Abstract
Aim
The first hepatitis C virus (HCV) recombinant, RF2k/1b, was initially described from Russia and has since then been identified from patients in Ireland, Estonia, Uzbekistan and Cyprus. Many of these patients originated from Georgia; however there is no information on its prevalence in Georgia or its susceptibility to antiviral treatment.
Methods
We retrospectively sequenced the non-structural region 5B (NS5B) of the HCV genome in samples from 72 Georgian patients, 36 of those had been treated with Pegylated Interferon and Ribavirin.
Results
The HCV genotype was determined using the Versant HCV Genotype v2 kit. Based on this typing, 32 patients (44.4%) were infected with genotype 1, 21 (29.1%) genotype 2 and 19 (26.3%) genotype 3. Partial NS5B of these strains was sequenced and analyzed for type, with concordant genotype results for all type 1 and 3 strains. Discrepant results were observed for genotyped 2 strains, with 16 (76%) having NS5B of subtype 1b. On phylogenetic analysis, 15 NS5B sequences of these strains were found in a clade formed by recombinant RF2k/1b stains. The remaining discordant sequence was found within a clade formed by 1b strains.
Conclusion
Our findings show that the RF2k/1b recombinant strain is common among Georgian patients previously assumed to be infected by genotype 2. Since genotyping is mainly performed to decide treatment strategies; there is a need to determine the genotype by analysis of at least two genomic regions in strains from Georgian patients considered infected by genotype 2 based on standard HCV genotyping methods.
Keywords: HCV sequence, phylogenetic analysis
Introduction
The World Health Organization (WHO) estimates that ~180 million people are infected with the hepatitis C virus (HCV) worldwide, with the highest prevalence rates reported in Africa and Asia1.In Western countries, HCV is the leading cause of end-stage liver disease and hepatocellular carcinoma, thereby the one of the major causes for liver transplantation globally2. It is estimates that as many as four million new hepatitis C infections occur annually and more than 350,000 individuals die from hepatitis C related liver diseases each year1.
Novel drugs known as direct-acting antiviral (DAAs) with or without Pegylated Interferon- Ribavirin (PEG/RBV) have revolutionized chronic hepatitis C treatment by providing shortened and simplified treatment regimens while minimizing related side effects and dramatically increasing sustained viral response (SVR) rates3, 4. Recent clinical guidelines include DAAs as a first-line treatment option for chronic hepatitis C5. However, due to the extremely high cost, dual therapy with PEG/RBV combination still continues to be the standard of care in resource limited countries, including Georgia. The success rate of this dual therapy regimen largely depends on the infecting HCV genotype. Clinical studies have shown that patients infected with genotype 2 or 3 are more likely to achieve SVR than patients infected with genotype 16, 7.
The antiviral treatment efficiency is not known for the natural intergenotypic recombinant forms that possess genotype 2 sequences in the structural regions and genotype 1, 5 or 6 sequences in the nonstructural regions8–12. The first of these recombinants, RF2k/1b, was primarily identified in Russia8, since then it has been described from patients in Ireland10, Estonia13, Uzbekistan14 and most recently in Cyprus15 and France16. Even if infection with this type is wide-spread, most of the diagnosed patients were infected in Georgia through either injection drug use (IDU) or sexual contact15–17, suggesting that this recombinant virus is circulating in Georgia.
Georgia has the highest HCV prevalence (6.7 %) in the general population18 in the Caucasus region, therefore, accurate HCV genotype determination is especially important for selection of correct treatment options. Taking into account that recombinant forms share components of different genotypes, HCV genotype interpretation based only on assays targeting one region, such as the 5'UTR/Core may mislead correct classification of HCV recombinants. Despite limited data reported on the susceptibility of RF2k/1b to PEG/RBV or to other antiviral compounds17, 19, it is important to correctly identify the genotype and recombinant forms, since they share interferon sensitivity-determining and interferon resistance-determining regions with the more difficult to treat genotype 120–22.
