Abstract
HIV-1 escapes antiretroviral agents by integrating into the host DNA and forming a latent transcriptionally silent HIV-1 provirus. Transcriptional activation is prerequisite for reactivation and the eradication of latent HIV-1 proviruses. dCas9-SunTag-VP64 transcriptional system has been reported that it can robustly activate the expression of an endogenous gene using a single guide RNA (sgRNA). Here, we systematically investigated the potential of dCas9-SunTag-VP64 with the designed sgRNAs for reactivating latent HIV-1. We found dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 targeted from −164 to −146 or −124 to −106 bp upstream of the transcription start sites of HIV-1 could induce high expression of luciferase reporter gene after screening of sgRNAs targeting different regions of the HIV-1 promoter. Further, we confirmed that dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 can effectively reactivate latent HIV-1 transcription in several latently infected human T-cell lines. Moreover, we confirmed that the reactivation of latent HIV-1 by dCas9-SunTag-VP64 with the designed sgRNA occurred through specific binding to the HIV-1 LTR promoter without genotoxicity and global T-cell activation. Taken together, our data demonstrated dCas9-SunTag-VP64 system can effectively and specifically reactivate latent HIV-1 transcription, suggesting that this strategy could offer a novel approach to anti-HIV-1 latency.
Introduction
Highly active antiretroviral therapy (HAART) has effectively suppressed the replication of human immunodeficiency virus-1 (HIV-1) and decreased the morbidity and mortality of HIV-infected patients during the last three decades.1,2 Unfortunately, HIV-1 infection remains incurable due to the persistence of a viral reservoir, which escaping antiretroviral agents by integrating into the host DNA and forming a latent transcriptionally silent HIV-1 proviruses. In such case, dormant viruses can bypass host immune system surveillance and antiretroviral drugs, followed by resuming active infection once HAART is interrupted. Therefore, the major barrier to the eradication of HIV-1 is the presence of latent reservoirs. Extensive efforts should be focused on identifying approaches to eliminating these dormant provirus.1,2 One strategy termed “shock and kill” has recently gained much attention. This approach involves reactivating latent HIV-1 by inducing the expression of the quiescent provirus and then stopping the spread of reactivated virus by HAART or clearing virus-producing cells by host immune responses or viral cytopathic effect.3,4,5 In devising the “shock and kill” strategy, focus has been placed on finding ways to reactivate latent HIV-1 without inducing global T-cell activation. A number of novel activators have been identified to reactivate latent HIV-1 by mechanism-directed approaches or a wide range of screening. However, several disadvantages: cytotoxicity, mutagenicity or a lack of target specificity existed when using these compounds, though some of them have already entered clinical testing in humans.6,7 Thus, better and more specific latency-reversing strategies are urgently needed in antiviral therapy.
Engineered transcription factors, generated by fusing activation or repression domains to DNA-binding domains, have been used to modulate desired gene expression through specifically targeting their promoters in many applications,8,9 including studying gene functions in complex biological processes and offering great potential in therapeutics. Zinc finger proteins (ZFPs) or transcription activator-like effectors (TALEs) coupled with functional domains are representative over the recent decades.8,9,10,11 Our group recently published related work on employing a synthetic ZFP and TALE specific for the HIV-1 5′-LTR (long terminal repeat) promoter were coupled with tetrameric herpes virus transcription activation domain VP16 (VP64) to activate latent HIV-1.10,11 However, due to either fixed DNA-sequence-binding requirements or their multistage DNA assembly protocols, engineered ZFP or TALE remains time-consuming and expensive to develop large-scale protein libraries for genome interrogation, thus severely limiting the potential use of them.12
The recently developed CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9) system is now frequently used for genome editing in human cells through sequence-specific sgRNA in complex with Cas9 proteins.12,13,14,15 This toolset greatly improves the ease of genome editing because of easy design and synthesis of sgRNA. Subsequently, a CRISPR/dCas9 system, mutant Cas9 protein without endonuclease activity (dead Cas9, dCas9) coupled with activator domain VP64 or repressor domain KRAB (Kruppel-associated box),16,17 is used to modulate eukaryotic transcription at native and synthetic promoters. Previous study shown that dCas9 fused with one copy of VP64 (dCas9-VP64) together with a designed sgRNA to increase transcription of interest gene usually resulted in less than twofold induction, thus limiting the potential application of this system.16,18,19 Subsequent study revealed that recruitment of multiple copies of dCas9-VP64 to native or artificial promoters via the combined use of nonoverlapping sgRNAs could improve the activation level.16,19,20,21,22 However, several sgRNAs needed to be transfected simultaneously into human cells. Recently, Tanenbaum et al. 18 demonstrated that dCas9-SunTag, a dCas9 protein fused with a repeating peptide array termed SunTag, can recruit up to 24 copies of an antibody-fusion VP64 to target promoter to robustly activate the expression of endogenous gene with a single sgRNA. The feature of this toolset (referred to as dCas9-SunTag-VP64) not only demonstrated in simplifying single gene activation but also opening possibilities to activate multiple genes simultaneously, potentially allowing functional re-engineering of cell behavior via precise modulation of gene expression.18
In this study, we systematically investigated the potential of dCas9-SunTag-VP64 with sgRNA for activating latent HIV-1. Our data suggested that the designed sgRNA with the dCas9-SunTag-VP64 could efficiently and specifically reverse HIV-1 from latency in latently infected human T cells, significantly better than the dCas9-VP64 system. Moreover, the dCas9-SunTag-VP64 system shows no cytotoxicity, genotoxicity, or global T-cell activation. Compared to current latency-reversing agents, a flexible and potent platform for reactivating latent HIV-1 through specifically binding to HIV-1 5′-LTR promoter was described in this article.
Results
Design and screen of sgRNAs for dCas9-SunTag-VP64-mediated HIV-1 LTR activation
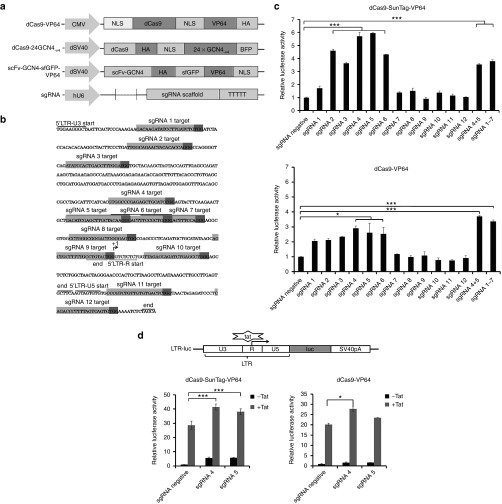
The HIV-1 LTR represents an attractive and ideal anti-HIV-1 target because it is highly conserved across all HIV-1 genomes. Here, we used the previously reported dCas9-VP64 system and the newly developed dCas9-SunTag-VP64 system together with sgRNAs targeting HIV-1 LTR to mediate gene transcription. The expression construct of dCas9-VP64 system, dCas9-SunTag-VP64 system (including dCas9-24GCN4-v4 and scFv-GCN4-sfGFP-VP64) and sgRNA were shown in Figure 1a. The detailed sgRNA cloning protocol was represented in Supplementary Figure S1. Initially, we detected the expression of dCas9-SunTag-VP64 and dCas9-VP64 system in human embryonic kidney 293T (HEK293T) cells. The expression of dCas9-24GCN4-v4 and scFv-GCN4-sfGFP-VP64 plasmid was visualized by fluorescence microscopy instead of western blot due to the large size of them (Supplementary Figure S2a). While the expression of dCas9-VP64 in HEK293T cells was detected by western blot (Supplementary Figure S2b).
Figure 1.
