Abstract
The Hedgehog (Hh) signaling pathway play critical roles in embryonic development and adult tissue homeostasis. A critical step in Hh signal transduction is how Hh receptor Patched (Ptc) inhibits the atypical G protein-coupled receptor Smoothened (Smo) in the absence of Hh and how this inhibition is release by Hh stimulation. It is unlikely that Ptc inhibits Smo by direct interaction. Here we discuss how Hh regulates the phosphorylation and ubiquitination of Smo, leading to cell surface and ciliary accumulation of Smo in Drosophila and vertebrate cells, respectively. In addition, we discuss how PI(4)P phospholipid acts in between Ptc and Smo to regulate Smo phosphorylation and activation in response to Hh stimulation.
Keywords: Hh signaling, phospholipid, phosphorylation, Smo, ubiquitination, signal transduction
Hh signaling
Hedgehog (hh) was first discovered as a segment polarity gene involved in Drosophila embryo development (Nüsslein-Volhard and Wieschaus, 1980). The Hh signaling pathway controls the most organs development and post developmental tissues homeostasis (Ingham and McMahon, 2001; Jiang and Hui, 2008; Briscoe and Thérond, 2013), malfunction of which causes birth defects as well as several types of cancer, including medulloblastoma, basal cell carcinoma (BCC), breast cancer, and lung cancer, thereby it has become an attractive therapeutic target.
The Hh signaling is transduced by a signaling cascade that is highly conserved among species, with the receptor complex consisting of two membrane protein, a 12-span transmembrane protein Patched (Ptc) and a seven-span transmembrane protein Smoothened (Smo). In the absence of Hh, Ptc inhibits Smo activity by promoting Smo endocytosis and turnover in intracellular compartments (Denef et al., 2000). Ptc likely inhibits Smo catalytically (Taipale et al., 2002), because substochiometric levels of Ptc are able to repress Smo activation (Taipale et al., 2002; Casali and Struhl, 2004). In Drosophila, the Hh signal is transduced through the receptor complex Ptc-Interference Hh (Ptc-Ihog) and the signal transducer Smo at the plasma membrane (Lum and Beachy, 2004; Camp et al., 2010; Zheng et al., 2010). Binding of Hh to Ptc-Ihog relieves the Ptc-mediated inhibition of Smo, which allows Smo to activate the Cubitus interruptus (Ci)/Gli family of zinc finger transcription factors and thereby induce the expression of Hh target genes, such as decapentaplegic (dpp), ptc, collier (col) and engrailed (en) (Hooper and Scott, 2005; Jia and Jiang, 2006) (Fig. 1A). Unlike Drosophila has single Ptc, Hh, and Gli genes, mammals have two Ptc receptors-Ptc1 and Ptc2, three Hh ligands-Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh), and three Gli transcription factors-Gli1, Gli2, and Gli3 (Jia and Jiang, 2006).
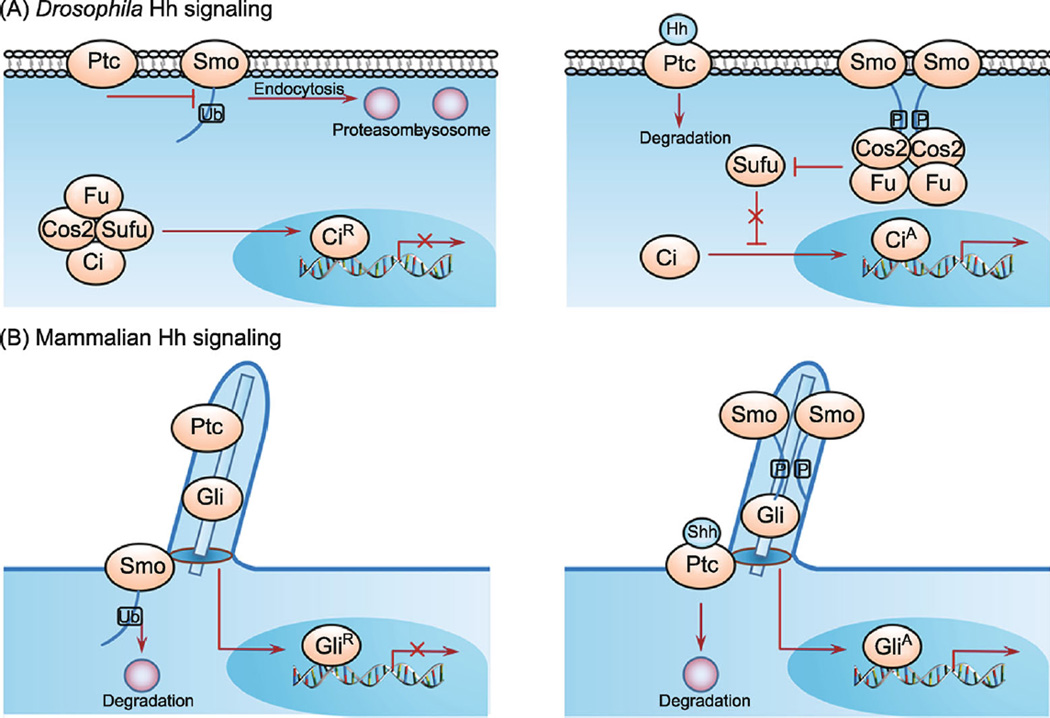
Figure 1.
The regulation of Smo phosphorylation and ubiquitination in Drosophila and mammalian Hh signaling pathways. (A) Schematic drawings of Drosophila Hh signaling. In the absence of Hh (left panel), Ptc inhibits Smo. Smo is ubiquitinated at multiple residues among its C-tail, resulting in endocytosis and degradation by both lysosome- and proteasome-dependent pathways. Full-length Ci (CiF) undergoes proteolytic processing to generate a truncated repressor form (CiR), which blocks the expression of Hh target genes. In the presence of Hh (right panel), binding of Hh to Ptc releases its inhibition on Smo and triggers phosphorylation of Smo by multiple kinases (PKA, CK1, CK2, Gprk2 and aPKC), leading to Smo cell surface accumulation, dimerization and activation, thus the conversion of CiF into the activator form CiA. (B) Schematic drawings of mammalian Hh signaling in the cilium. In the absence of Shh (left panel), Smo is excluded from the primary cilium, where Ptc resides to inhibit Hh signaling. In the presence of Shh (right panel), binding of Shh to Ptc leading its departure from primary cilium, where Smo is accumulated, phosphorylated, dimerized and activated. The active forms of Gli (GliA) induce Hh target gene expression.
Smo, an atypical G protein-coupled receptor (GPCR), is the key positive regulator of the pathway in both insects and vertebrates. Abnormal activation of Smo results in several cancers, thus Smo is an attractive therapeutic target (Xie et al., 1998; Yang et al., 2010). Currently, the most advanced drug is vismodegib that was approved by Food and Drug Administration (FDA) for treating cancers driven by Smo activation (Sekulic et al., 2012; Tang et al., 2012). However, cancer cells can acquire resistance and vismodegib becomes ineffective (Atwood et al., 2015; Sharpe et al., 2015). Several different mechanisms of resistance have been reported, including those through mutations in Smo (Wang et al., 2013), activation of Gli2 (Buonamici et al., 2010), mutations in the negative regulator Suppressor of fused (Sufu) (Kool et al., 2014), and activation of RAS/MAPK pathway (Zhao et al., 2015). Therefore, a better understanding the mechanisms of Smo regulation in the fundamental development processes is critical to developing more effective therapeutic treatments for cancers caused by Smo dysregulation.