The purpose of our study was to identify possible HCV recombinant forms among Georgian patients infected with HCV genotypes previously determined with an assay targeting the 5’UTR/Core region, and comparing these results with the genotypes obtained by sequencing the NS5B region of the strains.
Materials and methods
Study settings and population
The Infectious Diseases AIDS and Clinical Immunology Research Center (IDACIRC) is Georgia’s referral institution for diagnosis, and treatment of infectious diseases, including viral hepatitis. IDACIRC is the country’s largest provider of medical services related to hepatitis C infection. Patients referred to IDACIRC for hepatitis C care formed the study population. According to local protocols, after a positive screening for anti-HCV antibodies, patients are tested for HCV viremia followed by determination of HCV genotype. Overall, IDACIRC database contained specimens from 2,291 hepatitis C infected Georgian patients. All samples were genotyped from 2003 to 2011. From this cohort, serum samples for sequencing the NS5B region were selected from 100 patients, 36 of those had been treated with PEG/RBV. The selection was based on genotypes obtained by routine genotyping to achieve approximately equal proportions of each genotype in the subset as in the samples from all patients in IDACIRC HCV database. This database contains genotype data on HCV strains from 2,291 patients representing the majority of hepatitis C patients from 2003 to 2011 in Georgia. Analysis of the genotypes revealed that genotype 1 was predominant (42.0%) over genotypes 3 (32.9%) and 2 (24.9%). Genotype 4 was found among 3 patients (0.1 %), while other known genotypes were not identified. All samples had been stored at −20°C after routine HCV RNA quantification and genotyping.
IRB approval
Study was approved by Institutional Review Board of IDACIRC (OHRP3IRB00006106) and New York State Department of Health (#11-052).
HCV RNA quantification and genotyping
HCV RNA levels were determined by the COBAS TaqMan HCV Test, v.2 (Roche, Basel, Switzerland) with the quantification limit of 25 IU/ml. Patient specimens with detectable HCV viral load of more than 2,000 IU/ml were genotyped before initiation of PEG/RBV therapy.
HCV genotyping was performed by Versant HCV Genotype v2. Kit (Siemens, Ghent, Belgium). The kit is designed to reverse transcribe and amplify 240 and 270 base pairs of the 5' UTR and Core region. After amplification, PCR products were immobilized on a nitrocellulose strip, which resulted in a visible banding pattern. HCV genotyping results were then interpreted using the manufacturer’s protocol.
Personal error in baseline Versant HCV genotyping procedure was excluded, since all samples with discrepant results between Versant typing and sequencing, were re-tested in the Versant HCV genotyping assay.
HCV RNA Extraction and cDNA synthesis
HCV RNA was extracted from 0.5 mL plasma using High Pure Viral Nucleic Acid Kit with the elution volume of 75 ul. (Roche, Basel, Switzerland). cDNA synthesis was carried out with 100U of MuLV transcriptase and 0.1 U random hexamer primers (Qiagen, Valencia, CA) with the following steps: pre heat of 2ul random hexamer with 12ul RNA for 5 minutes at 85°C, followed by addition and heat of 28 ul mixture with 4 ul -10× PCR Buffer II, 8 ul – 25 mM MgCl2, 12 ul – 10 mM dNTP, 2 ul- Rnase Inhibitor and 2 ul -MulV RT at 42°C for 60 minutes and 85°C for 5 minutes.
Amplification and sequencing of the NS5B region
cDNA was amplified by nested PCR using the GeneAmp XL PCR kit (Applied Biosystems, Foster City, CA). For primary PCR, 5 µL of cDNA was amplified using primers P3 (TATGAYACCCGCTGYTTTGACTC) and P4 (GCNGARTAYCTVGTCATAGCCTC). Secondary PCR conditions were identical to the primary round, except that primer P4 was substituted with primer P5 (GCTAGTCATAGCCTCCGT).This amplification rendered a 260 bp amplicon, which was extracted from 1% agarose by using the QIAquick gel extraction kit (Qiagen, Valencia, CA) and sequenced at the Wadsworth Center’s Applied Genomic Technologies Core Facility.