Design and screen of sgRNAs for dCas9-SunTag-VP64-mediated HIV-1 LTR activation by luciferase assay. (a) Schematic representation of dCas9-VP64 and dCas9-SunTag-VP64 system. dCas9-VP64 system indicated that dCas9 protein was coupled with one copy of VP64, hemagglutinin (HA) epitope tag, and nuclear localization signal (NLS). The dCas9-SunTag-VP64 construct is composed of two expression cassettes. One is dCas9-SunTag, meaning dCas9 is coupled with 24 copies of the v4 version of the GCN4 peptides. The other cassette is scFv-GCN4-sfGFP-VP64, including a specific antibody-fusion VP64 to the GCN4 peptide. HA epitope tag and NLS were included in the two expression plasmids. The sgRNA expression cassette driven by human U6 promoter was used in two gene activation system. (b) Target sequence of sgRNAs in the HIV-1 5′-LTR promoter (from HXB2 reference strain, GenBank accession no. K03455). The sequence of the U3'R and U5 regions in the LTR promoter are underlined. The transcription start site is indicated with (+1). Individual sgRNA target site in the LTR promoter is shown in light gray. The protospacer-adjacent motif (PAM) sequence is indicated in dark gray. (c) Screen of sgRNAs for dCas9-SunTag-VP64 or dCas9-VP64-mediated HIV-1 LTR activation by luciferase assay. HEK293T cells were cotransfected indicated sgRNA with dCas9-SunTag-VP64 or dCas9-VP64, the LTR-luc reporter and internal control pRL-SV40 plasmid at the indicated time. The relative luciferase activity was measured using the dual-luciferase reporter assay system (Promega) at 72 hours post-transfection. The data was analyzed by normalizing individual sgRNA-transfected group to sgRNA negative vector-transfected group in two gene activation systems, respectively. Each data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test. (d) Analysis of transcription elongation of HIV-1 LTR when combining dCas9-VP64 or dCas9-SunTag-VP64 with Tat. HEK293T cells were transfected with the dCas9-VP64 or the dCas9-SunTag-VP64 expression cassette with indicated sgRNA in the presence or absence of Tat at the indicated time. The relative luciferase activity in each group was measured after 72 hours transfection. The data shown was normalized to sgRNA negative vector-transfected group in two gene activation systems, respectively. Each data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
To identify the binding sites most efficient for gene induction, 12 sgRNAs were designed targeting HIV-1 LTR promoter and tested their transactivation activity in HEK293T cells (Figure 1b and Supplementary Table S1). Towards this end, we cotransfected HEK293T cells with the dCas9-SunTag-VP64 or dCas9-VP64 expression vectors with individual sgRNA and a reporter vector comprising the luciferase gene driven by the HIV-1 LTR promoter (LTR-luc). The luciferase activity was measured at 72 hours post-transfection. As a control, a sgRNA-negative vector was used in both systems. The results showed that sgRNA 4, sgRNA 5, sgRNA 6 with dCas9-SunTag-VP64 or dCas9-VP64 yield significant activation of reporter gene expression relative to sgRNA negative control (Figure 1c). Especially, sgRNA 4 or sgRNA 5 yielded activation of reporter gene up to sixfold in dCas9-SunTag-VP64 system and threefold in dCas9-VP64 system (Figure 1c). To test whether a stronger activation could be generated by multiple sgRNAs with dCas9-SunTag-VP64 or dCas9-VP64, two most efficient sgRNAs (sgRNA 4+5) or seven sgRNAs (sgRNA 1–7) with dCas9-SunTag-VP64 or dCas9-VP64 were cotransfected into HEK293T cells, respectively. The results indicated that combined use of sgRNAs showed no robust induction in dCas9-SunTag-VP64, but a little higher than single sgRNA (sgRNA 4 or sgRNA 5) in dCas9-VP64 (Figure 1c). Among the sgRNAs designed, two sgRNAs (sgRNA 4 or sgRNA 5) binding within 200 bp region upstream of transcription start sites (TSS) induced significant activation with dCas9-SunTag-VP64. In the following experiments, we used sgRNA 4 or sgRNA 5 with dCas9-SunTag-VP64 system or dCas9-VP64 system for study.
We further explored transcription promotion effect of HIV-1 LTR induced by dCas9-SunTag-VP64 system or dCas9-VP64 system with Tat, a HIV-1-expressed protein responsible for viral transcription elongation through recruiting transcription elongation complexes to the transactivation response element of HIV-1 5′-LTR.23,24,25 For this purpose, HIV-1 LTR-luc reporter vector was cotransfected with dCas9-SunTag-VP64 or dCas9-VP64 and indicted sgRNA in the presence or absence of Tat expression vector into HEK293T cells. The results shown that cotransfection of Tat and dCas9-SunTag-VP64 with sgRNA negative vector induced around 27-fold increase of luciferase reporter expression, while around 40-fold induction of reporter gene expression was observed once coexpressing sgRNA 4 or sgRNA 5 with dCas9-SunTag-VP64 and Tat (Figure 1d). Our results indicated that dCas9-SunTag-VP64 system could promote HIV-1 LTR transcription elongation with Tat in a potent manner.
dCas9-SunTag-VP64 mediates activation of the HIV-1 5'-LTR in TZM-bl cells
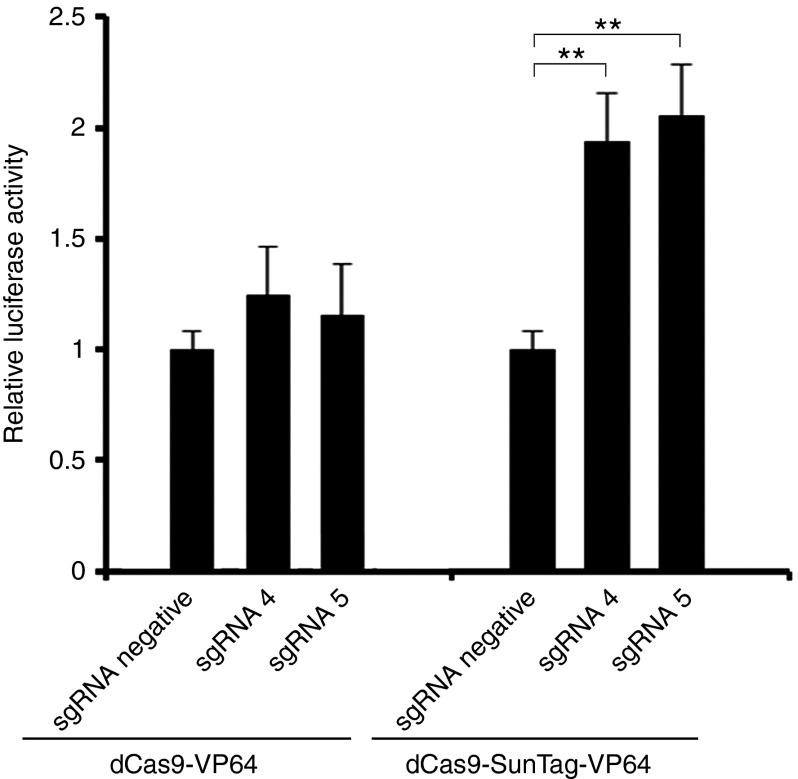
The regulation of gene expression in a transient transfection assay may be different from genomic expression because the reporter plasmid is not integrated into the genome as a chromosomal gene.26 To address this issue, we performed gene transcription activation experiment in TZM-bl cells, a HeLa cell line integrated with a luciferase reporter expression cassette driven by HIV-1 5′-LTR.27 We cotransfected dCas9-VP64 or dCas9-SunTag-VP64 with indicated sgRNA and HIV-1 LTR-luc expression cassette into TZM-bl cells. After 72 hours transfection, the luciferase level was measured. Our data showed that dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 yielded activation of reporter gene up to twofold in TZM-bl cells after normalized to sgRNA negative control. No gene expression induction was observed by dCas9-VP64 with sgRNA 4 or sgRNA 5 (Figure 2). These findings revealed that dCas9-SunTag-VP64 performed the same role of inducing integrated reporter gene expression as transient luciferase assay.
Figure 2.
dCas9-SunTag-VP64-mediated activation of the HIV-1 5′-LTR in TZM-bl cells. dCas9-VP64 and dCas9-SunTag-VP64 induced transcription of an integrated luciferase reporter in TZM-bl cells. TZM-bl cells were cotransfected with dCas9-VP64 or dCas9-SunTag-VP64 and indicated sgRNA at specified time. Cells were harvested and then subjected to luciferase activity assay after 3-day transfection. The data shown was normalized to sgRNA negative vector-transfected group in two gene activation systems, respectively. Each data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
dCas9-SunTag-VP64 can effectively reverse HIV-1 from latency in latently-infected cells
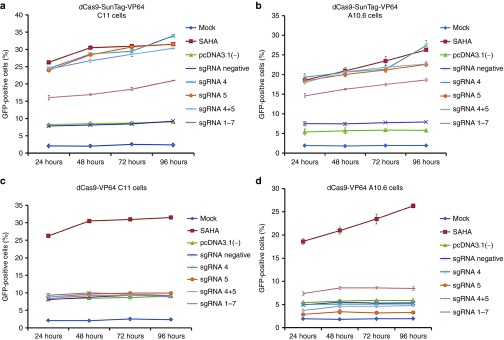
We further validated the effects of dCas9-SunTag-VP64 system in Jurkat T-cell-based latency models, C11 cells28,29,30 and J-Lat clone A10.6 cells.31 They both contained a green fluorescent protein (GFP) gene under the control of the HIV-1 LTR. The percentage of GFP-positive cells indicated the reversal of HIV-1 from latency. For this purpose, the C11 or J-Lat clone A10.6 cells were nucleofected with pcDNA3.1(-) vector or with dCas9-SunTag-VP64 or dCas9-VP64 expression vector and indicated sgRNA. Besides, in this experiment, C11 or J-Lat clone A10.6 cells untreated or treated with 0.5 μmol/l of suberoylanilide hydroxamic acid were used as mock and positive control, respectively. The percentage of GFP-positive cells was measured by flow cytometry to indicate the reactivation of latent HIV-1 at different time points. The results showed that dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 could significantly reactivate latent HIV-1 in C11 cells (33.89 and 31.23%, respectively) and J-Lat clone A10.6 (28.64 and 21.34%, respectively) at 72 hours post-transfection. Furthermore, the results showed that dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 dramatically activated HIV-1 expression at 24 hours post-transfection and persisted modest increasing activation over the next 3 days in both cell models (Figure 3a,b and Supplementary Figure S3a,b). Increased induction of latent HIV-1 expression was not obtained when combining multiple sgRNAs with dCas9-SunTag-VP64 in two HIV-1 latently infected cell models (Figure 3a,b and Supplementary Figure S3a,b). We wanted to test whether synergistic action could be observed at lower amount of indicated sgRNAs with dCas9-SunTag-VP64 system. To test this, sgRNA 4+5 or sgRNA 4–5 with dCas9-SunTag-VP64 was nucleofected into C11 cells. As shown in Supplementary Figure S4, no synergistic effects were observed by combined sgRNAs with dCas9-SunTag-VP64 on reactivating latent HIV-1 in C11 cells. While no significant induction of HIV-1 gene expression was observed by dCas9-VP64 with single or multiple sgRNAs in both cell lines at different time points (Figure 3c,d and Supplementary Figure S5a,b). However, the expression of HIV-1 in the cells transfected with pcDNA3.1(-) or with dCas9-SunTag-VP64 or dCas9-VP64 and sgRNA-negative vector was a little higher than mock, perhaps due to a partial activation of these cells by nucleofection.