Primary cilium is a microtubule-based short membrane protrusion found in most mammalian cells, and primary cilium dysfunction is the basis of a series of human pathologies, such as cancer, cystic kidney, obesity, blindness, cognitive disabilities, mental retardation, and various developmental malformations (Goetz and Anderson, 2010). In mammals, most of the Hh signaling components are located at or nearby the primary cilium, which is an absolute requirement for proper Hh signaling (Briscoe and Thérond, 2013). In the absence of Hh, Ptc1 is concentrated in the primary cilium, while in the presence of Hh Ptc1 is removed from the cilium, whereas Smo accumulates in the cilium that is required for its activation (Corbit et al., 2005; Rohatgi et al., 2007; Rohatgi et al., 2009; Wang et al., 2009) (Fig. 1B). Kim et al. identified that Ptc contains the ciliary localization sequence in the C-terminal domain, which is necessary to mediate protein localization to the primary cilium (Kim et al., 2015). Surprisingly, Ptc must accumulate in the cilium to inhibit Smo in the absence of Hh, but Ptc ciliary removal is secondary because Ptc mutants that are unable to exit the cilium not only repress Smo activity but also enabled Hh to neutralize its repression and elevate Smo activity in the presence of Hh (Kim et al., 2015). Increased ciliary accumulation of Smo is associated with activation of Gli transcription factors and expression of Hh target genes. Gli proteins are the transcriptional effectors of Hh pathway, the balance between activator and repressor forms of Gli2 and Gli3 is critical to the activity of the pathway. In addition, the base of the cilium acts as a gate to the basal body, which mediates the exchange of Hh signaling components in and out of the cilium. The cAMP-activated kinase, PKA, functions at the base of the cilium and is key in negative regulating Hh signaling by mediating the processing of Gli into GliR (Jiang and Struhl, 1998; Wang et al., 2000a; Tuson et al., 2011). Therefore, cilium is fundamentally function as a signaling center for the Hh pathway in mammals (Wilson and Chuang, 2010). However, it remains unclear how Smo cell surface/ciliary accumulation and intracellular trafficking are controlled.
Two products of human disease genes responsible for the Ellis-van Creveld syndrome, Evc and Evc2, are reported to have the positive role in Hh signaling (Dorn et al., 2012; Yang et al., 2012; Pusapati et al., 2014). The loss of Evc/Evc2 does not affect Shh-induced Smo phosphorylation and ciliary localization but blocks Hh pathway activation mediated by constitutively active forms of Smo, suggesting that Evc and Evc2 are recruited to activate Gli proteins by antagonizing Sufu in the primary cilia (Yang et al., 2012). Evc/Evc2 are optional for the constitutive Gli activity in Sufu (−/−) cells, suggesting that Evc/Evc2 act upstream of Sufu to promote Gli activation. In addition the EFCAB7-IQCE complex anchors the EVC-EVC2 at the base of cilium, and Smo-Evc signaling complex is required for Hh signal transmission (Dorn et al., 2012; Pusapati et al., 2014). Primary cilium also serve as specialized compartments for calcium signaling, containing a heteromeric PKD1L1-PKD2L1 channel in mice and humans, which regulates ciliary calcium concentration thereby regulates ciliary Smo-mediated Gli1 expression and Gli2 translocation (DeCaen et al., 2013; Delling et al., 2013). Further studies could determine whether other components in the Hh pathway are calcium signaling dependent, and respond to changes in the environment during development.
Drosophila cells do not have cilium during development, hence cilium-mediated Hh signaling is restricted to vertebrates. Kuzhandaivel et al. identifed and characterized a ciliamediated Hh pathway in Drosophila olfactory sensory neurons (Kuzhandaivel et al., 2014). They demonstrated that Costal2 (Cos2) and Fused (Fu) are required for the Smo ciliary transport and cilium mediate the expression of the Hh signaling target genes. The ciliary Hh pathway in olfactory sensory neurons is conserved, at least in part, from Drosophila to vertebrates.
Smo phosphorylation regulated by different levels of Hh signaling activity
Studies of the Hh signaling in the past have identified many core components that control the Hh signaling pathway (Wilson and Chuang, 2010), most of which are regulated by phosphorylation (Chen and Jiang, 2013; Jia, 2012). Smo has homologyto GPCRs (Alcedo et al., 1996), and Smo signals need Gαi, one of the G-proteins in Drosophila, to activate low threshold of Hh target genes expression (Ogden et al., 2008). Like many GPCRs, phosphorylation of Smo controls the switch between on and off of the signaling states.
In Drosophila, studies have shown that Hh induces cell surface accumulation and phosphorylation of Smo by multiple kinases, including protein kinase A (PKA) and casein kinase 1 (CK1) (Jia et al., 2004; Zhang et al., 2004; Apionishev et al., 2005), casein kinase 2 (CK2) (Jia et al., 2010), G protein-coupled receptor kinase 2 (Gprk2) (Chen et al., 2010) and atypical PKC (aPKC) (Jiang et al., 2014), which activate Smo by inducing differential phosphorylation, thus the gradual conformational change in the protein (Jia et al., 2004; Jiang et al., 2014; Apionishev et al., 2005; Zhang et al., 2004; Zhao et al., 2007; Chen et al., 2010; Jia et al., 2010; Fan et al., 2012;).
Smo forms homodimer and undergoes a conformational change when it is phosphorylated by PKA and CK1 upon Hh stimulation (Zeng et al., 2005; Zhao et al., 2007). Zhao et al. using fluorescence resonance energy transfer (FRET) analysis, propose a dimerization model to explain that Smo conformational states rely on the level of Hh activity and Smo phosphorylation, which can be manipulated by substitution of various Ser/Thr residues into either Ala or Asp (Zhao et al., 2007). PKA and CK1 play an essential role in activating Smobyphosphorylating Smoat three clusters of serine residues in its C-tail (Zhao et al., 2007; Aikin et al., 2008), but it would be interesting to understand how phosphorylation of Smo at the three clusters of residues contributes to its activity. Fan et al. demonstrated that Smo is differentially phosphorylated in response to different levels of Hh signaling activity (Fan et al., 2012). They also found in addition to the number of phosphorylated sites, the position of the phosphorylated residues is critical for establishing the stoichiometry of Smo activity. Thereupon a zipper-lock model, in which phosphorylation at the three clusters of serine residues promotes a gradual conformational change to activate Smo, is proposed (Fan et al., 2012).
