Abstract
Background
Recently, microRNA-21 (miR-21) has been reported to be associated with prognosis of pancreatic ductal adenocarcinoma (PDAC). The present studies aimed to evaluate the prognostic value of miR-21 for PDAC with meta-analysis.
Methods
A systematic search in the PubMed and other databases was conducted to identify eligible studies. The pooled hazard ratios (HRs) with 95 % confidence interval (CI) were calculated. The meta-analysis was conducted using the STATA 12.0 software.
Results
A total of 12 articles (13 studies) which included 963 cases were selected for the meta-analysis. Elevated miR-21 expression was significantly predictive of poor overall survival (HR = 2.05, 95 % CI 1.71–2.46, P < 0.001). In the subgroup analyses, similar results were observed in Asian (HR = 2.09, 95 % CI 1.62–2.71, P < 0.001) and Caucasian (HR = 2.36, 95 % CI 1.53–3.65, P < 0.001); in tissue sample (HR = 2.14, 95 % CI 1.73–2.65, P < 0.001) and serum sample (HR = 1.84, 95 % CI 1.30–2.60, P = 0.001); with quantitative real-time polymerase chain reaction assay method (HR = 2.31, 95 % CI 1.86–2.86, P < 0.001); and in patients receiving adjuvant chemotherapy (HR = 2.37, 95 % CI 1.88–3.00, P < 0.001). The association between miR-21 expression level and lymph node metastasis was statistically significant (OR = 1.45, 95 % CI 1.02–2.06, P = 0.038). However, no significant relationship between miR-21 expression level and sex or vascular invasion or neural infiltration was observed (P > 0.05).
Conclusions
Our meta-analysis indicated that elevated miR-21 expression level can predict poor prognosis in patients with PDAC.
Keywords: MicroRNA-21, Prognosis, Pancreatic ductal adenocarcinoma, Meta-analysis
Background
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignancies and the fourth cause of cancer-related death [1]. Despite the advancement in medical and surgical managements, the outcome of PDAC remains disappointing. Only 15 % of patients can undergo radically surgical resection [2] and approximately 6 % of patients can survive for 5 years following diagnosis [1]. The poor prognosis of PDAC primarilyattributes to the facts including late clinical manifestation, aggressive local invasion, high metastatic potential, and chemotherapy or radiotherapy resistance. These challenges motivate researchers to seek better approaches for PDAC, including finding out new biomarkers which can identify different phenotypes with differences in clinical feature and prognosis.
MicroRNAs are a class of small non-coding RNA, approximately 20 nucleotides in length [3]. By base pairing with the 3′-untranslated region (UTR) of target messenger RNAs, microRNAs act as endogenous regulators of protein-coding genes at transcriptional or post-transcriptional level [3, 4]. Thus, microRNAs (miRNAs) are involved in various biological processes, such as differentiation, proliferation, cell cycle regulation, and metabolism [5, 6]. More importantly, mounting evidences suggest that microRNAs are involved in cancer development and progression including activation of oncogenes and inactivation of tumor suppressor genes [7, 8].
MiR-21 is the most frequently observed cancer-related microRNA. Previous studies have observed that miR-21 dysregulates in many cancers and acts as a key factor mediating the growth, development, and progression of tumors [9–11], which is expected to be a novel predictor and target. However, evidence for the prognostic role of miR-21 expression in PDAC was still lacking. Thus, we conduct this meta-analysis to evaluate the prognostic value of miR-21 for PDAC.
Methods
Search strategy
A systematic search in the PubMed, ISI Web of Science, and EMBASE databases was performed to identify studies published before February 2016. The detailed keywords for the PubMed searches were as follows:
miR-21.
miRNA-21.
microRNA-21.
pancreatic cancer.
pancreatic tumor.
pancreatic carcinoma.
prognos*.
survival.
outcomes.
#1 OR #2 OR #3
#4 OR #5 OR #6
#7 OR #8 OR #9
#10 AND #11 AND #12
All of the studies which fulfilled the following criteria were included in the meta-analysis: (1) studied PADC based on histopathological confirmation; (2) expression of miR-21 was measured; (3) the association between expression of miR-21 level and survival outcome was studied. Studies were excluded based on the following criteria: (a) reviews, letters or laboratory studies, unpublished studies with only the abstracts presented at national and international meetings; (b) studies had overlapping or duplicate data; (c) absence of necessary information to calculate HR. All references of retrieved articles were manually selected to identify all the potential studies. The language of the articles was limited to English. A flow diagram of the study selection process is presented in Fig. 1.