Figure 3.
dCas9-SunTag-VP64 reactivates HIV-1 in different latently infected cells. (a) C11 cells or (b) J-Lat clone A10.6 cells were nucleofected with pcDNA3.1(-) vector or dCas9-SunTag-VP64 with indicated sgRNA and were mock treated or treated with 0.5 μmol/l of suberoylanilide hydroxamic acid at the indicated time. The percentage of GFP-expressing cells was measured by flow cytometry at different time points. Fluorescence histograms represented the reactivation efficiency of latent HIV-1 by dCas9-SunTag-VP64. (c) and (d) Time-dependent effects of dCas9-VP64-mediated activation of latent HIV-1 in C11 cells or J-Lat clone A10.6 cells. The data represent the mean ± SD of three independent experiments.
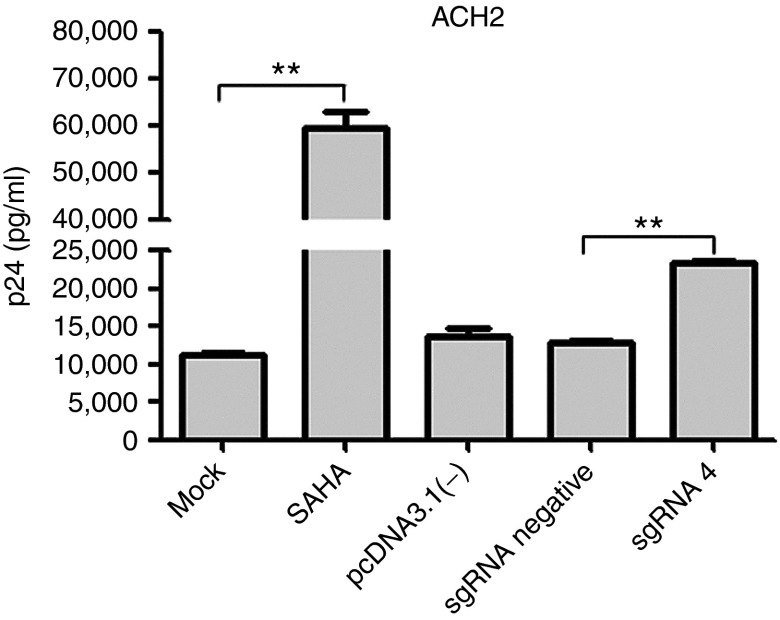
Additionally, the expression of p24 induced by dCas9-SunTag-VP64 system was detected in ACH2 cells, a chronically infected T-cell clone from the parental cell line A3.01, which contains a single copy of latent HIV-1 provirus.32 To this end, harvested supernatants from cells nucleofected with pcDNA3.1(-) plasmid or with dCas9-SunTag-VP64 and indicated sgRNA were subjected to p24 detection. The results indicated the expression of p24 from cells nucleofected with sgRNA 4 with dCas9-SunTag-VP64 was significantly higher than that from cells treated with sgRNA negative control with dCas9-SunTag-VP64 (Figure 4). Based on these data, our results revealed that dCas9-SunTag-VP64 system could efficiently induce HIV-1 expression in latently infected cells and could be further investigated as a novel anti-HIV-1 latency agent.
Figure 4.
dCas9-SunTag-VP64 system mediated the induction of latent HIV-1 in ACH2 cells. ACH2 cells were nucleofected with pcDNA3.1(-) plasmid or dCas9-SunTag-VP64 with indicated sgRNA at specified time. At 72 hours post-transfection, the supernatants were collected and analyzed for p24 antigen by enzyme-linked immunosorbent assay. ACH2 cells mock treated were used as the basal expression level of p24, and cells treated with 0.5 μmol/l suberoylanilide hydroxamic acid were shown as positive control. The data represent the mean ± SD of three independent experiments in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
Absence of dCas9-SunTag-VP64 system-related cytotoxicity, T-cell activation, and latent HIV-1 reactivation-induced cell death
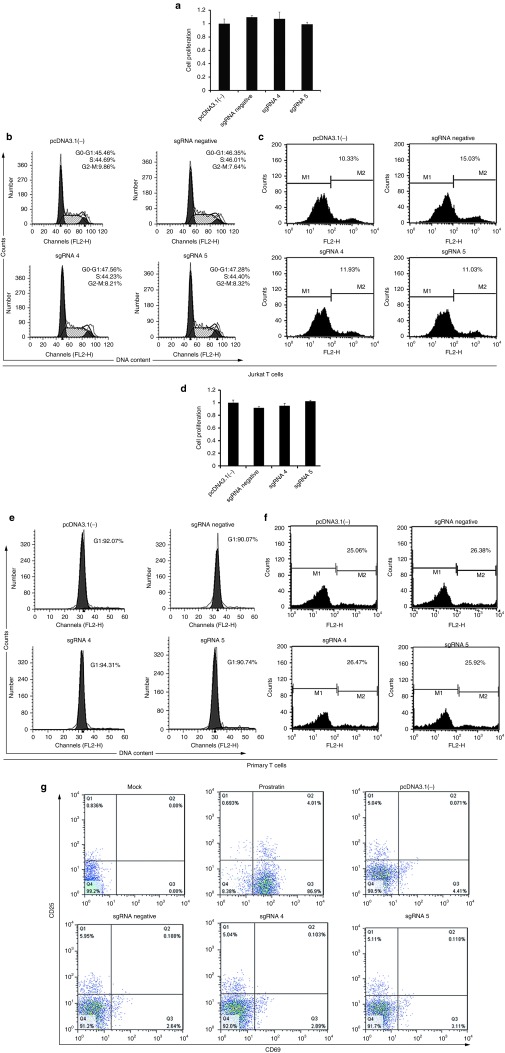
A critical concern of agents for activating HIV-1 latency is their safety. To address this issue, we evaluated the safety in the Jurkat T cells transfected with dCas9-SunTag-VP64 system. The CCK-8 assay was used to evaluate cell proliferation at 72 or 96 hours after Jurkat T cells transfected with pcDNA3.1(-) or with dCas9-SunTag-VP64 and indicated sgRNA. Our results suggested that dCas9-SunTag-VP64 with indicated sgRNAs did not affect cell proliferation (Figure 5a and Supplementary Figure S6a). The potential of dCas9-SunTag-VP64 to induce cell cycle progression was further investigated by staining these cells with propidium iodide at 72 or 96 hours post-transfection and analyzing the DNA content using flow cytometry. Compared with the control group, the transfection of the dCas9-SunTag-VP64 together with indicated sgRNA did not affect cell cycle progression and cell death (Figure 5b,c and Supplementary Figure S6b). Similar results were observed in primary cells treated with dCas9-SunTag-VP64 together with indicated sgRNA (Figure 5d–f).
Figure 5.
Absence of dCas9-SunTag-VP64 system-related cytotoxicity and T cell activation. (a) Jurkat T cells were transfected with pcDNA3.1(-) or dCas9-SunTag-VP64 and sgRNA 4 or sgRNA 5 or with a sgRNA negative vector at the indicated time. At 72 hours post-transfection, cell proliferation was measured using CCK-8 kit. The data shown are the cell viability of pcDNA3.1(-)-treated group divided by that of the dCas9-SunTag-VP64 with sgRNA-transfected groups. The data represent the mean ± SD of three independent experiments. (b) Jurkat T cells were nucleofected with pcDNA3.1(-) or with dCas9-SunTag-VP64 and sgRNA 4 or sgRNA 5 or with a sgRNA negative vector at the indicated time. Cell cycle progression was analyzed by flow cytometry after 72 hours transfection. Each experiment was replicated three times. (c) Jurkat T cells were nucleofected with pcDNA3.1(-) vector or dCas9-SunTag-VP64 along with indicated sgRNA. At 72 hours post-transfection, cells were harvested and then stained by propidium iodide and subjected to cell death assay. The results are presented as fluorescence histograms. (d) Cell proliferation assay, (e) cell cycle progression, (f) cell death detection were performed in primary CD4+ T cells as the methods described in Jurkat T cells. The data represent the mean ± SD of three independent experiments in triplicate. (g) Primary CD4+ T cells were nucleofected with pcDNA3.1(-) plasmid or dCas9-SunTag-VP64 with indicated sgRNA at specified time. Cells treated with prostratin (1 μmol/l) were used as positive control. Cells untreated were represented as mock. The expression of CD25 and CD69 was detected by flow cytometry using antibodies against CD25 and CD69. The results are representative of three independent experiments.