Smo phosphorylation has been reviewed elsewhere (Jia, 2012; Chen and Jiang, 2013), here we only discuss the roles of atypical protein kinase C (aPKC) in regulating Smo phosphorylation. Evidence shows that Shh increases the PKC activity and Shh-induced increase of Gli1 mRNA level requires PKC in mouse embryonic stem cells (Heo et al., 2007), indicate that PKC may play role in Hh signaling pathway. Jiang et al. indicated aPKC has a positive role in regulating Hh signaling by modulating both Smo and Ci (Jiang et al., 2014). Smo phosphorylation at Ser680 by aPKC promotes its activation, while Ci phosphorylation at Thr512 and Thr590 by aPKC promotes the DNA binding activity of Ci Zn finger DNA binding doma in. These findings indicate that aPKC is directly involved in Hh signaling beyond its roles in cell polarity regulation. In addition, inhibition of aPKCι/λ suppresses the Hh signaling and the growth of BCC cell lines, and aPKCι/λ phosphorylates Gli1 to regulate its DNA binding activity (Atwood et al., 2013). Interestingly, Hh signaling upregulates aPKC expression in a Ci dependent manner (Jiang et al., 2014), while aPKC-ι/λ is upregulated in BCCs, and Prkci is a direct target gene of Gli1(Atwood et al., 2013). These studies uncover a conserved role of aPKC in upregulating Hh signaling activity and a positive feedback regulation of aPKC itself. Moreover aPKC form a complex with Par6 to regulate the activity of Smo and Ci, which is independent of their function in apical/basal polarity, because inactivation of several other polarityproteins, such as Crb, Baz, Dlg5, Dlg1, and Lgl, shows polarity phenotypes but does not affect Smo accumulation and Hh signaling in Drosophila wings (Jiang et al., 2014). It is unclear whether these polarity proteins have additional roles in Hh signaling through polarity regulation.
Phosphorylation mediates the balance between transcriptional activator and repressor forms of Ci/Gli, which plays dual roles by two distinct forms (Lum and Beachy, 2004; Jia and Jiang, 2006). In the absence of Hh, full-length Ci (CiF) undergoes proteolytic processing to generate a truncated repressor form (CiR), which functions as a repressor to block the expression of Hh responsive genes such as dpp (Hooper and Scott, 2005; Jia and Jiang, 2006). Proteasome-mediated Ci processing requires cooperative and sequential phosphorylation by PKA, glycogen synthase kinase 3 (GSK3) and CK1 at multiple serine/threonine (Ser/Thr) residues in the C-terminal region of Ci (Jiang and Struhl, 1995; Jia et al., 2002; Price and Kalderon, 2002). In Drosophila, similar to the phosphorylation clusters in Smo, PKA and CK1 phosphorylate Ci at three clusters in the absence of Hh to inhibit pathway activation, while in the presence of Hh, PKA and CK1 phosphorylate Smo to stimulate its signaling activity. So how different substrates are chosen by the same set of kinases in different signaling states is important to the field. Recently, two studies demonstrated that Hh promotes the accumulation of Smo by inducing a switch from the association of PKA-Ci-Cos2 to a PKA-Smo-Cos2 complex, leading to the Smo phosphorylation and Hh signaling activation (Li et al., 2014; Ranieri et al., 2014).
Cos2 mainly acts as a negative regulator in Hh signal transduction, as it inhibits the transcriptional activator activity of CiF by inhibiting its nuclear translocation (Sisson et al., 1997; Chen et al., 1999;Wang et al., 2000b). Cos2 also acts as a scaffold protein to bring PKA, CK1, and GSK3 to CiF, thereby promoting sequential phosphorylation of CiF by these kinases (Zhang et al., 2005; Price, 2006). Cos2 has a negative role on Smo, because it blocks the phosphorylation and activation of Smo likely by masking its phosphorylation sites (Liu et al., 2007). In addition, Cos2 has a positive role in Hh-responding cells, and this correlates to its ability to form a complex with the C-terminal intracellular tail of Smo (Wang et al., 2000b; Jia et al., 2003; Lum et al., 2003; Ho et al., 2005). Cos2 can be phosphorylated at Ser572 and Ser931 by Fused (Fu), which likely inhibits the negative activity of Cos2 on Smo phosphorylation and leads Cos2 distribution from the cytoplasm to the plasma membrane, which also elevates Ci nuclear translocation and transcriptional activity (Ranieri et al., 2012).
One important question is how Hh regulates Smo phosphorylation and cell surface accumulation. Although Smo phosphorylation promotes its cell surface accumulation and signaling activity, the mechanisms controlling its cell surface accumulation are still unclear. Smo physically interacts with the Cos2-Fu complex to transduce Hh signaling (Robbins et al., 1997; Jia et al., 2003; Lum et al., 2003; Ruel et al., 2003). Accumulation of Smo to the cell surface recruits the large protein complex, which is thought to induce dissociation of the Cos2-Ci-Kinase complex and hence inhibition of Ci processing (Zhang et al., 2005). Fu-Cos2 protein complex have feedback regulation on Smo, because Cos2-Smo interaction blocks Hh-induced Smo phosphorylation, and Fu is essential for Hh-induced Smo phosphorylation and cell surface accumulation by antagonizing Cos2 (Liu et al., 2007). Fu is in the non-phosphorylated inactive state in the absence of Hh and a phosphorylated active state in the presence of Hh (Thérond et al., 1996; Lum et al., 2003). It has been shown that Hh induces Fu dimerization in a dose-dependent manner, which is mediated by a conformational change and dimerization of Smo (Shi et al., 2011; Zhang et al., 2011). In addition to its association with Cos2, forced dimerization of Fu induces its auto-phosphorylation and the activation of Hh signaling, regulates the ratio of CiA (an active form of CiF) and CiR by interfering with Ci-Sufu and Ci-Cos2-kinase complex formation (Shi et al., 2011; Zhang et al., 2011). Moreover, Thr158 in the activation loop of Fu was identified to be a critical residue phosphorylated upon Hh stimulation (Fukumoto et al., 2001). In Drosophila, Sufu forms a protein complex with Cos2, Fu and Ci, which is reported to regulate Ci subcellular localization and transcriptional activity in the nucleus (Méthot and Basler, 2000; Wang et al., 2000b). Sufu is phosphorylated in response to Hh and this phosphorylation event depends on Fu kinase activity (Lum et al., 2003; Dussillol-Godar et al., 2006), although it needs to be confirmed whether Fu is a direct kinase for Sufu. Recently, Oh et al. found Sufu phosphorylation is irrelevant for pathway regulation, meanwhile Fu and other components regulate Ci transcriptional activity independently of Sufu (Oh et al., 2015), leading to a possible additional layer of regulation.