Characterization of genotype by BLAST and phylogenetic analysis
All obtained HCV sequences were genotyped by BLAST comparison with sequences in the Los Alamos HCV sequence database http://hcv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html.
The sequences obtained were aligned with corresponding reference sequences representing HCV genotypes 1, 2, 3 and representative subtypes. The analyses also included published RF2k/1b sequences10, 14, 15, 23. In order to confirm the genotype classification, reference strains from the BLAST search were included, if there was a nucleotide similarity of >90% between sequences of known genotype and subtype in GenBank and those obtained in this study. The sequences were aligned using CLUSTAL W in BioEdit24. Phylogenetic analyses were conducted using MEGA version 6.025. The neighbor-joining method based on Kimura two-parameter model was used for tree construction. The robustness of the tree was estimated from 1,000 bootstrap replicates, with branches with more than 70 % bootstrap support considered significant.
Results
Serum samples from 100 patients, selected according to infecting HCV genotype were used for partial sequencing of the NS5B region. Sequences could be obtained for 72 of the 100 specimens. There was no difference regarding the genotype distribution between specimens group successfully sequenced and not sequenced (p-0.7) neither was the difference in terms of HCV viral load (p-0.13).
The majority of the 72 patients were male (84.7%). All were of white race and European ancestry (Table1). Based on the information available in medical charts probable sources of the hepatitis C acquisition were IDU for 50 (69.4%) patients, sexual contact for 8 (11.1%), and blood transfusion for 2 (2.7%) patients. The route of HCV transmission could not be determined for 12 (16.6%) patients (Table 1).
Table I.
Baseline characteristics of Georgian patients
| Characteristics | All patients (n=72) |
Genotype 1 (n=32) |
Genotype 2 (n=21) |
Genotype 3 (n=19) |
p value |
|---|---|---|---|---|---|
| Sex | |||||
| Male, n (%) | 61 (84.7%) | 25 (78.1%) | 19 (90.5%) | 17 (89.5%) | 0.49 |
| Female n (%) | 11 (15.3%) | 7 (21.9%) | 2 (9.5%) | 2 (10.5%) | |
| Possible route of HCV acquisition | |||||
| IDU | 50 (69.4%) | 23 (71.8%) | 15 (71.4%) | 12 (63.1%) | 0.68 |
| Sexual transmition | 8 (11.1%) | 4 (12.5%) | 2 (9.5%) | 2 (10.5%) | |
| Blood transfusion | 2 (2.7%) | 2 (6.3%) | 0 | 0 | |
| Unknown | 12 (16.6) | 3 (9.3%) | 4 (19.5%) | 5 (26.3%) | |
| Baseline HCV viral load | |||||
| >600 000 IU/ml, n (%) | 43 (59.7%) | 18 (56.3%) | 15 (71.4%) | 10 (52.6%) | 0.42 |
| <600 000 IU/ml, n (%) | 29 (40.3%) | 14 (43.7%) | 6 (28.6%) | 9 (47.4%) | |
Genotyping
Based on the Versant HCV Genotyping kit v.2, 32 of the 72 (44.4%) patients were infected with genotype 1, 21 (29.1%) with genotype 2 and 19 (26.3%) with genotype 3 (Table1).
When the NS5B region was sequenced and phylogenetically analyzed in these 72 strains, there was no genotype/subtype discordance observed between the Versant HCV Genotyping kit v.2 and NS5B genomic regions for the genotype 3 infected strains. There were discordances found for three genotype 1 strains, which were typed as 1a by Versant HCV Genotyping method, but had NS5B regions similar to subtype 1b.
