Abstract
The ubiquitin proteasome system (UPS) degrades misfolded proteins including those implicated in neurodegenerative diseases. We investigated the effects of tau accumulation on proteasome function in a mouse model of tauopathy and in a cross to a UPS reporter mouse (line Ub-G76V-GFP). Accumulation of insoluble tau was associated with a decrease in the peptidase activity of brain 26S proteasomes, higher levels of ubiquitinated proteins and undegraded Ub-G76V-GFP. 26S proteasomes from mice with tauopathy were physically associated with tau and were less active in hydrolyzing ubiquitinated proteins, small peptides and ATP. 26S proteasomes from normal mice incubated with recombinant oligomers or fibrils also showed lower hydrolyzing capacity in the same assays, implicating tau as a proteotoxin. Administration of an agent that activates cAMP–protein kinase A (PKA) signaling led to attenuation of proteasome dysfunction, probably through proteasome subunit phosphorylation. In vivo, this led to lower levels of aggregated tau and improvements in cognitive performance.
The UPS is the major pathway for protein degradation in eukaryotic cells1. Proteins are covalently tagged by the attachment of a polyubiquitin chain leading to rapid binding and hydrolysis by the 26S proteasome. This large (66-subunit) ATP-dependent proteolytic complex binds ubiquitinated proteins via receptor subunits on its 19S regulatory particle and then the ATPase complexes unfold and translocate the polypeptides into the 20S core particle where they are digested to small peptides by its six peptidase sites2–4. The proteasome’s ability to hydrolyze short peptides can be stimulated by agents that cause cAMP accumulation or by treatment with pure protein kinase A (PKA)5–7. The accumulation of ubiquitinated protein inclusions in neurodegenerative diseases8 suggests that defects exist in 26S proteasome-mediated clearance in affected neurons, and in support of this, tau from people with Alzheimer’s disease has been shown to be polyubiquitinated at several sites9–11 and several studies have implicated UPS dysfunction in response to tauopathy12–17. Herein, we demonstrate that pharmacological agents that raise cAMP in the brain and activate PKA can phosphorylate proteasome subunits, enhance proteasome activity, promote clearance of abnormal tau and improve cognition.
RESULTS
Tau aggregation and accumulation of ubiquitin conjugates
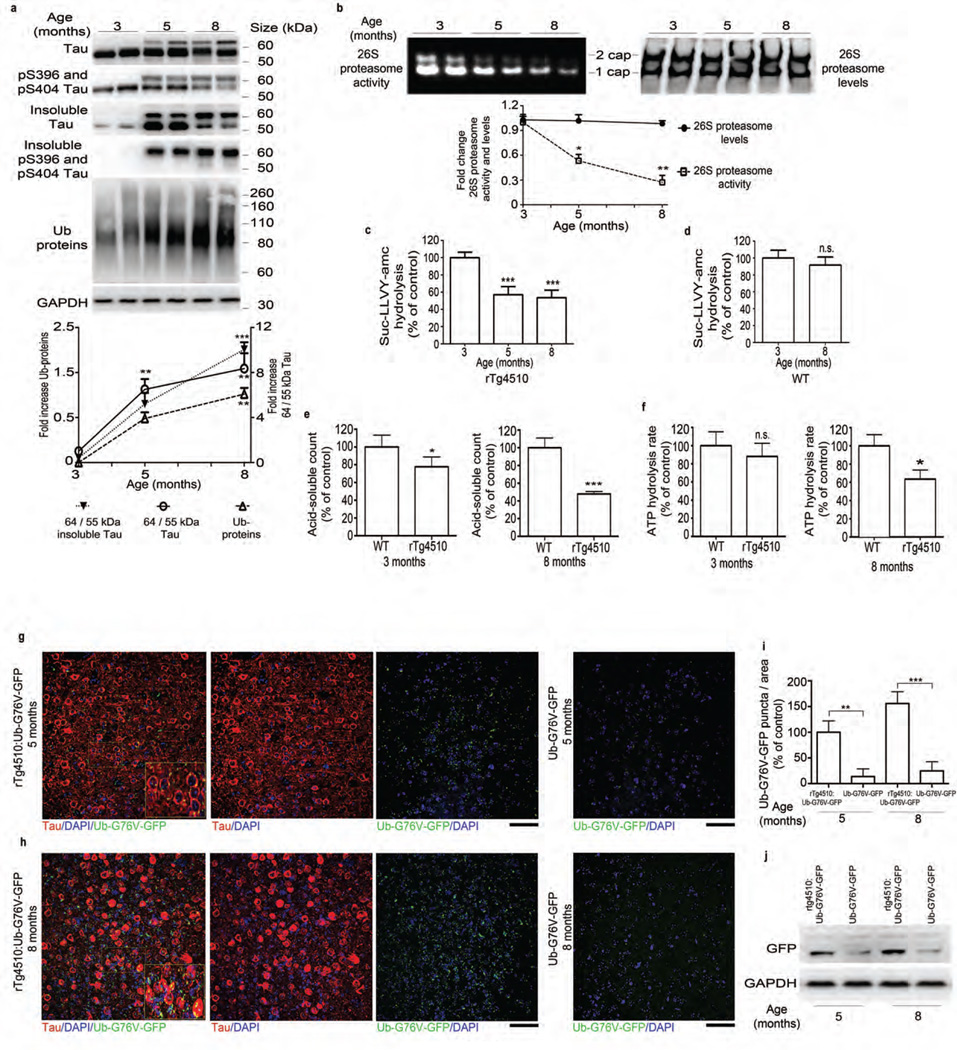
We first investigated the impact of progressive tauopathy on the UPS in the rTg4510 mouse, which expresses a pathogenic tau mutation (P301L) and exhibits progressive neurofibrillary pathology, neuronal loss and cognitive deficits18. At 3–4 months, these mice model early-stage disease; by 8 months they resemble a more severe stage of the human disease. By 5 months, soluble tau migrating at ~55 kDa converts to a disease-associated, hyperphosphorylated insoluble tau species that migrates at ~64 kDa (Fig. 1a). The ratio of 64-kDa to 55-kDa tau bands in cortical tissue (here referred to as the 64/55-kDa tau ratio) can be used to indicate the tauopathy stage of these mice. We observed the greatest change in the 64/55-kDa tau ratio in mice between 3 and 5 months of age, when the ratio increased fivefold. By 8 months, the 64/55 kDa tau ratio had increased further. Examination of additional time points (Supplementary Fig. 1a,b) identified 3.5–4.5 months as the time at which 64-kDa tau first began to accumulate. The shift to 64-kDa forms coincided with an increase in the amount of sarkosyl-insoluble total and phosphorylated tau, a concomitant decrease in soluble (heat-stable) tau (Fig. 1a and Supplementary Fig. 1c) and accumulation of total ubiquitinated proteins (Fig. 1a).
Figure 1.
Tauopathy is associated with a progressive decrease in proteasome function. (a) Top, immunoblot analysis of tau and pS396 and pS404 tau, Ub (ubiquitin) and GAPDH (for normalization) in total and sarkosyl-insoluble extracts from rTg4510 mice. Bottom, quantified densitometry of 64/55-kDa tau ratio in total and insoluble tau and ubiquitin, expressed as fold change relative to 3 months of age. (b) Native PAGE of 26S proteasome activity and levels (immunoprobing for Rpt6) and quantified densitometry (bottom). (c,d) Chymotrypsin-like activity of purified 26S proteasomes from rTg4510 (c) and WT (d) mice at indicated ages. (e) Degradation rate of 32P-labeled Ub5-DHFR by purified 26S proteasomes from rTg4510 and WT mice at indicated ages. (f) ATPase activity of purified 26S proteasomes from rTg4510 and WT (control) mice at indicated ages (g,h) Immunofluorescence labeling of human tau (red) and GFP signal (green, detected without antibody) in the frontal cortex of rTg4510:Ub-G76V-GFP and Ub-G76V-GFP mice at 5 (g) and 8 (h) months of age. Insets, high-magnification views of outlined areas. Scale bars, 50 µm. (i) Quantification of GFP puncta from analyses in g and h. Control, 5-month-old rTg4510:Ub-G76V-GFP. (j) Immunoblot analysis of GFP expression in rTg4510:Ub-G76V-GFP and Ub-G76V-GFP mice. For a,b and j at least three biological experiments (two mice per experiment, n = 6 mice) were performed. For c–f, n = 6 cortical brains per age group were used to elute 26S proteasomes, and at least three independent experiments were performed. Quantification of GFP signal for g and h was performed on slices from 6 mice per group. Error bars, mean ± s.e.m.; n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001 (one-way ANOVA followed by Tukey’s multiple comparison post hoc test).
