Abstract
Background
The dynamic changes that occur in protein expression after treatment of a cancer in vivo are poorly described. In this study we measure the effect of chemotherapy over time on the expression of a panel of proteins in ovarian cancer xenograft models. The objective was to identify phosphoprotein and other protein changes indicative of pathway activation that might link with drug response.
Methods
Two xenograft models, platinum-responsive OV1002 and platinum-unresponsive HOX424, were used. Treatments were carboplatin and carboplatin-paclitaxel. Expression of 49 proteins over 14 days post treatment was measured by quantitative immunofluorescence and analysed by AQUA.
Results
Carboplatin treatment in the platinum-sensitive OV1002 model triggered up-regulation of cell cycle, mTOR and DDR pathways, while at late time points WNT, invasion, EMT and MAPK pathways were modulated. Estrogen receptor-alpha (ESR1) and ERBB pathways were down-regulated early, within 24 h from treatment administration. Combined carboplatin-paclitaxel treatment triggered a more extensive response in the OV1002 model modulating expression of 23 of 49 proteins. Therefore the cell cycle and DDR pathways showed similar or more pronounced changes than with carboplatin alone. In addition to expression of pS6 and pERK increasing, components of the AKT pathway were modulated with pAKT increasing while its regulator PTEN was down-regulated early. WNT signaling, EMT and invasion markers were modulated at later time points. Additional pathways were also observed with the NFκB and JAK/STAT pathways being up-regulated. ESR1 was down-regulated as was HER4, while further protein members of the ERBB pathway were upregulated late. By contrast, in the carboplatin-unresponsive HOX 424 xenograft, carboplatin only modulated expression of MLH1 while carboplatin-paclitaxel treatment modulated ESR1 and pMET.
Conclusions
Thirteen proteins were modulated by carboplatin and a more robust set of changes by carboplatin-paclitaxel. Early changes included DDR and cell cycle regulatory proteins associating with tumor volume changes, as expected. Changes in ESR1 and ERBB signaling were also observed. Late changes included components of MAPK signaling, EMT and invasion markers and coincided in time with reversal in tumor volume reduction. These results suggest potential therapeutic roles for inhibitors of such pathways that may prolong chemotherapeutic effects.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2212-6) contains supplementary material, which is available to authorized users.
Keywords: Ovarian cancer, Carboplatin, Paclitaxel, Xenograft, Prognosis, Phosphoproteins
Background
Ovarian cancer is a leading cause of gynecologic cancer mortality worldwide. Platinum-based drugs, often combined with a taxane, are the main chemotherapy treatment that follows surgical debulking [1]. Although the majority of patients initially respond to this treatment, around 70 % relapse and present with recurrent disease between 6 and 24 months [2]. The overall 5-year survival is around 30 % [3], highlighting the need for improved patient targeting with appropriate treatment. Many treated chemo-resistant patients (30 %) suffer serious side effects needlessly, while their outcomes could be improved by guiding such patients to alternative therapies. Characterisation of the dynamic processes involved in response to chemotherapy in ovarian cancer models may help develop tools for the prediction of chemo-response and outcome and help identify key molecular changes occurring on treatment.
Cancer is the result of alterations in a limited number of pathways [4]. Recent genomic analyses of ovarian cancer have identified several deregulated pathways [5, 6]. For instance cell cycle associated defects are a hallmark of many cancer types including ovarian cancer [7]. Ovarian cancer is characterised by dysregulation of multiple cellular signaling pathways involved in cancer initiation and progression, such as EGFR/ERBB [8], MAPK [9], PI3K/AKT/mTOR [6, 10], WNT [11], JAK/STAT [12], NFκB [13], ERα [14]. Hypoxic response promotes invasiveness by inducing tumor migration and metastasis [15], while SNAIL [16], AKT, EGFR [17] and WNT pathway [18] mediate epithelial-to-mesenchymal (EMT) transition. Several DNA repair pathways, such as ataxia-telangiectasia-mutated (ATM) [19], Fanconi anemia (FA) [20], nucleotide excision repair (NER) pathways [21], are activated in response to platinum-induced DNA damage. A number of these pathways have been associated with chemo-resistance [22], while oncogenic pathway profiles have been associated with clinical outcome [23]. BRCA1/2 mutation status is also informative with respect to platinum response [24].
Activation states of cell signaling pathways can be inferred by measuring expression of representative proteins or phosphoproteins [25]. Quantitative proteomic approaches allow for the estimation of expression of multiple proteins in a large number of samples and have been used in ovarian cancer to identify factors associated with platinum response and to characterise histotype-dependent protein expression [26, 27]. Measurement of multiple proteins over time may reveal dynamic expression patterns of several pathways simultaneously. Clustering tumors according to activation patterns of oncogenic pathways can identify disease relevant deregulation patterns and support molecular classification [28].
In an earlier study, we analysed genome-wide dynamic gene expression changes in ovarian cancer models after chemotherapy treatment [29]. We observed deregulated DNA repair, cell cycle and apoptosis pathways and identified gene sets associated with survival [28]. Here we measured the dynamic effect of carboplatin and paclitaxel chemotherapy on the expression of a combined panel of 49 proteins and phosphoproteins using the same xenograft models. The models had distinct histological origin and diverse response patterns to carboplatin. As in the gene expression study, protein expression was assessed over a period of 14 days after treatment with drugs with concurrent monitoring of tumor volume. The selected proteins are frequently aberrant or belong to oncogenic pathways often deregulated in ovarian cancer [6, 30]. The targeted proteomics approach allowed the simultaneous detection of dynamic oncogenic pathway activation at the protein level in response to treatment, thus providing a complementary aspect to the transcriptomics study. We identified time-related changes in activated oncogenic pathways and we analysed those in respect to tumor volume reduction.