Fig. 1.
The PRISMA flowchart of literature review
Data extraction
Two reviewers (FT and KWJ) independently extracted the data following the guidelines of a critical review checklist of the Dutch Cochrane Centre proposed by Meta-analysis of Observational Studies in Epidemiology (MOOSE) [12] and decided upon the controversial issues through discussion. For disagreements, a consensus was reached by a third investigator (WW). The following information was extracted from each study: author, published year, country or area, ethnicity, sample size, sample type, TNM stage, detection method, cutoff value, follow-ups, adjuvant therapy, HRs with 95 % confidence intervals (CIs), and P value of miR-21 for overall survival (OS). If not available, data were extracted using the method described by Tierney et al. [13].
Statistical analysis
Pooled hazard ratio (HR) of miR-21 expression for OS was calculated. The heterogeneity was checked using I2 statistic described by Higgins [14]. A P < 0.05 or I2 > 50 % for Q test suggested significant heterogeneity among studies. If high heterogeneity exists among studies, pooled effect was calculated using the random-effects model (DerSimonian–Laird method) [15], Otherwise, the fixed-effects model (Mantel–Haenszel method) was used [16]. Publication bias was assessed using the funnel plot. Subgroup analysis of PADC patients with elevated miR-21 expression were examined with respect to gender (male vs. female), lymph node metastasis (positive vs. negative), vascular invasion (positive vs. negative), and neural infiltration (positive vs. negative). Data analyses were performed using STATA software version 12.0 (STATA Corporation, College Station, TX, USA). P < 0.05 was considered statistically significant.
Results
Summary of included study characteristics
A total of 206 published articles were initially retrieved in the databases of PubMed, EMBASE, and Web of Science. After abstract review, 189 articles were removed which left 17 articles for full-text evaluation. Finally, 12 articles (13 studies) remained which excluded 5 articles following full-text evaluation [17–28], Fig. 1. The main characteristics and results of the eligible studies are summarized in Table 1. These studies investigated a total of 963 cases from USA, China, Italy, UK, Korea, Japan, Greece, and Germany. MiR-21 expression was detected by either quantitative real-time polymerase chain reaction assay (qRT-PCR) or in situ hybridization (ISH) with 6 in FFPE, 2 in cancerous tissue, 2 in frozen tissue, and 3 in serum. The cutoff values of miR-21 varied in each study. Median and mean values were extracted from 9 studies; 2-fold values or score > 2 or score > 1 were considered in the remaining 4 studies and Liu et al. did not report the cutoff value [22]. The technical details in the detection of miR-21 expression are summarized in Table 2.
Table 1.
The characteristics of the pooled studies
| Study | Year | Country | Ethnicity | Sample size | Stage | Results | Survival analysis | HR (95 % CI) for prognostic outcomes | Adjuvant chemotherapy |
|---|---|---|---|---|---|---|---|---|---|
| Dillhoff | 2008 | USA | Caucasian | 80 | NR | OS | KM | OS, 4.08 (1.98, 8.38), P = 0.037 | NR |
| Giovannetti a | 2010 | Italy | Caucasian | 28 | I–III | OS, DFS | U | OS, 3.1 (1.4–7.3), P = 0.008 | Y |
| DFS, 4.4 (1.8–10.7), P = 0.001 | |||||||||
| Giovannetti b | 2010 | Italy | Caucasian | 31 | IV | OS, PFS | U | OS, 3.1 (1.4–7.1), P = 0.01 | Y |
| PFS, 2.4 (1.1–5.3), P = 0.03 | |||||||||
| Hwang | 2010 | Korea | Asian | 82 | II–IV | OS, DFS | M | OS, 2.26 (1.34, 3.80), P = 0.002 | Y |
| DFS, 2.793 (1.466–5.319), P = 0.002 | |||||||||
| Jamieson | 2012 | UK | Caucasian | 48 | II–IV | OS | M | OS, 3.22 (1.21–8.58), P = 0.019 | Y |
| Liu | 2012 | China | Asian | 38 | I–IV | OS | KM | OS, 2.89 (1.22–6.81), P = 0.02 | NR |
| Nagao | 2012 | Japan | Asian | 65 | I–IV | OS | U | OS, 2.32 (1.19–4.52), P = 0.045 | NR |
| Wang | 2013 | China | Asian | 177 | III–IV | OS | M | OS, 1.71 (1.15–2.54), P = 0.008 | Y |
| Kadera | 2013 | USA | Caucasian | 145 | I–IV | OS | U | OS, 1.1 (0.7–1.6), P = 0.7 | NR |
| Papaconstantinou | 2013 | Greece | Caucasian | 88 | I–IV | OS | M | OS, 1.72 (1.25–12.3), P = 0.019 | NR |
| Ma | 2013 | China | Asian | 78 | I–IV | OS | M | OS, 2.60 (1.15–5.87), P = 0.021 | NR |
| Dhayat | 2015 | Germany | Caucasian | 91 | II | OS, RFS | M | OS, 3.06 (1.37–6.85), P = 0.0064 | Y |
| RFS, 2.25 (1.06–4.77), P = 0.0338 | |||||||||
| Khan | 2015 | UK | Caucasian | 12 | IV | OS, PFS | KM | OS, 1.46 (0.41–5.21), P = 0.564 | Y |
| PFS, 4.7 (1.1–19.7), P = 0.032 |
OS overall survival, DFS disease-free survival, RFS relapse-free survival, PFS progression-free survival, KM Kaplan–Meier analysis, U univariate analysis, M multivariate analysis, Y yes, NR not reported
Table 2.