Tauopathy decreases 26S proteasome activity
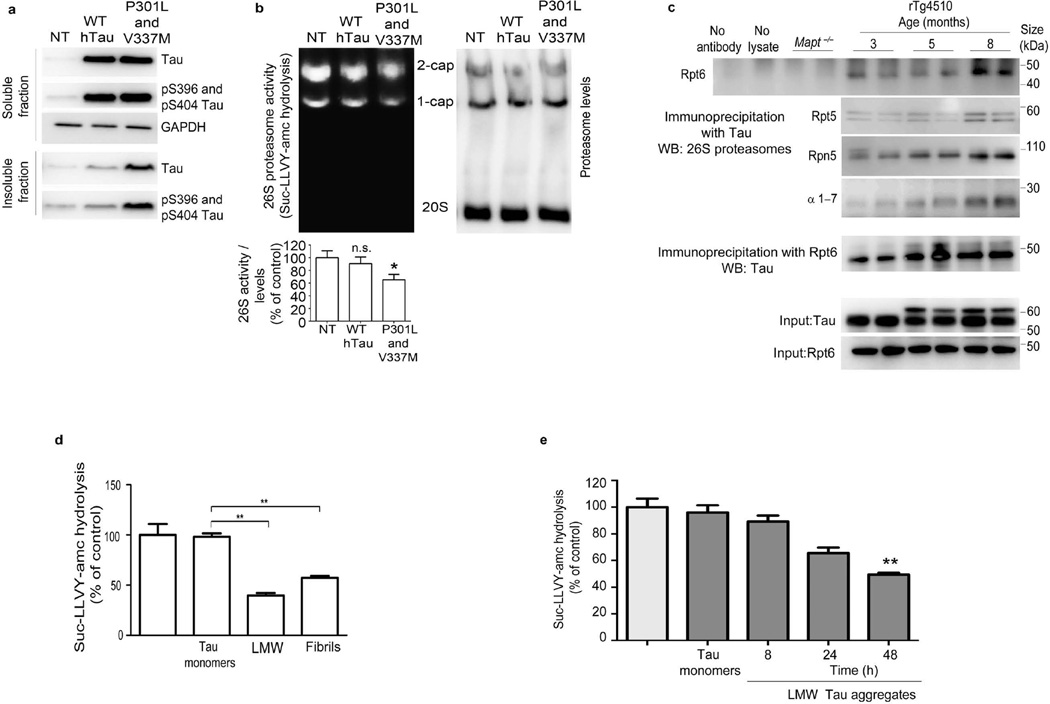
To assess whether worsening tauopathy impairs 26S proteasome function, we first measured the chymotrypsin-like activity of the 26S proteasomes. In older mice with a higher 64/55-kDa tau ratio, peptidase activity in the cortical brain extracts decreased. The activity of both singly (1-cap) and doubly (2-cap) capped 26S proteasomes decreased under these assay conditions; the free 20S particles showed no activity (Fig. 1b). This decrease was not due to reduced 26S or 20S proteasome levels, as there was no change in the levels of the 26S proteasome regulatory subunit Rpt6 (Fig. 1b) or the 20S subunit (Supplementary Fig. 2a). Wild-type (WT) mice showed no decrease in 26S proteasome activity over this period (Supplementary Fig. 2b). To assay proteasome function more rigorously, we purified 26S proteasomes from mouse cortex by affinity chromatography using the ubiquitin-like domain (UBL)19. Purified proteasomes also showed a decrease in peptidase activity in mice between 3 and 5 months of age, coinciding with the period when levels of ~64 kDa tau increased. All three of the particle’s peptidase activities—chymotrypsin- (Fig. 1c), trypsin- and caspase-like activities (Supplementary Fig. 2c,e) decreased during this period. Peptidase activity of WT proteasomes was not affected up to 8 months of age (Fig. 1d and Supplementary Fig. 2d,f). Proteasomes from rTg4510 mice showed lower 26S peptidase activity than proteasomes from age-matched WT mice (Supplementary Fig. 2g–i) but no changes in 26S (2-cap and 1-cap) levels, as assessed by immunoblotting of native PAGE gel for the Rpt6 subunit (Supplementary Fig. 2g), indicating that the decrease in 26S peptidase activity was related to tauopathy. Ubiquitinated proteins are the primary substrates of 26S proteasomes in cells. Therefore, to obtain a more physiologically relevant measure of proteasome function in rTg4510 mice, we measured the degradation rates of two well-characterized substrates, ubiquitinated DHFR (Ub5-DHFR) and ubiquitinated Sic1 (Ubn-Sic1)20,21. Proteasomes from rTg4510 mice aged 5 and 8 months showed a reduced capacity to hydrolyze these ubiquitin conjugates, compared to proteasomes from 3-month-old rTg4510 or age-matched WT mice (Fig. 1e and Supplementary Fig. 2j). ATP hydrolysis by the six AAA+ ATPases of the 19S regulatory particles is crucial for substrate degradation20,22,23. To examine the function of ATPase subunits, we measured ATP hydrolysis20,23 of purified proteasomes from WT and rTg4510 mice. Proteasomes from 8-month-old rTg4510 mice showed reduced ATP hydrolysis, as compared to age-matched WT proteasomes (Fig. 1f), but the difference between rTg4510 and WT was not significant at 3 months. To obtain additional evidence that worsening tauopathy is associated with proteasome dysfunction, we generated a transgenic mouse by crossing rTg4510 mice to a UPS reporter line expressing a 26S-targeted fragment fused to GFP24. Normally, the fusion protein is ubiquitinated and hydrolyzed rapidly by the 26S proteasome via the ubiquitin-fusion degradation (UFD) pathway20. GFP is therefore undetectable (or present at very low levels) in normal cells, but it is stabilized and accumulates when there are defects in the UPS. The transgenic (rTg4510:Ub-G76V-GFP) mice showed accumulation of GFP puncta, which increased with worsening tauopathy from 5 (Fig. 1g) to 8 months (Fig. 1h). In contrast, the brains of littermate mice expressing Ub-G76V-GFP but not mutant tau (Ub-G76V-GFP mice) accumulated very low amounts of GFP at both 5 (Fig. 1g) and 8 (Fig. 1h) months, although there was a slight (nonsignificant) increase with age. Quantitative analysis of GFP by immunohistochemistry (Fig. 1i) and immunoblotting (Fig. 1j) showed more GFP in rTg4510:Ub-G76V-GFP mice than in age-matched Ub-G76V-GFP controls. The GFP signal was often localized in the cell body and appeared to overlap with human tau, which is predominantly somatodendritic at 5–8 months. However, in the rTg4510:Ub:G76V-GFP mice, the intensity of tau staining resulting from the high level of tau expression made it difficult to observe GFP signal in the same cells. We therefore examined a second mouse line (JNPL3, which carry a human tau with a P301L mutation25, crossed to Ub-G76V-GFP mice). Hindbrain neurons from the resulting JNPL3:Ub-G76V-GFP mice showed colocalization of tau and GFP (Supplementary Fig. 3), probably owing to co-sequestration of aggregated tau and GFP into inclusion bodies. To confirm that the accumulation of tau aggregates and not soluble tau impairs proteasome function, we generated stable lines of human embryonic kidney (HEK) cells expressing WT tau or highly aggregable mutant tau. The cell lines expressed similar amounts of soluble tau, but the mutant line showed higher amounts of insoluble tau (Fig. 2a). The chymotrypsin-like activity of 26S proteasomes from tau mutant– expressing cells was lower than that in WT-expressing cells, which was not different from that in non-transfected, control cells (Fig. 2b). Protein aggregates that cause neurodegenerative diseases have been proposed to physically associate with proteasomes and interfere with substrate degradation26. Immunoprecipitation of human tau from cortical brain extracts of rTg4510 mice aged 3, 5 or 8 months (Fig. 2c) co-precipitated several proteasome subunits. To test whether aggregates of tau can directly cause proteasome dysfunction, we incubated proteasomes with recombinant monomeric tau or aggregates27. Incubation of 26S proteasomes from WT mice with low–molecular weight (LMW) oligomeric or fibrillar tau reduced peptidase activity, whereas incubation of 26S proteasomes with monomeric tau had no effect (Fig. 2d). To examine the effect of LMW forms of tau, we incubated recombinant tau for 8, 24 or 48 h and then incubated equivalent amounts of each oligomers preparation with purified proteasomes. Tau monomers had no effect, but oligomers incubated for 48 h impaired proteasome function significantly (Fig. 2e).
Figure 2.
Aggregated tau directly inhibits 26S proteasomes and associates with proteasomes in brains of mice with tauopathy. (a) Immunoblot analysis for tau and phosphorylated (pS396 and pS404) tau epitopes from soluble and insoluble fractions in HEK293 cells nontransfected (NT) or stably transfected with WT human tau (hTau) or double-mutant P301L and V337M tau. (b) Native PAGE assay (top) and quantification (bottom) of 26S proteasome activity and levels (assessed by immunoblotting for β5 subunit) in NT HEK293 cells and HEK293 cells transfected with WT hTau or P301L and V337M tau. The densitometric quantification of 26S proteasome activity normalized to 26S proteasome levels (c) Immunoprecipitation with antibody to human tau of cortical brain lysates from Mapt−/− (negative control) and rTg4510 mice at indicated ages and immunoblot analysis of Rpt6, Rpt5, Rpn5 and 20S α-subunits 1–7 (α1–7) expression and a reciprocal experiment using anti-Rpt6 for immunoprecipitation and anti-hTau for immunoblotting. The total extracts (input) for tau and Rpt6 are shown as loading controls. (d,e) Rate of succinyl-Leu-Leu-Val-Tyr-amc (Suc-LLVY-amc) hydrolysis by 26S proteasomes, incubated with tau monomers, LMW aggregates and fibrils generated from recombinant tau (d), or incubated with tau monomers and LMW aggregates generated by incubating recombinant tau at indicated time points (e). Untreated WT 26S proteasomes were used as a control for d and e. At least three biological replicates were performed with stably transfected clones (a,b). For immunoprecipitation (c), two animals per experiment were analyzed in three biological replicates (n = 6 mice). Purified proteasomes from n = 6 brains from 3-month-old WT were used for d and e, and at least three independent experiments were performed (d,e). Error bars, means ± s.e.m. *P < 0.05, **P < 0.01 (one-way ANOVA followed by Tukey’s multiple comparison post hoc test).
