of obligate heterodimer FokI domains.72,73,74,75 Using these engineered variants, two pairs of ZFNs or TALENs can be expressed at the same time without mis-pairing. As CRISPR/Cas9 possesses inherent endonuclease activity and facile reprogramming via RNA guides, it is well suited for multiplexing by simply providing additional guides. Additionally, Cas9 orthologs and engineered variants are currently being developed to accommodate different PAM sequences to enable distinct engineering operations at different loci.120,121
Several groups have also attempted to increase the efficiency of gene editing via additional factors that perturb and bias DNA repair pathways. For example, It has also been demonstrated that a significant number of DNA breaks made by 3' overhang meganucleases can be precisely repaired by the nonhomologous end-joining pathway.122,123 These breaks can be forced to a mutagenic resolution by coexpressing the 3' exonuclease Trex2, resulting in very high gene disruption rates.122,123,124 While this exonuclease and other DNA repair proteins have been shown to increase the gene disruption rates of other gene-editing platforms,123,125,126 it tends to be to a lesser extent. Additional efforts to influence break-processing for gene editing are underway and include siRNAs, small molecule inhibitors, and expressing viral proteins to skew the DNA repair environment towards the desired editing outcome.127,128,129,130
Lastly, one potential important distinction between gene-editing platforms with respect to efficiency is the biology at the break point. For example, the polarity of DNA overhangs (megaTALs leave a 3' overhang break,17 ZFNs and TALENs leave 5' overhangs14 and S. pyogenes CRISPR/Cas9 leaves blunt ends15) could lead to recruitment of different DNA repair proteins and thus editing outcome.131 The biological impact of different break overhangs on gene editing has yet to be fully elucidated, though some observations on repair pathway choice and translocation formation have been documented.102,126,132 Likewise, Cas9 has been shown to exhibit an extremely long half-life at its target sites in vitro,133 which presents the possibility that the endonuclease itself could alter the detection or processing of the break-site by the host DNA-repair machinery.127 The continued mining of putative natural RNA-guided endonucleases will likely yield unique architectures with distinct biophysical and biochemical properties (for example the recently discovered Cpf1 CRISPR family which generates breaks with 4 or 5 bp 5' overhangs134), that may begin to shed more light on the impact of break biology on gene-editing outcomes.
Conclusions
Much progress has been made in our ability to precisely modify genomes, and these efforts are already being explored clinically. Access to reagents to induce targeted DNA edits has been greatly expanded and they are rapidly becoming common laboratory methods. From the standpoint of the researcher, the CRISPR/Cas9 and TALEN systems have proven to be accessible and robust enough for routine use. In the clinical setting where gene-editing efficiency and specificity will be most important, ZFNs are leading the way with TALENs near to initial clinical applications and meganuclease and CRISPR/Cas 9 systems close behind. It is likely that all four systems will see specific clinical applications and in the long-term, it is possible that multiple commercial products will be developed to treat patients with a variety of genetic and acquired diseases. The initial paradigm that gene therapy would one day advance to repair and replace defects using gene surgery may be an attainable goal.
Acknowledgments
The authors would like to thank Jordan Jarjour for providing the figure and Jordan Jarjour and Phillip Gregory for critical reading of this manuscript. The authors are employees of and own stock in bluebird bio, which is actively involved in commercial development of gene therapy technology including gene editing using meganuclease/MegaTALs.