The technical details in detection of miR-21 expression
| Study | Sample size | Sample | Sample technique | Method | Endogenous control | Cutoff |
|---|---|---|---|---|---|---|
| Dillhoff | 80 | FFPE | MDS | ISH | RNU6 | Score > 1 |
| Giovannetti a | 28 | Tissue | MDS | RT-PCR | RNU43 | Median |
| Giovannetti b | 31 | Tissue | MDS | RT-PCR | RNU44 | Median |
| Hwang | 82 | FFPE | MDS | RT-PCR | RNU66 or RNU43 | Median |
| Jamieson | 48 | FT | NR | RT-PCR | RNU6 | Median |
| Liu | 38 | Serum | RT-PCR | NR | NR | |
| Nagao | 65 | FFPE | NR | RT-PCR | RNU6 | Mean |
| Wang | 177 | Serum | RT-PCR | RNU7 | Median | |
| Kadera | 145 | FFPE | NR | ISH | RNU6 | Median |
| Papaconstantinou | 88 | FFPE | NR | RT-PCR | RNU6 | Mean |
| Ma | 78 | FT | NR | RT-PCR | RNU6 | ≥2-fold change |
| Dhayat | 91 | FFPE | NR | RT-PCR | NR | Mean |
| Khan | 12 | Serum | ISH | NR | Median |
FFPE formalin-fixed paraffin-embedded, FT frozen tissue, MD microdissected sample, ISH in situ hybridization, qRT-PCR quantitative real-time polymerase chain reaction assay, NR not reported
Correlation between miR-21 expression and survival outcome
Elevated miR-21 expression was significantly predictive of poor OS (HR = 2.05, 95 % CI 1.71–2.46, P < 0.001), Fig. 2. In the subgroup analyses by ethnicity, no matter the cases were Asian or Caucasian, the high-level miR-21 expression was still a significantly poor predictor for OS (Asian, HR = 2.09, 95 % CI 1.62–2.71, P < 0.001; Caucasian, HR = 2.36, 95 % CI 1.53–3.65, P < 0.001), Fig. 3. We also observed that the high-level miR-21 expression was associated with poor OS both in tissue sample (HR = 2.14, 95 % CI 1.73–2.65, P < 0.001) and serum sample (HR = 1.84, 95 % CI 1.30–2.60, P = 0.001), Fig. 4. Further analyses of studies revealed that the high-level miR-21 expression detected by the qRT-PCR method predicted poor OS (HR = 2.31, 95 % CI 1.86–2.86, P < 0.001), while similar result did not arise in high-level miR-21 expression detected by the ISH method (HR = 1.86, 95 % CI 0.73–4.74, P = 0.191), Fig. 5. In patients receiving adjuvant chemotherapy, high expression of miR-21 is also associated with poor prognosis (HR = 2.37, 95 % CI 1.88–3.00, P < 0.001), Fig. 6.
Fig. 2.