Enhanced proteasome activity via cAMP-PKA attenuates tauopathy
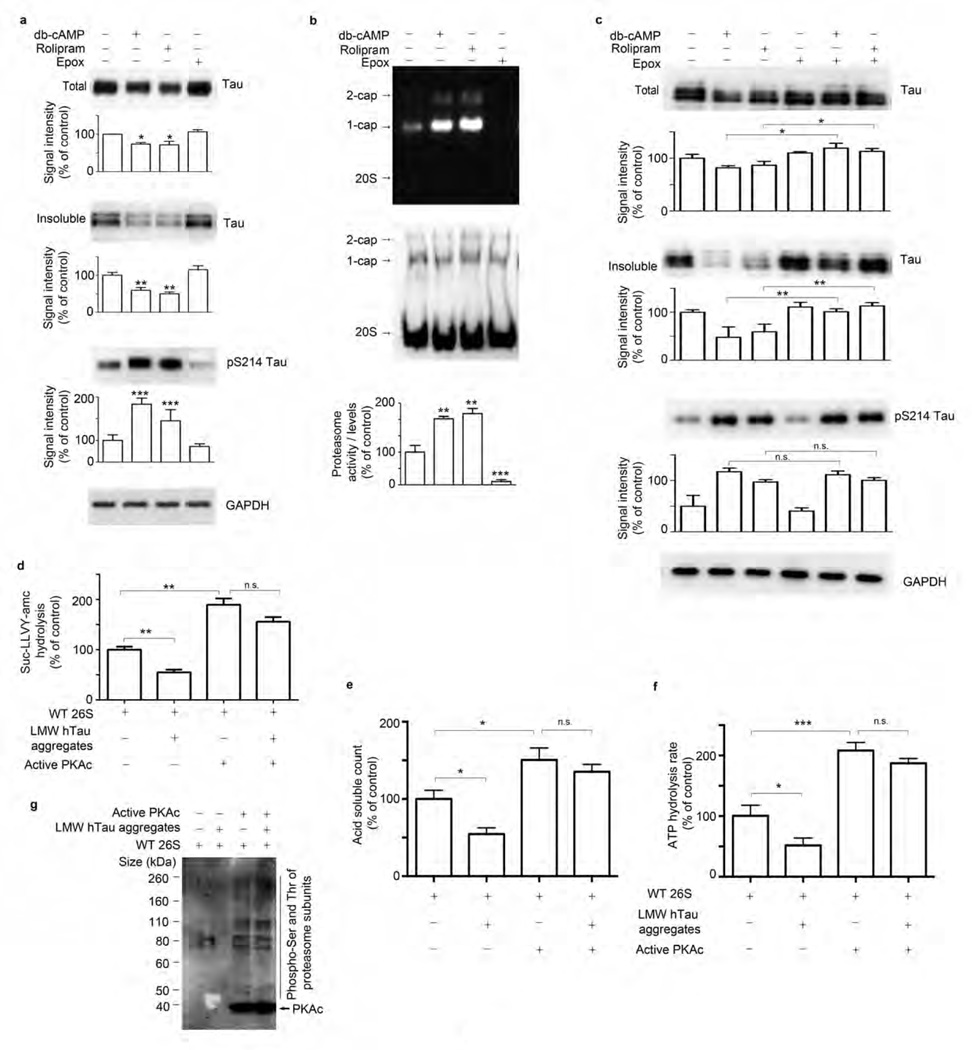
PKA enhances proteasome peptidase activity5,6. Therefore, we tested whether raising levels of cAMP and activating PKA would stimulate proteasome activity ex vivo (Fig. 3) and in vivo (Figs. 4 and 5). Rolipram is a specific phosphodiesterase type 4 (PDE4) inhibitor that increases cAMP levels in multiple tissues in vivo28. To assess whether activation of cAMP-PKA enhances proteasome function and reduces the accumulation of tau, we prepared cortical brain slices from rTg4510 mice aged 3–4 months (early-stage mice) and treated them with vehicle, dibutyryl-cAMP (db-cAMP), rolipram or epoxomicin (a specific inhibitor of the proteasome’s chymotrypsin-like activity). Exposure of slices from the early-stage mice to rolipram or db-cAMP for 8 h reduced amounts of total and insoluble tau (Fig. 3a), including phosphorylated tau (Supplementary Fig. 4a,b). In contrast, epoxomicin treatment increased levels of phosphorylated tau (Supplementary Fig. 4), although the amount of total tau remained unchanged (Fig. 3a). PKA is known to phosphorylate tau at Ser214 (pS214)29. We therefore monitored the amount of pS214 tau to determine whether rolipram and db-cAMP activated PKA. The amount of pS214 tau was higher in db-cAMP– and rolipram-treated samples than in control (DMSO) and epoxomicin-treated slices (Fig. 3a). The chymotrypsin-like activity of the 26S proteasomes in crude extracts was elevated after administration of db-cAMP or rolipram but was blocked by epoxomicin (Fig. 3b). To verify that these effects were mediated through enhancement of proteasomal activity and not some other effect of cAMP-PKA, we treated organotypic slices from early-stage rTg4510 mice with epoxomicin and db-cAMP or rolipram (Fig. 3c). Epoxomicin prevented the decrease in total and insoluble tau seen with db-cAMP and rolipram treatment alone. pS214 tau has been shown to reduce recombinant tau aggregation in vitro30,31. However, in acute organotypic slices, hyper phosphorylation of Ser214 by db-cAMP or rolipram occurred in the presence of epoxomicin but without a reduction in tau aggregates (Fig. 3c).
Figure 3.
Activation of PKA stimulates hydrolyzing activity of the proteasome in slices-ex vivo and in vitro. (a–c) Immunoblot analysis and corresponding densitometric quantification of total and insoluble extracts for tau, pS214 tau epitope and GAPDH (a,c) and native PAGE analysis and quantification of 26S proteasome activity and level using antibody to the β5 subunit (b) in acute organotypic cortical slices from 3- to 4-month-old rTg4510 mice treated with vehicle, db-cAMP, rolipram or epoxomicin (Epox) alone or a combination of epoxomicin and db-cAMP or rolipram. (d–f) Hydrolysis rate of succinyl-Leu-Leu-Val-Tyr-amc (Suc-LLVY-amc) (d), 32P-labeled Ub5-DHFR substrate (e) and ATP (f) by purified 26S proteasomes from WT mice untreated (WT 26S), incubated with LMW tau aggregates and/or pre-incubated with PKA. (g) Immunoblot analysis of phosphorylated serine and threonine (pSer and pThr) epitopes of proteasome subunits by PKA in purified 26S proteasomes treated as in d–f. For a–c, two animals per experiment were analyzed in three biological replicates (n = 6 mice). For d–g, purified proteasomes from n = 6 brains from 3-month-old WT mice were used for each hydrolyzing assay and at least three independent experiments were performed. Error bars, mean ± s.e.m.; n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001 (one-way ANOVA followed by Tukey’s multiple comparison post hoc test).
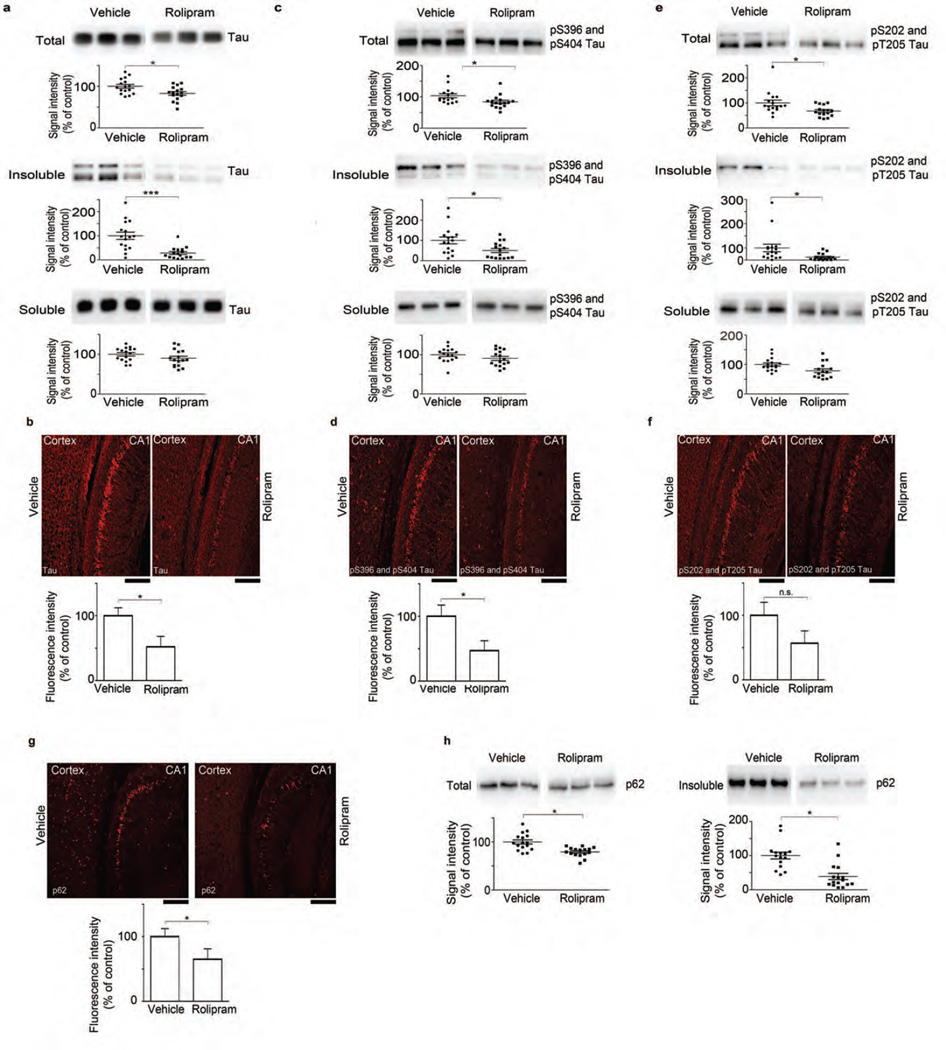
Figure 4.