Methods
Xenograft dataset
The platinum sensitive OV1002 ovarian cancer xenograft model was derived from a high grade serous adenocarcinoma, while the HOX424 xenograft model was of clear cell/endometrioid origin and had reduced responsiveness to platinum. Both cell lines were established at the Institute of Genetics and Molecular Medicine, University of Edinburgh as described previously [31]. Ovarian tumor fragments were implanted subcutaneously into adult (12 week old) female CD-1 nu/nu mice and were allowed to grow to 4–6 mm in diameter in negative pressure isolators (La Calhene, Cambridge, UK). Mice were then stratified into treatment groups such that mean tumor volumes were equivalent across the groups. Tumor size was monitored twice weekly and relative tumor volumes (RTV) were calculated for each individual tumor by dividing the tumor volume on day t (Vt) by the tumor volume on day 0 (V0). Mice were grouped into three groups: untreated controls, carboplatin treated (50 mg/kg) and carboplatin (50 mg/kg) + paclitaxel treated (10 mg/kg). Treatment was administered as a single intraperitoneal dose on day 0. Groups of both treated and untreated control mice were sacrificed on Day 1, 2, 4, 7 and 14 after treatment. Tumors were placed into formalin, fixed and paraffin embedded. Tissue sections were cut and stained with a range of antibodies as described below against 49 protein targets (Table 1). The xenograft dataset consisted of 99 OV1002 and 67 HOX424 xenograft samples. Phenotypic and raw data are shown in Additional file 1. There were 3–8 biological replicates at each time point and treatment, except at day 2 for HOX424, where there were 2 and 1 replicates for each treatment. Agreement between sample expression levels was good as measured by the correlation coefficients (r) among replicates of each condition (mean r 0.94, 95 % confidence interval 0.934–0.955, Additional file 2, column E). The xenograft studies were undertaken under a UK Home Office Project Licence in accordance with the Animals (Scientific Procedures) Act 1986 and studies were approved by the University of Edinburgh Animal Ethics Committee.
Table 1.
Protein targets
| Entrez Gene ID | Gene symbol | Protein (phospho-protein) | Gene name | Function |
|---|---|---|---|---|
| 1017 | CDK2 | CDK2, pCDK2 | cyclin-dependent kinase 2 | CELL CYCLE |
| 891 | CCNB1 | CyclinB1 | cyclin B1 | CELL CYCLE |
| 595 | CCND1 | CyclinD1 | cyclin D1 | CELL CYCLE |
| 4609 | MYC | Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | CELL CYCLE |
| 1026 | CDKN1A | P21 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CELL CYCLE |
| 1027 | CDKN1B | P27 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CELL CYCLE |
| 5925 | RB1 | pRb | retinoblastoma 1 | CELL CYCLE |
| 672 | BRCA1 | BRCA1, pBRCA1 | breast cancer 1, early onset | DNA DAMAGE REPAIR |
| 2067 | ERCC1 | ERCC1 | excision repair cross-complementing rodent repair deficiency, complementation group 1 | DNA DAMAGE REPAIR |
| 4292 | MLH1 | MLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | DNA DAMAGE REPAIR |
| 4436 | MSH2 | MSH2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | DNA DAMAGE REPAIR |
| 2956 | MSH6 | MSH6 | mutS homolog 6 (E. coli) | DNA DAMAGE REPAIR |
| 1111 | CHEK1 | pChk1 | CHK1 checkpoint homolog (S. pombe) | DNA DAMAGE REPAIR |
| 3014 | H2AFX | pH2AX | H2A histone family, member X | DNA DAMAGE REPAIR |
| 3021 | H3F3B | pHH3 | H3 histone, family 3B (H3.3B) | DNA DAMAGE REPAIR |
| 5395 | PMS2 | PMS2 | PMS2 postmeiotic segregation increased 2 (S. cerevisiae) | DNA DAMAGE REPAIR |
| 7157 | TP53 | pP53 | tumor protein p53 | DNA DAMAGE REPAIR |
| 8202 | NCOA3 | AIB1 | nuclear receptor coactivator 3 | GROWTH SIGNALING:MAPK signaling |
| 207 | AKT1 | AKT, pAKT | v-akt murine thymoma viral oncogene homolog 1 | GROWTH SIGNALING:PI3K/AKT signaling |
| 1499 | CTNNB1 | BCatenin, pBCatenin | catenin (cadherin-associated protein), beta 1, 88 kDa | GROWTH SIGNALING:Wnt signaling |
| 1956 | EGFR | EGFR | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | GROWTH SIGNALING:ERBB signaling |
| 2099 | ESR1 | ER,pER | estrogen receptor 1 | GROWTH SIGNALING:ESR signaling |
| 5594 | MAPK1 | ERK, pERK | mitogen-activated protein kinase 1 | GROWTH SIGNALING:MAPK signaling |
| 2064 | ERBB2 | HER2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | GROWTH SIGNALING:ERBB signaling |
| 2065 | ERBB3 | HER3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | GROWTH SIGNALING:ERBB signaling |
| 2066 | ERBB4 | HER4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | GROWTH SIGNALING:ERBB signaling |
| 3486 | IGFBP3 | IGFBP3 | insulin-like growth factor binding protein 3 | GROWTH SIGNALING: IGF signaling |
| 3488 | IGFBP5 | IGFBP5 | insulin-like growth factor binding protein 5 | GROWTH SIGNALING: IGF signaling |
| 6194 | RPS6 | S6, pS6 | ribosomal protein S6 | GROWTH SIGNALING:mTOR signaling, hypoxia |
| 5241 | PGR | PgR | progesterone receptor | GROWTH SIGNALING:PI3K/AKT signaling |
| 5604 | MAP2K1 | pMEK | mitogen-activated protein kinase kinase 1 | GROWTH SIGNALING:MAPK signaling |
| 4790 | NFKB1 | pNFkB | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | GROWTH SIGNALING:NFkB signaling |
| 6774 | STAT3 | pSTAT3 | signal transducer and activator of transcription 3 | GROWTH SIGNALING:Jak/STAT signaling |
| 5728 | PTEN | PTEN | phosphatase and tensin homolog | GROWTH SIGNALING:PI3K/AKT signaling |
| 1366 | CLDN7 | Claudin7 | claudin 7 | ETM/INVASION |
| 999 | CDH1 | Ecadherin | cadherin 1, type 1, E-cadherin (epithelial) | ETM/INVASION |
| 3091 | HIF1A | HIF1alpha | hypoxia inducible factor 1, alpha subunit | ETM/INVASION |
| 4233 | MET | MET, pMET | met proto-oncogene | ETM/INVASION |
| 5054 | SERPINE1 | PAI | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | ETM/INVASION |
| 6615 | SNAI1 | Snail | snail homolog 1 (Drosophila) | ETM/INVASION |
| 7431 | VIM | Vimentin | vimentin | ETM/INVASION |
Details and function of 49 protein form targets (41 proteins) and their pathways tested in the xenograft protein expression experiment. There were 16 phosphorylated proteins, 8 of which were tested for both phosphorylated and unphosphorylated forms
Protein expression
Protein expression was measured by immunofluorescence using methods previously described [25]. Tissue microarrays were prepared from xenograft material that had been formalin-fixed at the time of collection and had been processed into paraffin blocks. Microarray slides were deparaffinized and antigen-retrieved by pressure-cooking. Endogenous peroxidases were blocked with 2.5 % hydrogen peroxide for 15 min and non-specific binding blocked with serum-free protein block for 15 min. Slides were then incubated with primary antibodies (Table 2) with AE1/AE3 mouse monoclonal cytokeratin antibody at room temperature for 1 h. After washing, sections were incubated for 1 h at room temperature with secondary antibodies, which included an Alexa 555-conjugated goat anti-mouse antibody diluted 1:100, and prediluted goat anti-rabbit antibody conjugated to a horseradish peroxidase decorated dextran-polymer backbone (EnVision, Dako). Slides were then incubated for 10 min with Cy5-tyramide, which is activated by horseradish peroxidase, to visualise target protein expression. 4', 6-Diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, Ore) was used to stain the nuclear compartment. For analysis, pan-cytokeratin antibody was used to identify tumor cells and normal epithelial cells, DAPI counterstain to identify nuclei, and Cy-5-tyramide detection for target protein in compartmentalised (tissue and subcellular) analysis of tissue sections. Monochromatic images of each 0.6 mm TMA core were captured at 20× objective using an Olympus AX-51 epifluorescence microscope, and high-resolution digital images analysed by the AQUAnalysis software. AQUA scores, which represent the sum of the Cy5-tyramide score for the target protein divided by the area of the cellular compartment (cytoplasm or nucleus), were then generated for each sample. A detailed description of the AQUA analysis has been reported elsewhere [32].