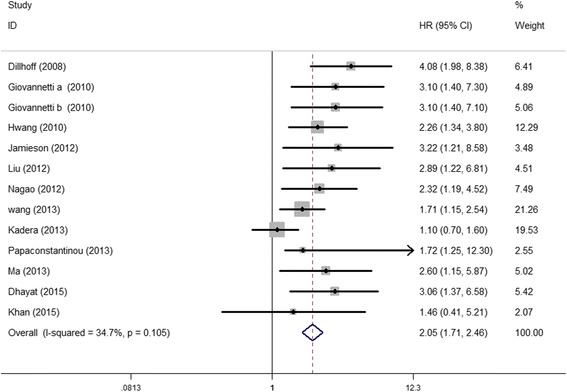
The relationship between elevated miR-21 level and OS
Fig. 3.
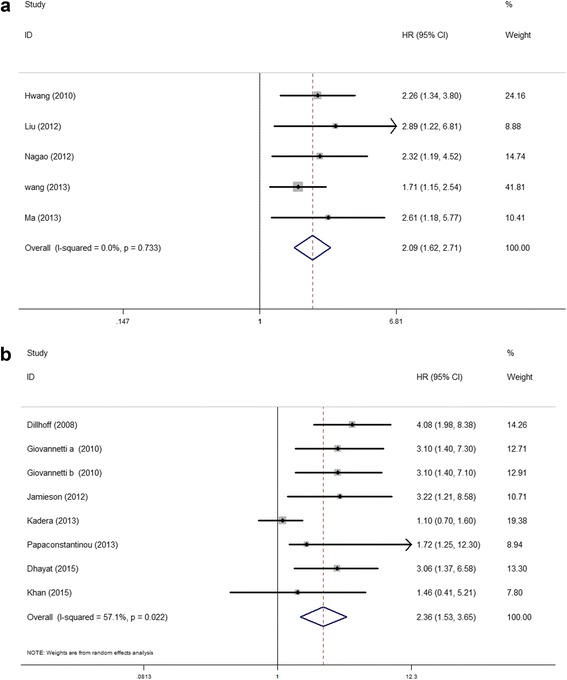
Subgroup analyses of relationship between elevated miR-21 level and OS by ethnicity. a Asian. b Caucasian
Fig. 4.
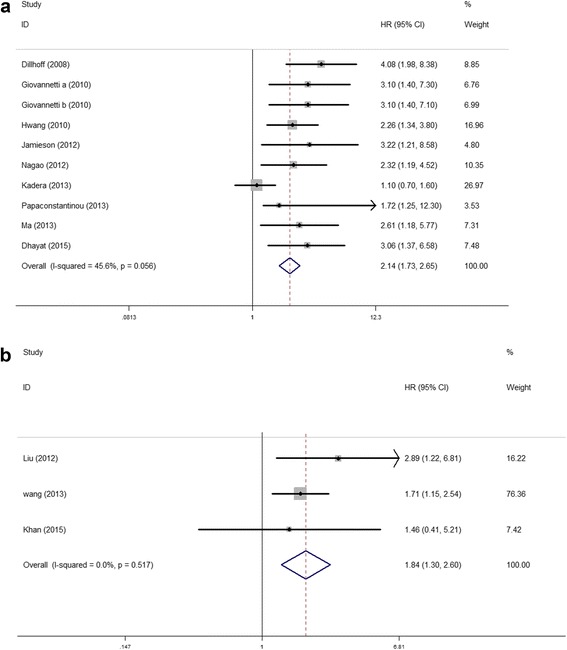
Subgroup analyses of relationship between elevated miR-21 level and OS by sample type. a Tissue. b Serum
Fig. 5.
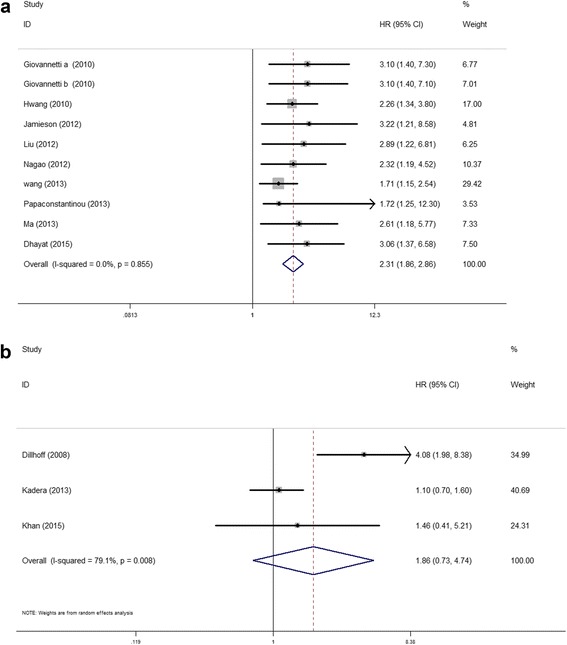
Subgroup analyses of relationship between elevated miR-21 level and OS by detection method. a qRT-PCR. b ISH
Fig. 6.
