Summary
Although proteasomes are critical in cell regulation and cancer therapy, little is known about the factors regulating proteasome content or activity. In this issue, Zhang et al report that miR-101 suppresses the expression of chaperone POMP and 20S assembly, and certain cancers raise proteasome content by losing miR-101.
In the ubiquitin proteasome pathway, protein half-lives are determined by the hundreds of different ubiquitinating enzymes that determine which proteins are degraded and when. Ubiquitinated proteins are then either deubiquitinated and escape degradation or are rapidly digested by the 26S proteasome. It is generally assumed that ubiquitination is the only regulated step in this pathway. However, there is growing evidence that protein breakdown rates are also regulated through changes in proteasome activity or content. 20S proteasomes can associate with alternative activating complexes (PA200 and PA28), whose physiological roles are still unclear. Several protein kinases, including PKA and CaMKII, can alter proteasome localization and activity, and the phosphatase UBLCP1 represses activity of nuclear proteasomes. The functional consequences of proteasome phosphorylation are only now emerging. Also, when proteasomes are inhibited or stalled, they auto-ubiquitinate a subunit that binds ubiquitinated proteins, Rpn13, and thus prevents substrate binding to non-functional proteasomes (Besche et al., 2014).
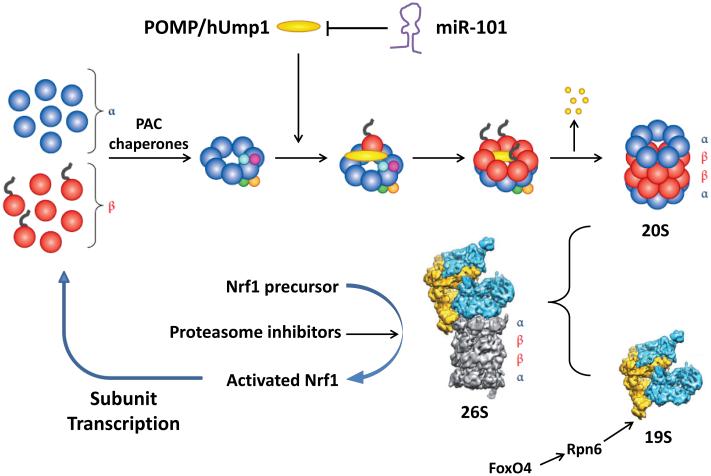
These intriguing mechanisms have the potential to rapidly alter proteolysis and cell composition, but their in vivo importance has not been extensively studied, nor has the control of proteasome content. In yeast, proteasome expression can change dramatically through a simple feedback mechanism involving the rapidly degraded transcription factor Rpn4. However, mammalian cells lack this system, and their proteasome content changes only little, even in conditions like muscle in fasting, where overall proteolysis rises and transcription of proteasome subunits increases (Piccirillo and Goldberg, 2012). Proteasome content varies several fold between cell types, and changes with differentiation,, and in some physiological states (e.g. denervated muscle (Piccirillo and Goldberg, 2012)). Proteasome activity falls during stem cell differentiation, because of decreased FoxO4-mediated transcription of Rpn6 (Vilchez et al., 2012), which can limit the assembly of the 26S. Interestingly, overexpressing Rpn6 in C. elegans increases longevity, presumably by raising 26S proteasome levels. Proteasomes also regulate their own levels. When their function is partially inhibited, proteasomes increase the proteolyitc processing of NrF1 to its active form (Figure 1), which stimulates transcription of all 26S subunits (Sha and Goldberg, 2014).
Figure 1. Different Strategies to Regulate 26S Abundance.
In this issue of Molecular Cell, Zhang et al describe the regulation of 20S assembly by controlling levels of a limiting chaperone, POMP/hUMP1 by the miR-101. This mode of regulation appears to be disrupted in at least some cancer types. Stem cells maintain high levels of 26S proteasomes by increasing transcription of a limiting subunit, Rpn6, under control of FoxO4. Cells that are treated with proteasome inhibitors and cells in certain types of proteotoxic stress compensate by inducing via the transcription factor Nrf1 all proteasome subunits plus POMP and other 20S assembly factors.
In this issue of Molecular Cell, Zhang et al. describe a novel mechanism controlling proteasome content that can limit tumor cell growth. Assembling the 28-subunit 20S core proteasome is a complex process catalyzed by several chaperones (Tomko and Hochstrasser, 2013). Forming this particle involves special challenges since it contains six proteolytic sites with the potential to digest most cell proteins. Consequently, 20S formation must be precisely regulated. It’s proteolytic sites function by an unusual catalytic mechanism involving an N-terminal threonine residue, whose generation requires the autolytic removal of a N-terminal propeptide. Two pairs of chaperones, Pac1-Pac2 and Pac3-Pac4, help assemble the outer α-ring and someβ-subunit precursors to form two-ring hemi-proteasomes. Another chaperone, POMP/hUMP1 then promotes the assembly of the four-ring mature particle by incorporating remaining β-subunits and promoting proteolytic processing of the catalytic subunits (Figure 1). In this process, POMP becomes trapped within the proteasome and becomes its first victim (Tomko and Hochstrasser, 2013). POMP thus functions non-catalytically. Several other chaperones catalyze 19S base assembly, but are not consumed during proteasome formation. POMP thus limits 26S production, since its overexpression enhances proteasome content and resistance to stressors, while its downregulation reduces proteasome content (Heink et al., 2005).