Table 2.
Sources of antibodies and the dilutions used in this study
| Protein name | Gene name | Source | Catalog No. | Dilution |
|---|---|---|---|---|
| AIB1 | NCOA3 | BD Biosciences | 61105 | 1 in 50 |
| AKT1 | AKT | Cell Signaling | Ab4685 | 1 in 200 |
| pAKT | AKT | Cell Signaling | 9271 | 1 in 50 |
| BRCA1 | BRCA1 | Eurogentec | 75460 | 1 in 70 |
| pBRCA1 | BRCA1 | Cell Signaling | 9009 | 1 in 25 |
| CyclinB1 | CCNB1 | Epitomics | 1495-1 | 1 in 50 |
| CyclinD1 | CCND1 | Dako | M3635 | 1 in 100 |
| E-Cadherin | CDH1 | BD sciences | 610181 | 1 in 1500 |
| CDK2 | CDK2 | Cell Signaling | 2546 | 1 in 100 |
| pCDK2 | CDK2 | Cell Signaling | 2561 | 1 in 1500 |
| CDKN1A | P21 | Cell Signaling | 2946 | 1 in 100 |
| CDKN1B | P27 | Cell Signaling | 2552 | 1 in 100 |
| CLDN7 | Claudin7 | Abcam | AB27487 | 1 in 200 |
| B-Catenin | CTNNB1 | BD sciences | 610153 | 1 in 500 |
| pB-Catenin | CTNNB1 | Cell Signaling | 9561 | 1 in 25 |
| EGFR | EGFR | Invitrogen | 28-0005 | 1 in 50 |
| ERBB2 | HER2 | Dako | A0485 | 1 in 400 |
| ERBB3 | HER3 | Dako | M7297 | 1 in 50 |
| ERCC1 | ERCC1 | Labvision | MS-671-P0 | 1 in 600 |
| ER1 | ESR1 | Neomarkers | RM-9101-S1 | 1 in 100 |
| pER1 | ESR1 | Cell Signaling | 2511 | 1 in 50 |
| pH2AX | H2AFX | Cell Signaling | 2577 | 1 in 50 |
| HER2 | HER2 | Dako | M7297 | 1 in 400 |
| HER3 | HER3 | Cell Signaling | 4754 | 1 in 50 |
| HER4 | HER4 | Cell Signaling | 4792 | 1 in 300 |
| HIF1A | HIF1a | Cell Signaling | 3176 | 1 in 25 |
| IGFBP3 | IGFBP3 | Abcam | Ab4248 | 1 in 100 |
| IGFBP5 | IGFBP5 | Abcam | Ab4255 | 1 in 50 |
| MAPK1 | ERK | Cell Signaling | 9107 | 1 in 250 |
| pMEK | MAP2K1 | Cell Signaling | 9154 | 1 in 400 |
| MET | MET | Eurogentec | 75551 | 1 in 400 |
| pMET | MET | Eurogentec | 65559 | 1 in 25 |
| MLH1 | MLH1 | Leica | NCL-L-MLH1 | 1 in 100 |
| MSH2 | MSH2 | Invitrogen | 33-7900 | 1 in 50 |
| MSH6 | MSH6 | Leica | NCL-L-MSH6 | 1 in 250 |
| MYC | MYC | Eurogentec | 75355 | 1 in 70 |
| pNFKB | NFKB1 | Cell Signaling | 3037 | 1 in 25 |
| PAI | SERPINE1 | BD Sciences | P612024 | 1 in 200 |
| PgR | PGR | Dako | 3569 | 1 in 50 |
| PMS2 | PMS2 | Leica | NCL-L-PMS2 | 1 in 250 |
| pP53 | TP53 | Cell Signaling | 9286 | 1 in 100 |
| PTEN | PTEN | Cell Signaling | 9559 | 1 in 200 |
| pRB | RB1 | Cell Signaling | 9308 | 1 in 50 |
| S6 | RPS6 | Cell Signaling | 2217 | 1 in 100 |
| pS6 | RPS6 | Cell Signaling | 2211 | 1 in 200 |
| SERPINE1 | PAI | BD Sciences | P612024 | 1 in 200 |
| SNAIL | SNAI1 | Abcam | Ab17732 | 1 in 800 |
| pSTAT3 | STAT3 | Cell Signaling | 9145 | 1 in 25 |
| Vimentin | VIM | Sigma | V6630 | 1 in 400 |
Protein expression analysis and statistics
AQUA scores were log-transformed averaged from replicate cores. Means of differentially expressed proteins log fold change values for each time point were hierarchically clustered using Cluster 3.0 in order to identify significant proteins with similar temporal expression profiles. Heatmaps were visualized using TreeView [33]. Bioconductor package limma [34] was used for differential expression calculations. Significant genes had FDR adjusted p-values below 0.05. Treated samples were contrasted to pooled untreated control from all time points in each xenograft. Differentially expressed proteins were classified as expressed early (Days 1–4) or late (Days 7, 14), and transient (expressed significantly in 1 time point) or continuous (expressed significantly at least 2 continuous time points).