Smo ciliary accumulation is regulated by phosphorylation. Primary cilium is critical for mammalian Hh signaling (Huangfu et al., 2003; Huangfu and Anderson, 2005; Rohatgi et al., 2007), which can be used as a platform for investigating the mechanism how activators or repressors regulate the signaling pathway. Since Shh or Smo oncogenic mutations promote Smo activation by inducing its ciliary localization (Corbit et al., 2005; Rohatgi et al., 2007), it is interesting whether Smo phosphorylation could regulate its ciliary accumulation. Mammalian Smo does not contain the three PKA/CK1 phosphorylation clusters found in Drosophila Smo C-tail, however, Chen et al. found that phosphorylation of mammalian Smo by CK1α and GRK2 at multiple residues in its C-tailpromotes the activation and ciliary localization of Smo (Chen et al., 2011). Differential phosphorylation of vertebrate Smo correlates with the gradient of Hh activity. Similarly, Smo phosphorylation is also induced by oncogenic mutation and the agonist SAG (Zhao et al., 2007), but not by the antagonist cyclopamine that promotes Smo ciliary localization but blocks Smo phosphorylation, conformational change and Hh pathway activation (Zhao et al., 2007; Rohatgi et al., 2009; Wilson et al., 2009). These results suggest that Smo ciliary localization is insufficient for potent Smo activation, whereas Hh induced phosphorylation of Smo is required for the full activation of Smo. Since other components like intraflagellar transport (IFT) proteins (Huangfu et al., 2003; Wang et al., 2009) and β-arrestins (Kovacs et al., 2008) have roles in mediating Smo ciliary localization, the relationship between Smo phosphorylation and its interaction with these components will be an interesting question to investigate.
In mammalian Hh signaling, aPKCι/λ phosphorylates Gli1, promotes Gli1 binding to the target DNA and elevates Gli1 activity, therefore activates Hh signaling (Atwood et al., 2013). Drosophila aPKC-Par6 complexalso has a positive role in Hh signaling by phosphorylating Smo, prompting Smo basolateral accumulation and regulating the activity of Ci (Jiang et al., 2014). Considering the similarity between Smo membrane accumulation in Drosophila and Smo translocation to cilium in mammalian cells, it is possible that malfunction of aPKC will lead to aberrant Smo ciliary accumulation and activation. Par3/Par6/aPKC polarity complex localizes to cilium and regulates intraflagellar transport and ciliogenesis (Fan et al., 2004; Prulière et al., 2011; He et al., 2012), suggesting an important role for aPKC to associate with microtubule motors in cilium formation and ciliogenesis. Further investigation on the functions of aPKC in the cilium may result in a novel therapeutic approach for the treatment of Hh-mediated tumorigenesis.
Hh may also regulate the dephosphorylation of Hh signaling components that is mediated by their phosphatases. Protein phosphatase 2A (PP2A) was identified as a positive regulator in Hh signaling (Nybakken et al., 2005; Rorick et al., 2007; Casso et al., 2008), although the relevant substrate remain unidentified. Jia et al. found protein phosphatase 4 (PP4) and PP2A are the phosphatases in regulating dephosphorylation of Smo and Ci, respectively (Jia et al., 2009). The other independent study found that PP2A may also play a role in dephosphorylating Smo (Su et al., 2011). As discussed earlier, multiple Ser/Thr kinases regulate Hh signaling by phosphorylating Smo and Ci, it is not surprisingly that other phosphatases are also involved in dephosphorylating Smo and Ci, as well as other Hh signaling components.
Smo ubiquitination regulated by Hh
As one of the posttranslational modification, protein modification by a small protein called ubiquitin is also involved in Hh signal transduction, which can modulate the stability, activity and sub-cellular localization of the Hh signaling components (Jiang, 2006; Hsia et al., 2015). Unlike the positive role for phosphorylation to regulate Smo, ubiquitination has a negative role in promoting Smo endocytosis, trafficking and degradation in the cell (Li et al., 2012; Xia et al., 2012).
Two independent studies have demonstrated that Smo is poly- and mono-ubiquitinated, leading to the endocytosis and degradation by the lysosome-mediated pathway, which is regulated by Hh stimulation (Li et al., 2012; Xia et al., 2012). Smo ubiquitination also promotes Smo degradation through the proteasome-mediated pathway. Using RNAi screen, Xia et al. identify that ubiquitin-specific protease 8 (USP8) as a deubiquitinase (DUB) required for Hh-induced deubiquitination and cell surface accumulation of Smo. Hh promotes the formation of a Smo-USP8 complex, and USP8 further promotes the accumulation of Smo at the cell surface and prevents localization to the early endosomes by deubiquitinating Smo, leading to increased Hh signaling activity. Importantly, evidence also showed that USP8-mediated elevation of Smo is still inhibited by Ptc, and that phosphorylation and dimerization is required for Smo activation even though Smo can be stabilized by deubiquitination (Xia et al., 2012). Both studies found that Smo ubiquitination is regulated by both Hh and PKA/CK1-mediated Smo phosphorylation. Inactivation of the ubiquitin activating enzyme-Uba1 promotes Smo accumulation on the cell surface and Hh signaling activation, and USP8 decreases Smo ubiquitination to promote its cell surface accumulation in the absence or presence of Hh (Li et al., 2012; Xia et al., 2012).
Since the endosomal sorting complex required for transport (ESCRT) facilitates the ubiquitinated proteins trafficking from endosomes to lysosomes via MVBs (Williams and Urbé, 2007; Wollert and Hurley, 2010), some ESCRT homolog members were found to regulate ubiquitinated Smo in endosomal sorting, like HGF-regulated tyrosine kinase substrate (Hrs) (Li et al., 2012; Fan et al., 2013), Tumor susceptibility gene 101 (Tsg101) (Li et al., 2012) and Vacuolar protein sorting 36 (Vps36) (Yang et al., 2013). Hrs promotes Smo ubiquitination, mediates Smo trafficking in the late endosome (Fan et al., 2013). The N-terminus of Hrs directly interacts with the PKA/CK1 phosphorylation clusters to prevent Smo phosphorylation and activation, indicating an ubiquitin-independent regulation of Smo by Hrs, unlike in many cases that Hrs requires substrate ubiquitination. Knockdown of Tsg101 accumulates Smo in wing discs that is co-localized with Hrs and late endosome markers-Rab7 and Lamp1, indicate that Tsg101 and Hrs mediate Smo trafficking in the late endosomes (Fan et al., 2013). In mammals, Shh treatment induced a substantial decrease in Smo ubiquitination, suggest a conserved mechanism that regulates Smo (Xia et al., 2012), although more details are still lacking regarding Smo regulation by ubiquitination in mammals.
Although the role of ubiquitination in regulating Smo activity has been documented recently (Hsia et al., 2015), the ubiquitin-protein ligase(s) that directly regulate Smo are still unknown. In an RNAi screen targeting the E3 ligases in the fly genome, Du et al. identified neddylation genes involved in Hh signaling (Du et al., 2011), however, none of the genes in this collection are directly involved in Smo ubiquitination. It is possible that multiple E3 ligases are involved in Smo ubiquitination and degradation by the proteasome- and lysosome-mediated pathways.