References
- Hinnen, A, Hicks, JB and Fink, GR (1978). Transformation of yeast. Proc Natl Acad Sci USA 75: 1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, TL, Szostak, JW and Rothstein, RJ (1981). Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA 78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell, DA and Kolb, AF (2005). Targeted modification of mammalian genomes. Biotechnol Adv 23: 431–469. [DOI] [PubMed] [Google Scholar]
- Pâques, F and Duchateau, P (2007). Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther 7: 49–66. [DOI] [PubMed] [Google Scholar]
- Szostak, JW, Orr-Weaver, TL, Rothstein, RJ and Stahl, FW (1983). The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Kostriken, R, Strathern, JN, Klar, AJ, Hicks, JB and Heffron, F (1983). A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 35: 167–174. [DOI] [PubMed] [Google Scholar]
- Jacquier, A and Dujon, B (1985). An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41: 383–394. [DOI] [PubMed] [Google Scholar]
- Colleaux, L, D'Auriol, L, Galibert, F and Dujon, B (1988). Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA 85: 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet, P, Smih, F and Jasin, M (1994). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard, BL (2011). Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus, MH and Carroll, D (2005). Gene targeting using zinc finger nucleases. Nat Biotechnol 23: 967–973. [DOI] [PubMed] [Google Scholar]
- Urnov, FD, Rebar, EJ, Holmes, MC, Zhang, HS and Gregory, PD (2010). Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Bogdanove, AJ and Voytas, DF (2011). TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846. [DOI] [PubMed] [Google Scholar]
- Scharenberg, AM, Duchateau, P and Smith, J (2013). Genome engineering with TAL-effector nucleases and alternative modular nuclease technologies. Curr Gene Ther 13: 291–303. [DOI] [PubMed] [Google Scholar]
- Doudna, JA and Charpentier, E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Jurica, MS and Stoddard, BL (1999). Homing endonucleases: structure, function and evolution. Cell Mol Life Sci 55: 1304–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard, BL (2014). Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grizot, S, Duclert, A, Thomas, S, Duchateau, P and Pâques, F (2011). Context dependence between subdomains in the DNA binding interface of the I-CreI homing endonuclease. Nucleic Acids Res 39: 6124–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme, SB, Boissel, SJ, Arshiya Quadri, S, Nolan, T, Baker, DA, Park, RU et al. (2014). Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res 42: 2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, R, Lambert, AR, Mak, AN, Jacoby, K, Dickson, RJ, Gloor, GB et al. (2011). Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci USA 108: 13077–13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, K, Metzger, M, Shen, BW, Certo, MT, Jarjour, J, Stoddard, BL et al. (2012). Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space. Nucleic Acids Res 40: 4954–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour, J, West-Foyle, H, Certo, MT, Hubert, CG, Doyle, L, Getz, MM et al. (2009). High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res 37: 6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, R, Choi, M and Stoddard, BL (2014). Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc Natl Acad Sci USA 111: 4061–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli, RR and Barbas, CF 3rd (2002). Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol 20: 135–141. [DOI] [PubMed] [Google Scholar]
- Klug, A (2010). The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem 79: 213–231. [DOI] [PubMed] [Google Scholar]
- Isalan, M, Choo, Y and Klug, A (1997). Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci USA 94: 5617–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan, M, Klug, A and Choo, Y (1998). Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry 37: 12026–12033. [DOI] [PubMed] [Google Scholar]
- Isalan, M, Klug, A and Choo, Y (2001). A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol 19: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, YG, Cha, J and Chandrasegaran, S (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee, ES, Santagata, S and Aggarwal, AK (2001). FokI requires two specific DNA sites for cleavage. J Mol Biol 309: 69–78. [DOI] [PubMed] [Google Scholar]
- Boch, J and Bonas, U (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48: 419–436. [DOI] [PubMed] [Google Scholar]