Results
Carboplatin dynamically activates phosphoproteins in ovarian cancer xenografts
Protein candidates were selected from pathways identified in a recent analysis of somatic mutations and copy number changes in ovarian cancer [5] and known pathways of interest in this disease [25]. A total of 49 protein forms were investigated which are known to be involved in DDR (BRCA1/pBRCA1, ERCC1, MLH1, MSH2, MSH6, pCHK1, pH2AX, PHH3, PMS2, pP53), cell cycle regulation (CDK2/pCDK2, Cyclin B, Cyclin D1, MYC, p21, p27, pRb), EMT/invasion (Claudin 7, E-cadherin, HIF1α, MET/pMET, PAI1, SNAIL, Vimentin) and growth signaling including WNT (CTNNB1/pCTNNB1), PI3K/AKT (AKT/pAKT, PTEN), mTOR (S6/pS6), NFκB (NFκB1), MAPK (ERK1/pERK1, pMEK), EGFR/ERBB (EGFR, HER2, HER3, HER4), IGF (IGFBP3, IGFBP5), JAK/STAT (pSTAT3) and ERα (ESR1/pESR1, PGR, AIB1) pathways. Further details of protein targets are shown in Table 1. Examples of stained images are illustrated in Fig. 1. There were 41 unique proteins in the set, 8 of which were in both native and phosphorylated forms and 8 only in phosphorylated form. Activated pathways would feature increased expression of phosphorylated proteins, usually coupled with a concurrent drop in expression of the native form.
Fig. 1.
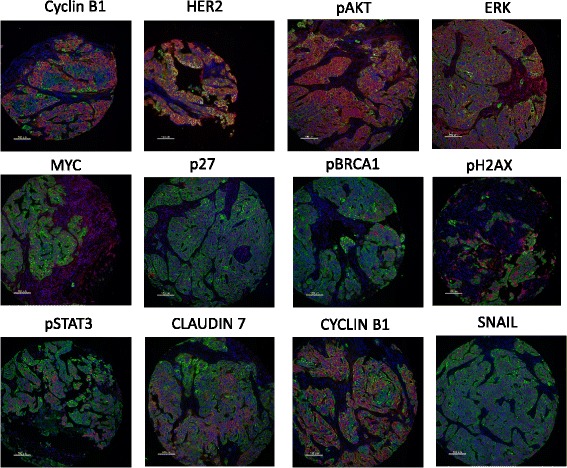
Stained tissue images for AQUA quantitative image analysis. Representative immunofluorescence images of examples of the phosphoproteins or total proteins used within the study. Blue = DAPI nuclear counterstain, Green = cytokeratin tumor mask, Red = target protein. Formaldehyde fixed paraffin-embedded xenograft tissue was stained and analysed as described in Methods
We have previously shown significant relative tumor volume reduction within the period up to 14 days after a single treatment in the platinum responsive OV1002 and a minimal response in platinum resistant HOX424 ovarian cancer xenograft models [29] (Fig. 2). This same sample set was analysed for the above-named proteins. Carboplatin was administered at 50 mg/kg and tumors collected on days 1, 2, 4, 7 and 14 after treatment. Over this time period, analysis of the protein set identified 13 differentially expressed protein forms (ESR1, βCatenin, Claudin 7, MET, Cyclin B1, HER4, S6, pBRCA1, pERK, pCDK2, pCHK1, pH2AX, pS6) in carboplatin treated OV1002 xenografts, of which 6 were phosphoproteins (Additional file 3). Proteins were grouped by hierarchical clustering into 3 clusters (which we have identified as downregulated early, upregulated late, and upregulated overall, corresponding to their dynamic changes) and a singleton (ESR1 - which was downregulated) (Fig. 3a). Differential expression was mostly late and transient (Fig. 4). Native S6 was reduced early and substituted by its late expressed phosphorylated form (pS6) over time. pCDK2, pERK, pBRCA1 were upregulated at late time points, while expression of their unphosphorylated counterparts was not significantly changed (Fig. 5). Phosphorylated pH2AX was differentially expressed at all five time points, while pCHK1 only at day 14.
Fig. 2.
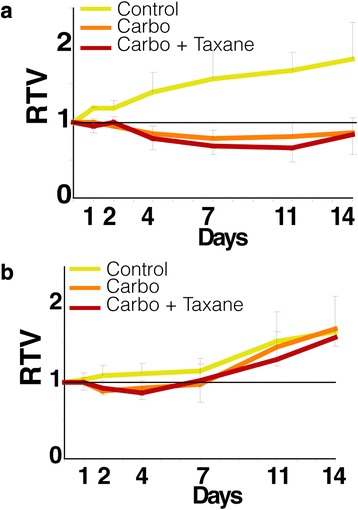
Relative Tumor Volume graphs. Post-treatment tumor volume growth relative to day of treatment (Day 0) for the OV1002 (a) and HOX424 (b) xenografts. (Carbo: Carboplatin; Taxane: Paclitaxel). Modified from [29]
Fig. 3.
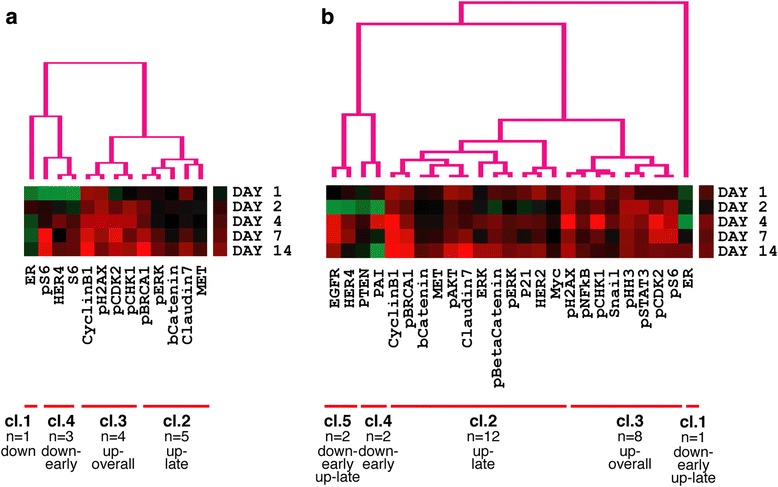
Hierarchical clustering of post-treatment dynamic phosphoprotein expression. Heatmaps illustrating clusters of proteins with differentially increased (red) or decreased (green) expression in response to carboplatin (a) or carboplatin-paclitaxel (b) treatment in OV1002 xenografts
Fig. 4.