Phosphoinositides in Hh signaling
Intraflagellar transport proteins are essential for the assembly and maintenance of cilium required for vertebrate Hh signaling (Rosenbaum and Witman, 2002; Huangfu et al., 2003; Liu et al., 2005). However, the negative role of IFT-A complex subunits in Hh signaling suggests a role of IFT-A complex in additional to regulating ciliogenesis. Tubby-like protein 3 (Tulp3) and IFT-A promote trafficking of the GPCR 161 (GPR161), thus mediate the PKA-dependent basal repression of Hh signaling (Mukhopadhyay et al., 2010; Mukhopadhyay et al., 2013). Endogenous GPR161 is absence in the cilium when the cells are treated with Shh, whereas overexpression of GPR161 resulted in increased cAMP production and therefore PKA activation (Mukhopadhyay et al., 2013). Interestingly, in the in vitro protein-lipid overlay assay, GST-TULP3 binds most strongly to PI (4,5)P2 on lipid-dotted strips (Mukhopadhyay et al., 2010), raising the possibility that phosphoinositides (PIs) may have function in the Hh signaling pathway. Although some regulators of Hh signaling are known to be accumulated in the primary cilium, much less is known about the lipids.
PIs are a group of lipids found in various cellular membranes, which have critical functions in regulating numerous biologic processes, including gene expression, protein and membrane trafficking, cell signaling, ion channels, as well as endocytic and exocytic processes (Di Paolo and De Camilli, 2006; Balla, 2013). The inositol ring of phosphatidylinositol, the precursor of PI, can be reversibly phosphorylated at the third, fourth and fifth positions in all combinations, resulting in generating seven different PI species: phosphatidylinositol 3-phosphate (PI(3)P), PI(4)P, PI (5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2 and PI(3,4,5)P3. PIs are enriched on specific subcellular membranes, as PI(4,5)P2 and PI(3,4,5)P3 are concentrated at the plasma membrane, PI(3,4) P2 is mostly found at the plasma membrane and in the early endocytic pathway, PI(4)P is enriched at the Golgi complex, but also present at the plasma membrane, PI(3)P is concentrated in early endosomes, while PI(3,5)P2 is formed at the multivesicular body and PI(3,5)P2 concentrated on late endosomal pathway (Di Paolo and De Camilli, 2006; Balla, 2013).
Inositol polyphosphate 5-phosphatase E (INPP5E) is known to remove the 5-phosphate from PI(4,5)P2, PI(3,4,5) P3, and PI(3,5)P2. INPP5E has a proline-rich sequence and a COOH-terminal CAAX box in addition to the 5-phosphatase domain, which locates mostly in the Golgi and partially in the plasma membrane. Evidence showed that mutations in the phosphatase domain of INPP5E promoted premature destabilization of cilia in response to stimulation, hence are responsible for Joubert syndrome (Bielas et al., 2009). INPP5E mutation also affect its ciliary localization and cilium stability in a family with MORM syndrome (Jacoby et al., 2009). The physical targeting of INPP5E to the cilium is dependent on a protein-protein interaction network included the ADP-ribosylation factor-like 13B (ARL13B), phosphodiesterase 6D (PDE6D), and the centrosomal protein 164 (CEP164) (Humbert et al., 2012). All of them have been linked to ciliopathies, kind of disease associated with ciliary dysfunction (Hildebrandt et al., 2011). These findings indicate a role for PIs to regulate the function of cilium. Recently, two studies show that INPP5E restricts the ciliary PIs trafficking and accumulation of negative regulators of Hh signaling (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Chávez et al. identified that the main PI in neural stem cell primary cilium membrane is PI(4)P, and that PI(4,5)P2 is limited to the ciliary base (Chávez et al., 2015). They also showed that the INPP5E regulates PI distribution, as in INPP5E knockout neuronal stem cells primary cilium PI(4,5) P2 is detected and PI(4)P is absence, and that this regulation depends on its phosphatase activity. INPP5E affects recruitment of the PI(4,5)P2-interacting protein Tulp3, as well as Hh signaling repressor IFT-A components and Gpr161, thus regulates Hh signaling (Chávez et al., 2015). The same conclusion was made by Garcia-Gonzalo et al. in mouse embryonic fibroblasts (MEFs) with a slightly different approach (Garcia-Gonzalo et al., 2015), meanwhile they found INPP5E promotes Hh signal at a step subsequent to Smo ciliary localization and prior to Gli3 accumulation at the ciliary tip.
Ptc is not physically associated with Smo because they do not appear to be enriched at similar subcellular locations in vivo (Jia and Jiang, 2006; Jiang and Hui, 2008). Ptc acts indirectly to inhibit Smo activity, instead functions as a permease to facilitate transmembrane movement of a small molecule(s) that acts as Smo agonist or antagonist (Taipale et al., 2002). Evidence indicates that, in Drosophila, downregulation of the Stt4 kinase or upregulation of the Sac1 phosphatase keep the PI(4)P at low level in cells, which results in a phenotype similar to that of loss of Ptc (Yavari et al., 2010). Taken together of the recent published studies (Yavari et al., 2010; Chávez et al., 2015; Garcia-Gonzalo et al., 2015), it is still unknown whether phospholipids directly regulate the key Hh signaling regulators, Ptc and Smo.
In a most recent study, Jiang et al. found that Hh promotes the production of PI(4)P in both wing discs and cultured cells (Jiang et al., 2016) (Fig. 2). In both Drosophila S2 cells and mammalian NIH3T3 cells, PI(4)P was detected by the sensitive direct mass-spec assay and the levels of PI(4)P are elevated by Hh stimulation. Also in cultured cells, PI(4)P stimulates the phosphorylation and dimerization of Smo through direct interaction with an arginine motif in Smo, which is conserved between Drosophila Smo and mammalian Smo, as such motifs have been mapped in Smo species (Zhao et al., 2007). Mutation in the R4 arginine motif abolishes PI (4)P-Smo direct interaction, indicated that Smo conformation change due to the binding of PI(4)P to the specific arginine motif is critical. The position for PI(4)P binding is also critical to trigger Smo activation, because fusion of a pleckstrin homology (PH) domain to either the 3rd intracellular loop or the C terminus of Smo attracts PI(4)P to different locations inside Smo, thus blocking Smo activation by PI(4)P. Surprisingly phospho-mimetic Smo mutations (SmoSD123 and mSmoSD) are still regulated by PI(4)P, suggesting that PI (4)P either promotes phosphorylation at other residues or has additional role(s) in activating Smo (Jiang et al., 2016).