Rolipram administration reduces accumulation of tau species and p62 in vivo. (a,c,e) Immunoblot analysis and corresponding densitometric quantification of total, insoluble and soluble extracts of tau (a) and pS396 and pS404 (c) and pS202 and pT205 (e) tau epitopes from cortical tissue of rTg4510 mice treated with vehicle or rolipram (b,d,f,g). Immunofluorescence labeling and quantification of fluorescence intensity for tau (b), pS396 and pS404 (d) and pS202 and pT205 (f) tau epitopes, and p62 (g) in the CA1 region of the hippocampus of rTg4510:Ub-G76V-GFP mice treated with vehicle or rolipram. Scale bars, 200 µm. (h) Immunoblot analysis and densitometric quantification of p62 in total and insoluble extracts from cortical tissue. Scatter plots represent quantification of immunoreactivity normalized to GAPDH. Statistical analyses of rolipram (n = 16) and vehicle-treated (n = 16) mice for a,c and e were performed in two sets. For quantification of immunofluorescence signal (b,d,f,g), slices from 6 mice per treatment group were analyzed. Error bars, mean ± s.e.m.; n.s., not significant; *P < 0.05, ***P < 0.001 (unpaired two-tailed Student’s t-test). Vehicle treatment served as control for all experiments.
Figure 5.
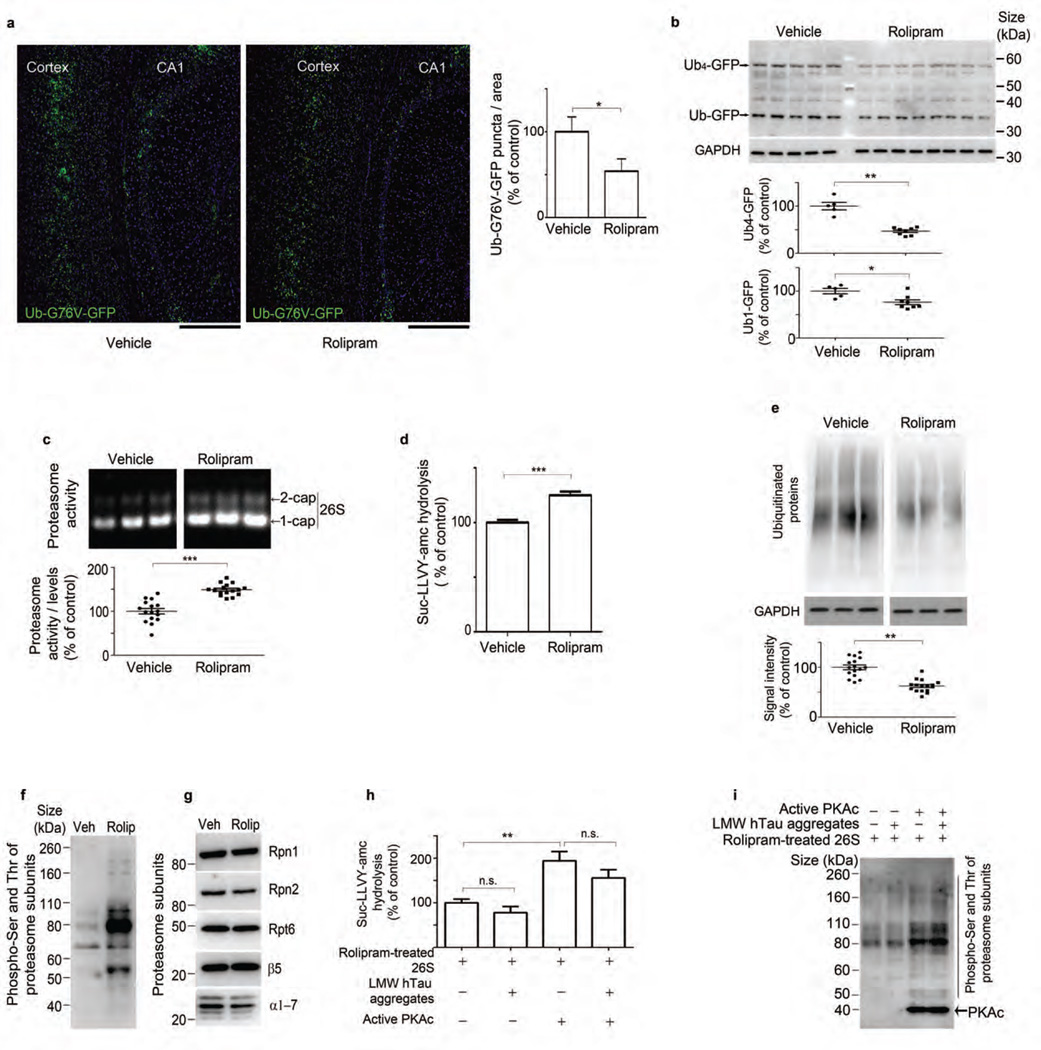
Rolipram treatment increases proteasome function and reduces ubiquitinated protein accumulation in vivo. (a) GFP signal and quantification of GFP puncta from hippocampus CA1 and the cortex of rTg4510:Ub-G76V-GFP mice. Scale bars, 200 µm. (b) Immunoblot analysis and densitometric quantification of GFP and GAPDH in vehicle- and rolipram-treated rTg4510:Ub-G76V-GFP mice. (c) Native PAGE of 26S proteasome and densitometric quantification of activity normalized to 26S proteasome levels in vehicle- and rolipram-treated rTg4510 mice. (d) Rate of hydrolysis by 26S proteasomes from vehicle- and rolipram-treated mice, incubated with succinyl-Leu-Leu-Val-Tyr-amc (Suc-LLVY-amc). (e) Immunoblot analysis and corresponding densitometric quantification of ubiquitinated protein and GAPDH in rTg4510 mice treated with vehicle or rolipram. (f,g) Immunoblot analysis of phosphorylated serine and threonine (pSer and pThr) (f) and proteasome subunits Rpn1, Rpn2, Rpt6, β5 and α-subunits 1–7 (α1–7) (g) in purified 26S proteasomes from vehicle- and rolipram-treated rTg4510 mice. (h) Rate of Suc-LLVY-amc hydrolysis by 26S proteasomes purified from rolipram-treated rTg4510 mice (control) or incubated with LMW tau aggregates and/or pre-incubated with PKA. (i) Immunoblot analysis of serine and threonine phosphorylation in rolipram-treated 26S proteasomes treated as in h. Statistical analyses for rTg4510 (rolipram n = 16 and vehicle n = 16) (c and e) and for rTg4510:Ub-G76V-GFP (rolipram n = 8 and vehicle n = 5) mice (b) and for quantification of GFP signal (n = 3 mice per treatment group) (a) were carried out using unpaired two-tailed Student’s t-test between groups. Purified proteasomes were pooled from n = 6 brains (d, f–i), and at least three independent experiments were performed. Statistical analyses employed two-tailed Student’s t-test between groups (d) and one-way ANOVA followed by Tukey’s multiple comparison post hoc test (h). Error bars, mean ± s.e.m.; n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001. Veh, vehicle; rolip, rolipram.
Phosphorylation of proteasomes protects against tau toxicity
To further confirm that the impact of enhanced PKA activity was on proteasome function and phosphorylation, we incubated purified 26S proteasomes from WT mouse brains with active PKA for 60 min and assayed their chymotrypsin-like activity. PKA treatment enhanced proteasome activity almost twofold (Fig. 3d). Proteasomes incubated with oligomeric tau showed a reduced rate of hydrolysis of the small fluorogenic peptide (Fig. 3d), but proteasomes pre-treated with PKA were protected from the inhibitory effect of tau oligomers, as the decrease in activity in PKA-treated proteasomes after the addition of oligomers was not statistically significant (Fig. 3d). WT proteasomes incubated with LMW oligomeric tau showed a reduced rate of hydrolysis of Ub5-DHFR (Fig. 3e) and a decrease in ATP hydrolysis (Fig. 3f), as compared to proteasomes without added oligomers. Pre-treatment of proteasomes with PKA enhanced degradation of Ub5-DHFR (Fig. 3e) and ATP hydrolysis (Fig. 3f) and protected against the inhibitory effect of oligomers on these activities (Fig. 3e,f). Thus PKA treatment appeared to protect proteasomes from the inhibitory effects of aggregated tau in vitro. PKA treatment led to an increase in phosphorylation of proteasome particles (Fig. 3g), supporting the idea that proteasome activation by PKA is mediated by phosphorylation of proteasome subunits. Analyses of the mechanisms involved in proteasome activation by PKA are presented in Lokireddy et al.32
Rolipram enhances proteasome activity and reduces tauopathy
To test whether raising cAMP with rolipram could reduce tauopathy and cognitive impairment, we administered rolipram to rTg4510 and rTg4510:Ub-G76V-GFP mice. Rolipram treatment during early-stage tauopathy in rTg4510 mice reduced amounts of total (Fig. 4a and Supplementary Fig. 5a) and phosphorylated tau (Fig. 4c,e and Supplementary Fig. 5b,c) in both the crude extracts and the insoluble fraction. The soluble (heat-stable) tau species also decreased slightly but not significantly. Rolipram treatment increased the total amount of pS214 tau in the extracts, confirming that PKA activity was enhanced (Supplementary Fig. 5d). To assess whether rolipram treatment attenuated UPS deficits in vivo, we also treated rTg4510:Ub-G76V-GFP mice. Rolipram-treated rTg4510:Ub-G76V-GFP mice showed reduced immunofluorescence labeling of total and phosphorylated tau (Fig. 4b,d,f). Because tau aggregates were reduced, we also measured p62 protein, which associates with insoluble ubiquitinated proteins after proteasome inhibition33. Rolipram treatment significantly decreased p62 levels in rTg4510:Ub-G76V-GFP mice, as assessed by immunofluorescence staining (Fig. 4g), and in rTg4510 mice, as assessed by quantitative immunoblotting (Fig. 4h and Supplementary Fig. 5e). GFP (an indicator of UPS dysfunction) was reduced as assessed by detection of fluorescence without antibody (Fig. 5a and Supplementary Fig. 6a) and by immunoblotting (Fig. 5b) in the CA1 region of the hippocampus and in the cortex of rTg4510: Ub-G76V-GFP mice treated with rolipram. Age-matched control mice (Ub-G76V-GFP) showed no change in the amount of GFP between vehicle- and rolipram-treated animals, as assessed by immunoblotting (Supplementary Fig. 6b), and no accumulation of GFP puncta when analyzed by confocal microscopy for GFP signal (Supplementary Fig. 6c). These findings indicate that rolipram treatment improved UPS-mediated degradation in mice with tauopathy in vivo. In support of these findings, when 26S proteasomes from the brains of these mice were resolved by native PAGE, both doubly capped and singly capped 26S particles showed increased activity in rolipram-treated mice (Fig. 5c and Supplementary Fig. 7a). The levels of proteasome proteins were unchanged (Supplementary Fig. 7b). Purified proteasomes from rolipram-treated mice had increased chymotrypsin- (Fig. 5d), trypsin- and caspase-like activities (Supplementary Fig. 8a,b), and the amount of ubiquitinated proteins was lower in extracts from rolipram-treated mice, consistent with enhanced proteasome activity (Fig. 5e and Supplementary Fig. 7c).