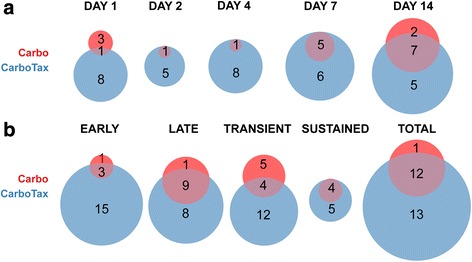
Differential expression analysis of treated OV1002 xenografts. Venn diagrams of differentially expressed protein form counts in each time point (a) and according to their temporal characteristics (b) in carboplatin and carboplatin-paclitaxel treated OV1002 xenografts. Protein expression was analysed in 5 time points post treatment (Days 1, 2, 4, 7 and 14). Early (late) genes were differentially expressed in Days 1, 2 and 4 (Days 7 and 14). Transient (sustained) were differentially expressed in a single time point (more than one time points)
Fig. 5.
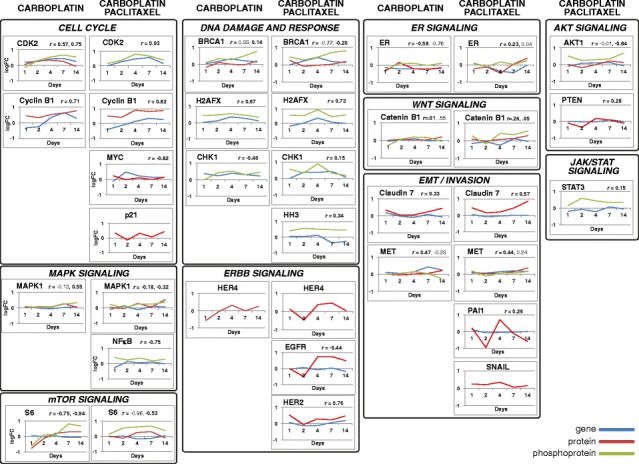
Post-treatment dynamic phosphoprotein/native protein differential expression graphs. Mean log fold differential expression of phosphoproteins (orange)/native proteins (red) in treated OV1002 xenografts as compared to untreated controls. Proteins are significantly expressed in at least one time point. Corresponding gene expression from [29] (GEO accession number GSE49577) is shown in blue where available, and its correlation to protein expression is indicated
In contrast to the increased phosphoprotein expression in treated xenografts, a range of unphosphorylated protein expression responses was observed. Expression of HER4, S6 and ER was inhibited one day post treatment, while MET, βCatenin, Claudin7 and CyclinB1 were upregulated late (Fig. 5). Four of these proteins were found differentially expressed also in the gene level: CDK2, Cyclin B1, CHK1 and MET [29].
The HOX424 response to treatment was very limited. Only one protein (MLH1) was differentially expressed in carboplatin-treated xenografts (Additional file 3). This minimal response is consistent with the very limited tumor growth response to carboplatin (Additional file 3).
Carboplatin-paclitaxel dynamically trigger a broader and stronger phosphoprotein expression in ovarian cancer xenografts
The combined carboplatin-paclitaxel treatment triggered a more extensive response than carboplatin alone in the OV1002 model. In contrast to only 13 proteins showing differential expression with carboplatin, a total of 23 proteins (25 protein forms) (EGFR, HER4, PTEN, PAI, Cyclin B1, pBRCA1, CTNNB1, MET, pAKT, Claudin 7, ERK, pCTNNB1, pERK, p21, HER2, MYC, pH2AX, pNFκB, pCHK1, SNAIL, pHH3, pSTAT3, pCDK2, pS6, ESR1) were differentially expressed at both early and late time points, of which 11 were phosphoproteins (Figs. 4 and 5, Additional file 3). These were clustered by hierarchical clustering into 4 groups (identified as upregulated late, upregulated overall, downregulated early, down- early/upregulated late) and a singleton (ESR1 - downregulated) (Fig. 3b). Differentially expressed proteins included all but one (S6) that were found significant after carboplatin only treatment. Longitudinal expression profiles between treatments were similar, especially among phosphoproteins. Some distinct expression patterns were observed after carboplatin-paclitaxel treatment. Expression of pBRCA1 was increased immediately after treatment (day 1), while pCHK1 had a higher log fold change across all time points compared to carboplatin only treatment. Five additional phosphoproteins were differentially expressed only after combined treatment. Expression of pAKT and pNFκB were triggered shortly after treatment (day 1), followed by pSTAT3 and pHH3 which were differentially expressed from day 2. Phospho-βCatenin was expressed late, from 7 days after treatment (Fig. 5). Expression of four phosphoproteins was changed at least 2-fold: pH2AX, pCHK1, pCDK2 and pBRCA1 (Fig. 5).
In contrast to carboplatin only treatment, early HER4 and ERα down-regulation were followed by up-regulation at late time points after combination treatment. Additionally, up-regulation of Cyclin B1 and Claudin 7 started immediately after treatment. Another eight proteins were differentially expressed after carboplatin-paclitaxel treatment. Expression of PAI and PTEN was inhibited at day 2. Expression of HER2 and MYC was upregulated at day 1 only, while EGFR and ERK were upregulated late. Snail and p21 were upregulated at early and late time points (Fig. 4). CDK2, Cyclin B1, BRCA1, CHK1, ESR1, HH3 and STAT3 were differentially expressed in the corresponding treatment to control comparison in the gene expression study [29]. All were upregulated in both gene and protein level, except the last two which were downregulated in the gene level and positively regulated in the protein level. Protein expression of HH3 and STAT3 was assessed only in the phosphorylated form and this may explain the discrepancy in expression between gene and protein.
The HOX424 response to combination treatment, as with carboplatin alone treatment, was very limited. Only two proteins (ESR1 and pMET) were differentially expressed in carboplatin-paclitaxel treated xenografts (Additional file 3). Again this is consistent with the very minimal growth response shown to these drugs (Fig. 2).
Discussion
In this study, we characterised a dynamic protein response to chemotherapy in two ovarian cancer models over a period of 14 days using a set of protein markers of pathways that are frequently deregulated in cancer and may be associated with survival. Complementing an earlier gene expression study, we found multiple proteins differentially expressed after treatment in either or both their native and phosphorylated forms allowing assessment of relative expression of each form over time. An increase of the activated (phosphorylated) forms was observed indicating the activation of several pathways in response to treatment. Certain proteins showed late change in expression, detected here due to the relatively prolonged period of expression monitoring (14 days). There is potential to collect serial samples during treatment of ovarian cancer, e.g., during neoadjuvant therapy which is being increasingly considered prior to surgery [35] or during adjuvant therapy by studying peritoneal ascitic cells removed from patients before and after therapy. Measurement of the dynamic markers identified within this study could provide further information to aid in the assessment of whether the tumor is sensitive to treatment or not.