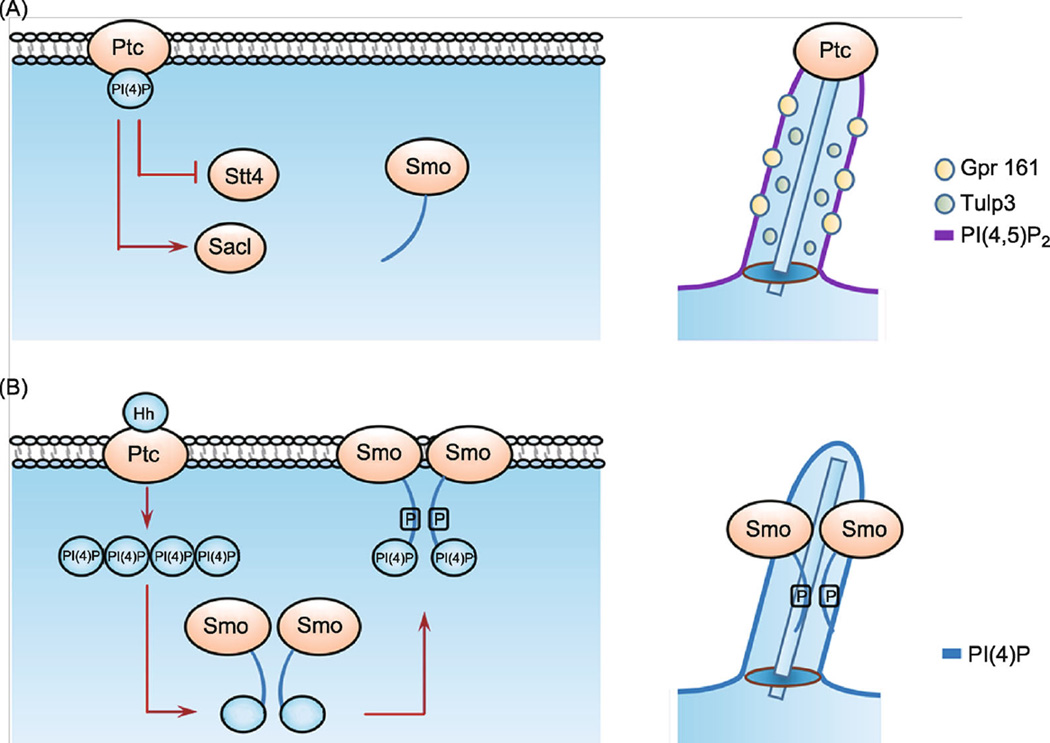
Figure 2.
PI(4)P regulates the phosphorylation and ciliary accumulation of Smo. Models are based on studies from recent publications. (A) Smo regulation in the absence of Hh. In Drosophila (left panel), Stt4 has low whereas Sac1 has high activity, resulting in low levels of PI(4)P that interacts with Ptc. In vertebrate cilium (right panel), Ptc1/2 and Grp161 localize in the cilium to inhibit Hh signaling. PI(4,5)P2 accumulates in the cilium but PI(4)P does not. (B) Smo regulation in the presence of Hh. In Drosophila (left panel), Hh promotes the production of PI(4)P by activating Stt4 and inactivating Sac1. PI(4)P is also released from Ptc upon Hh stimulation, allowing PI(4)P to bind and activate Smo through promoting Smo phosphorylation and dimerization. In the vertebrate cilium (right panel), PI(4)P levels are elevated by Shh, which promote Smo phosphorylation and ciliary accumulation. PI(4)P elevated by Shh inhibits the ciliary localization of Ptc1/2.
Gprk2 is positively involved in Hh signaling by directly phosphorylating Smo, and by forming a dimer to promote Smo dimerization and activation in a kinase activity-independent manner (Chen et al., 2010). But how Gprk2 promotes Smo dimerization and activation is not clear. Jiang et al. demonstrate that the PH domain of Gprk2 is responsible to enrich PI(4)P, which promotes Smo phosphorylation and dimerization. The findings that lipids bind to the PH domain promotes protein dimerization (Swanson et al., 2008; Zwolak et al., 2013), leading to the possibility for PI(4)P to promote Gprk2 dimerization, eventually promoting Smo phosphorylation and dimerization (Chen et al., 2010).
Hh likely promotes the production of PI(4)P by regulating the activity of PI(4)P kinase Stt4 and phosphatase Sac1 (Jiang et al., 2016), which may account for the mechanism by which Ptc acts catalytically to inhibit Smo. In this recent study, an additional layer of regulation has been identified, in which Hh may regulate the release of PI(4)P from Ptc in order for PI(4)P to activate Smo (Jiang et al., accepted). The sterol-sensing domain (SSD)of Ptc strongly interacts with PI(4)P, raising the possibility that Ptc may control the pool of phospholipids in regulating the accessibility of Smo to PI(4)P. Indeed, Hh treatment increases Smo-PI(4)P interaction however decreases Ptc-PI(4)P interaction, therefore binding of Hh to Ptc may cause the conformation change, leading to the release of PI(4)P from Ptc. The regulation of Smo by PI(4)P is likely conserved between Drosophila and vertebrate. PI(4)P promotes vertebrate Smo phosphorylation, activation and localization in the cilium. Meanwhile, PI(4)P prevents the ciliary accumulation of Ptc1 and Ptc2. This interesting discovery should facilitate further studies to better understand the direct role of phospholipids in Hh signaling pathway, ciliary function and how related regulators are controlled in this process.
Concluding remarks
Many studies regarding Smo regulation demonstrate that the mechanisms are highly conserved among species, although Drosophila and mammalian Smo have big difference in sequence homology in the carboxyl-terminal tail (Arensdorf et al., 2015). Smo also has other functional domains as GPCR, including N-terminal cysteine rich domain (CRD), three extracellular, three intracellular loops, and seven transmembrane domains. Recent structural analysis of the Smo N-terminal have suggested that Smo CRD may serve as binding site for exogenous ligand, such as cyclopamine and oxysterols (Myers et al., 2013; Nachtergaele et al., 2013; Rana et al., 2013). It is unknown whether the N-terminal of Smo also has posttranslational modifications. A recent study found that loss of N-glycosylation disrupted Smo trafficking and attenuated its signaling capability (Marada et al., 2015), however, more evidence may be needed for a better understanding.
Studies have shown that USP8 also interacts with and deubiquitinate Hrs (Zhang et al., 2014; Pradhan-Sundd and Verheyen, 2015), demonstrating multiple roles of USP8 in both cargo de-ubiquitination and ESCRT-0 stability during development, which is helpful to address the mechanisms of Hh signaling. Besides, it is interesting to test whether USP8 or other DUBs are involved in reversing the ubiquitination of other Hh signaling components. Balmer et al. performed an RNAi screen in Drosophila non-ciliated cells to test whether cilium-associated proteins have cilium-independent functions, and the results show there is no effect on Hh signaling (Balmer et al., 2015), indicating that cilium-associated regulators need to reach the primary cilium to modulate Hh signaling. Whether the DUBs required to be localized in the cilium in order to modulate Hh signaling is unknown.
Studies on the function of endogenous sterol derivatives in Hh signaling also suggest that lipids synthesis and trafficking may regulate not only Hh secretion and spreading, but the activation and trafficking of Hh signaling components (Callejo et al., 2008; Eaton, 2008; Khaliullina et al., 2009). Because recent work has shown that PI(4)P can modulate Hh signaling through regulating Smo, and because PIs have critical roles in regulating cell physiology, signaling, metabolism and membrane trafficking, it is possible for other phospholipids to positively or negatively regulate Hh signaling activity. PI(4)P acts likely in between Ptc and Smo to promote Hh signaling. Further investigation of the effects for phospholipids in the regulating Smo and other Hh components may yield important insights into the intracellular trafficking and activation of Smo that is the key for Hh signaling.
Acknowledgments
We apologize to colleagues whose works were not cited because of space limited. Our work is supported by funding from the National Institute of Health (R01GM079684).
Footnotes
Compliance with ethics guidelines
This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
Kai Jiang and Jianhang Jia declare that they have no conflict of interest.