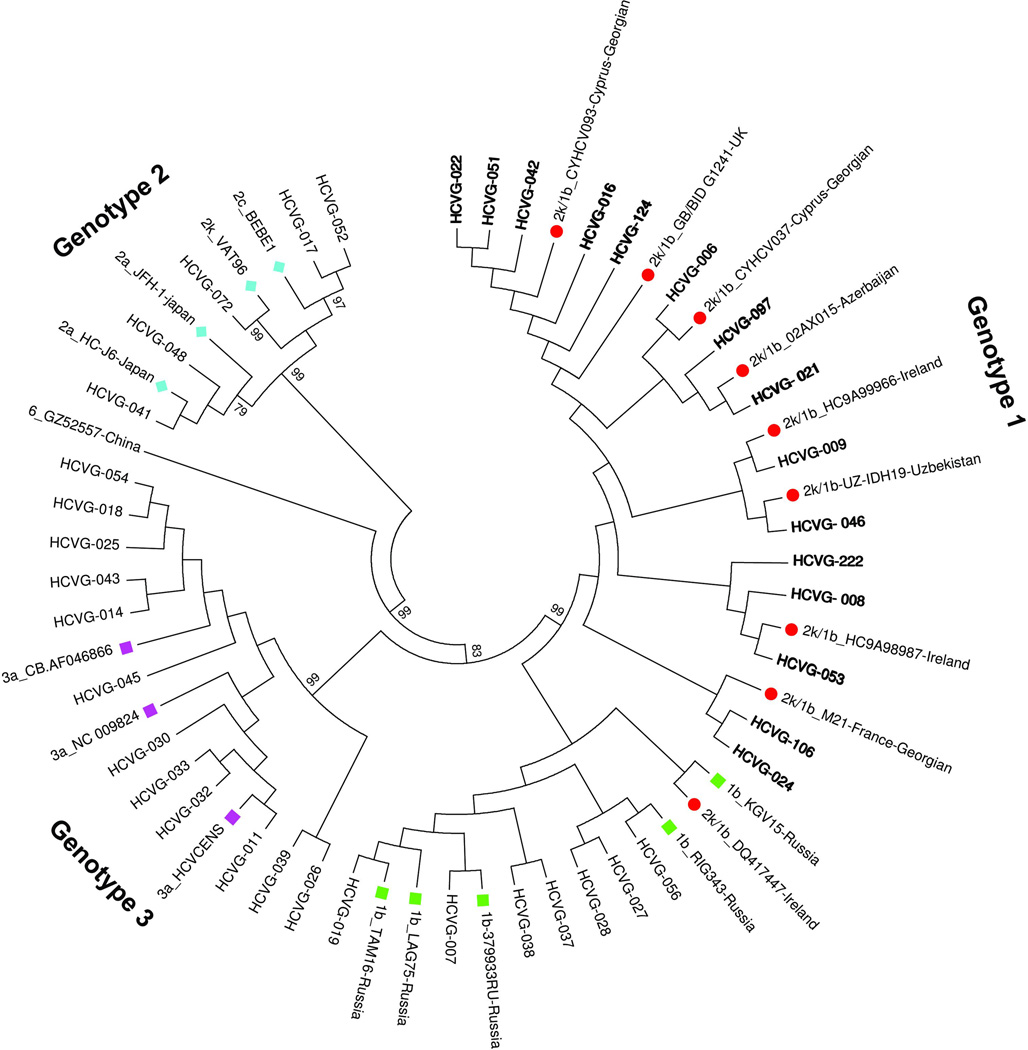
Discordant results were found for 16 (76.2%) of the 21 strains previously typed as either genotype 2 or 2a/2c. The NS5B sequences were similar to subtype 1b, 15 of them formed a clade together with the RF2k/1b sequences from EU, Russia and Azerbaijan in the phylogenetic tree (Figure 1).The remaining sample, HCVG 056, was found in a clade formed by 1b strains together with 1b strains from Russia. Sample HCV G072 considered as genotype 2a/2c based on Versant HCV Genotype v2 kit, was found to be genotype 2k in the NS5B region. The remaining four 2a/2c sequences were all identified either as 2a or 2c based on sequencing of the NS5B region (Figure 1).