To assess whether the mechanism of enhanced serine and threonine phosphorylation of proteasome subunits could occur in rolipram-treated mice in vivo as observed in 26S proteasomes treated with purified PKA (Fig. 3e), we purified 26S particles from vehicle- (control) and rolipram-treated rTg4510 mouse brains and analyzed them by quantitative immunoblotting with an antibody to phosphorylated serines and threonines. Increased phosphorylation of serine and threonine epitopes of several subunits was evident in the rolipram-treated group (Fig. 5f), without any change in protein levels of proteasomes (Fig. 5g).
To assess whether rolipram treatment could protect proteasomes from tau-mediated inhibition, we isolated 26S proteasomes from rolipram-treated mice and incubated them with tau oligomers. Inhibition was not significant after rolipram-treated 26S proteasomes were incubated with tau oligomers (Fig. 5h), unlike the marked inhibition seen in proteasomes from untreated WT mice incubated with oligomers (Fig. 3d). Pre-incubation of proteasomes from rolipram-treated rTg4510 mice with PKA further increased proteasome phosphorylation (Fig. 5i), and peptidase activity was increased (Fig. 5h), suggesting that further activation was possible even in the presence of tau oligomers. cAMP-PKA signaling has been shown to suppress autophagy induction and autophagy-dependent neurite degeneration in primary neuronal cells bearing an LRRK2 mutation34. In our study, rolipram administration in rTg4510 mice had no effect on the levels of proteins associated with autophagic induction or on proteins responsible for elongation and maturation of the autophagosome (Supplementary Fig. 9).
Rolipram improves cognition in early-stage tauopathy
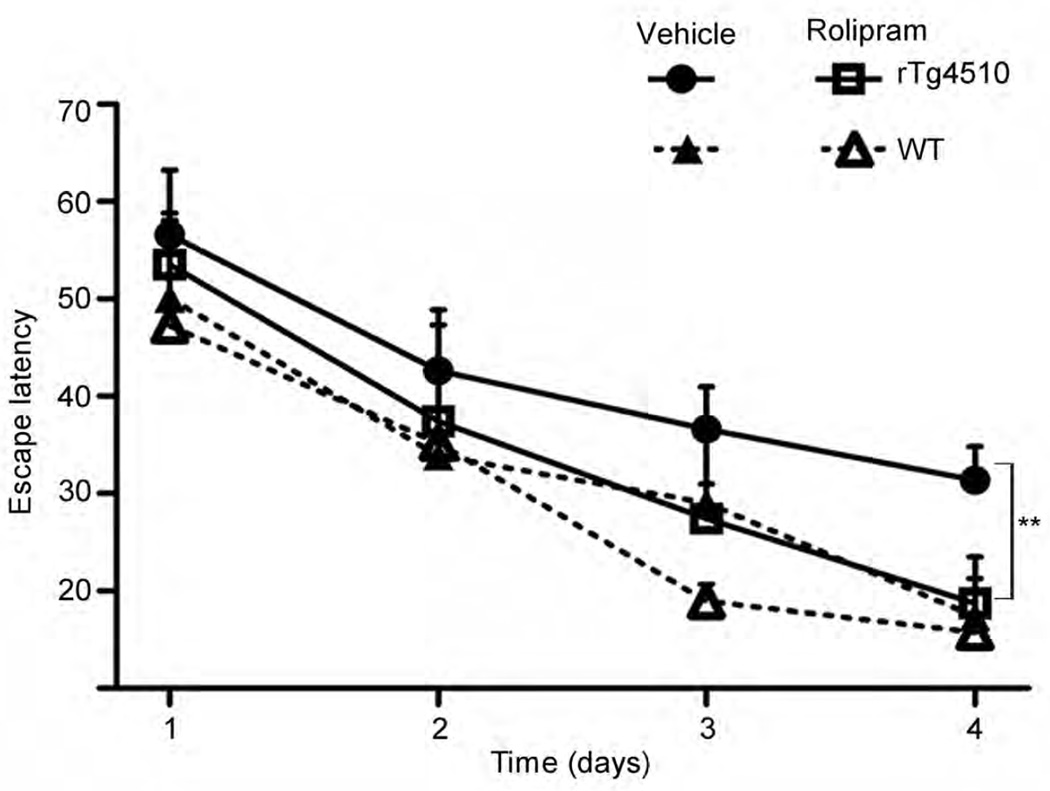
To assess the effect of rolipram on tauopathy-associated cognitive impairment, we examined spatial reference memory in rTg4510, rTg4510:Ub-G76V-GFP and WT mice in the Morris water maze. We observed a difference between WT and rTg4510 mice (genotype effect) and between treated and untreated (vehicle or rolipram) rTg4510 mice (treatment effect) on spatial learning over time (Fig. 6 and Supplementary Data). Escape latency at day 3 was not different across treatment groups for any strains tested. However, by day 4 of testing, cognitive impairment was detectable in vehicle-treated rTg4510 mice but not in rolipram-treated rTg4510 mice or WT groups, and the performance of rolipram-treated rTg4510 mice did not differ from that of WT mice. Rolipram treatment had the same effect in rTg4510:Ub-G76V-GFP mice (data not shown). This effect could not be attributed to a general improvement in cognition or performance, as rolipram treatment had no effect on the performance of WT mice.
Figure 6.
Rolipram treatment improves cognition in rTg4510 mice with early-stage disease. Cognitive performance, assessed by Morris water maze escape latency, of rTg4510 and WT mice treated with vehicle or rolipram. n = 16 (vehicle) or 15 (rolipram) in rTg4510 mice group; and n = 7 (vehicle) or n =7 (rolipram) in WT mice group. Error bars, mean ± s.d. **P < 0.01 (repeated-measures ANOVA test (days 1–3) plus one-way ANOVA with Bonferroni correction (day 4)).
Rolipram treatment is not effective in late-stage disease
Rolipram was not as effective at later stages of tauopathy (age 8–10 months in rTg4510 mice) as at 3–4 months, either ex vivo (Supplementary Fig. 10) or in vivo (Supplementary Fig. 11). In contrast to our observations in slices from early-stage mice, acute slices from cortex of mice with advanced tauopathy showed only a slight (not significant) decrease in total (Supplementary Fig. 10a) and phosphorylated (Supplementary Fig. 10b,c) tau levels after treatment with db-cAMP or rolipram. There was a trend toward increased proteasome activity upon exposure to db-cAMP or rolipram, but this was not statistically significant (Supplementary Fig. 10d), and activity was lower than that evoked in slices from early-stage brains. PKA activity (indicated by the level of pS214 tau) was increased after db-cAMP or rolipram treatment, but the change reached significance only after db-cAMP treatment (Supplementary Fig. 10e). In agreement with our findings ex vivo, rolipram administration in late-stage mice did not reduce the levels of total (Supplementary Fig. 11a) or phosphorylated tau (Supplementary Fig. 11b,c). The levels of ubiquitinated proteins were not significantly different in late-stage mice treated with vehicle and rolipram (Supplementary Fig. 11e). Furthermore, neither the activity of proteasomes nor the levels of proteasome proteins (Supplementary Fig. 11f) were significantly different. In vivo phosphorylation of 26S proteasome was lower in late-stage rolipram-treated rTg4510 mice than in early-stage, rolipram-treated mice (Supplementary Fig. 11g). Spatial learning was not improved in late-stage rTg4510 mice after rolipram treatment (Supplementary Fig. 12a and Supplementary Data). As PKA-mediated phosphorylation and activation of cAMP response element–binding protein (CREB) has been shown to lead to memory improvement35, we wanted to assess whether CREB was phosphorylated by rolipram. We assessed the amount of phosphorylated and total CREB in tissue lysates from vehicle- and rolipram-treated rTg4510 mice, and found that phosphorylated CREB was increased in early- and late-stage rolipram-treated mice (Supplementary Fig. 12b,c). As we observed cognitive improvement only in rolipram-treated early-stage mice, it is unlikely that the activation of CREB signaling by cAMP-PKA accounted for the improved cognition.