We observed distinct responses to treatment in each cell line xenograft model. The serous derived OV1002 xenograft was highly responsive to carboplatin-based treatment as judged by a decrease in RTV and marked differential protein expression analysis. Differential protein expression in the slow growing and mainly unresponsive clear cell/endometrioid derived HOX424 was very limited after treatment, while the drop in RTV was smaller in duration and magnitude. This observation coincided with the post-treatment differential expression pattern of these models at the gene level [29]. Correlation of gene and protein expression was higher when both were differentially expressed [36]. These results suggest that responses to treatment among ovarian cancer models of distinct histological origin vary which is consistent with clinical experience. Testing more models of diverse origin would be valuable in helping to assess the range of responses to treatment in ovarian cancer.
Carboplatin treatment in the platinum-sensitive OV1002 model triggered up-regulation of cell cycle (Cyclin B1, pCDK2, pCHK1), mTOR (S6, pS6) and DRR (pH2AX, pBRCA1, pCHK1) pathways, while at late time points WNT (βCatenin), invasion (Claudin 7), EMT (c-MET) and MAPK (pERK) pathways were modulated. Estrogen receptor-alpha (ESR1) and ERBB (HER4) pathways were down-regulated early, within 24 h, as was S6. The early increase in pH2AX at 24 h was maintained through to Day 7 after which it fell. Cell cycle changes (pCDK2, Cyclin B1) increased progressively over the first 7 days and at the latest time point indicators of WNT, EMT, Invasion and MAPK pathways were increased. The changes in DNA damage response and repair proteins and in cell cycle associated proteins are in line with the DNA lesions produced by carboplatin and consistent with its mode of action [37]. In platinum-sensitive ovarian cancer cells in vitro, we have previously shown that markers of DDR such as pBRCA1 and pɣH2AX are known to be induced over a 3–4 day period comparable to that seen in vivo [38]. The changes observed in the current study are also consistent with differential expression patterns observed in the gene expression study, where several cell cycle and DDR proteins used here were significant (Fig. 5). These variations were anticipated and match the changes in tumor volume after treatment.
Phosphorylation of H2AX on Ser139 is consistent with formation of DNA strand breaks [39], while BRCA1 is phosphorylated by ATM in response to double-stranded DNA breaks [40]. CHK1 Ser 317 phosphorylation is then consistent with checkpoint activation [41]. βCatenin has been proposed as a target in this disease and may be associated with chemoresistance [18]. Claudin 7 is frequently overexpressed in ovarian cancer and has been linked to increased migration [42]. High expression of c-MET in ovarian cancer has been associated with poor survival and the use of c-MET inhibitors could enhance the effects of platinum-taxane therapy [43, 44]. The early changes in ESR1 and HER4 were unexpected as were the later changes in invasion and EMT proteins. High expression of ESR1 has been associated with outcome for ovarian cancer after platinum-based therapy [45] and also decreased on carboplatin treatment. Expression of HER4 (ERBB4) has been associated with chemoresistance [46] in ovarian cancer and this decreased on carboplatin treatment. Several of the later changes may reflect rapid regrowth, for example the increases in pERK and pAKT. These proteins are key in activation of their corresponding ERK and AKT pathways which are complex, interacting cascades playing key roles in normal and malignant cell growth [47].
Further responses were observed when carboplatin-paclitaxel combination therapy was compared with carboplatin treatment alone. The dynamic response to the carboplatin-paclitaxel combination involved similar or greater responses in the same pathways but with more proteins demonstrating a response while additional pathway molecules were also modulated (Fig. 6). Therefore the cell cycle (Cyclin B1, pCDK2, pCHK1 and also pHH3, MYC and p21) and DDR (pH2AX, pBRCA1, pCHK1) pathways showed similar or more pronounced changes than with carboplatin alone and with more of the tested proteins demonstrating changed expression. Consistent to our findings, p21 (CDKN1A) is reported to increase after paclitaxel treatment in ovarian cancer cells contributing to cell cycle regulation [48]. In addition to the mTOR molecule pS6 increasing, components of the AKT pathway (pAKT, PTEN) were modulated with pAKT increasing, while its tumor suppressor regulator PTEN was down-regulated early on. The MAPK pathway was again modulated with pERK expression increasing. WNT signaling (both βCatenin and phospho-βCatenin), EMT markers (MET, SNAI1), invasion (Claudin 7 and PAI) were again modulated generally at later time points. Increased SNAI1 expression has been reported to relate to paclitaxel resistance in this disease [49]. Additional pathways were also observed with the NFκB (pNFκB) and JAK/STAT (pSTAT3) pathways being up-regulated. NFκB has been shown to possess a biphasic role in ovarian cancer [50] while paclitaxel treatment of ovarian cancer cells has been reported to activate the STAT3 pathway [51]. The ER-α pathway was again down-regulated as was HER4, however further protein members of the ERBB pathway, EGFR and HER2, were also upregulated late. HER2 expression has been reported to relate to paclitaxel sensitivity in ovarian cancer cells in vitro [52]. These findings are consistent with observations from the gene expression study where several of the proteins used here, mostly within the cell cycle and DDR functional groups, were differentially expressed (Fig. 5).
Fig. 6.
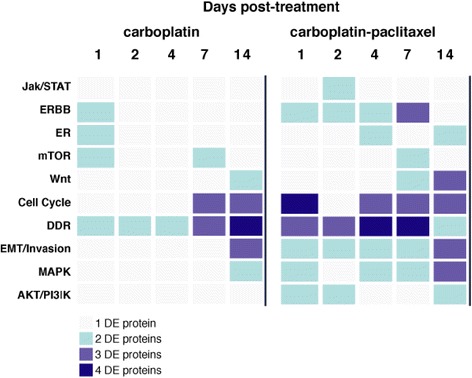
Heatmap of post-treatment dynamic pathway activation in OV1002 xenografts. The number of differentially expressed (DE) protein members of a pathway (Jak/STAT signaling, ERBB signaling, ERα signaling, mTOR signaling, WNT signaling, cell cycle, DDR, EMT/Invasion, MAPK signaling and AKT signaling) in each time point is colour coded in shades of blue, for each treatment: carboplatin and carboplatin-paclitaxel
Conclusion
The signaling changes induced by carboplatin and carboplatin-paclitaxel indicate pathways activated by the cytotoxic agents and may suggest new targets for therapeutic intervention and potential ways to enhance chemotherapy effects. The changed expression also provides predictive biomarkers of response. If such dynamic changes can be identified in clinical samples during treatment, they may help indicate the responsiveness of the tumor to chemotherapy. Early changes included DDR and cell cycle regulatory proteins associating with tumor volume changes, which were expected. Therefore inhibitors of the cell cycle or the DDR signaling pathway, e.g., CHK inhibitors [53], may merit further testing and assessment. Changes in ESR1 and HER signaling were also observed, as were late changes in EMT and invasion markers, and these require further investigation to assess their generality.