Zhang et al. discovered that tumor cells can have increased levels of proteasomes and demonstrated an important new factor determining proteasome content and assembly, miR-101. This micro-RNA may function as a tumor suppressor, because its levels are low in a number of malignancies, and its overexpression prevents cell proliferation. These workers were initially investigating whether miR-101 might activate the tumor suppressor p53, but, to their surprise, found that overproducing miR-101 causes p53 and other short-lived proteins to accumulate in ubiquitinated forms (Zhang et al., 2015). In studying how miR-101 suppresses conjugate degradation generally, they discovered that one target of miR-101 is POMP (Figure 1). Overexpressing miR-101 depleted POMP and eventually reduced 20S and 26S content (Zhang et al., 2015).
However, it is unclear how much proteasome levels have to be reduced to suppress proteolysis. Neurons, and presumably other cells, contain a large excess of 26S, which are not actively engaged in proteolysis (Asano et al., 2015), and may only function in stressful conditions. Many cells also have abundant free 20S whose possible involvement in normal protein turnover remains controversial. It had been proposed that cancers are inherently more active in protein degradation due to more rapid proliferation and the presence of many abnormal proteins resulting from aneuploidy. Therefore, malignant and normal cells may differ in their reservoirs of unused proteasomes and their susceptibility to miR-101.
Although genetic loss of miR-101 occurs in some cancers (Varambally et al., 2008), little is known concerning the normal regulation of miR-101 content. Different types of T cells vary markedly in their levels of miR-101, which regulates immune responses through other targets (Yu et al., 2007). However, since these T cells probably have different amounts of POMP, they may also differ in proteasome content and proteolytic capacity, which could be important in determining their immune function.
Because miR-101 has many targets, it was unclear if POMP is truly important in miR-101’s anti-proliferative activity. Therefore, these workers mutated its binding sites in the 3’-UTR of POMP mRNA and demonstrated that miR-101 requires POMP knockdown to slow proliferation of U2OS (Zhang et al., 2015). Interestingly, miR-101 retards proliferation of SKBr3 breast cancer cells primarily by downregulating a histone methyltransferase, EZH2 (Varambally et al., 2008). Thus, miR-101 may slow proliferation via distinct targets in different cells.
These observations emphasize the therapeutic importance of studying proteasomal expression and assembly in normal and malignant cells. Proteasome inhibitors (e.g., bortezomib) have dramatically improved the treatment of multiple myeloma, a cancer of plasma cells. These cells are particularly sensitive to proteasome inhibition because they express and degrade exceptionally high loads of aberrant proteins (mutant immunoglobins), and require proteasomal production of NFκB for growth and invasiveness. Elevated 20S levels were observed in the bortezomib-resistant U2OS cells, and miR-101 overexpression re-sensitized these cells to this drug (Zhang et al., 2015). Therefore, expression of miR-101 by myeloma cells will be important to measure and targeting POMP represents a potentially valuable strategy to inhibit proteasome function and cancers generally. Inhibiting POMP and 20S assembly without blocking subunit expression must significantly increase the cellular content of aggregation-prone free subunits, further challenging the cancer cell’s already compromised protein degradative machinery. 26S proteasomes respond to proteasome inhibitors by Nrf1-dependent production of new proteasomes, which can enhance their cellular resistance to the drugs. However, inhibiting 20S formation could avoid this compensatory mechanism, and would also block formation of immunoproteasomes, the specialized forms found in immune cells. Thus far, proteasome inhibitors have not proven useful in treating solid tumors, in part due to drug binding to large numbers of proteasomes present in red cells. Inhibiting proteasome assembly, which does not occur in these non-nucleated cells, may avoid this pharmacokinetic problem.
This important new role for mir-101 in the regulation of POMP opens many intriguing questions for study. It will now be important to determine how miR-101 and particle assembly are regulated during differentiation, with physiological stimuli, and in different tumors, and whether mir-101 or other miRNAs also regulate proteasome expression or assembly.
Acknowledgements
The authors are grateful to the NIGMS (RO-051923-17) and Target ALS for research support and to Megan LaChance for assistance in preparing this manuscript. We regret that space limitations did not allow us to cite all the investigators whose contributions were discussed.
References
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L, Goldberg AL. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci U S A. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 2012;31:3334–3350. doi: 10.1038/emboj.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr., Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]