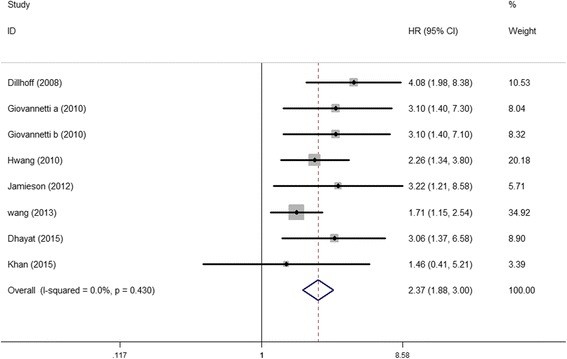
The relationship between elevated miR-21 level and OS in patients with chemotherapy
Correlation between miR-21 expression and clinical characteristics
There were 4 studies that reported correlations between miR-21 expression and some clinical characteristics (sex, lymph node metastasis, vascular invasion, and neural infiltration). The association between miR-21 expression level and lymph node metastasis was statistically significant (OR = 1.45, 95 % CI 1.02–2.06, P = 0.038). However, no significant relationship between miR-21 expression level and sex or vascular invasion or neural infiltration was observed, Table 3.
Table 3.
Correlation between miR-21 expression and clinical characteristics
| Variables | Number of studies | Model | OR(95 % CI), P value | Heterogeneity (I2, P-value) |
|---|---|---|---|---|
| Gender (male vs. female) | 4 | Fixed | 1.05(0.75–1.47), 0.78 | 0.0 %, 1.00 |
| Lymph node metastasis (positive vs. negative) | 4 | Fixed | 1.45(1.02–2.06),0.04 | 0.0 %, 0.70 |
| Vascular infiltration (yes vs. no) | 3 | Fixed | 0.91(0.47–1.76),0.78 | 0.0 %,0.79 |
| Neural infiltration (yes vs. no) | 3 | Fixed | 1.30(0.80–2.10),0.29 | 0.0 %,0.67 |
Publication bias and sensitivity analysis
The publication bias of the pooled studies was evaluated by funnel plots. Visual inspection of the funnel plots was almost symmetric in studies reported OS, Fig. 7. Sensitivity analysis was performed by exclusion of the highest weighted study and there was no individual study dominantly influenced overall HR.
Fig. 7.
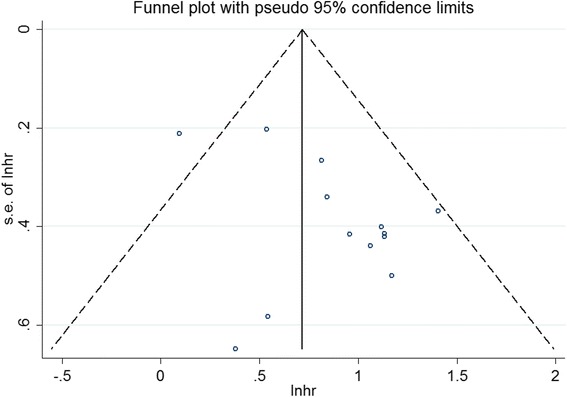
Funnel plot for publication bias analysis based on OS
Discussion
Recently, miR-21 has gain wide attentions in cancer research for its crucial role in gene regulation and cancer development. The potential role of miR-21 as a novel diagnostic or prognostic biomarker has proved by accumulated evidence in various types of cancer, such as colon cancer, gastric cancer, breast cancer, and lung cancer [29–32]. Regarding PDAC, although the potential diagnostic effect of miR-21 had already been reported [22, 33, 34], the prognostic role is still contingent. In addition, the relation between the expression of miR-21 and patients’ clinical characteristics still needs to be defined.