- Boch, J, Scholze, H, Schornack, S, Landgraf, A, Hahn, S, Kay, S et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Moscou, MJ and Bogdanove, AJ (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Christian, M, Cermak, T, Doyle, EL, Schmidt, C, Zhang, F, Hummel, A et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino, C, Morbitzer, R, Lütge, F, Dannemann, N, Lahaye, T and Cathomen, T (2011). A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, JC, Tan, S, Qiao, G, Barlow, KA, Wang, J, Xia, DF et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Mak, AN, Bradley, P, Cernadas, RA, Bogdanove, AJ and Stoddard, BL (2012). The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, D, Yan, C, Pan, X, Mahfouz, M, Wang, J, Zhu, JK et al. (2012). Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak, T, Doyle, EL, Christian, M, Wang, L, Zhang, Y, Schmidt, C et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon, D, Tsai, SQ, Khayter, C, Foden, JA, Sander, JD and Joung, JK (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk, JL, Schmidt, T, Kaiser, V, Höning, K and Hornung, V (2013). A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol 31: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, AW, Rios, X, Chari, R, Yang, L, Zhang, F, Mali, P et al. (2012). Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res 40: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath, D, Amlinger, L, Rath, A and Lundgren, M (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117: 119–128. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy, J and Davidson, AR (2014). To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends Microbiol 22: 218–225. [DOI] [PubMed] [Google Scholar]
- Barrangou, R and Marraffini, LA (2014). CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, R and Wiedenheft, B (2014). A CRISPR method for genome engineering. F1000Prime Rep 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA and Charpentier, E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas, G, Barrangou, R, Horvath, P and Siksnys, V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, SW, Kim, S, Kim, JM and Kim, JS (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, East, A, Cheng, A, Lin, S, Ma, E and Doudna, J (2013). RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X and Gonçalves, MA (2015). Engineered viruses as genome editing devices. Mol Ther. [DOI] [PMC free article] [PubMed]
- Cox, DB, Platt, RJ and Zhang, F (2015). Therapeutic genome editing: prospects and challenges. Nat Med 21: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio, I and Gonçalves, MA (2015). Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol 33: 280–291. [DOI] [PubMed] [Google Scholar]
- Yin, H, Kanasty, RL, Eltoukhy, AA, Vegas, AJ, Dorkin, JR and Anderson, DG (2014). Non-viral vectors for gene-based therapy. Nat Rev Genet 15: 541–555. [DOI] [PubMed] [Google Scholar]
- Goubau, D, Deddouche, S and Reis e Sousa, C (2013). Cytosolic sensing of viruses. Immunity 38: 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F, Pruett-Miller, SM, Huang, Y, Gjoka, M, Duda, K, Taunton, J et al. (2011). High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers, M, Maggio, I, Henriques, SF, Janssen, JM, Cathomen, T and Gonçalves, MA (2014). Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat Methods 11: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Boissel, S, Jarjour, J, Astrakhan, A, Adey, A, Gouble, A, Duchateau, P et al. (2014). megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res 42: 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z, Yang, H and Colosi, P (2010). Effect of genome size on AAV vector packaging. Mol Ther 18: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas, P, Stein, D, Tang, WW, Frank, I, Wang, SQ, Lee, G et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L, Guell, M, Byrne, S, Yang, JL, De Los Angeles, A, Mali, P et al. (2013). Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41: 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paper: First Clinical Application of Talen Engineered Universal CAR19 T Cells in B-ALL. https://ash.confex.com/ash/2015/webprogram/Paper81653.html.
- Gori, JL, Hsu, PD, Maeder, ML, Shen, S, Welstead, GG and Bumcrot, D (2015). Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther 26: 443–451. [DOI] [PubMed] [Google Scholar]
- Ran, FA, Cong, L, Yan, WX, Scott, DA, Gootenberg, JS, Kriz, AJ et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard, BL (2005). Homing endonuclease structure and function. Q Rev Biophys 38: 49–95. [DOI] [PubMed] [Google Scholar]
- Anders, C, Niewoehner, O, Duerst, A and Jinek, M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu, H, Ran, FA, Hsu, PD, Konermann, S, Shehata, SI, Dohmae, N et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton, J, Daboussi, F, Leduc, S, Molina, R, Redondo, P, Macmaster, R et al. (2012). 5'-Cytosine-phosphoguanine (CpG) methylation impacts the activity of natural and engineered meganucleases. J Biol Chem 287: 30139–30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton, J, Dupuy, A, Daboussi, F, Thomas, S, Maréchal, A, Macmaster, R et al. (2012). Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem 287: 38427–38432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek, M, Brondani, V, Büchel, J, Serrano, L, Segal, DJ and Cathomen, T (2007). Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793. [DOI] [PubMed] [Google Scholar]