Figure 1.
Genotype reference sequences are shown with different colors. Nodes with bootstrap support over 70% are noted.
Clinical data on patients infected by recombinant forms
Based on the medical records, 36 of the 72 patients studied had been previously treated with PEG/RBV. Sustained viral response e.i negative HCV RNA at 24 weeks after treatment completion (SVR24) was achieved in 5 (38.5%) patients infected with genotype 1b, 7 (58.3%) infected with genotype 2, and 8 (72.7%) genotype 3 infected patients, with total SVR rate of 55.6 % (Table 2), when genotypes were determined by the Versant HCV Genotyping kit.
Table II.
Interferon therapy and SVR rates
| Treatment information | Total treated (n=36) |
Genotype 1 (n=13) |
Genotype 2 (n=12) |
Genotype 3 (n=11) |
|---|---|---|---|---|
| SVR †, (%) | 20 (55.6%) | 5 (38.5%) | 7 (58.3%) | 8 (72.7%) |
| Relapse/no response (%) | 16 (44.4%) | 8 (61.5%) | 5 (41.6%) | 3 (27.2%) |
SVR †- Sustained viral response
SVR was reached among four of five confirmed genotype 2 patients (80.0%) and three out of seven patients with RF2k/1b strain (42.8%) (p-0.29).
Among the seven genotype 2 infected patients with SVR, three (HCVG-097, HCVG-124, and HCVG-050) were found to be infected with the recombinant RF2k/1b virus. Five of the genotype 2 patients did not respond to treatment; 4 of those (HCVG-016, HCVG-021, HCVG -024, and HCVG-222) were infected by the RF2k/1b recombinant virus (Table 2).
Treatment information on the remaining 8 patients was not available, since they were lost to follow up.
Discussion
In this study HCV strains infecting 72 Georgian patients were retrospectively genotyped by sequencing partial NS5B. There was a 100% genotype concordance between two different typing methods for genotype 1 and 3 patients. However, only few specimens that were previously typed as genotype 2 were confirmed by NS5B region sequencing. The discrepancy was mainly due to the misclassification of RF2k/1b during initial genotype identification by Versant HCV genotyping kit, as this assay is limited to only amplifying structural parts of the HCV genome.
A high prevalence of this recombinant RF2k/1b strains was thus revealed, with 76.1% of the strains initially classified as genotype 2 found to be recombinants. This recombinant strain has been reported among ethnic Georgian persons residing in EU15–17 as well as among intravenous drug users in countries from Former Soviet Union, where it was originally discovered. Further studies are needed on more genotype 2 samples from Georgia by sequencing the 5’UTR/Core region for confirming the Versant HCV genotyping and the NS2 breakpoint for confirming that the samples contained the recombinant strain. However it is most probable that the patients in this study were infected by the RF2k/1b recombinants, since all strains gave discrepant results in the genotyping assays, and formed clades in the phylogenetic tree with confirmed recombinant strains from different countries. Our study shows that this strain prevalent in Georgia, where it may have originated, since it is circulating in the country and not a rare event in the hepatitis C infected population.
As stated by Viazov et al, that recombination between HCV strains is a rare event and that the resulting recombinants are usually not viable may hold true for many recombinant strains26. In addition, the necessary step of co-infection of two different strains may be rare event even in highly exposed settings26. However, the RF2k/1b recombinant has in this study shown to be highly viable and frequent in Georgia where it is spreading nosocomially, by intravenous drug use and sexually.
The high prevalence of the recombinant in Georgia has clinical importance with regard to treatment, as shown in this study with unsuccessful 24 week PEG/RBV therapy in four out of seven RF2k/1b infected patients, although only numerically lower compare to confirmed genotype 2 patients (p-0.29). This finding may provide a possible explanation of lower SVR rates among Georgian genotype 2 infected patients compare to SVR rates reported in different countries27–29.