In this meta-analysis, the pooled HR from 963 patients in 13 studies for OS was 2.05 (95 % CI 1.71 to 2.46, P < 0.01), which was considered strongly predictive [35]. Our present meta-analysis found strongly predictive value of miR-21 both in Asian and Caucasian patients; thus, we assume that it may be applied as a general prognostic marker regardless of ethnicity. An ideal prognostic biomarker should be convenient, sensitive, and credible. The majority of pooled studies detected the miR-21 expression in the tissue with qRT-PCR method. Elevated expression of miR-21 in the serum sample also predicted poor prognosis as tissue sample, which may be favored in the clinical management of PDAC. Conversely, in the subgroup analysis of ISH method, elevated expression of miR-21 did not imply poor outcomes. This result should be elaborated causing limited pooled studies using ISH method with a different cutoff value. Therefore, further studies with consistent normalization are warranted. Though we did not report the pooled relationship of miR-21 with disease-free survival, relapse-free survival, and progression-free survival due to limited studies for analysis, several studies demonstrated the significant prediction of miR-21 on these prognostic outcomes [18, 19, 27, 28]. Moreover, the patients’ clinical characteristics, including male gender, positive vascular infiltration, and positive neural infiltration showed no correlation with high miR-21 expression, but positive lymph node metastasis showed significant correlations. This important feature was consistent with several other malignancies [36, 37]. Kadera et al. reported that pancreatic cancer cells induce the fibroblasts with high expression of miR-21 and increase the invasive potential as manifested by lymph node metastasis [21].
As potential mechanisms of enhanced PDAC invasion, miR-21 has been identified to target on the phosphatase and tensin homolog (PTEN). The PTEN gene in humans generally acts as a tumor suppressor gene through the actions of its phosphatase protein product [38], which plays a critical role in cell cycle arrest, cell migration, cell spreading, and cell invasions. Aberrant expression of PTEN was associated with the development and progression of multiple tumors [39, 40]. Meng et al. found that inhibition of miR-21 in cultured HCC cells increased the expression of PTEN [41]. Giovannetti et al. reported that transfection with pre-miR-21 resulted in the reduction of PTEN expression [18], suggesting that PTEN was a direct target of miR-21. The reversion-inducing-cysteine-rich protein with Kazal motifs (RECK) is another potential target gene of miR-21 identified recently. RECK functions as a metastasis suppressor by membrane-bound inhibiting MMPs, which play an important role in extracellular matrix remodeling during tumor progression [42, 43]. Zhao et al. reported that miR-21 modulated the RECK expression through directly binding to RECK 3′-UTR [44]. Down-regulation of RECK expression was observed in tumors including PDAC when the expression of miR-21 is elevated [45, 46].
Several clinical trials have approved that adjuvant chemotherapy can effectively improve the outcome of PDAC [47, 48]. The gemcitabine or 5-fluorouracil-based chemotherapy was considered as the most effective regimen till now, while the response to the adjuvant chemotherapy remains unsatisfactory. The dense fibrotic bulk of PDAC appears to impede the delivery of chemotherapeutic drugs to cancer cells. More importantly, miR-21 expression is likely to be correlated with chemoresistance as growing evidence suggests that aberrant miR-21 expression strongly cripples the response to the chemotherapy [49–51]. The present meta-analysis revealed that elevated expression of miR-21 was associated with shorter OS in patients with chemotherapy as well. Higher expression of miR-21 was detected in PDAC cells with the higher 50 % inhibitory concentration values of gemcitabine or 5-fluorouracil [18, 19]. Meanwhile, antisense inhibition of miR-21 induced the reduction of cancer cell proliferation, invasion, and chemoresistance against gemcitabine and 5-fluorouracil [52].
Apoptotic evasion is considered to be one of the main causes of chemoresistance. Previous studies have revealed that overexpression of miR-21 can down-regulate PTEN expression and consequently activate the PI3K/Akt signaling pathway, while anti-miR-21 strategy strongly reduced phospho-AKT levels and enhanced apoptosis when used in combination with gemcitabine [18]. Furthermore, Toste et al. identified an inverse correlation between miR-21 and p85α [53], a major negative regulatory subunit of PI3K/Akt signaling [54], which was likely a potential mechanism for the known relationship between miR-21 expression and PI3K/Akt signaling. Beyond this, Wang et al. reported that miR-21 directly regulates the FasL/Fas pathway by binding the 3′-UTR region of the FasL messenger RNA (mRNA). Additionally, ectopic expression of FasL significantly abrogated the miR-21-induced chemoresistance [26]. These evidences may also partially explain that high expression of miR-21 predicts poor prognosis.