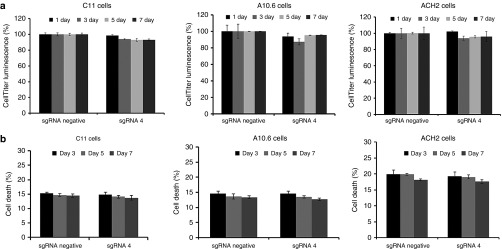
The major disadvantage of some current therapeutic agents is their propensity to nonspecifically activate bystander T cells. We therefore investigated the induction of global T-cell activation markers by dCas9-SunTag-VP64 treatment. Our results shown that no induction of CD25 or CD69 expression was found in the pcDNA3.1(-) or dCas9-SunTag-VP64-treated primary CD4+ T cells (Figure 5g). Nevertheless, cells treated with 1 μmol/l prostratin stimulated the expression of activation markers on the surface of primary CD4+ T cells, consistent with previous study.33 These data suggested that dCas9-SunTag-VP64 system is a potentially safe tool for anti-HIV-1 latency therapy. Our results showed that dCas9-SunTag-VP64 system did not have cytotoxicity in Jurkat T cells and primary T cells. We further wanted to test the virus-induced suicide death effects in latency models after reactivation by dCas9-SunTag-VP64 system. To this end, dCas9-SunTag-VP64 with indicated sgRNA was nucleofected into the three latently infected cell lines (C11, A10.6, and ACH2 cells) at indicated time. Compared to sgRNA negative-treated group, dCas9-SunTag-VP64 with sgRNA 4 did not affect cell viability at different time. Further cell death assay also showed no difference between dCas9-SunTag-VP64 with sgRNA negative and sgRNA 4-treated group at indicated time (Figure 6a,b).
Figure 6.
Analysis of effects of dCas9-SunTag-VP64 system in HIV-1 latently infected cell models. (a) The cell viability of C11, A10.6, and ACH2 cells nucleofected with dCas9-SunTag-VP64 with indicated sgRNA was measured by CellTiter-Glo luminescent assay kit at different time post-transfection. The data from dCas9-SunTag-VP64 with sgRNA 4-treated group was relative to sgRNA negative control in such three latently infected cell lines at indicated time, respectively. The data represent the mean ± SD of three independent experiments. (b) Cell death assay were performed as the methods described in Jurkat T cells. The data represent the mean ± SD of three independent experiments in triplicate.
dCas9-SunTag-VP64 reactivates latent HIV-1 through specific binding to the HIV-1 LTR promoter
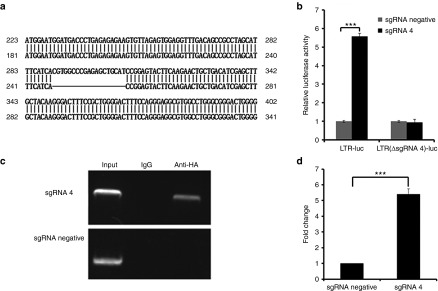
We investigated whether dCas9-SunTag-VP64 system reactivated HIV-1 through direct binding to the HIV-1 5′-LTR promoter. To address this issue, we constructed a HIV-1 LTR-luc reporter vector lacking the designed sgRNA 4 target site, termed LTR(ΔsgRNA4)-luc. The deletion of the sgRNA 4 target site in the LTR-luc reporter vector was aligned with wild type of LTR-luc vector (Figure 7a). Then, we transfected HEK293T cells with the dCas9-SunTag-VP64 with sgRNA 4 and HIV-1 LTR-luc or LTR(ΔsgRNA4)-luc expression vector. The results shown a more than fivefold induction of luciferase expression in LTR-luc group (consistent with previous results), but failed to generate induction in LTR(ΔsgRNA4)-luc group when coexpressing dCas9-SunTag-VP64 with sgRNA 4 in HEK293T cells (Figure 7b), indicating that the induction of HIV-1 by dCas9-SunTag-VP64 system occurred through specific binding to the HIV-1 5′-LTR promoter.
Figure 7.
dCas9-SunTag-VP64 specifically binds to the LTR to reactivate latent HIV-1. (a) Schematic representation of the deletion of the sgRNA 4 target site in the LTR-luc reporter vector aligned with wild type of LTR-luc vector. (b) Cotransfection of dCas9-SunTag-VP64 with sgRNA 4 or with sgRNA negative vector and the LTR-luc or LTR(ΔsgRNA4)-luc reporter plasmids into HEK293T cells at the indicated time. The luciferase activity induced by dCas9-SunTag-VP64 with sgRNA 4 was normalized to that of sgRNA-negative vector in each group at 72 hours post-transfection. The data represent the mean ± SD of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test. (c) Analysis of dCas9-SunTag-VP64 with sgRNA 4-specific binding to the HIV-1 5′-LTR promoter by a chromatin immunoprecipitation assay. C11 cells were nucleofected with dCas9-SunTag-VP64 and indicated sgRNA at the indicated time. Chromatin fragments were immunoprecipitated with anti-HA antibody or normal mouse IgG and then amplified by primers specific for HIV-1 5′-LTR promoter. (d) Analysis of the fold change of immunoprecipitated chromatin fragments from cells treated with dCas9-SunTag-VP64 with sgRNA 4 or with sgRNA negative vector group by quantitative polymerase chain reaction. The signal was normalized to that of input in each group. The data represent the mean ± SD of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
To further validate the direct binding of dCas9-SunTag-VP64 with sgRNA 4 to the HIV-1 5′-LTR in vivo, we performed a chromatin immunoprecipitation (ChIP) assay to identify this relationship. Briefly, C11 cells transfected with dCas9-SunTag-VP64 and sgRNA 4 or sgRNA negative vector were crosslinked with formaldehyde, and the chromatin fragments were immunoprecipitated with an antibody that recognizes the hemagglutinin (HA) tag at the C-terminus of dCas9-SunTag or with negative antibody IgG. DNA was isolated from the immunoprecipitates and then analyzed by polymerase chain reaction (PCR) using primers specific for HIV-1 LTR. We observed the immunoprecipitated HIV-1 LTR promoter fragment from dCas9-SunTag-VP64 with sgRNA 4-transfected cells rather than with sgRNA negative-transfected cells using the anti-HA antibody (Figure 7c). No immunoprecipitated DNA fragments were observed in negative antibody IgG (Figure 7c). Moreover, we calculated the fold change of immunoprecipitated DNA fragments from cells transfected with dCas9-SunTag-VP64 and sgRNA 4 relative to sgRNA negative-transfected group is more than fivefold induction (Figure 7d). Our results confirmed that the observed reactivation of HIV-1 was through a direct interaction between dCas9-SunTag-VP64 together with sgRNA 4 and HIV-1 5′-LTR.
Specific targeting of HIV-1 LTR without off-target modifications by CRISPR/Cas9
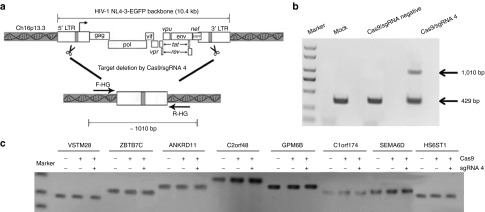
Next, we wanted to test the specific targeting of HIV-1 LTR promoter without off-target modification by CRISPR/Cas9. In order to test the specific targeting, wild-type Cas9 with sgRNA 4 was selected to determine whether HIV-1 proviral DNA could be deleted from HIV-infected cells C11, which having been found to carry a single integrated HIV-1 vector at position Ch16p13.3 (Figure 8a). To probe for deletion of the provirus, we used primers located at outsides of the integrated HIV genome normally separated by 10.4 kb when the provirus is integrated into the host genome (Figure 8a). The PCR product size we observed after Cas9 with sgRNA 4 treatment was approximately 1,010 bp (Figure 8b), a band size expected if the DNA segments between the two sgRNA 4 target sites were deleted from the chromosome. No 1,010-bp band in the control groups were observed (Figure 8b), indicating that deletion occurred only in the presence of Cas9 with sgRNA 4. To ensure that the 1,010-bp band was produced by rejoining DNA at the endonuclease cut sites, we cloned the PCR product and performed sequence analysis. Indeed, the sgRNA 4 target site at the 5′-LTR locus is directly linked to the same target site at 3′-LTR locus, indicating that the integrated HIV 5′- and 3′-LTR sites had been joined and the intervening 9.8 kb DNA segment had been deleted (Supplementary Figure S7).