Availability of supporting data
All protein expression values analysed here are available as Additional file 1 (Protein expression matrix). Additionally, calculated log fold change values of differentially expressed proteins for each condition are available as Additional file 3 (Differential protein expression).
Gene expression values shown in Fig. 5 are taken from [29] and are available from the Gene Expression Omnibus (GEO) with accession number GSE49577 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49577).
Acknowledgements
The authors thank Medical Research Scotland and the Scottish Funding Council.
This work was supported by Medical Research Scotland [FRG353 to V.A.S.]; the FP7-Directorate-General for Research and Innovation of the European Commission [EU HEALTH-F4-2012-305033 to Coordinating Action Systems Medicine - D.J.H.]; the Chief Scientist Office of Scotland [D.J.H.], the Scottish Funding Council [D.J.H. and S.P.L.]. Health Canada Scholarship (Indspire) [KEF], Scottish Overseas Research Student Award Scheme (University of Edinburgh)[KEF] and the Three Fires Award (Wikwemikong Board of Education)[KEF].
Abbreviations
- AQUA
automated quantitative analysis
- DDR
DNA damage response
- EMT
epithelial to mesenchymal
- RTV
relative tumor volume
Additional files
Protein expression matrix. Protein expression matrix of raw data of 49 proteins/phosphoproteins assayed in 166 xenograft samples of the dataset. (XLSX 224 kb)
Assessment of biological replicate concordance. Number of replicates for all tested conditions (xenograft/treatment/day) is shown. Correlation coefficients were calculated for all possible pairs of biological replicates and averaged (mean replicate r) using expression values from all detected proteins within each condition. (XLSX 42 kb)
Differential protein expression. Log fold changes of differentially expressed proteins over 14 days are shown in 4 worksheets, each for a xenograft-treatment condition: carboplatin treated OV1002; carboplatin treated HOX424; carboplatin-paclitaxel treated OV1002; carboplatin-paclitaxel treated HOX424. (XLS 57 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AK and SPL wrote the manuscript and AK performed all computational analyses. SPL, IU, KEF and CK performed all biological experiments. VAS, SPL and DJH conceived the study. All authors read and commented on the manuscript and agreed upon a final version together.
Contributor Information
Antonis Koussounadis, Email: ak73@st-andrews.ac.uk.
Simon P. Langdon, Email: Simon.Langdon@ed.ac.uk
Inhwa Um, Email: ihu@st-andrews.ac.uk.
Charlene Kay, Email: charkay22@gmail.com.
Kyle E. Francis, Email: K.E.Francis@sms.ed.ac.uk
David J. Harrison, Email: david.harrison@st-andrews.ac.uk
V. Anne Smith, Phone: +44 (0)1334-463368, Email: anne.smith@st-andrews.ac.uk.
References
- 1.Covens A, Carey M, Bryson P, Verma S, Fung Kee Fung M, et al. Systematic review of first-line chemotherapy for newly diagnosed postoperative patients with stage II, III, or IV epithelial ovarian cancer. Gynecol Oncol. 2002;85:71–80. doi: 10.1006/gyno.2001.6552. [DOI] [PubMed] [Google Scholar]
- 2.Moss C, Kaye SB. Ovarian cancer: progress and continuing controversies in management. Eur J Cancer. 2002;38:1701–7. doi: 10.1016/S0959-8049(02)00161-2. [DOI] [PubMed] [Google Scholar]
- 3.Levi F, Lucchini F, Negri E, Boyle P, La Vecchia C. Cancer mortality in Europe, 1995–1999, and an overview of trends since 1960. Int J Cancer. 2004;110:155–69. doi: 10.1002/ijc.20097. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Kinzler K. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 5.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham JM, Vierkant RA, Sellers TA, Phelan C, Rider DN, Liebow M, et al. Cell cycle genes and ovarian cancer susceptibility: a tagSNP analysis. Br J Cancer. 2009;101:1461–8. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maihle NJ, Baron AT, Barrette BA, Boardman CH, Christensen TA, et al. EGF/ErbB receptor family in ovarian cancer. Cancer Treat Res. 2002;107:247–58. doi: 10.1007/978-1-4757-3587-1_11. [DOI] [PubMed] [Google Scholar]
- 9.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 10.Dobbin ZC, Landen CN. The importance of the PI3K/AKT/MTOR pathway in the progression of ovarian cancer. Int J Mol Sci. 2013;14:8213–27. doi: 10.3390/ijms14048213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbolina MV, Burkhalter RJ, Stack MS. Diverse mechanisms for activation of WNT signalling in the ovarian tumor microenvironment. Biochem J. 2011;437:1–12. doi: 10.1042/BJ20110112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 13.Alvero AB. Recent insights into the role of NF-kappaB in ovarian carcinogenesis. Genome Med. 2010;2:56. doi: 10.1186/gm177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Stefano I, Zannoni GF, Prisco MG, Fagotti A, Tortorella L, Vizzielli G, et al. Cytoplasmic expression of estrogen receptor beta (ERβ) predicts poor clinical outcome in advanced serous ovarian cancer. Gynecol Oncol. 2011;122:573–9. doi: 10.1016/j.ygyno.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi A, Hayashi T, Kikuchi N, Hayashi A, Fuseya C, Shiozawa T, et al. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1α to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int J Cancer. 2012;131:1755–67. doi: 10.1002/ijc.27448. [DOI] [PubMed] [Google Scholar]
- 16.Kim MK, Kim MA, Kim H, Kim YB, Song YS. Expression profiles of epithelial-mesenchymal transition-associated proteins in epithelial ovarian carcinoma. Biomed Res Int. 2014;2014:e495754. doi: 10.1155/2014/495754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, et al. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arend RC, Londoño-Joshi AI, Straughn JM, Jr, Buchsbaum DJ. The WNT/β-catenin pathway in ovarian cancer: a review. Gynecol Oncol. 2013;131:772–9. doi: 10.1016/j.ygyno.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 19.Yazlovitskaya EM, Persons DL. Inhibition of cisplatin-induced ATR activity and enhanced sensitivity to cisplatin. Anticancer Res. 2003;23:2275–9. [PubMed] [Google Scholar]
- 20.D’Andrea AD. The Fanconi Anemia/BRCA signaling pathway: disruption in cisplatin-sensitive ovarian cancers. Cell Cycle. 2003;2:290–2. [PubMed] [Google Scholar]
- 21.Basu A, Krishnamurthy S. Cellular responses to cisplatin-induced DNA damage. J Nucleic Acids. 2010;201367. [DOI] [PMC free article] [PubMed]