Although miR-21 holds a great promise as a novel prognostic biomarker for PADC, several limitations need to be addressed in practical application. Firstly, statistical heterogeneity among pooled studies, which may be derived from different ethnicities (Caucasian vs. Asian), sample types (tissue vs. serum), treatment (without chemotherapy vs. with gemcitabine or 5-fluorouracil), different detection methods (qRT-PCR vs. ISH), decreased the plausibility of the results. Secondly, only studies published in English were pooled in this meta-analysis which may omit some valuable articles on this issue. Thirdly, miR-21 as a novel prognostic marker of PDAC just looms in recent years, and still limited research work was done. So, the study size obtained in this meta-analysis was relatively small.
Conclusions
In conclusion, the current meta-analysis demonstrated that miR-21 might serve as a potential prognostic marker of PDAC. Due to some limitations of this study, more complementary researches are awaited to validate our results.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WW and GYH designed the study. GYH, FT, and KWJ collected the data and performed the statistical analysis. GYH and FT drafted the manuscript. WW revised and proofread the manuscript. All authors read and approved the final manuscript.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Camacho D, Reichenbach D, Duerr GD, Venema TL, Sweeney JF, Fisher WE. Value of laparoscopy in the staging of pancreatic cancer. JOP. 2005;6:552–561. [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Redis RS, Berindan-Neagoe I, Pop VI, Calin GA. Non-coding RNAs as theranostics in human cancers. J Cell Biochem. 2012;113:1451–1459. doi: 10.1002/jcb.24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Blelloch R. Cell cycle regulation by microRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al: A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. [DOI] [PMC free article] [PubMed]
- 8.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Zhang W, Lv Q, Zhu D. Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncol Rep. 2015;33:3108–3116. doi: 10.3892/or.2015.3931. [DOI] [PubMed] [Google Scholar]
- 10.Ma X, Conklin DJ, Li F, Dai Z, Hua X, Li Y, Xu-Monette ZY, Young KH, Xiong W, Wysoczynski M, et al: The oncogenic microRNA miR-21 promotes regulated necrosis in mice. Nat Commun. 2015;6:7151. [DOI] [PMC free article] [PubMed]
- 11.Lou Y, Yang X, Wang F, Cui Z, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 2010;26:819–827. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 12.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB: Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–12. [DOI] [PubMed]
- 13.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 17.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, et al: MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–38. [DOI] [PubMed]
- 19.Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, et al: Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. [DOI] [PMC free article] [PubMed]
- 20.Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter CR, Sansom OJ, Evans TR, McKay CJ, Oien KA: MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18:534–45. [DOI] [PubMed]
- 21.Kadera BE, Li L, Toste PA, Wu N, Adams C, Dawson DW, Donahue TR: MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS One. 2013;8:e71978. [DOI] [PMC free article] [PubMed]
- 22.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, et al: Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–8. [DOI] [PubMed]
- 23.Ma MZ, Kong X, Weng MZ, Cheng K, Gong W, Quan ZW, Peng CH: Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. [DOI] [PMC free article] [PubMed]
- 24.Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu S, Hamada T, Fukuyama T, Nakano R, Uchiyama A, Kawamoto M, Yamaguchi K, Hashimoto H: Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol. 2012;25:112–21. [DOI] [PubMed]
- 25.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D: Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. [DOI] [PubMed]
- 26.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z: The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334–45. [DOI] [PMC free article] [PubMed]
- 27.Dhayat SA, Abdeen B, Kohler G, Senninger N, Haier J, Mardin WA. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin Epigenetics. 2015;7:132. doi: 10.1186/s13148-015-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan K, Cunningham D, Peckitt C, Barton S, Tait D, Hawkins M, Watkins D, Starling N, Rao S, Begum R, et al: miR-21 expression and clinical outcome in locally advanced pancreatic cancer: exploratory analysis of the pancreatic cancer erbitux, radiotherapy and UFT (PERU) trial. Oncotarget. 2016. doi:10.18632/oncotarget.7208. [DOI] [PMC free article] [PubMed]