Figure 8.
Off-target analysis for Cas9/sgRNA4 in latently infected cell clone C11. (a) Schematic representation of Cas9/sgRNA4-mediated genome deletions. The gray box in the 5′- and 3′-LTR of HIV-1 genome (10.4 kb) indicate CRISPR/Cas9 target sites. F-HG and R-HG (arrows) located at outsides of the integrated HIV-1 genome were used for amplifying genome deletion events. (b) Polymerase chain reaction products validated Cas9/sgRNA4-induced HIV-1 proviral genome deletions. C11 cells were untransfected (mock) or nucleofected with Cas9 and indicated sgRNA at a mass ration of 3:1. Genomic DNA was extracted 3 days post-transfection and then subjected to polymerase chain reaction using the F-HG and R-HG primers. (c) Off-target analysis for Cas9/sgRNA4. Eight potential off-target sites were amplified from Cas9/sgRNA4-treated cells and the mock cells. The closest gene name is used to indicate the amplicon presented on top of the lines. These amplicons were subjected to T7E1 assay to reveal any off target sites between the targeted cells and the mock cells by gel electrophoresis.
To further determine the off-target sites in genome, the sequence of sgRNA 4 target was BLAST-searched in the NCBI database of the human genome. We got eight homologous sequences (Supplementary Table S2) and amplified from cells untransfected or transfected with Cas9 and indicated sgRNA using the corresponding primers (Supplementary Table S3). The amplicons were purified and then treated with T7 endonuclease 1 (T7E1) as previously described.34,35,36 Our results showed that no mutation was detected by T7E1 analysis (Figure 8c), suggesting high specificity of the the designed sgRNA 4-guided Cas9.
Discussion
Up to date, the major strategy to eradication of latent HIV-1 reservoirs has been focused on purging the pool of latently infected cells in the presence of HAART by reactivating dormant virus. A number of small molecule activators have been shown to stimulate HIV-1 transcription in latently infected cells. However, there are major safety concerns with these compounds: toxicity, a lack of target specificity and the development of acquired drug resistance.3,4,5,6,7 Therefore, better and more specific latency activators are needed to explore. Previous studies have reported ZFP-VP64 and TALE-VP64 could induce HIV-1 gene expression.10,11 However, because of either user-defined DNA sequence requirements or a repetitive composition and size, using them is laborious and requires specialized expertise to interrogate the genome.16,19,20 As a result, it remains challenging to use DNA-binding proteins for modulating multiple gene expression simultaneously and implementing wide-range of genetic programs. The currently repurposed CRISPR system provides a potential platform for targeted gene regulation in gene therapy. In our study, we used dCas9-SunTag-VP64 toolset to activate HIV-1 latency with a single sgRNA. Our findings indicated that dCas9-SunTag-VP64 system could efficiently reactivate latent HIV-1 in different HIV-1 latently infected cells, significantly better than the previously developed dCas9-VP64 system.
Optimal design of sgRNAs on promoter region is critical to achieve high modulation efficiency using dCas9-SunTag-VP64 system.19,20 In our study, we designed 12 sgRNAs spanning HIV-1 LTR promoter, including 9 sgRNAs upstream of TSS and 3 sgRNAs downstream of TSS. The results revealed that sgRNA 4 or sgRNA 5, located within −164 to −146 or −124 to −106 bp upstream of TSS, showed the most efficient gene activation effect with dCas9-SunTag-VP64. Using additional sgRNAs targeting upstream of TSS had a little lower activity of inducing gene expression, suggesting that the position of sgRNAs targeting given promoter are important to activate gene expression. Previous study had demonstrated that efficient activation of endogenous genes could be achieved by sgRNAs binding within 200 bp region upstream of TSS,37 termed sgRNAs “hotspot”. However, the binding sites of sgRNA 7 and sgRNA 8 were also located at this region, no induction of HIV-1 expression were observed in our results. This was probably because the binding site of sgRNA 7 targeting at LTR promoter has been occupied by NF-kB, having been shown to induce transcription of HIV-1 gene by binding in the promoter region.38 While the binding sites of sgRNA 8 at LTR promoter has been occupied by Sp1 cis-regulatory sites, having been shown to stimulate HIV-1 gene expression in a synergistic manner that was dependent on the presence of both NF-kB and Spl elements.38 These reasons perhaps resulted in that dCas9-SunTag-VP64 with sgRNA 7 or sgRNA 8 complex were inaccessible to their target sites to further induce gene expression. Besides, our data also indicated that those sgRNAs downstream of TSS (sgRNA10, 11, 12) did not activate the expression of gene. This result is consistent with a previous report on CRISPR interference (CRISPRi)-mediated transcriptional repression.22 It is possible that binding of dCas9-SunTag-VP64 to downstream of TSS sterically hinders transcription by blocking polymerase.
In our data, the reactivation efficiency of latent HIV-1 by dCas9-SunTag-VP64 with sgRNA 4 or sgRNA 5 was modest in two latently infected cell models, similar to previous reactivation efficiency by ZFP-VP64 and TALE-VP64-induced. Actually, our activation efficiency was not as high as previous report regarding the efficiency of inducing other endogenous gene expression.18 This was probably attributed to the transfection strategy that we used. Three plasmids including dCas9-24GCN4-v4, scFv-GCN4-VP64, and sgRNA expression plasmid were simultaneously transfected into cells, whereas the former two plasmids were rather difficult for transfection, the co-transfection efficiency will be compromised. Combined use of multiple sgRNAs with dCas9-SunTag-VP64 system did not enhance the induction of latent HIV-1. One reason is likely due to the lower transfection efficiency and high variants for more plasmids to get into one target cell. The other reason we speculate that there may be the steric hindrance of dCas9-SunTag-VP64 with multiple sgRNAs at respective target site. Besides, our result is also consistent with a previous report that dCas9p300 Core was capable of robustly activating gene expression with a single sgRNA at promoters and characterized enhancers instead of with multiple sgRNAs.39 Previous study shown that significant gene activation of endogenous gene was based on virus-delivered methods.18 We proposed that a higher efficiency could be achieved by adenovirus-delivered dCas9-SunTag-VP64 to induce HIV-1 expression. The application of lentivirus-based gene delivery in the study may be limited: the risk of lentiviral random insertional mutagenesis, the chance of homologous recombination (HR) occurring at the sequence of LTR integrated in latent HIV-1 cells and LTR derived from the reverse transcription of lentiviral RNA genome into a double-stranded DNA and potential magnified off-target induced by stable expression of dCas9 in host cells. Furthermore, it was recently reported that SpCas9 (Streptococcus pyogenes Cas9) orthologue from Staphylococcus aureus, SaCas9, which is smaller than the widely used SpCas9 but with similar gene-editing efficiency,40 might be used in the dCas9-SunTag-VP64 system to increase gene activation efficiency. Moreover, a recent work reported that Cas9 could be engineered to recognize several types of PAM,41 which could be benefit for dCas9-SunTag-VP64 system to provide wider selection range of sgRNAs on promoters. The SunTag system is based on high affinity of scFv antibodies recognizing short peptides. However, a major drawback of antibodies is relatively large size and poorly expressed in the cytoplasm. DARPins, which are small, highly stable proteins, may be developed into a second-generation SunTag system by evolving in vitro to bind their epitope with high affinity.42
In spite of this newly developed technology, the safety concerns with dCas9-SunTag-VP64 activation system should be considered severely. To address this issue, we performed cytotoxicity assay to asses this toolset. Our results revealed that no cellular cytotoxicity induced by dCas9-SunTag-VP64 system in Jurkat T cells and primary T cells. Moreover, our results showed that no expression of CD25 or CD69 in T cells treated with dCas9-SunTag-VP64 with indicated sgRNA. Besides, the evidence of HIV-1 reactivation-induced cell death by dCas9-SunTag-VP64 system was not observed in three HIV-1 latently infected cells. Previous study showed that env, nef, tat, and vpr played a key role in the modulation of apoptosis by HIV-1 infection.43 While in our study, C11 cells contained HIV-1 NL4-3 genome driven by LTR promoter but inactivated env, nef, and vpr gene.28,29,30 A10.6 cells only included tat driven by LTR promoter.31 Viral protein such as Env, Vpr, and Nef were not expressed in C11 and A10.6 cells, indicating that no evidence of cell death was observed after reactivation by dCas9-SunTag-VP64 system in such two cells. As far as ACH2 cells, perhaps one reason was the low reactivation level. This result was similar to the previous study that the lower activation efficiency of latent HIV-1 in Jurkat-derived 2D10 or E4 cell lines resulted in less cell death.44 The other reason may be the provirus reactivation response and mechanisms varied with different latent cells.44 In this regard, the group of Siliciano has recently reported that virus reactivation with vorinostat occurs in latently infected primary CD4+ T cells generated in vitro, but is not associated with death induced directly by viral protein, i.e., viral cytopathic effect.45 In further study, we verified dCas9-SunTag-VP64 together with indicated sgRNA reactivating latent HIV-1 through specifically binding to HIV-1 LTR promoter by transient transfection assay and a ChIP experiment. The targeting specificity of this strategy is its major advantage over existing small molecule activators and also supposed to be a potent antiretroviral approach in future.