- Miller, JC, Holmes, MC, Wang, J, Guschin, DY, Lee, YL, Rupniewski, I et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Söllü, C, Pars, K, Cornu, TI, Thibodeau-Beganny, S, Maeder, ML, Joung, JK et al. (2010). Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res 38: 8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y, Vo, TD, Mendel, MC, Greenberg, SG, Wang, J, Xia, DF et al. (2011). Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- Sternberg, SH, LaFrance, B, Kaplan, M and Doudna, JA (2015). Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak, V, Lin, S, Guilinger, JP, Ma, E, Doudna, JA and Liu, DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Scott, DA, Weinstein, JA, Ran, FA, Konermann, S, Agarwala, V et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y, Foden, JA, Khayter, C, Maeder, ML, Reyon, D, Joung, JK et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick, TJ, Fine, EJ, Antico, CJ and Bao, G (2013). CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41: 9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Aach, J, Stranges, PB, Esvelt, KM, Moosburner, M, Kosuri, S et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H, Xiao, T, Chen, CH, Li, W, Meyer, CA, Wu, Q et al. (2015). Sequence determinants of improved CRISPR sgRNA design. Genome Res 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger, JP, Thompson, DB and Liu, DR (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, SQ, Wyvekens, N, Khayter, C, Foden, JA, Thapar, V, Reyon, D et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker, IM, Gao, L, Zetsche, B, Scott, DA, Yan, WX and Zhang, F (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres, A, Gosis, BS, Ding, Q, Collins, R, Ragavendran, A, Brand, H et al. (2014). Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C, Gore, A, Yan, W, Abalde-Atristain, L, Li, Z, He, C et al. (2014). Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15: 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V, Shen, B, Zhang, W, Hodgkins, A, Keane, T, Huang, X et al. (2015). Off-target mutations are rare in Cas9-modified mice. Nat Methods 12: 479. [DOI] [PubMed] [Google Scholar]
- Certo, MT, Ryu, BY, Annis, JE, Garibov, M, Jarjour, J, Rawlings, DJ et al. (2011). Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods 8: 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, CL, Certo, MT, Mussolino, C, Goodwin, MJ, Cradick, TJ, McCaffrey, AP et al. (2012). Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 40: 5560–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L and Maizels, N (2011). DNA nicks promote efficient and safe targeted gene correction. PloS One 6: e23981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L and Maizels, N (2014). Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci USA 111: E924–E932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Hsu, PD, Lin, CY, Gootenberg, JS, Konermann, S, Trevino, AE et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan-Curay, J, O'Reilly, M, Kohn, DB, Cannon, PM, Bao, G, Bushman, FD et al. (2015). Genome editing technologies: defining a path to clinic. Mol Ther 23: 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, R, Lombardo, A, Arens, A, Miller, JC, Genovese, P, Kaeppel, C et al. (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823. [DOI] [PubMed] [Google Scholar]
- Tsai, SQ, Zheng, Z, Nguyen, NT, Liebers, M, Topkar, VV, Thapar, V et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto, N, Mitra, A, Silva, MJ, Bienko, M, Dojer, N, Wang, Q et al. (2013). Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10: 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock, RL, Hu, J, Meyers, RM, Ho, YJ, Kii, E and Alt, FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D, Bae, S, Park, J, Kim, E, Kim, S, Yu, HR et al. (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12: 237–43, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- Perez, EE, Wang, J, Miller, JC, Jouvenot, Y, Kim, KA, Liu, O et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei, D and Foiani, M (2008). Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9: 297–308. [DOI] [PubMed] [Google Scholar]