DISCUSSION
The degradation of ubiquitin conjugates by the 26S proteasome is coupled to ATP hydrolysis36, and ATP enhances multiple steps in this process, including conjugate binding, deubiquitination, substrate unfolding, gate opening in the 20S particle22,37 and substrate trans-location into the 20S central chamber21. Protein aggregates, such as tau, that bind the 26S particle but resist dissociation or translocation through the ATPases, could disrupt this multistep process and obstruct the degradation of other proteins. In support of this, defects in proteasomal degradation of ubiquitinated proteins have been observed in the brains of mice infected with the prion protein PrPSc and microaggregates of PrPSc were shown to directly bind to 20S core particles and interfere with ATPase-induced gate opening for substrate entry26, leading to reduced hydrolysis of peptides, as we have shown for tau. It has generally been assumed that the rate-limiting step in the UPS is substrate ubiquitination and that the proteasome digests all ubiquitinated proteins that bind to it. However, recent findings indicate that many ubiquitinated proteins bind the proteasome but are released (or deubiquitinated and released) without degradation2,36,38. In proteotoxic diseases, a larger fraction of ubiquitin conjugates may fail to be digested. Similarly to a Huntington’s disease model39, PKA-mediated phosphorylation in the tauopathy model may enhance the fraction of ubiquitinated proteins that bind and are degraded36,40. Our in vitro studies showed that enhancement of proteasome activity by cAMP-PKA caused a rapid drop in ubiquitin conjugate levels, strongly suggesting that PKA-induced enhancement of conjugate degradation was responsible for the enhanced tau clearance. Thus, the rate of protein breakdown in cells appears to be regulated by proteasome phosphorylation and not just by substrate ubiquitination32. Of particular note in the present study was the ability of a pharmacological agent to promote proteasome activity, reduce tau and ameliorate disease-associated cognitive defects through cAMP-PKA signaling in mice during early-stage disease. However, short-term cAMP-PKA activation in mice at later stages of disease did not have the same effect. It is possible that this was a result of the short (3-week) exposure to rolipram or of suboptimal dosing of the mice, which have a very robust tauopathy phenotype. Alternatively, the form of tau cleared by the UPS might be the form involved in cell-to-cell propagation of tauopathy, and prevention of propagation of this form of tau by rolipram might be apparent only in a mouse model, where propagation of the disease can be quantified41,42. We have shown that enhanced cAMP-PKA signaling in vivo leads to phosphorylation of several proteins in purified proteasomes leading to and that phosphorylation of critical proteasome subunits (particularly Rpn6) enhanced peptide and conjugate breakdown and ATPase function32. cAMP and PKA activate many other cellular processes, but enhancement of protein breakdown by phosphorylation of the proteasome is a new action for cAMP signaling. Together, these findings point to enhancement of proteasomal function through agents that phosphorylate proteasome subunits as a potentially promising therapeutic approach to treatment of proteotoxic diseases.
Online Methods
Mouse models
Double-transgenic rTg451018 mice express human tau with four microtubule–binding domain repeats (4R0N) and the P301L mutation (line FVB-Tg(tetO-MAPT*P301L human)#Kha/JlwsJ) under the control of mouse calcium-calmodulin kinase II–driven tetracycline-controlled transcriptional activator (tTa) (line Tg(Camk2a-tTA). Strain of origin is FVB. Ub-G76V-GFP mice express the transgene under the control of ACTB promoter with a cytomegalovirus immediate-early enhancer24 (line Tg(CAG-Ub*G76V/GFP) (The Jackson Laboratory, Stock No. 008112); strain of origin is CBA × C57BL/6. JNPL3 mice express human 4R0N P301L mutation, and expression is driven by the mouse prion promoter Tg(Prnp-MAPT*P301L)JNPL325 strain of origin is FVB. Two novel transgenic crosses were generated. The triple-transgenic rTg4510:Ub-G76V-GFP crosses were obtained by cross-breeding rTg4510 with Ub-G76V-GFP. Resulting offspring were FVB:C57BL/6F1. Breeding was between one male rTg4510 and two female Ub-G76V-GFP mice, which we found resulted in the largest litters. The double-transgenic JNPL3:Ub-G76V-GFP mice were generated by cross-breeding JNPL3 and Ub-G76V-GFP mice; resulting offspring were FVB:C57BL/6 F1. Control mice were nontransgenic or Ub-G76V-GFP littermates. For time-course studies, male rTg4510 mice aged 3–8 months were used (n = 6 per age group). Male, early-stage (3–4 months old) and late-stage disease (8–10 months old) mice were used for ex vivo studies. For in vivo studies, rTg4510 and nontransgenic littermate males were used, as female mice from this line have more variability in tau expression. Studies with rTg4510:Ub-G76V-GFP mice and their control littermates (Ub-G76V-GFP) used male and female mice. Males from the double-transgenic cross JNPL3:Ub-G76V-GFP were used. No animal or extracted sample was excluded from any of the analyses. For the in vivo rolipram trials, mice from the same litters were randomly assigned to each experimental group (vehicle or rolipram). Drug administration and testing was performed in two sets of mice. Mice were housed in 12 h light–12 h dark cycles with free access to food and water. Rolipram (0.03 mg/kg) or DMSO (0.05%) in 0.9% normal saline solution was administered intraperitoneally twice daily for 21 days. Animal experiments were in full compliance with the US National Institutes of Health Institutional Animal Care and Use Committee guidelines and overseen by Columbia University Medical Center.
To assess whether rolipram could prevent overt tauopathy development or reverse it after pathology had already developed, rolipram was administered to mice at an early stage of the disease (3–4 months of age) or a late stage (8–10 months). As a control, WT and Ub-G76V-GFP mice (3–4 months old) were also administered rolipram. The mice were tested for cognitive performance in the Morris water maze 6 h after the final administration and immediately sacrificed. Proteasome activity and tauopathy were assessed in brain extracts. Western blots for all rTg4510 mice from two sets of administrations of 3–4 months of age and from 8–10 months are shown in Supplementary Figures 5 and 9, respectively. Investigators were blinded to mouse genotype during cognitive testing.
Antibodies
Monoclonal antibodies to total human tau (CP27), pS396 and pS404 (PHF1) and pS202 and pT205 (CP13) were gifts from P. Davies. Polyclonal rabbit anti-pS214 tau was from Life Technologies (#44-742G); phospho-(Ser and Thr) PKA substrate (#9621), ULK1 (#4776), pS757 (#6888) and pS317 (#6887) ULK1, beclin 1 (#3788), Atg12 (#2010) were from Cell Signaling; anti-LC3 was from Novus Biological (NB600-1384); anti-GFP was from Abcam (#ab6556); rabbit anti-ubiquitin and rabbit anti-human tau were from Dako (Z0458 and A0024, respectively). Monoclonal mouse anti-CREB and phospho-S133 CREB (clone E306, #04-218 and clone 634-2, #05-807, respectively) were from Millipore; anti-GAPDH was from Sigma (clone GADH-71.1, #G8795); anti-p62 was from Abnova (clone 1C9, #H0000878-001); anti-Rpt6/S8 (clone EPR13565(B), #PW9265) and polyclonal rabbit anti-β5 (PW8895) were from BIOMOL; mono-clonal mouse anti-proteasome 20S (α1–α7) (clone MCP231, #BML-PW8195) was from Enzo. Anti-Rpn 1 (clone yC-19, #sc-26454), Rpn2 (clone 112-1,#sc-58007) and Rpn5 (clone N-12, #sc-107976) were from Santa Cruz. Secondary antibodies were from Jackson Immunoresearch, anti-mouse (115-035-003) and anti-rabbit (115-036-003).
Acute organotypic slice cultures
We prepared slice cultures using a method modified from Duff et al.43. Brains from rTg4510 mice at 3 and 8 months of age were harvested and hemispheres free of cerebellum and brainstem were sectioned into 350-µm-thick transverse slices and separated in ice-cold oxygenated Neurobasal A medium (Life Technologies). Slices were transferred to a 12-well plate connected to 95% O2/5% CO2 tank, containing 3.5 ml of Neurobasal A medium. Submerged slices were maintained for 8 h under continuous flow of 95% O2/5% CO2 (carbogen gas) at 37 °C bath temperature. We performed at least three biological experiments. For each experiment, hemispheres from two animals were randomly distributed in the wells, with each well containing 6–7 slices. Following a 15-min incubation, slices were treated with 2 mM db-cAMP (EMD Chemical), 20 µM rolipram, 500 nM epoxomicin (Sigma) or DMSO (final concentration of DMSO in the medium was 0.008%). In the co-treatment experiments, epoxomicin was added at the same time as db-cAMP and rolipram. Slices were collected and kept on dry ice until homogenization and protein extraction.
Cell lines
HEK293 cell lines were mock transfected (transfected with an empty expression plasmid) or stably transfected with one of two tau constructs: human 4R WT tau or human 4R P301L and V331M mutant tau. Transfected cells were subject to kanamycin selection of tau-expressing cells for native PAGE assay and western blotting. Prior to conducting assays, we tested cells for mycoplasma by microbiological culture method, where cell culture supernatant is cultivated on a mycoplasma agar medium and incubated at 37 °C for 2 weeks. Cells were grown in DMEM containing 10% FBS, 100 U/ml kanamycin and 1% nonessential amino acids.
Morris water maze
The Morris water maze test was carried out as previously described44. After nonspatial training, young (3–4 months of age) rTg4510 (15 rolipram- and 16 vehicle-treated mice, analyzed in two experiments) and age-matched WT mice (7 rolipram- and 7 vehicle-treated) underwent place-discrimination testing for 4 d with two trials per day. A second test was performed on rolipram- (n = 9) and vehicle-treated (n = 7) rTg4510 mice, aged 8–10 months, under identical conditions. Statistical methods were not employed to predetermine sample size; however, compared to other reported in vivo studies with transgenic animals, our studies included similar (old) or greater (young) numbers of animals. A third test was performed on young rTg4510:Ub-G76V-GFP (n = 8) and Ub-G76V-GFP (n = 5) mice. Sample sizes were smaller in this experiment owing to the complexity of generating a triple-transgenic cross. Mice from the same litter (4–5 mice per litter) were randomly assigned to each experimental group (vehicle or rolipram). Investigators carrying out tests were blinded to treatment group allocation and genotype. No animal was excluded from any of the analyses. We did not apply formal statistical tests for normality or equality of variances but assumed an approximation to the normal distribution when appropriate. Statistical analyses for the Morris water maze test were done by repeated-measures ANOVA or one-way ANOVA with Bonferroni correction, as described below.
