Abstract
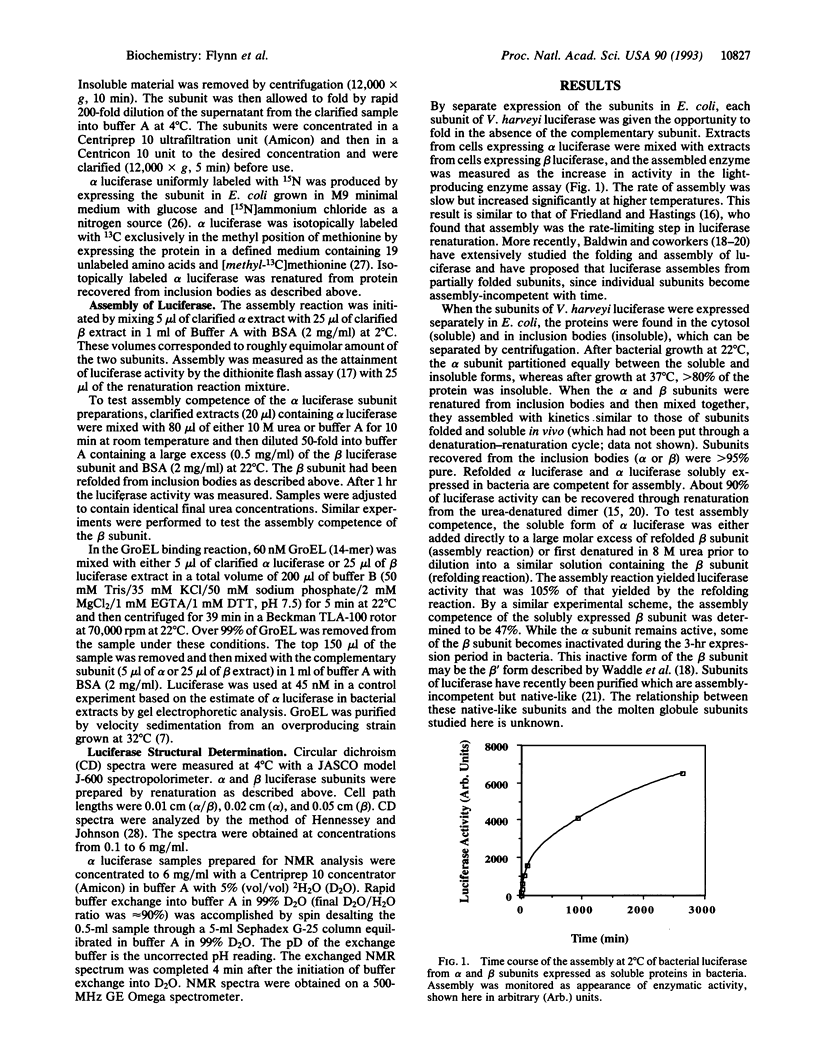
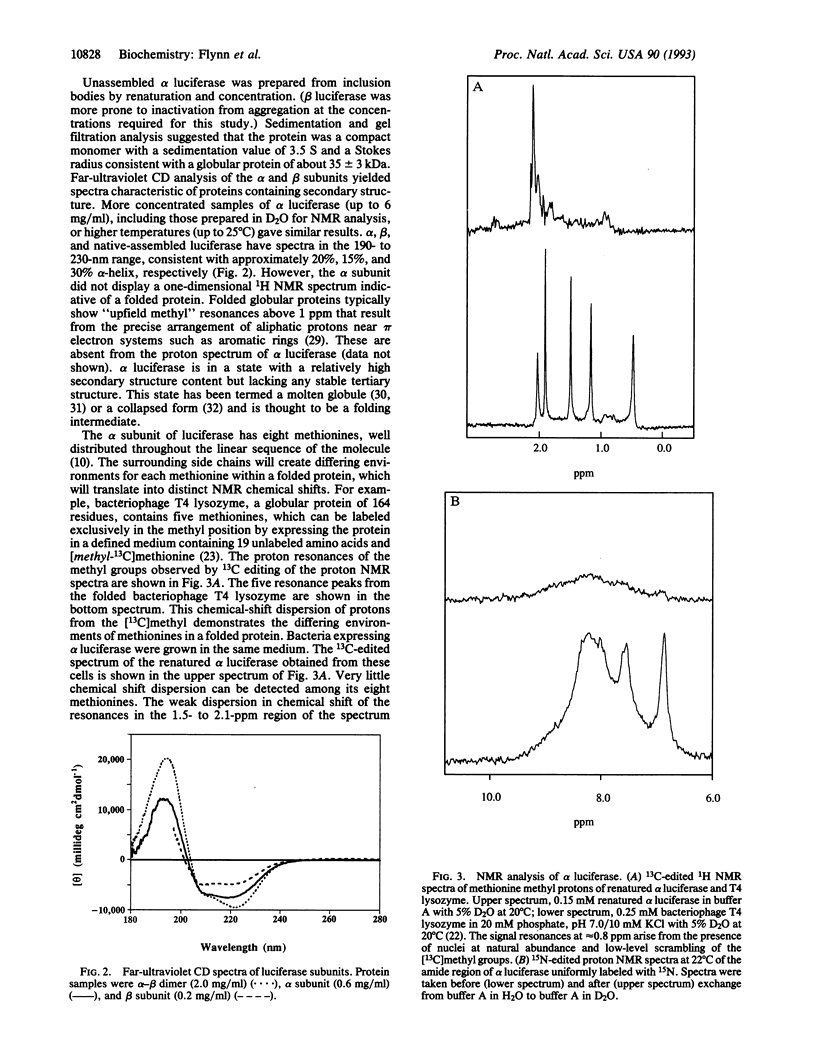
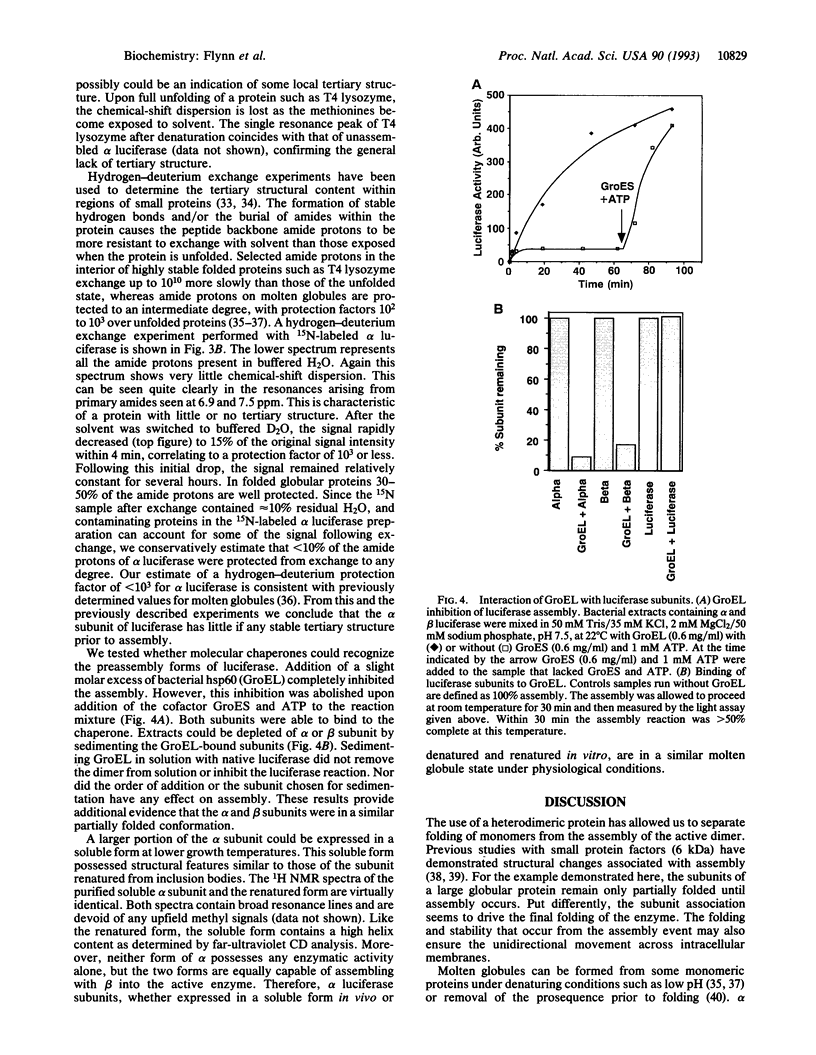
We have studied the assembly of a large heterodimeric protein, bacterial luciferase, by mixing purified subunits expressed separately in bacteria. The individual subunits alpha and beta contain much (66% and 50%, respectively) of the alpha-helix content of the native heterodimer as measured by circular dichroism, yet the alpha subunit lacks observable tertiary structure as measured by NMR. These results are consistent with the alpha subunit existing in a molten globule or collapsed form prior to assembly. The molecular chaperone GroEL binds reversibly to both subunits prior to assembly. Since these observations were obtained under physiological conditions, we propose that the molten globule exists as a stable form during folding or assembly in the cell. Either the molten globule form of the subunits is an authentic folding intermediate or it is in rapid equilibrium with one. GroEL may function by facilitating assembly through stabilization of these incompletely folded subunits.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker D., Sohl J. L., Agard D. A. A protein-folding reaction under kinetic control. Nature. 1992 Mar 19;356(6366):263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- Baldwin T. O., Nicoli M. Z., Becvar J. E., Hastings J. W. Bacterial luciferase. Binding of oxidized flavin mononucleotide. J Biol Chem. 1975 Apr 25;250(8):2763–2768. [PubMed] [Google Scholar]
- Baldwin T. O., Ziegler M. M., Chaffotte A. F., Goldberg M. E. Contribution of folding steps involving the individual subunits of bacterial luciferase to the assembly of the active heterodimeric enzyme. J Biol Chem. 1993 May 25;268(15):10766–10772. [PubMed] [Google Scholar]
- Bochkareva E. S., Lissin N. M., Flynn G. C., Rothman J. E., Girshovich A. S. Positive cooperativity in the functioning of molecular chaperone GroEL. J Biol Chem. 1992 Apr 5;267(10):6796–6800. [PubMed] [Google Scholar]