Currently, no recommendation exists on the duration or optimal regimen for RF2k/1b due to limited information on its susceptibility to antiviral treatment. Two studies have discrepant results regarding interferon treatment of RF2k/1b and SVR17, 19, with one study19 showing higher susceptibility of the RF2k/1b strain to PEG/RBV treatment than the other17. Our results are consistent with one of these studies from France on treatment of a Georgian patient double infected with 3a and RF2k/1b strains17. The recombinant RF2k/1b strain emerged and became dominant after eradication of the previously dominant 3a subtype by 24 weeks PEG/RBV therapy. The recombinant strain was thus less responsive to a 24 week PEG/RBV regimen than genotype 3a. Recent study revealed lower SVR rates among patients infected with recombinant forms on DAA therapy30. This result is also consistent with our finding in this study, with the RF2k/1b as difficult-to-treat as genotype 1. Patients infected by this strain may therefore be considered for longer treatment duration or DAA regimen, as is now performed for genotype 1 infected patients.
Since 2003 the IDACIRC has been using the Versant HCV Genotype v2 method for routine HCV genotyping. Several studies have shown that this method is accurate for HCV genotype identification in specific populations31, 32. However, the method cannot be used for identifying recombinant forms since it is based on reverse transcription and amplification in the 5'UTR/Core region of the HCV genome only, so the sequence variation in the NS5B region is missed. Our finding of missed RF2k/1b strains by using this technique is in agreement with the recent proposal by Morel et al on new genotyping schemes for HCV using at least two different genomic regions33. Since the results were concordant for genotype 1 and 3 an approach for sequencing only genotype 2 strains will save cost and resources. Based on our results, we therefore suggest that all strains from Georgian patients infected by genotype 2 as determined by Versant HCV genotyping assay should also be sequenced in the NS5B region for either confirmation of genotype 2 or identification of RF2k/1b strains. This scheme will be implemented at IDACIRC, and it is recommended also for neighboring countries where this recombinant form may also circulate. If strains from patients infected with this recombinant for will not be typed also by sequencing the single typing can cause that these patients receive too short duration of treatment and are not offered alternative treatment regimens because of cost reasons.
Studies on more genotype 2 infected patients treated with PEG/RIB will confirm our finding on non-responders and relapsers among patients infected by RF2k/1b. The high prevalence of this recombinant form in Georgia stresses the need of further studies, since this strain may widely circulate in the country and thus, may alter treatment options and/or duration compared to genotype 2 infected, since it shares non-structural regions with genotype 1 and may be as difficult to treat as this genotype. Future studies will also reveal if the IL28B genotypes in patients infected with RF2k/1b strain will influence on the antiviral response. None of these patients in this study were on triple therapy with DAA that may be considered as a treatment option. Despite the development of highly potent DAA drugs, the pricing policy still remains a challenge in developing countries. DAAs are not affordable for patients in Georgia, thus; dual therapy with PEG/RBV will remain the standard of care unless the price policy changes. Recent initiatives supported by the Global Fund and the Georgian Government increased access to free PEG/RBV among HIV/HCV co-infected patients and prisoners. In addition, the cost of PEG/RBV therapy was decreased by 60.0% in 2014 for 10 000 patients, which also triggered increased demand. However, for the remaining patients the overall treatment costs are not covered by the health system or insurance companies, but have to be paid by the patients, and remains very expensive. As the relatively affordable option with PEG/RBV treatment is genotype dependent, the correct identification of infecting HCV genotype is important for SVR prediction or correct treatment selection.
Acknowledgments
We thank Infectious Diseases, AIDS and Clinical Immunology Research Center. The project described was supported by Award Number D43TW000233 and D43TW007384 from the Fogarty International Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health.