Native PAGE assay for proteasome activity
Frozen brain tissues were processed for proteasome activity according to published methods45. Briefly, frozen tissue was homogenized in a buffer containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 5 mM ATP, 1 mM DTT and 10% glycerol, which preserves 26S proteasome assembly, and centrifuged at 20,000 × g for 18 min at 4 °C. The supernatant was normalized for protein concentration. Samples were loaded on a 4% nondenaturing gel as described45 and run for 200 min at 150 V. Activity of the 26S proteasome was measured by 400 µM Suc-LLVY-amc (BACHEM Bioscience) diluted in homogenizing buffer. 26S proteasome bands were detected by UV light (365 nm) and photographed by iPhone 4S camera. Following activity assessment, native gels were transferred for immunoblot analysis to detect proteasome levels using the anti-β5 (1:1,000) or anti-Rpt6 (1:1,000) antibody. At least three biological experiments were performed in HEK293 cell lines and acute slices (n = 6). For in vivo studies, proteasome activity was measured in extracts from young rTg4510 (15 rolipram- and 16 vehicle-treated mice, analyzed in two experiments) and from old rTg4510 (9 rolipram- and 7 vehicle-treated) mice.
Immunoblot analysis
Samples (5–20 µg protein) were typically run on 4–12% Bis-Tris gels (Life Technologies; WG1403BOX10) using MOPS buffer (NP0001) with antioxidant (NP0005) and on Tris-Glycine gels (EC64985BOX) for LC3. Proteins were analyzed after electrophoresis on SDS-PAGE and transferred onto 0.2-µm nitrocellulose membranes (Whatman). Blots were blocked and incubated with primary and secondary antibodies at concentrations listed below. Membranes were developed with enhanced chemiluminescent reagent (Immobilon Western HRP substrate and Luminol reagent (WBKLS0500, Millipore) using a Fujifilm LAS3000 imaging system. ImageJ (http://rsb.info. nih.gov/ij) was used to quantify the signal. Relative intensity (fold change or fold increase, no units) is the ratio of the value for each protein to the value of the respective loading control. At least three biological experiments from two animals were performed in ex vivo and tauopathy in vivo studies. For in vivo study across tauopathy stages (Supplementary Figs. 1 and 2b,g) n = 6 mice were used. Dilutions for western blotting were as follows: mouse monoclonal anti-tau (CP27, 1:5,000), pS396 and pS404 (PHF1, 1:5,000) and pS202 and pT205 (CP13,1:5,000). anti-pS214 tau (1:3,000); anti–phospho-(Ser and Thr) PKA substrate (1:1,000); anti-ULK1 (1:1,000), pS757 (1:1,000) and pS317 (1:1,000) ULK1; anti-Beclin 1 (1:1,000); anti-Atg12 (1:1,000); anti-CREB and phospho-S133 CREB (1:1,000); anti-LC3 (1:1,000); anti-GFP (1:1,000); anti-ubiquitin (1:1,500) anti-rabbit Tau (1:5,000); anti-GAPDH (1:5,000); anti-p62 (1:3,000); anti-proteasome, 19S regulatory particle: anti-Rpt6 (1:1,000) Rpn1 (1:500) and Rpn2 (1:500) and Rpn5 (1:500); 20S proteasome: β5 (1:1,000) and α1–α7 (1:1,000). Secondary antibodies were from Jackson ImmunoResearch: anti-mouse (1:3,000) and anti-rabbit (1:3,000) diluted in blocking buffer containing 5% milk.
Immunofluorescence
Mouse brains were isolated after transcardial perfusion with PBS and drop-fixed in 4% PFA overnight then subject to cryoprotection treatment in 30% sucrose in PBS for 24 h. Free-floating brain sections (35 µm) from brains sectioned in the sagittal plane were used. The sections were incubated at 4 °C overnight with primary antibody diluted in PBS containing 0.3% Triton X-100 and 5% normal goat serum blocking solution (Vector Laboratories, #S-1000). Antibodies were as follows: anti-mouse monoclonal human tau (CP27,1:1,000); pS396 and pS404 (PHF1, 1:1,000); pS202 and pT205 (CP13,1:1,000); p62 (1:1,000). GFP signal was obtained without antibody staining. Following washes, sections were incubated with goat anti-mouse IgG Alexa 594 (ThermoFisher Scientific #A-11005, 1:500). Staining was visualized by confocal microscopy, FV1200 FluoView (FV) 1200 laser-scanning confocal microscope (Olympus) and Zeiss AxioVision Imager Z1 microscope. Images were processed using FLUOVIEW software and AxioVision 4.8 image software. Quantification of the immunofluorescence signal for total and phosphorylated epitopes of tau, p62 and GFP signal was obtained by ImageJ.
Immunoprecipitation
The immunoprecipitation assay was performed with the Pierce Crosslink IP kit as described by manufacturer’s protocol (#26147 Thermo Scientific). Briefly, 10 µg mouse monoclonal human tau (clone # CP27) or 10 µg Rpt6 mouse monoclonal antibody (clone EPR13565B) per sample was incubated with Pierce Protein A/G Plus agarose for 60 min, followed by cross-linking with DSS cross-linker reagent for 60 min. 500 µg protein in 400 µl immunoprecipitation (IP) lysis buffer was incubated overnight at 4 °C. Antigen elution was performed the next day. At least three independent experiments from two animals of each age group were performed (n = 6). Immunoprecipitation for tau and Rpt6 was performed with anti-mouse antibodies followed by immuno-blotting with anti-rabbit antibodies at the following dilutions: Rpt6 (1:1,000), Rpt5 (1:1,000), Rpn5 (1:500) and 20S (α1–α7) (1:1,000) and rabbit anti-human tau(1:3,000). Secondary antibodies (anti-mouse and anti-rabbit) were diluted at 1:3,000. The specificity of co-precipitation of proteasome subunits was confirmed, as other large complexes (for instance, ribosomes or mitochondria) were not co-precipitated with tau (data not shown).
Preparation of recombinant tau aggregates
Preparation of tau aggregates from recombinant tau was performed as described27. Briefly, recombinant tau monomers were purchased from rPeptide (Tau-441, (2N4R) catalog #T-1001–2). Tau LMW aggregates were prepared by incubating tau solution (8 µM) at room temperature with 100 mM sodium acetate. Incubation times for LMW tau aggregates varied from 8 h to 2 d. For fibril assembly, tau solution (8 µM) was incubated with DTT (Invitrogen), heparin (Invitrogen), and sodium azide (0.02%), Invitrogen) for 5 d at room temperature, centrifuged at 14,000 × g and resuspended in 100 mM sodium acetate.
Purification of 26S proteasomes
Cortices from 6 mice were pooled each time proteasome purification was carried out, and 26S proteasomes were affinity purified using a UBL domain as the ligand19. Briefly, brain homogenates were spun for 1 h at 100,000 × g. The soluble extracts were incubated at 4 °C with 2 mg/ml glutathione-S-transferase–ubiquitin-like domain (GST-UBL) and a corresponding amount of glutathione–Sepharose 4B (GSH-Sepharose). The slurry containing 26S proteasomes bound to GST-UBL was poured into an empty column and washed, then incubated with 2 mg/ml His10-ubiquitin–interacting motif (10× His-UIM). The eluate was collected and incubated with Ni2+-NTA-agarose for 20 min at 4 °C. The Ni2+-NTA-bound 10× His-UIM was removed by filtration. The resulting flow-through (~0.6 mL) contained purified 26S proteasomes. The molarity of 26S proteasome particles was calculated assuming a molecular weight of 2.5 MDa. To assess peptidase capacity of 26S proteasome subunits, we incubated 10 nM of proteasome with 50 µM Suc-LLVY-amc fluorogenic peptide for chymotrypsin-like activity (β5 activity); 50 µM of Bz-VRG-amc (Bz-Val-Gly-Arg-amc) for trypsin-like activity (β2 activity); and 50 µM of Z-LLE-amc (Z-Leu-Leu-Glu-amc) for caspase-like activity (β1 activity). Kinetic reactions were carried out for a period of 90 min. The rate of kinetic reaction (slope) over time was calculated. Fluorescence signal was captured at 380 nm excitation, 460 nm emission by Infinite 200 PRO multimode reader (TECAN).
In vitro phosphorylation of purified proteasomes with PKA and recombinant tau
Purified 26S proteasomes (10 nM) were incubated with 300 nM (13.8 ng/µL; MW, 45.92 kDa) recombinant tau. The assay was carried out in the presence of proteasome purification buffer: 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 0.01 mg/ml BSA. To measure chymotrypsin-like activity of proteasomes, 50 µM of the fluorogenic substrate Suc-LLVY-amc was added to purified proteasomes and monomeric or aggregated recombinant tau mix. Emitted fluorescence was monitored by Infinite 200 PRO multimode reader (TECAN) with excitation and emission wavelengths at 380 nm and 460 nm, respectively, for a period of 90 min. The rate of kinetic reaction (slope) was calculated. To confirm the effect on proteasome activity, 26S proteasomes (10 nM) were pre-treated with 400 U (1 uL) catalytic subunit of PKA (New England BioLabs #P6000S) for 60 min at 30 °C. Activated proteasomes were then incubated with recombinant tau forms (300 nM or 13.8 ng/µL) and 50 µM Suc-LLVY-amc peptide, and the fluorescence monitored as described above.
Degradation assay of radiolabeled ubiquitinated proteins
Ub5-DHFR (a gift from Millennium Pharmaceuticals) was radiolabeled using PKA (Sigma) and [γ-32P]ATP40. Sic1PY was radiolabeled using casein kinase II and [γ-32P]ATP (NEB), followed by ubiquitination using Rsp5 as described previously (Ubn-Sic1)46. Degradation of these substrates (50 nM) by 26S proteasomes (1 nM) was assayed in the presence of 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 0.01 mg/ml BSA (Sigma) as reported47,48 and measured by after the conversion of the substrate to trichloroacetic acid (TCA)-soluble 32P-labeled peptides. The reaction of 26S was carried out at 37 °C for 0, 10, 20 or 30 min with Ub5-DHFR and at 0, 3 or 9 min with Ubn-Sic1.