- 22.Helleman J, Smid M, Jansen MP, van der Burg ME, Berns EM. Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol. 2010;117:170–6. doi: 10.1016/j.ygyno.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Trinh XB, Tjalma WA, Dirix LY, Vermeulen PB, Peeters DJ, Bachvarov D, et al. Microarray-based oncogenic pathway profiling in advanced serous papillary ovarian carcinoma. PLoS One. 2011;6:e22469. doi: 10.1371/journal.pone.0022469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Yang D, Sun Y, Xue F, Sood AK, Zhang W. Differing clinical impact of BRCA1 and BRCA2 mutations in serous ovarian cancer. Pharmacogenomics. 2012;13:1523–35. doi: 10.2217/pgs.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faratian D, Um I, Wilson DS, Mullen P, Langdon SP, Harrison DJ. Phosphoprotein pathway profiling of ovarian carcinoma for the identification of potential new targets for therapy. Eur J Cancer. 2011;47:1420–31. doi: 10.1016/j.ejca.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Carey MS, Agarwal R, Gilks B, Swenerton K, Kalloger S, Santos J, et al. Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin Cancer Res. 2010;16:2852–60. doi: 10.1158/1078-0432.CCR-09-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegand KC, Hennessy BT, Leung S, Wang Y, Ju Z, McGahren M, et al. A functional proteogenomic analysis of endometrioid and clear cell carcinomas using reverse phase protein array and mutation analysis: protein expression is histotype-specific and loss of ARID1A/BAF250a is associated with AKT phosphorylation. BMC Cancer. 2014;14:120. doi: 10.1186/1471-2407-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 29.Koussounadis A, Langdon SP, Harrison DJ, Smith VA. Chemotherapy-induced dynamic gene expression changes in vivo are prognostic in ovarian cancer. Br J Cancer. 2014;110:2975–84. doi: 10.1038/bjc.2014.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creixell P, Reimand J, Haider S, Wu G, Shibata T, Vazquez M, et al. Pathway and network analysis of cancer genomes. Nat Methods. 2015;12:615-21. [DOI] [PMC free article] [PubMed]
- 31.Faratian D, Zweemer AJ, Nagumo Y, Sims AH, Muir M, Dodds M, et al. Trastuzumab and pertuzumab produce changes in morphology and estrogen receptor signaling in ovarian cancer xenografts revealing new treatment strategies. Clin Cancer Res. 2011;17:4451–61. doi: 10.1158/1078-0432.CCR-10-2461. [DOI] [PubMed] [Google Scholar]
- 32.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–8. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth GK. Limma: linear models for microarray data. In: Gentleman RRW, Carey V, Dudoit S, Irizarry R, Huber WRW, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 35.Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239–45. doi: 10.1038/nrclinonc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koussounadis A, Langdon SP, Um I, Harrison DJ, Smith VA. Differential gene expression has meaning: gene and protein expression correlate better across experimental manipulation for differentially expressed genes. Sci Rep. 2015;5:10775. doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–83. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 38.Huang R, Langdon SP, Tse M, Mullen P, Um IH, Faratian D, et al. The role of HDAC2 in chromatin remodelling and response to chemotherapy in ovarian cancer. Oncotarget. 2015;7:4695–711. doi: 10.18632/oncotarget.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 40.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–6. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 41.Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27:2572–81. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahiya N, Becker KG, Wood WH, 3rd, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One. 2011;6:e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawada K, Radjabi AR, Shinomiya N, Kistner E, Kenny H, Becker AR, et al. c-Met overexpression is a prognostic factor in ovarian cancer and an effective target for inhibition of peritoneal dissemination and invasion. Cancer Res. 2007;67:1670–9. doi: 10.1158/0008-5472.CAN-06-1147. [DOI] [PubMed] [Google Scholar]
- 44.Marchion DC, Bicaku E, Xiong Y, Bou Zgheib N, Al Sawah E, Stickles XB, et al. A novel c-Met inhibitor, MK8033, synergizes with carboplatin plus paclitaxel to inhibit ovarian cancer cell growth. Oncol Rep. 2013;2:2011–8. doi: 10.3892/or.2013.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamagni C, Wirtz RM, De Iaco P, Rosati M, Veltrup E, Rosati F. Oestrogen receptor 1 mRNA is a prognostic factor in ovarian cancer patients treated with neo-adjuvant chemotherapy: determination by array and kinetic PCR in fresh tissue biopsies. Endocr Relat Cancer. 2009;16:1241–9. doi: 10.1677/ERC-08-0342. [DOI] [PubMed] [Google Scholar]
- 46.Gilmour LM, Macleod KG, McCaig A, Gullick WJ, Smyth JF, Langdon SP. Expression of erbB-4/HER-4 growth factor receptor isoforms in ovarian cancer. Cancer Res. 2001;6:2169–76. [PubMed] [Google Scholar]
- 47.Steelman LS, Chappell WH, Abrams SL, Kempf CR, Long J, Laidler P, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192–222. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitsuuchi Y, Johnson SW, Selvakumaran M, Williams SJ, Hamilton TC, Testa JR. The phosphatidylinositol 3-kinase/AKT signal transduction pathway plays a critical role in the expression of p21WAF1/CIP1/SDI1 induced by cisplatin and paclitaxel. Cancer Res. 2000;60:5390–4. [PubMed] [Google Scholar]
- 49.Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53 mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–68. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 50.Yang G, Xiao X, Rosen DG, Cheng X, Wu X, Chang B, et al. The biphasic role of NF-kappaB in progression and chemoresistance of ovarian cancer. Clin Cancer Res. 2011;17:2181–94. doi: 10.1158/1078-0432.CCR-10-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abubaker K, Luwor RB, Zhu H, McNally O, Quinn MA, Burns CJ, et al. Inhibition of the JAK2/STAT3 pathway in ovarian cancer results in the loss of cancer stem cell-like characteristics and a reduced tumor burden. BMC Cancer. 2014;14:317. doi: 10.1186/1471-2407-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aigner A, Hsieh SS, Malerczyk C, Czubayko F. Reversal of HER-2 over-expression renders human ovarian cancer cells highly resistant to taxol. Toxicology. 2000;144:221–8. doi: 10.1016/S0300-483X(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 53.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142:1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]