Our study demonstrated that dCas9-SunTag-VP64 system could efficiently and specifically reactivate latent HIV-1 in a potentially safe manner. In order to achieve the full potential of this toolset, a number of questions and challenges needed to be addressed, such as the optimal methods for delivering this technology and the evaluation of specificity and toxicity of it in vivo.46,47,48,49 Taken together, the promising CRISPR/dCas9-based activation strategy would constantly develop and provide much broader use in the field of HIV-1 gene therapy.
Materials and Methods
Cell culture. C11 cells are a type of HIV-1 latently infected cells constructed in our laboratory.28,29,30 J-Lat clone A10.6 cells were kindly provided by the NIH AIDS Research and Reference Reagent Program (Dr. Eric Verdin).31 ACH2 cells were also obtained from the NIH AIDS Research and Reference Reagent Program. The three HIV-1 latently infected cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, Grand Island, NY) and 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Shanghai, China) at 37 °C under 5% CO2. Human embryonic kidney 293T (HEK293T) cells were purchased from American Type Culture Collection (Manassas, VA). TZM-bl cells obtained from the NIH AIDS Research and Reference Reagent Program were generated by introducing separate integrated copies of the luciferase and β-galactosidase genes driven by the HIV-1 promoter.27 HEK293T and TZM-bl cells were maintained in Dulbecco′s modified Eagle′s medium supplemented with 10% FBS (Gibco), 1% penicillin-streptomycin at 37 °C with 5% CO2.
Transfection. HEK293T cells and TZM-bl cells were transfected with ViaFect reagent (Promega, Madison, WI) according to the manufacturer's instructions. C11 cells, J-Lat clone A10.6 cells, and ACH2 cells were nucleofected with pcDNA3.1(-) or with dCas9-VP64 or dCas9-SunTag-VP64 expression plasmid together with individual sgRNA using the Amaxa Cell Line Nucleofector Kit V (Lonza, Gaithersburg, MD). dCas9-VP64 expression plasmid was transfected into target cells at a mass ratio of 3:1 to the individual sgRNA expression plasmid.16 The dCas9-SunTag-VP64 plasmid, including an equal transfection mass ratio between the dCas9-24×GCN4-v4 and scFv-GCN4-sfGFP-VP64 expression plasmid, was also transfected at a mass ratio of 3:1 to the individual sgRNA expression plasmid.
Imaging. To detect the expression of dCas9-SunTag-VP64 system, HEK293T cells were cotransfected with dCas9-24GCN4-v4-BFP and scFv-GCN4-sfGFP-VP64 in 96-well glass bottom dishes at indicated time. After 48 hours transfection, Cells were imaged on a Zeiss LSM510 laser scanning confocal microscope.
Vector constructs. Plasmids encoding dCas9-24×GCN4-v4 and scFv-GCN4-sfGFP-VP64 were obtained from Addgene (plasmid 60910 and plasmid 60904).18 dCas9-VP64 expression plasmid was obtained from Addgene (plasmid 47107).16 sgRNA expression plasmid was obtained from Addgene (plasmid 47108).16 Because the reactivation of HIV-1 from latency was visualized by GFP under the control of the HIV-1 LTR promoter in latently infected cells C11 and A10.6, we needed to delete sfGFP from the scFv-GCN4-sfGFP-VP64 expression plasmid using BstBI and BamHI restriction sites and then ligated into the plasmid. The individual sgRNA clone vector included a BbsI restriction site facilitating rapid insert of a single sgRNA sequence. The multiple sgRNAs clone vector including NheI and XhoI restriction sites facilitated the insertion of different sgRNAs sequence. The sgRNA information and cloning protocol are detailed in Supplementary Table S1 and Supplementary Figure S1.
To obtain the HIV-1 LTR-luciferase reporter expression plasmid (LTR-luc), a full-length LTR fragment was amplified from HIV-1NL4-3-EGFP backbone using the forward primer F-LTR (5′-CGGGGTACCTGGAAGGGCTAATTCACTCCCAAAG-3′) and the reverse primer R-LTR (5′-CCGCTCGAGTGCTAGAGATTTTCCACACTGACTA-3′), followed by purification and digestion by KpnI and XhoI and then ligation into the KpnI-XhoI clone site of the pGL3-basic plasmid (Promega). HIV-1 LTR-luc reporter vector lacking the sgRNA 4 target site LTR(ΔsgRNA4)-luc was constructed using two-step fusion PCR: forward primer amplifying upstream of 5′-LTR (5′-AGGTACCTGGAAGGGCTAATTCACTCCCAA-3′) and reverse primer amplifying upstream of the 5′-LTR (5′-AGTTCTTGAAGTACTCCGGTGATGAAATGCTAGGCG-3′); forward primer amplifying downstream of 5′-LTR (5′-CGCCTAGCATTTCATCACCGGAGTACTTCAAGAACT-3′) and reverse primer amplifying downstream of 5′-LTR (5′-ACTCGAGTGCTAGAGATTTTCCACACTGAC-3′). Finally, a full-length HIV-1 5′-LTR lacking sgRNA 4 target site fragment was amplified using the forward primer of amplifying upstream of the 5′-LTR (5′-AGGTACCTGGAAGGGCTAATTCACTCCCAA-3′) and the reverse primer of amplifying downstream of the 5′-LTR (5′-ACTCGAGTGCTAGAGATTTTCCACACTGAC-3′). The purified LTR(ΔsgRNA4)-luc PCR product was then ligated into digested pGL3-basic plasmid using KpnI and XhoI sites. All the expression plasmids were confirmed by sequencing.
Luciferase reporter assay. To examine the effects of dCas9-SunTag-VP64 or dCas9-VP64 on the HIV-1 LTR, dCas9-SunTag-VP64 or dCas9-VP64 (600 ng) with indicated sgRNA (200 ng) were cotransfected into HEK293T cells with LTR-luc (100 ng) and internal control pRL-SV40 (50 ng) using ViaFect reagent (Promega) according to the manufacturer's instructions. Transcription elongation assay between dCas9-SunTag-VP64 or dCas9-VP64 and Tat were performed as described below: the transfection mixture included 100 ng of LTR-Luc plasmid, dCas9-24×GCN4-v4 construct (300 ng), the scFv-GCN4-sfGFP-VP64 (300 ng) or dCas9-VP64 (600 ng) expression plasmid together with individual sgRNA (200 ng) or sgRNA negative vector (200 ng) and 50 ng of pRL-SV40 with or without 40 ng of the Tat expression plasmid, after 72 hours transfection, the cells were harvested and lysed, and the collected supernatant was subjected to detect luciferase activity using Dual-Luciferase Reporter Assay system (Promega). Each experiment was performed in triplicate.
Since the genome of the TZM-bl cells was integrated with a luciferase reporter driven by the HIV-1 5′-LTR promoter, we only needed to transfect dCas9-24×GCN4-v4 construct (300 ng), scFv-GCN4-sfGFP-VP64 (300 ng), or dCas9-VP64 (600 ng) together with indicated sgRNA (200 ng) into TZM-bl cells using ViaFect (Promega) according to manual instructions. Cells were harvested at 72 hours post-transfection, and the lysate was assayed for luciferase activity. Triplicate cultures were measured for each experiment.
Western blot. HEK293 T Cells were preseeded in 60-mm dish and then transfected with 8 μg of EGFP vector or with dCas9-VP64 vector using ViaFect reagent (Promega) according to the manufacturer's instructions, respectively. After 3 days transfection, cells were harvested, lysed, and subject to SDS PAGE, then transferred on N.C membrane, followed by the incubation with primary antibody against HA. Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc XRS+ System and processed using ImageLab software (Bio-Rad).
Isolation of primary CD4+ T cells. Peripheral blood mononuclear cells (from two blood units, 400 ml) isolated from healthy donors were purchased from the Shanghai Blood Center (Shanghai, China). CD4+ T cells were further purified from peripheral blood mononuclear cells by negative selection according to the manufacturer's instructions (Miltenyi). The CD4+ T cells were maintained in RPMI 1640 medium containing 2 mmol/l L-glutamine supplemented with 10% FBS (Gibco), 1% penicillin-streptomycin, and 5 ng/ml recombinant human interleukin-2 (Roche Applied Science) at 37 °C under 5% CO2.