- Kuhar, R, Gwiazda, KS, Humbert, O, Mandt, T, Pangallo, J, Brault, M et al. (2014). Novel fluorescent genome editing reporters for monitoring DNA repair pathway utilization at endonuclease-induced breaks. Nucleic Acids Res 42: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane, JD, Lee, G, Zheng, Z, Mendel, M, Abate-Daga, D, Bharathan, M et al. (2015). Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol Ther 23: 1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot, L, Philip, B, Schiffer-Mannioui, C, Le Clerre, D, Chion-Sotinel, I, Derniame, S et al. (2015). Multiplex genome-edited T-cell manufacturing platform for “Off-the-Shelf” adoptive T-cell immunotherapies. Cancer Res 75: 3853–3864. [DOI] [PubMed] [Google Scholar]
- Mandal, PK, Ferreira, LM, Collins, R, Meissner, TB, Boutwell, CL, Friesen, M et al. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris, JA, Thompson, DB, Shu, Y, Guilinger, JP, Bessen, JL, Hu, JH et al. (2015). Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S, Staahl, BT, Alla, RK and Doudna, JA (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3: e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S, Kim, D, Cho, SW, Kim, J and Kim, JS (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, K, Lin, S, Boyer, E, Simeonov, DR, Subramaniam, M, Gate, RE et al. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA 112: 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel, A, Bak, RO, Clark, JT, Kennedy, AB, Ryan, DE, Roy, S et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese, P, Schiroli, G, Escobar, G, Di Tomaso, T, Firrito, C, Calabria, A et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. W. et al. Gene Editing of CCR5 in Hematopoietic Stem Cells in a Nonhuman Primate Model of HIV/AIDS. Blood 124, 4802–4802 (2014). [Google Scholar]
- Yin, H, Xue, W, Chen, S, Bogorad, RL, Benedetti, E, Grompe, M et al. (2014). Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32: 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H, Haurigot, V, Doyon, Y, Li, T, Wong, SY, Bhagwat, AS et al. (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q, Strong, A, Patel, KM, Ng, SL, Gosis, BS, Regan, SN et al. (2014). Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res 115: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahiny, AJ, Dewerth, A, Mays, LE, Alkhaled, M, Mothes, B, Malaeksefat, E et al. (2015). In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat Biotechnol 33: 584–586. [DOI] [PubMed] [Google Scholar]
- Sharma, R, Anquela, XM, Doyon, Y, Wechsler, T, DeKelver, RC, Sproul, S et al. (2015). In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfs, JM, DaSilva, M, Meister, SE, Wang, X, Schild-Poulter, C and Edgell, DR (2014). MegaTevs: single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res 42: 8816–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, KM, Mali, P, Braff, JL, Moosburner, M, Yaung, SJ and Church, GM (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, BP, Prew, MS, Tsai, SQ, Topkar, VV, Nguyen, NT, Zheng, Z et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo, N, Gunn, A, Cheng, A, Hasty, P and Stark, JM (2009). Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet 5: e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo, MT, Gwiazda, KS, Kuhar, R, Sather, B, Curinga, G, Mandt, T et al. (2012). Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods 9: 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacôte, F, Perez, C, Guyot, V, Duhamel, M, Rochon, C, Ollivier, N et al. (2013). High frequency targeted mutagenesis using engineered endonucleases and DNA-end processing enzymes. PLoS One 8: e53217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo, T, Kaneko, T, Sakuma, T, Kobayashi, J, Kunihiro, Y, Voigt, B et al. (2013). Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci Rep 3: 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, MC, Yanez, DA and Stark, JM (2014). An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells. Nucleic Acids Res 42: 7720–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C, Liu, Y, Ma, T, Liu, K, Xu, S, Zhang, Y et al. (2015). Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, VT, Weber, T, Wefers, B, Wurst, W, Sander, S, Rajewsky, K et al. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Maruyama, T, Dougan, SK, Truttmann, MC, Bilate, AM, Ingram, JR and Ploegh, HL (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, F, Barbeau, M, Éthier, S, Dostie, J and Pelletier, J (2015). Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, LS and Gautier, J (2011). Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Ghezraoui, H, Piganeau, M, Renouf, B, Renaud, JB, Sallmyr, A, Ruis, B et al. (2014). Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell 55: 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, SH, Redding, S, Jinek, M, Greene, EC and Doudna, JA (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B, Gootenberg, JS, Abudayyeh, OO, Slaymaker, IM, Makarova, KS, Essletzbichler, P et al. (2015). Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]