- 29.Xia X, Yang B, Zhai X, Liu X, Shen K, Wu Z, Cai J: Prognostic role of microRNA-21 in colorectal cancer: a meta-analysis. PLoS One. 2013;8:e80426. [DOI] [PMC free article] [PubMed]
- 30.Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of microRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668–1674. doi: 10.12659/MSM.892096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Zhang Y, Pan C, Ma F, Zhang S. Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One. 2015;10:e0118647. doi: 10.1371/journal.pone.0118647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu K, Wei L, Chu M: The prognostic value of miR-21 and miR-155 in non-small-cell lung cancer: a meta-analysis. Jpn J Clin Oncol. 2013;43:813–20. [DOI] [PubMed]
- 33.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang JY, Sun YW, Liu DJ, Zhang JF, Li J, Hua R. MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer. Am J Cancer Res. 2014;4:663–673. [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–392. doi: 10.1023/A:1014778713034. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Sun J, Xu J, Li Q, Guo Y, Zhang Q. miR-21 is a promising novel biomarker for lymph node metastasis in patients with gastric cancer. Gastroenterol Res Pract. 2012;2012:640168. doi: 10.1155/2012/640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY: MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. [DOI] [PMC free article] [PubMed]
- 38.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Chu EC, Tarnawski AS. PTEN regulatory functions in tumor suppression and cell biology. Med Sci Monit. 2004;10:Ra235–241. [PubMed] [Google Scholar]
- 40.Dong Y, Richards JA, Gupta R, Aung PP, Emley A, Kluger Y, Dogra SK, Mahalingam M, Wajapeyee N: PTEN functions as a melanoma tumor suppressor by promoting host immune response. Oncogene. 2014;33:4632–42. [DOI] [PubMed]
- 41.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, Nishimura S, Imamura Y, Kitayama H, Alexander DB, et al: The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell. 2001;107:789–800. [DOI] [PubMed]
- 43.Reis ST, Pontes-Junior J, Antunes AA, Dall’Oglio MF, Dip N, Passerotti CC, Rossini GA, Morais DR, Nesrallah AJ, Piantino C, et al: miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012;12:14. [DOI] [PMC free article] [PubMed]
- 44.Zhao W, Dong Y, Wu C, Ma Y, Jin Y, Ji Y. MiR-21 overexpression improves osteoporosis by targeting RECK. Mol Cell Biochem. 2015;405:125–133. doi: 10.1007/s11010-015-2404-4. [DOI] [PubMed] [Google Scholar]
- 45.Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang N, Zhang CQ, He JH, Duan XF, Wang YY, Ji X, Zang WQ, Li M, Ma YY, Wang T, Zhao GQ: MiR-21 down-regulation suppresses cell growth, invasion and induces cell apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal carcinoma. Dig Dis Sci. 2013;58:1863–70. [DOI] [PubMed]
- 47.Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, et al: Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–77. [DOI] [PubMed]
- 48.Valle JW, Palmer D, Jackson R, Cox T, Neoptolemos JP, Ghaneh P, Rawcliffe CL, Bassi C, Stocken DD, Cunningham D, et al: Optimal duration and timing of adjuvant chemotherapy after definitive surgery for ductal adenocarcinoma of the pancreas: ongoing lessons from the ESPAC-3 study. J Clin Oncol. 2014;32:504–12. [DOI] [PubMed]
- 49.Chan JK, Blansit K, Kiet T, Sherman A, Wong G, Earle C, Bourguignon LY: The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol. 2014;132:739–44. [DOI] [PubMed]
- 50.Xu L, Huang Y, Chen D, He J, Zhu W, Zhang Y, Liu X: Downregulation of miR-21 increases cisplatin sensitivity of non-small-cell lung cancer. Cancer Genet. 2014;207:214–20. [DOI] [PubMed]
- 51.Oue N, Anami K, Schetter AJ, Moehler M, Okayama H, Khan MA, Bowman ED, Mueller A, Schad A, Shimomura M, et al: High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. 2014;134:1926–34. [DOI] [PMC free article] [PubMed]
- 52.Zhao Y, Zhao L, Ischenko I, Bao Q, Schwarz B, Niess H, Wang Y, Renner A, Mysliwietz J, Jauch KW, et al: Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target Oncol. 2015;10(4):535–48. [DOI] [PubMed]
- 53.Toste PA, Li L, Kadera BE, Nguyen AH, Tran LM, Wu N, Madnick DL, Patel SG, Dawson DW, Donahue TR: p85alpha is a microRNA target and affects chemosensitivity in pancreatic cancer. J Surg Res. 2015;196:285–93. [DOI] [PMC free article] [PubMed]
- 54.Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JH, Patel SG, Wu N, Hindoyan A, Farrell JJ, et al: Integrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352–63. [DOI] [PMC free article] [PubMed]