Nucleofection and flow cytometry assay. To assess whether the two-activation toolsets could reverse HIV-1 from latency, C11 cells or J-Lat clone A10.6 cells were nucleofected with 3 μg of pcDNA3.1(-) plasmid or with dCas9-SunTag-VP64 (1.2 μg of dCas9-24GCN4-v4 and 1.2 μg of scFv-GCN4-VP64) or dCas9-VP64 (1.8 μg) and 0.6 μg of indicated sgRNA using Amaxa Cell Line Nucleofector Kit V (Lonza). The GFP-expressing cells indicated a reversal of HIV-1 from latency. Cells were washed with 1× phosphate-buffered saline (PBS) at 1,000 rpm centrifugation for 5 minutes at ambient temperature. The percentage of GFP-positive cells was measured by flow cytometry to determine the reversal level of HIV-1 from latency at 24, 48, 72, and 96 hours post-transfection. GFP expression was measured using a FACScan flow cytometer (Becton Dickinson, Mountain View, CA), and the data were analyzed using CellQuest software (Macintosh, Sunnyvale, CA).
To test the synergistic effects induced by dCas9-SunTag-VP64 system with combined sgRNAs, we nucleofected dCas9-SunTag-VP64 (1.2 μg of dCas9-24GCN4-v4 and 1.2 μg of scFv-GCN4-VP64) with individual sgRNA (100 or 200 ng) or combined sgRNA 4+5 (100 ng + 100 ng or 200 ng + 200 ng) or combined sgRNA 4–5 (133 or 233 ng) into C11 cells at indicated time. sgRNA empty plasmid was added to bring the total amount of plasmids to be equal between the single sgRNA and combined sgRNAs. pcDNA3.1(-) plasmid was used to fill up a total amount of 3 μg. At 72 hours post-transfection, the percentage of GFP-positive cells was measured by flow cytometry.
Enzyme-linked immunosorbent assay detection of antigen p24 levels. ACH2 cells were nucleofected with pcDNA3.1(-) plasmid or with dCas9-SunTag-VP64 and indicated sgRNA using the Amaxa Human T Cell Line Nucleofector kit V (Lonza). After 3-day transfection, viral reactivation were monitored via quantifying the amounts of p24 produced in supernatant by using HIV-1 p24 Antigen ELISA kit (ZeptoMetriX) according to the manufacturer's instructions. ACH2 cells were untreated or treated with 0.5 μmol/l of suberoylanilide hydroxamic acid in this study as mock and positive control, respectively.
Analysis of dCas9-SunTag-VP64 system-related cellular toxicity, global T-cell activation and latent HIV-1 reactivation-induced suicide death. Our study wanted to examine whether potent activation induced by dCas9-SunTag-VP64 system had impacts on cell proliferation, cell cycle progression, and cell death. Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) was used to measure cell proliferation after dCas9-SunTag-VP64 treatment. Briefly, Jurkat T cells or primary CD4+ T cells were seeded approximately 4 × 105 cell per well in 96-well plates and transfected with dCas9-SunTag-VP64 constructs and indicated sgRNA expression plasmid at the following day. After 72 hours transfection, 10 μl of CCK-8 solution was added to each cell and incubated for 4 hours at 37 °C. The absorbance at 450 nm was measured using a microplate reader. Each experiment was performed independently in triplicate.
In order to test the effects of suicide death on C11, J-Lat A10.6 and ACH2 cells after reactivation by dCas9-SunTag-VP64 system, respectively. The cells were nucleofected with dCas9-SunTag-VP64 with indicated sgRNA at specified time. The CellTiter-Glo luminescent cell viability assay, which is a homogeneous and sensitive method, is used to quantitate ATP produced by metabolically active cells related to the number of viable cells. Briefly, Cells were grown in 96-well plates and added with 100 μl of CellTiter-Glo reagent for 2 minutes in an orbital shaker, followed by 10 minutes at ambient temperature to stabilize luminescence signal. The luminescence in each well was measured using 96 microplate Luminometer (Promega).
To determine cell cycle distribution, 1 × 106 Jurkat T cells or primary CD4+ T cells were harvested and fixed with 500 μl precold 70% ethanol at −20 °C for 2 hours or 4 °C overnight. The following day cells were washed twice with cold phosphate-buffered saline at 1,000 rpm for 5 minutes and then stained with propidium iodide (50 μg/ml propidium iodide and 100 μg/ml RNase A in phosphate-buffered saline) at 37 °C for 30 minutes. The cell cycle analysis was performed using a FACScan flow cytometer. All experiments were performed independently in triplicate.
To determine the cell death effects induced by dCas9-SunTag-VP64, 1 × 106 Jurkat T cells, latently infected cells (C11, A10.6, and ACH2) or primary CD4+ T cells with treatment of dCas9-SunTag-VP64 system and indicated sgRNA were performed by staining propidium iodide at a final concentration of 10 μg/ml at room temperature in the dark for 5 minutes. The cells were washed with 1× PBS and analyzed using a BD FACSCanto II (Becton Dickinson) system.
To analyze the effect induced by dCas9-SunTag-VP64 on the expression of T-cell marker CD25 and CD69, 1 × 106 cells transfected with pcDNA3.1(-) plasmid or dCas9-SunTag-VP64 with indicated sgRNA were collected and incubated with 20 μl of fluorescently labeled antibodies against CD25 and CD69 in 100 μl PBS containing 1% FBS for 45 minutes on ice. Subsequently, cells were washed three times with 1× PBS and resuspended in 1 ml PBS containing 1% FBS and 10,000 cells were acquired using FACScan with Cell Quest software.
ChIP. A ChIP experiment was performed as previously described.28,30 2 × 106 C11 cells were harvested and nucleofected with dCas9-SunTag-VP64 and sgRNA 4- or sgRNA-negative vector using the Amaxa Cell Line Nucleofector Kit V. After 72 hours transfection, cells were then crosslinked with formaldehyde to a final concentration of 1% for 30 minutes at 37 °C, washed with cold 1× PBS on ice twice and then suspended in 200 µl sodium dodecyl sulfate (SDS) lysis buffer and incubated on ice for 20 minutes. Lysates were subjected to sonication to generate DNA fragments within 500–1,000 bp in length. Then 10% of the total sonicated chromatin DNA was used as the input DNA in different groups. The remaining sonicated chromatin DNA was subsequently used for incubation with antibodies against the hemagglutinin HA tag (Sigma-Aldrich) or negative antibody IgG overnight at 4 °C after preclearing samples with Protein G agarose for 30 minutes at 4 °C with slow rotation. The following day Protein G agarose beads were added to each group for 2 hours rotation at 4 °C. After incubation, the mixtures were centrifuged and washed for 5 minutes at 4 °C. Immune complexes were eluted with elution buffer, and the collected supernatants were isolated and incubated for 6 hours at 65 °C to reverse crosslinking. Meanwhile, input group was also treated to reverse crosslinking. Proteinase K was added to immunoprecipitated samples and input group, and the samples were incubated for 2 hours at 45 °C. DNA was purified and resuspended in 30 µl of water. The purified DNA product was analyzed by PCR using primers specific for HIV-1 LTR: Forward primer 5′-AGACTGCTGACATCGAGCTTTCT-3′ and reverse primer 5′-GTGGGTTCCCTAGTTAGCCAGAG-3′. The amount of PCR product amplified was measured using quantitative reverse transcription PCR on the ABI 7900 HT system with FAST SYBR Green Master Mix (Applied Biosystems).
PCR and sequencing analysis. To detect HIV-1 genomic deletions in human cells by Cas9/sgRNA4 treatment, PCR analysis were performed. Briefly, genomic DNA was extracted using a Blood & Cell Culture DNA Midi Kit (Qiagen) according to the manufacturer's instructions and then subjected to PCR analysis. The primers F-HG (5′-TGCCACCCGAAACTATTCACAAG-3′) and R-HG (5′-CCGGCATGGATTCCAGTTCTTAG-3′) were used for DNA template from HIV-1 latently infected cell clone 11 (C11) untransfected or transfected with Cas9/sgRNA negative or Cas9/sgRNA4. PCR products were analyzed by agarose gel electrophoresis and further sequencing.
Off-target analysis induced by CRISPR/Cas9. Off-target analysis was performed using bioinformatics-based search tools to determine the potential off-target sites in the human genome by CRISPR/Cas9 with sgRNA 4. Eight potential off-target sites (Supplementary Table S2) were identified by BLAST search in the NCBI database of the human genome and http://crispr.mit.edu. The primers (Supplementary Table S3) for amplifying the eight off-target sites resulted in 225–524 bp amplicons in cells mock treated or transfected with Cas9 and indicated sgRNA. The eight loci was amplified, melted, and annealed to form heteroduplex DNA. The annealed DNA was treated with five units (10U/μl) of the mismatch-sensitive T7 endonuclease 1 (T7E1) (New England BioLabs) for 15 minutes at 37 °C and then precipitated by addition of 2 μl of 0.25 M EDTA solution. The precipitated DNA was analyzed by agarose gel electrophoresis
Statistical analysis. Data are representative of three independent experiments, and error bars represent standard errors (SD). Paired samples t-tests were performed with use of SPSS version 13.0 (SPSS, Chicago, IL), and statistical significance was indicated at *P < 0.05, **P < 0.01, or ***P < 0.001.