Footnotes
Infectious Diseases, AIDS and Clinical Immunology Research Center, Address: 16 Al Kazbegi Ave, Tbilisi, Georgia 0160
Department of Infectious Diseases, Institute of Biomedicine, University of Gothenburg, Address: Västergatan 25 43430, Gothenburg, Sweden
David Axelrod Institute, Wadsworth Center, Address: 120 New Scotland Ave, Albany, NY, US
References
- 1.World Health Organization. Hepatitis C fact sheet. 2013 [cited 2013; Available from: http://www.who.int/mediacentre/factsheets/fs164/en/
- 2.Lauer GM, Walker BD. Hepatitis C virus infection. The New England journal of medicine. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 3.Lawitz E, Lalezari JP, Hassanein T, Kowdley KV, Poordad FF, Sheikh AM, et al. Sofosbuvir in combination with peginterferon alfa-2a and ribavirin for non-cirrhotic, treatment-naive patients with genotypes 1, 2, and 3 hepatitis C infection: a randomised, double-blind, phase 2 trial. Lancet Infect Dis. 2013;13(5):401–408. doi: 10.1016/S1473-3099(13)70033-1. [DOI] [PubMed] [Google Scholar]
- 4.Lawitz E, Gane EJ. Sofosbuvir for previously untreated chronic hepatitis C infection. The New England journal of medicine. 2013;369(7):678–679. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- 5.EASL Recommendations on Treatment of Hepatitis C 2014. J Hepatol. 2014;61(2):373–395. doi: 10.1016/j.jhep.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Ferenci P. Predictors of response to therapy for chronic hepatitis C. Seminars in liver disease. 2004;24(Suppl 2):25–31. doi: 10.1055/s-2004-832925. [DOI] [PubMed] [Google Scholar]
- 7.Zeuzem S. Heterogeneous virologic response rates to interferon-based therapy in patients with chronic hepatitis C: who responds less well? Annals of internal medicine. 2004;140(5):370–381. doi: 10.7326/0003-4819-140-5-200403020-00033. [DOI] [PubMed] [Google Scholar]
- 8.Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. Journal of virology. 2002;76(8):4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kageyama S, Agdamag DM, Alesna ET, Leano PS, Heredia AM, Abellanosa-Tac-An IP, et al. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. Journal of medical virology. 2006;78(11):1423–1428. doi: 10.1002/jmv.20714. [DOI] [PubMed] [Google Scholar]
- 10.Moreau I, Hegarty S, Levis J, Sheehy P, Crosbie O, Kenny-Walsh E, et al. Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland. Virology journal. 2006;3:95. doi: 10.1186/1743-422X-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legrand-Abravanel F, Claudinon J, Nicot F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, et al. New natural intergenotypic (2/5) recombinant of hepatitis C virus. Journal of virology. 2007;81(8):4357–4362. doi: 10.1128/JVI.02639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. Journal of virology. 2006;80(15):7569–7577. doi: 10.1128/JVI.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tallo T, Norder H, Tefanova V, Krispin T, Schmidt J, Ilmoja M, et al. Genetic characterization of hepatitis C virus strains in Estonia: fluctuations in the predominating subtype with time. Journal of medical virology. 2007;79(4):374–382. doi: 10.1002/jmv.20828. [DOI] [PubMed] [Google Scholar]
- 14.Kurbanov F, Tanaka Y, Avazova D, Khan A, Sugauchi F, Kan N, et al. Detection of hepatitis C virus natural recombinant RF1_2k/1b strain among intravenous drug users in Uzbekistan. Hepatology research : the official journal of the Japan Society of Hepatology. 2008;38(5):457–464. doi: 10.1111/j.1872-034X.2007.00293.x. [DOI] [PubMed] [Google Scholar]
- 15.Demetriou VL, Kyriakou E, Kostrikis LG. Near-full genome characterisation of two natural intergenotypic 2k/1b recombinant hepatitis C virus isolates. Advances in virology. 2011;2011:710438. doi: 10.1155/2011/710438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramiere C, Tremeaux P, Caporossi A, Trabaud M, Lebosse F, Bailly F, et al. Recent evidence of underestimated circulation of hepatitis C virus intergenotypic recombinant strain RF2k/1b in the Rhone-Alpes region, France, January to August 2014: implications for antiviral treatment. Euro Surveill. 2014;19(43) doi: 10.2807/1560-7917.es2014.19.43.20944. [DOI] [PubMed] [Google Scholar]