References
- Aikin RA, Ayers KL, Thérond PP. The role of kinases in the Hedgehog signalling pathway. EMBO Rep. 2008;9(4):330–336. doi: 10.1038/embor.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86(2):221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7(1):86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- Arensdorf AM, Marada S, Ogden SK. Smoothened Regulation: A Tale of Two Signals. Trends Pharmacol Sci. 2015;37(1):62–72. doi: 10.1016/j.tips.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature. 2013;494(7438):484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, Ally MS, Kim J, Yao C, Chang AL, Oro AE, Tang JY. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93(3):1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer S, Dussert A, Collu GM, Benitez E, Iomini C, Mlodzik M. Components of intraflagellar transport complex A function independently of the cilium to regulate canonical Wnt signaling in Drosophila. Dev Cell. 2015;34(6):705–718. doi: 10.1016/j.devcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, Kayserili H, Swistun D, Scott LC, Bertini E, Boltshauser E, Fazzi E, Travaglini L, Field SJ, Gayral S, Jacoby M, Schurmans S, Dallapiccola B, Majerus PW, Valente EM, Gleeson JG. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41(9):1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, Yu Q, Ostrom L, Fordjour P, Anderson DL, Monahan JE, Kelleher JF, Peukert S, Pan S, Wu X, Maira SM, García-Echeverría C, Briggs KJ, Watkins DN, Yao YM, Lengauer C, Warmuth M, Sellers WR, Dorsch M. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51):51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejo A, Culi J, Guerrero I. Patched, the receptor of Hedgehog, is a lipoprotein receptor. Proc Natl Acad Sci USA. 2008;105(3):912–917. doi: 10.1073/pnas.0705603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp D, Currie K, Labbé A, van Meyel DJ, Charron F. Ihog and Boi are essential for Hedgehog signaling in Drosophila. Neural Dev. 2010;5(1):28. doi: 10.1186/1749-8104-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A, Struhl G. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature. 2004;431(7004):76–80. doi: 10.1038/nature02835. [DOI] [PubMed] [Google Scholar]
- Casso DJ, Liu S, Iwaki DD, Ogden SK, Kornberg TB. A screen for modifiers of hedgehog signaling in Drosophila melanogaster identifies swm and mts. Genetics. 2008;178(3):1399–1413. doi: 10.1534/genetics.107.081638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez M, Ena S, Van Sande J, de Kerchove d’Exaerde A, Schurmans S, Schiffmann SN. Modulation of ciliary phosphoinositide content regulates trafficking and sonic Hedgehog signaling output. Dev Cell. 2015;34(3):338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98(3):305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23(2):186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, Jia J, Jiang J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010;24(18):2054–2067. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, Jiang J. Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9(6):e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504(7479):315–318. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504(7479):311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N, Neubüser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102(4):521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23(4):823–835. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhang J, Su Y, Liu M, Ospina JK, Yang S, Zhu AJ. In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling. PLoS ONE. 2011;6(9):e24168. doi: 10.1371/journal.pone.0024168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussillol-Godar F, Brissard-Zahraoui J, Limbourg-Bouchon B, Boucher D, Fouix S, Lamour-Isnard C, Plessis A, Busson D. Modulation of the Suppressor of fused protein regulates the Hedgehog signaling pathway in Drosophila embryo and imaginal discs. Dev Biol. 2006;291(1):53–66. doi: 10.1016/j.ydbio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9(6):437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- Fan J, Jiang K, Liu Y, Jia J. Hrs promotes ubiquitination and mediates endosomal trafficking of smoothened in Drosophila hedgehog signaling. PLoS ONE. 2013;8(11):e79021. doi: 10.1371/journal.pone.0079021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Liu Y, Jia J. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Dev Biol. 2012;366(2):172–184. doi: 10.1016/j.ydbio.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14(16):1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Fukumoto T, Watanabe-Fukunaga R, Fujisawa K, Nagata S, Fukunaga R. The fused protein kinase regulates Hedgehog-stimulated transcriptional activation in Drosophila Schneider 2 cells. J Biol Chem. 2001;276(42):38441–38448. doi: 10.1074/jbc.M105871200. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G, 3rd, Abedin M, Schurmans S, Inoue T, Reiter JF. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev Cell. 2015;34(4):400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Wang G, Dasgupta S, Dinkins M, Zhu G, Bieberich E. Characterization of an apical ceramide-enriched compartment regulating ciliogenesis. Mol Biol Cell. 2012;23(16):3156–3166. doi: 10.1091/mbc.E12-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JS, Lee MY, Han HJ. Sonic hedgehog stimulates mouse embryonic stem cell proliferation by cooperation of Ca2+/protein kinase C and epidermal growth factor receptor as well as Gli1 activation. Stem Cells. 2007;25(12):3069–3080. doi: 10.1634/stemcells.2007-0550. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KS, Suyama K, Fish M, Scott MP. Differential regulation of Hedgehog target gene transcription by Costal2 and Suppressor of Fused. Development. 2005;132(6):1401–1412. doi: 10.1242/dev.01689. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6(4):306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Hsia EY, Gui Y, Zheng X. Regulation of Hedgehog signaling by ubiquitination. Front Biol (Beijing) 2015;10(3):203–220. doi: 10.1007/s11515-015-1343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102(32):11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426(6962):83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA. 2012;109(48):19691–19696. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M, Pernot E, Kisseleva MV, Compère P, Schiffmann SN, Gergely F, Riley JH, Pérez-Morga D, Woods CG, Schurmans S. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat Genet. 2009;41(9):1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. J Biol Chem. 2010;285(48):37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Liu Y, Yan W, Jia J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development. 2009;136(2):307–316. doi: 10.1242/dev.030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J. Phosphorylation regulation of Hedgehog signaling. Vitam Horm. 2012;88:253–272. doi: 10.1016/B978-0-12-394622-5.00011-0. [DOI] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416(6880):548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell Mol Life Sci. 2006;63(11):1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17(21):2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432(7020):1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle. 2006;5(21):2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80(4):563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391(6666):493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Jiang K, Liu Y, Fan J, Epperly G, Gao T, Jiang J, Jia J. Hedgehog-regulated atypical PKC promotes phosphorylation and activation of Smoothened and Cubitus interruptus in Drosophila. Proc Natl Acad Sci USA. 2014;111(45):E4842–E4850. doi: 10.1073/pnas.1417147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Liu Y, Fan J, Zhang J, Li X, Evers BM, Zhu H, Jia J. PI (4)P promotes phosphorylation and conformational change of Smoothened through interaction with its C-terminal tail. PLoS Biol. 2016;14(1):e1002375. doi: 10.1371/journal.pbio.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaliullina H, Panáková D, Eugster C, Riedel F, Carvalho M, Eaton S. Patched regulates Smoothened trafficking using lipoprotein-derived lipids. Development. 2009;136(24):4111–4121. doi: 10.1242/dev.041392. [DOI] [PubMed] [Google Scholar]