ATPase assay of 26S proteasomes
ATPase function of 26S proteasomes subunits was performed using a malachite green system that detects the formation of inorganic phosphate (Pi) during ATP hydrolysis. The assay buffer contains 25 mM HEPES-KOH, pH 8, 2.5 mM MgCl2, 125 mM potassium acetate, 0.025% Triton X-100, 1 mM ATP, 1 mM DTT, 0.1 mg/ml BSA. The reaction with 5 nM of 26S proteasomes was carried out at 37 °C for 0, 20, 40 or 60 min. After incubation, samples were transferred to a 96-well plate on ice, and 170 µl malachite green hydrochloride/ammonium molybdate tetrahydrate solution (3:1) was added to the plate Immediately afterward, 20 µl 34% of citric acid was added to each well. Final concentration of the assay solution was 6 mM ammonium molybdate tetrahydrate, 120 µM malachite green hydrochloride and 3.4% sodium citrate. Incubation at room temperature for 30 min is required for color development. The absorbance was read at 650 nm by infinite 200 PRO multimode reader (TECAN). The amount of Pi released was calculated on the basis of the absorbance standard curve established by KH2PO4.
Tissue fractionation and protein extraction
Mice were sacrificed by cervical dislocation, and the brains were immediately dissected on wet ice and stored on dry ice. Briefly, frozen hemispheres free of cerebellum and brainstem were weighed and homogenized without thawing in RIPA buffer (10× volume/weight) (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl (PMSF), 1 mM sodium orthovanadate, 1 mM sodium fluoride (NaF), 1 µl/ml protease inhibitor mix (Sigma-Aldrich)). Homogenates were centrifuged for 10 min at 3,000 × g at 4 °C. Protein assay was performed on the clear supernatants representing the total extract used for analysis of the total protein levels. Sample volumes were adjusted with RIPA buffer containing 100 mMDTT and NuPAGE LDS Sample Buffer 4× buffer (Life Technologies) and boiled for 5 min. The sarkosyl-insoluble extracts, which are highly enriched in aggregated tau species, were generated when 200 µg aliquots from the total protein extracts were normalized into 200 µl final volume containing 1% sarkosyl, followed by ultra-centrifugation at 100,000 × g for 1 h at 20 °C. Without disturbing the pellet, the supernatant was transferred to new tubes. The pellet was resuspended in 100 µL RIPA buffer con-taining DTT and NuPAGE LDS Sample Buffer 4× buffer, followed by vortexing for 1 min and 5 min heating at 95 °C. The heat-stable extract, which contains soluble tau, was obtained when the supernatant was further processed, first by heating for 5 min at 95 °C followed by 30 min centrifugation at 20,000 × g. Extracts were transferred to new tubes containing NuPAGE LDS Sample Buffer 4× buffer (4:1 (extracts/buffer) ratio).
Statistical analysis
Statistical analyses for immunoblots, proteasome assays, ATPase activity and Ub5-DHFR and Ubn-Sic1 degradation assays were performed with Prism5 (Graphpad Software, San Diego, CA) using unpaired two-tailed Student’s t-test between groups with unequal variance or one-way ANOVA followed by Tukey’s multiple comparison post hoc test. Data are mean ± s.e.m. The Morris water maze was employed to assess the effect of rolipram in vivo. Statistical analyses were performed with Stata v.12 on the mean time to find the hidden platform (latency) per day for each day of testing (4 d total) using repeated-measures ANOVA. Treatment (rolipram vs. vehicle) and genotype (rTg4510 vs. WT, or rTg4510:Ub-G76V-GFP vs. Ub-G76V-GFP) were employed as between-subject factors and day of testing as the within-subject factor. We reported (Supplementary Data) the F test statistic, degrees of freedom and P value for the main effects on escape latency of treatment, genotype, and day of testing, plus each interaction. Escape latency on the final day of testing (day 4), plus day 3, was also subjected to one-way ANOVA with a Bonferroni correction. Data are mean ± s.d. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank J. Lewis and K. Ashe for providing rtg4510 and JNPL3 mice and P. Davies for the generous gift of tau antibodies. We thank L. Liu for assisting with mice perfusion. This work was supported by grants from the US National Institute of Neurological Disorders and Stroke NS074593 (K.E.D.), curePSP Foundation (N.M.), the US National Institute of General Medical Sciences GM051923 (to A.L.G.), the Fidelity Biosciences Research Initiative (A.L.G.) and the multiple myeloma Research Foundation (N.V.K.).
Footnotes
Author contributions
N.M. performed all experiments except for the degradation assay of ubiquitinated proteins. C.L.C. performed STATA analyses for behavioral studies; S.E. assisted with IHC and water maze experiments; N.V.K. performed degradation assays. N.M., W.H.Y., A.L.G. and K.E.D. designed the studies. N.M., A.L.G. and K.E.D. wrote the manuscript. C.L.C., N.V.K. and W.H.Y. contributed to manuscript preparation. W.H.Y. contributed transfected cell lines. All authors reviewed and commented on the manuscript.
References
- 1.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asai M, et al. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. Journal of molecular and cellular cardiology. 2009;46:452–462. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, et al. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. The Journal of biological chemistry. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 7.Myeku N, Wang H, Figueiredo-Pereira ME. cAMP stimulates the ubiquitin/proteasome pathway in rat spinal cord neurons. Neuroscience letters. 2012;527:126–131. doi: 10.1016/j.neulet.2012.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. Journal of neurochemistry. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 9.Cripps D, et al. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. The Journal of biological chemistry. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 10.Thomas SN, Cripps D, Yang AJ. Proteomic analysis of protein phosphorylation and ubiquitination in Alzheimer's disease. Methods in molecular biology. 2009;566:109–121. doi: 10.1007/978-1-59745-562-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris M, et al. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nature neuroscience. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tai HC, et al. The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. The American journal of pathology. 2012;181:1426–1435. doi: 10.1016/j.ajpath.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MJ, Lee JH, Rubinsztein DC. Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Progress in neurobiology. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 14.David DC, et al. Proteasomal degradation of tau protein. Journal of neurochemistry. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 15.Han DH, et al. Direct cellular delivery of human proteasomes to delay tau aggregation. Nature communications. 2014;5:5633. doi: 10.1038/ncomms6633. [DOI] [PubMed] [Google Scholar]
- 16.Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. Journal of neurochemistry. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 17.Metcalfe MJ, Huang Q, Figueiredo-Pereira ME. Coordination between proteasome impairment and caspase activation leading to TAU pathology: neuroprotection by cAMP. Cell death & disease. 2012;3:e326. doi: 10.1038/cddis.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santacruz K, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besche HC, Goldberg AL. Affinity purification of mammalian 26S proteasomes using an ubiquitin-like domain. Methods in molecular biology. 2012;832:423–432. doi: 10.1007/978-1-61779-474-2_29. [DOI] [PubMed] [Google Scholar]
- 20.Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. The Journal of biological chemistry. 2013;288:7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma R, McDonald H, Yates JR, 3rd, Deshaies RJ. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Molecular cell. 2001;8:439–448. doi: 10.1016/s1097-2765(01)00308-2. [DOI] [PubMed] [Google Scholar]
- 22.Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochimica et biophysica acta. 2012;1823:67–82. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsten K, Menendez-Benito V, Masucci MG, Dantuma NP. A transgenic mouse model of the ubiquitin/proteasome system. Nature biotechnology. 2003;21:897–902. doi: 10.1038/nbt851. [DOI] [PubMed] [Google Scholar]
- 25.Lewis J, et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nature genetics. 2000;25:402–405. doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- 26.Deriziotis P, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. The EMBO journal. 2011;30:3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JW, et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. The Journal of biological chemistry. 2013;288:1856–1870. doi: 10.1074/jbc.M112.394528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu B, et al. Protein kinase A phosphorylation of tau-serine 214 reorganizes microtubules and disrupts the endothelial cell barrier. American journal of physiology. Lung cellular and molecular physiology. 2010;299:L493–L501. doi: 10.1152/ajplung.00431.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 31.Sadik G, et al. Phosphorylation of tau at Ser214 mediates its interaction with 14-3-3 protein: implications for the mechanism of tau aggregation. Journal of neurochemistry. 2009;108:33–43. doi: 10.1111/j.1471-4159.2008.05716.x. [DOI] [PubMed] [Google Scholar]
- 32.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E7176–E7185. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myeku N, Figueiredo-Pereira ME. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. The Journal of biological chemistry. 2011;286:22426–22440. doi: 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherra SJ, 3rd, et al. Regulation of the autophagy protein LC3 by phosphorylation. The Journal of cell biology. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell. 2014;157:163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Molecular cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sha Z, Peth A, Goldberg AL. Keeping proteasomes under control--a role for phosphorylation in the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18573–18574. doi: 10.1073/pnas.1115315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin JT, et al. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington's disease. Molecular and cellular biology. 2013;33:1073–1084. doi: 10.1128/MCB.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. The Journal of biological chemistry. 2013;288:29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, et al. Trans-synaptic spread of tau pathology in vivo. PloS one. 2012;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Calignon A, et al. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duff K, Noble W, Gaynor K, Matsuoka Y. Organotypic slice cultures from transgenic mice as disease model systems. Journal of molecular neuroscience : MN. 2002;19:317–320. doi: 10.1385/JMN:19:3:317. [DOI] [PubMed] [Google Scholar]
- 44.Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. Journal of neuroscience methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 45.Myeku N, Metcalfe MJ, Huang Q, Figueiredo-Pereira M. Assessment of proteasome impairment and accumulation/aggregation of ubiquitinated proteins in neuronal cultures. Methods in molecular biology. 2011;793:273–296. doi: 10.1007/978-1-61779-328-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeki Y, Isono E, Toh EA. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods in enzymology. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 47.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Molecular cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam YA, Huang JW, Showole O. The synthesis and proteasomal degradation of a model substrate Ub5DHFR. Methods in enzymology. 2005;398:379–390. doi: 10.1016/S0076-6879(05)98031-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.