- Bowie J. U., Sauer R. T. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989 Sep 5;28(18):7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F. X., Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991 Feb 12;30(6):1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- Cline T. W., Hastings J. W. Mutationally altered bacterial luciferase. Implications for subunit functions. Biochemistry. 1972 Aug 29;11(18):3359–3370. doi: 10.1021/bi00768a008. [DOI] [PubMed] [Google Scholar]
- Cohn D. H., Mileham A. J., Simon M. I., Nealson K. H., Rausch S. K., Bonam D., Baldwin T. O. Nucleotide sequence of the luxA gene of Vibrio harveyi and the complete amino acid sequence of the alpha subunit of bacterial luciferase. J Biol Chem. 1985 May 25;260(10):6139–6146. [PubMed] [Google Scholar]
- Flynn G. C., Chappell T. G., Rothman J. E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989 Jul 28;245(4916):385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Friedland J., Hastings J. W. Nonidentical subunits of bacterial luciferase: their isolation and recombination to form active enzyme. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2336–2342. doi: 10.1073/pnas.58.6.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland J., Hastings J. W. The reversibility of the denaturation of bacterial luciferase. Biochemistry. 1967 Sep;6(9):2893–2900. doi: 10.1021/bi00861a033. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989 Dec 21;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Hennessey J. P., Jr, Johnson W. C., Jr Information content in the circular dichroism of proteins. Biochemistry. 1981 Mar 3;20(5):1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- Holzman T. F., Baldwin T. O. Isolation of bacterial luciferases by affinity chromatography on 2,2-diphenylpropylamine-Sepharose: phosphate-mediated binding to an immobilized substrate analogue. Biochemistry. 1982 Nov 23;21(24):6194–6201. doi: 10.1021/bi00267a026. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Wright P. E., Baldwin R. L. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990 Sep 28;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- Johnston T. C., Thompson R. B., Baldwin T. O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J Biol Chem. 1986 Apr 15;261(11):4805–4811. [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- Landry S. J., Jordan R., McMacken R., Gierasch L. M. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992 Jan 30;355(6359):455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992 Apr 23;356(6371):683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Lu J., Dahlquist F. W. Detection and characterization of an early folding intermediate of T4 lysozyme using pulsed hydrogen exchange and two-dimensional NMR. Biochemistry. 1992 May 26;31(20):4749–4756. doi: 10.1021/bi00135a002. [DOI] [PubMed] [Google Scholar]
- Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate. Nature. 1991 Jul 4;352(6330):36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Mayo K. H., Barker S., Kuranda M. J., Hunt A. J., Myers J. A., Maione T. E. Molten globule monomer to condensed dimer: role of disulfide bonds in platelet factor-4 folding and subunit association. Biochemistry. 1992 Dec 8;31(48):12255–12265. doi: 10.1021/bi00163a040. [DOI] [PubMed] [Google Scholar]
- McIntosh L. P., Wand A. J., Lowry D. F., Redfield A. G., Dahlquist F. W. Assignment of the backbone 1H and 15N NMR resonances of bacteriophage T4 lysozyme. Biochemistry. 1990 Jul 10;29(27):6341–6362. doi: 10.1021/bi00479a003. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Hastings J. W. Binding site determination from kinetic data. Reduced flavin mononucleotide binding to bacterial luciferase. J Biol Chem. 1971 Dec 25;246(24):7666–7674. [PubMed] [Google Scholar]
- Meighen E. A., Nicoli M. Z., Hastings J. W. Functional differences of the nonidentical subunits of bacterial luciferase. Properties of hybrids of native and chemically modified bacterial luciferase. Biochemistry. 1971 Oct 26;10(22):4069–4073. doi: 10.1021/bi00798a009. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Nicoli M. Z., Hastings J. W. Hybridization of bacterial luciferase with a variant produced by chemical modification. Biochemistry. 1971 Oct 26;10(22):4062–4068. doi: 10.1021/bi00798a008. [DOI] [PubMed] [Google Scholar]
- Mendoza J. A., Rogers E., Lorimer G. H., Horowitz P. M. Chaperonins facilitate the in vitro folding of monomeric mitochondrial rhodanese. J Biol Chem. 1991 Jul 15;266(20):13044–13049. [PubMed] [Google Scholar]
- Miyamoto C. M., Boylan M., Graham A. F., Meighen E. A. Organization of the lux structural genes of Vibrio harveyi. Expression under the T7 bacteriophage promoter, mRNA analysis, and nucleotide sequence of the luxD gene. J Biol Chem. 1988 Sep 15;263(26):13393–13399. [PubMed] [Google Scholar]