- Kim J, Hsia EY, Brigui A, Plessis A, Beachy PA, Zheng X. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal. 2015;8(379):ra55. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jäger N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, König R, Beroukhim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimling A, Scheurlen W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schüller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM the ICGC PedBrain Tumor Project. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25(3):393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz RJ. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320(5884):1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhandaivel A, Schultz SW, Alkhori L, Alenius M. Cilia-mediated hedgehog signaling in Drosophila. Cell Reports. 2014;7(3):672–680. doi: 10.1016/j.celrep.2014.03.052. [DOI] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol. 2012;10(1):e1001239. doi: 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma G, Wang B, Jiang J. Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci Signal. 2014;7(332):ra62. doi: 10.1126/scisignal.2005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132(13):3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cao X, Jiang J, Jia J. Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev. 2007;21(15):1949–1963. doi: 10.1101/gad.1557407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304(5678):1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12(5):1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Marada S, Navarro G, Truong A, Stewart DP, Arensdorf AM, Nachtergaele S, Angelats E, Opferman JT, Rohatgi R, McCormick PJ, Ogden SK. Functional divergence in the role of N-linked glycosylation in Smoothened signaling. PLoS Genet. 2015;11(8):e1005473. doi: 10.1371/journal.pgen.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méthot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127(18):4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24(19):2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152(1–2):210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, Bazan JF, Beachy PA. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell. 2013;26(4):346–357. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, Whalen DM, Mydock LK, Zhao Z, Malinauskas T, Krishnan K, Ingham PW, Covey DF, Siebold C, Rohatgi R. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife. 2013;2:e01340. doi: 10.7554/eLife.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37(12):1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456(7224):967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Kato M, Zhang C, Guo Y, Beachy PA. A comparison of Ci/Gli activity as regulated by Sufu in Drosophila and mammalian Hedgehog response. PLoS ONE. 2015;10(8):e0135804. doi: 10.1371/journal.pone.0135804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan-Sundd T, Verheyen EM. The Myopic-Ubpy-Hrs nexus enables endosomal recycling of Frizzled. Mol Biol Cell. 2015;26(18):3329–3342. doi: 10.1091/mbc.E15-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MA. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20(4):399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108(6):823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Prulière G, Cosson J, Chevalier S, Sardet C, Chenevert J. Atypical protein kinase C controls sea urchin ciliogenesis. Mol Biol Cell. 2011;22(12):2042–2053. doi: 10.1091/mbc.E10-10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati GV, Hughes CE, Dorn KV, Zhang D, Sugianto P, Aravind L, Rohatgi R. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev Cell. 2014;28(5):483–496. doi: 10.1016/j.devcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana R, Carroll CE, Lee HJ, Bao J, Marada S, Grace CR, Guibao CD, Ogden SK, Zheng JJ. Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat Commun. 2013;4:2965. doi: 10.1038/ncomms3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri N, Ruel L, Gallet A, Raisin S, Thérond PP. Distinct phosphorylations on kinesin costal-2 mediate differential hedgehog signaling strength. Dev Cell. 2012;22(2):279–294. doi: 10.1016/j.devcel.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Ranieri N, Thérond PP, Ruel L. Switch of PKA substrates from Cubitus interruptus to Smoothened in the Hedgehog signalosome complex. Nat Commun. 2014;5:5034. doi: 10.1038/ncomms6034. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Thérond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90(2):225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106(9):3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rorick AM, Mei W, Liette NL, Phiel C, El-Hodiri HM, Yang J. PP2A:B56epsilon is required for eye induction and eye field separation. Dev Biol. 2007;302(2):477–493. doi: 10.1016/j.ydbio.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3(11):813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Thérond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5(10):907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, Marmur E, Rudin CM, Chang AL, Low JA, Mackey HM, Yauch RL, Graham RA, Reddy JC, Hauschild A. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, Tsui V, Durham AB, Dlugosz AA, Haverty PM, Bourgon R, Tang JY, Sarin KY, Dirix L, Fisher DC, Rudin CM, Sofen H, Migden MR, Yauch RL, de Sauvage FJ. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138(19):4219–4231. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90(2):235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Su Y, Ospina JK, Zhang J, Michelson AP, Schoen AM, Zhu AJ. Sequential phosphorylation of smoothened transduces graded hedgehog signaling. Sci Signal. 2011;4(180):ra43. doi: 10.1126/scisignal.2001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KD, Tang Y, Ceccarelli DF, Poy F, Sliwa JP, Neel BG, Eck MJ. The Skap-hom dimerization and PH domains comprise a 3′-phosphoinositide-gated molecular switch. Mol Cell. 2008;32(4):564–575. doi: 10.1016/j.molcel.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418(6900):892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, Coppola C, Chanana AM, Marji J, Bickers DR, Epstein EH., Jr Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366(23):2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thérond PP, Knight JD, Kornberg TB, Bishop JM. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc Natl Acad Sci USA. 1996;93(9):4224–4228. doi: 10.1073/pnas.93.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138(22):4921–4930. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000a;100(4):423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497(7449):338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 2000b;14(22):2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc Natl Acad Sci USA. 2009;106(8):2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS ONE. 2009;4(4):e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137(13):2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its sub-cellular localization. PLoS Biol. 2012;10(1):e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yang C, Chen W, Chen Y, Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res. 2012;22(11):1593–1604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29(4):469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- Yang X, Mao F, Lv X, Zhang Z, Fu L, Lu Y, Wu W, Zhou Z, Zhang L, Zhao Y. Drosophila Vps36 regulates Smo trafficking in Hedgehog signaling. J Cell Sci. 2013;126(Pt 18):4230–4238. doi: 10.1242/jcs.128603. [DOI] [PubMed] [Google Scholar]
- Yavari A, Nagaraj R, Owusu-Ansah E, Folick A, Ngo K, Hillman T, Call G, Rohatgi R, Scott MP, Banerjee U. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev Cell. 2010;19(1):54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438(7069):873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc Natl Acad Sci USA. 2004;101(52):17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Du J, Lei C, Liu M, Zhu AJ. Ubpy controls the stability of the ESCRT-0 subunit Hrs in development. Development. 2014;141(7):1473–1479. doi: 10.1242/dev.099564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell. 2005;8(2):267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mao F, Lu Y, Wu W, Zhang L, Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011;21(10):1436–1451. doi: 10.1038/cr.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, Li W, Atwood SX, Whitson RJ, Chang AL, Li J, Oro AE, Chan JA, Kelleher JF, Segal RA. RAS/MAPK Activation Drives Resistance to Smo Inhibition, Metastasis, and Tumor Evolution in Shh Pathway-Dependent Tumors. Cancer Res. 2015;75(17):3623–3635. doi: 10.1158/0008-5472.CAN-14-2999-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450(7167):252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- Zheng X, Mann RK, Sever N, Beachy PA. Genetic and biochemical definition of the Hedgehog receptor. Genes Dev. 2010;24(1):57–71. doi: 10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolak A, Yang C, Feeser EA, Ostap EM, Svitkina T, Dominguez R. CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nat Commun. 2013;4:2523. doi: 10.1038/ncomms3523. [DOI] [PMC free article] [PubMed] [Google Scholar]