SUPPLEMENTARY MATERIAL Figure S1. sgRNA expression vector constructs. Figure S2. The expression of dCas9-SunTag-VP64 and dCas9-VP64 construct. Figure S3. Reactivation of HIV-1 in latently infected cells by dCas9-SunTag-VP64. Figure S4. Analysis of the synergistic effects of dCas9-SunTag-VP64 with combined sgRNAs in latently infected cells. Figure S5. dCas9-VP64 mediated the reversal of HIV-1 from latency in HIV-1 latently infected cells. Figure S6. Analysis of the cytotoxicity effects of dCas9-SunTag-VP64 in Jurkat T cells at 96 h post-transfection. Figure S7. DNA sequences of PCR products from latently infected C11 cells treated with CRISPR/Cas9. Table S1. The sequence and position of the designed sgRNAs targeting the HIV-1 5′-LTR promoter are listed. Table S2. Off-target analysis for CRISPR/Cas9-mediated targeting in C11 cells. Table S3. Primers information for CRISPR/Cas9 off-target analysis.
Acknowledgments
The authors thank the NIH AIDS Research and Reference Reagent Program for providing the J-Lat clone A10.6 cells. The authors also thank Jianqing Xu (Shanghai Medical College of Fudan University) for providing the pNL4-3-EGFP plasmid. This work was supported by National High-tech Research and Development Program of China (2015AA020307), the National Grand Program on Key Infectious Disease (2014ZX10001003). All authors have contributed to their respective portions of the manuscript and reviewed the manuscript. The authors have no conflict of interest.
Supplementary Material
References
- Liu, H, Ma, Y, Su, Y, Smith, MK, Liu, Y, Jin, Y et al. (2014). Emerging trends of HIV drug resistance in Chinese HIV-infected patients receiving first-line highly active antiretroviral therapy: a systematic review and meta-analysis. Clin Infect Dis 59: 1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis-Cremer, N (2014). The emerging profile of cross-resistance among the nonnucleoside HIV-1 reverse transcriptase inhibitors. Viruses 6: 2960–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Margolis, DM, Delaney M, Greene WC, Hazuda, D and Pomerantz, RJ (2009). The challenge of finding a cure for HIV infection. Science 323: 1304–1307. [DOI] [PubMed] [Google Scholar]
- Coiras, M, López-Huertas, MR, Pérez-Olmeda, M and Alcamí, J (2009). Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7: 798–812. [DOI] [PubMed] [Google Scholar]
- Deeks, SG, Autran, B, Berkhout, B, Benkirane, M, Cairns, S, Chomont, N et al. (2012). Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarbanti, M and Battistini, A (2013). Therapeutics for HIV-1 reactivation from latency. Curr Opin Virol 3: 394–401. [DOI] [PubMed] [Google Scholar]
- Xing, S and Siliciano, RF (2013). Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discov Today 18: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, KH, Kwon, YD, Shin, HC, Hwang, MS, Ryu, EH, Park, KS et al. (2003). Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol 21: 275–280. [DOI] [PubMed] [Google Scholar]
- Beerli, RR, Dreier, B and Barbas, CF 3rd (2000). Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA 97: 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P, Qu, X, Wang, X, Zhu, X, Zeng, H, Chen, H et al. (2014). Specific reactivation of latent HIV-1 with designer zinc-finger transcription factors targeting the HIV-1 5'-LTR promoter. Gene Ther 21: 490–495. [DOI] [PubMed] [Google Scholar]
- Wang, X, Wang, P, Fu, Z, Ji, H, Qu, X, Zeng, H et al. (2015). Designed transcription activator-like effector proteins efficiently induced the expression of latent HIV-1 in latently infected cells. AIDS Res Hum Retroviruses 31: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashant, M, Luhan, Y, Esvelt, KM, John, A, Marc, G, Dicarlo, JE, et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, HK, Gu, Y, Diaz, A, Marlett, J, Takahashi, Y, Li, M et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6: 6413. [DOI] [PubMed] [Google Scholar]
- Jinek, M, East, A, Cheng, A, Lin, S, Ma, E and Doudna, J (2013). RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera, P, Kocak, DD, Vockley, CM, Adler, AF, Kabadi, AM, Polstein, LR et al. (2013). RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress, BF, Toparlak, ÖD, Guleria, S, Lebovich, M, Stieglitz, JT, Englaender, JA et al. (2015). CRISPathBrick: Modular Combinatorial Assembly of Type II-A CRISPR Arrays for dCas9-Mediated Multiplex Transcriptional Repression in E. coli. ACS Synth Biol 4: 987–1000. [DOI] [PubMed] [Google Scholar]
- Tanenbaum, ME, Gilbert, LA, Qi, LS, Weissman, JS and Vale, RD (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S, Brigham, MD, Trevino, AE, Joung, J, Abudayyeh, OO, Barcena, C et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Lei, Y, Wong, WK, Liu, S, Lee, KC, He, X et al. (2014). Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res 42: 4375–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, AW, Wang, H, Yang, H, Shi, L, Katz, Y, Theunissen, TW et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, LS, Larson, MH, Gilbert, LA, Doudna, JA, Weissman, JS, Arkin, AP et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak, RA, Calnan, BJ, Frankel, AD, and Sharp, PA (1990). HIV-1 Tat protein trans-activates transcription in vitro. Cell 63: 791–802. [DOI] [PubMed] [Google Scholar]
- Wei, P, Garber, ME, Fang, SM, Fischer, WH and Jones, KA (1998). A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462. [DOI] [PubMed] [Google Scholar]
- Van Herreweghe, E, Egloff, S, Goiffon, I, Jády, BE, Froment, C, Monsarrat, B et al. (2007). Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 26: 3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy, SR, Goncalves, J, Coelho, S, Segal, DJ, Berkhout, B and Barbas, CF 3rd (2006). Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol 80: 2873–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, EJ, Bilska, M, Kozak, SL, Kabat, D and Montefiori, DC (2009). Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83: 8289–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D, Qu, X, Li, L, Zhou, X, Liu, S, Lin, S et al. (2013). Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology 440: 182–189. [DOI] [PubMed] [Google Scholar]
- Qu, X, Wang, P, Ding, D, Li, L, Wang, H, Ma, L et al. (2013). Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res 41: 7771–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P, Qu, X, Wang, X, Liu, L, Zhu, X, Zeng, H et al. (2013). As2O3 synergistically reactivate latent HIV-1 by induction of NF-κB. Antiviral Res 100: 688–697. [DOI] [PubMed] [Google Scholar]
- Jordan, A, Bisgrove, D and Verdin, E (2003). HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh, EJ, Maury, WJ, Folks, TM, Fauci, AS and Rabson, AB (1989). Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA 86: 5974–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancotto, A, Grivel, JC, Gondois-Rey, F, Bettendroffer, L, Vigne, R, Brown, S et al. (2004). Dual role of prostratin in inhibition of infection and reactivation of human immunodeficiency virus from latency in primary blood lymphocytes and lymphoid tissue. J Virol 78: 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Scott, DA, Weinstein, JA, Ran, FA, Konermann, S, Agarwala, V et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, EE, Wang, J, Miller, JC, Jouvenot, Y, Kim, KA, Liu, O et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Horlbeck, MA, Adamson, B, Villalta, JE, Chen, Y, Whitehead, EH et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, ND, Edwards, NL, Duckett, CS, Agranoff, AB, Schmid, RM and Nabel, GJ (1993). A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12: 3551–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, IB, D'Ippolito, AM, Vockley, CM, Thakore, PI, Crawford, GE, Reddy, TE et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Cong, L, Yan, WX, Scott, DA, Gootenberg, JS, Kriz, AJ et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, BP, Prew, MS, Tsai, SQ, Topkar, VV, Nguyen, NT, Zheng, Z et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz, HK, Amstutz, P, Kohl, A, Stumpp, MT, Briand, C, Forrer, P et al. (2004). High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol 22: 575–582. [DOI] [PubMed] [Google Scholar]
- Roshal, M, Zhu, Y and Planelles, V (2001). Apoptosis in AIDS. Apoptosis 6: 103–116. [DOI] [PubMed] [Google Scholar]
- Zhang, Y, Yin, C, Zhang, T, Li, F, Yang, W, Kaminski, R et al. (2015). CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep 5: 16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L, Deng, K, Shroff, NS, Durand, CM, Rabi, SA, Yang, HC et al. (2012). Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, F and Grimm, D (2015). CRISPR genome engineering and viral gene delivery: a case of mutual attraction. Biotechnol J 10: 258–272. [DOI] [PubMed] [Google Scholar]
- Fine, EJ, Appleton, CM, White, DE, Brown, MT, Deshmukh, H, Kemp, ML et al. (2015). Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes. Sci Rep 5: 10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman, SM, Lazar, DC, Scott, TA, Hart, JR, Takahashi, M, Burnett, JC, et al. (2015). Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther 23: 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsirichai, P, Gaj, T and Schaffer, DV (2015). CRISPR-mediated activation of latent HIV-1 expression. Mol Ther 23: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.