- 17.Morel V, Descamps V, Francois C, Fournier C, Brochot E, Capron D, et al. Emergence of a genomic variant of the recombinant 2k/1b strain during a mixed Hepatitis C infection: a case report. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2010;47(4):382–386. doi: 10.1016/j.jcv.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Sharvadze L, Nelson KE, Imnadze P, Karchava M, Tsertsvadze T. Prevalence of HCV and genotypes distribution in general population of Georgia. Georgian medical news. 2008;(165):71–77. [PubMed] [Google Scholar]
- 19.Kurbanov F, Tanaka Y, Chub E, Maruyama I, Azlarova A, Kamitsukasa H, et al. Molecular epidemiology and interferon susceptibility of the natural recombinant hepatitis C virus strain RF1_2k/1b. The Journal of infectious diseases. 2008;198(10):1448–1456. doi: 10.1086/592757. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. The New England journal of medicine. 1996;334(2):77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 21.El-Shamy A, Nagano-Fujii M, Sasase N, Imoto S, Kim SR, Hotta H. Sequence variation in hepatitis C virus nonstructural protein 5A predicts clinical outcome of pegylated interferon/ribavirin combination therapy. Hepatology. 2008;48(1):38–47. doi: 10.1002/hep.22339. [DOI] [PubMed] [Google Scholar]
- 22.Raghwani J, Thomas XV, Koekkoek SM, Schinkel J, Molenkamp R, van de Laar TJ, et al. Origin and evolution of the unique hepatitis C virus circulating recombinant form 2k/1b. Journal of virology. 2012;86(4):2212–2220. doi: 10.1128/JVI.06184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman RM, Kuntzen T, Weiner B, Berical A, Charlebois P, Kuiken C, et al. Whole genome pyrosequencing of rare hepatitis C virus genotypes enhances subtype classification and identification of naturally occurring drug resistance variants. The Journal of infectious diseases. 2013;208(1):17–31. doi: 10.1093/infdis/jis679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 98/98/NT. Nucleic Acids Symposium Series. 1999;31:95–98. [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viazov S, Ross SS, Kyuregyan KK, Timm J, Neumann-Haefelin C, Isaeva OV, et al. Hepatitis C virus recombinants are rare even among intravenous drug users. Journal of medical virology. 2010;82(2):232–238. doi: 10.1002/jmv.21631. [DOI] [PubMed] [Google Scholar]
- 27.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 28.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. The New England journal of medicine. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 29.Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Annals of internal medicine. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 30.Hedskog C, Doehle B, Chodavarapu K, Gontcharova V, Crespo Garcia J, De Knegt R, et al. Characterization of hepatitis C virus inter-genotypic recombinant strains and associated virologic response to sofosbuvir/ribavirin. Hepatology. 2014 doi: 10.1002/hep.27361. [DOI] [PubMed] [Google Scholar]
- 31.Verbeeck J, Stanley MJ, Shieh J, Celis L, Huyck E, Wollants E, et al. Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0. J Clin Microbiol. 2008;46(6):1901–1906. doi: 10.1128/JCM.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouchardeau F, Cantaloube JF, Chevaliez S, Portal C, Razer A, Lefrere JJ, et al. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol. 2007;45(4):1140–1145. doi: 10.1128/JCM.01982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morel V, Fournier C, Francois C, Brochot E, Helle F, Duverlie G, et al. Genetic recombination of the hepatitis C virus: clinical implications. J Viral Hepat. 2011;18(2):77–83. doi: 10.1111/j.1365-2893.2010.01367.x. [DOI] [PubMed] [Google Scholar]