- Miyamoto C. M., Graham A. D., Boylan M., Evans J. F., Hasel K. W., Meighen E. A., Graham A. F. Polycistronic mRNAs code for polypeptides of the Vibrio harveyi luminescence system. J Bacteriol. 1985 Mar;161(3):995–1001. doi: 10.1128/jb.161.3.995-1001.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday R. S., Englander S. W., Kallen R. G. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972 Jan 18;11(2):150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- Muchmore D. C., McIntosh L. P., Russell C. B., Anderson D. E., Dahlquist F. W. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- Ohgushi M., Wada A. 'Molten-globule state': a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983 Nov 28;164(1):21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- Roder H., Elöve G. A., Englander S. W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988 Oct 20;335(6192):700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P. E., Krieg U. C., Hwang S. T., Vestweber D., Schatz G. A precursor protein partly translocated into yeast mitochondria is bound to a 70 kd mitochondrial stress protein. EMBO J. 1990 Dec;9(13):4315–4322. doi: 10.1002/j.1460-2075.1990.tb07880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J. F., Waddle J. J., Waddill E. F., Baldwin T. O. Purified native subunits of bacterial luciferase are active in the bioluminescence reaction but fail to assemble into the alpha beta structure. Biochemistry. 1993 May 18;32(19):5036–5044. doi: 10.1021/bi00070a010. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- Waddle J. J., Johnston T. C., Baldwin T. O. Polypeptide folding and dimerization in bacterial luciferase occur by a concerted mechanism in vivo. Biochemistry. 1987 Aug 11;26(16):4917–4921. doi: 10.1021/bi00390a004. [DOI] [PubMed] [Google Scholar]
- Ziegler M. M., Goldberg M. E., Chaffotte A. F., Baldwin T. O. Refolding of luciferase subunits from urea and assembly of the active heterodimer. Evidence for folding intermediates that precede and follow the dimerization step on the pathway to the active form of the enzyme. J Biol Chem. 1993 May 25;268(15):10760–10765. [PubMed] [Google Scholar]




