Abstract
Protein arrays are typically made by random absorption of proteins to the array surface, potentially limiting the amount of properly oriented and functional molecules. We report the development of a DNA encoded antibody microarray utilizing site-specific antibody–oligonucleotide conjugates that can be used for cell immobilization as well as the detection of genes and proteins. This technology allows for the facile generation of antibody microarrays while circumventing many of the drawbacks of conventionally produced antibody arrays. We demonstrate that this method can be used to capture and detect SK-BR-3 cells (Her2+ breast cancer cells) at concentrations as low as 102 cells/mL (which is equivalent to 10 cells per 100 µL array) without the use of microfluidics, which is 100- to 105-fold more sensitive than comparable techniques. Additionally, the method was shown to be able to detect cells in a complex mixture, effectively immobilizing and specifically detecting Her2+ cells at a concentration of 102 SK-BR-3 cells/mL in 4 × 106 white blood cells/mL. Patients with a variety of cancers can have circulating tumor cell counts of between 1 and 103 cells/mL in whole blood, well within the range of this technology.
Graphical abstract
INTRODUCTION
Protein microarrays have been used in many biomedical applications including the detection of proteins in serum, the analysis of protein–protein interactions, and the study of posttranslational modifications.1 Specifically, antibody-based proteomics can identify and validate cancer biomarkers as well as provide a diagnostic approach for identification of different tumor types.2,3 Recently, antibody microarrays have been used to identify metastatic breast cancer as well as distinguish patients with pancreatic cancer from healthy controls.4,5 Additionally, because antibody microarrays also have the ability to capture cells, they allow the possibility of detecting rare cells such as circulating tumor cells (CTCs).6,7
Although there are numerous applications for antibody arrays, construction of these protein arrays is a significantly greater challenge compared with conventional DNA microarrays. The generation of antibody microarrays requires immobilization of the antibody on either hydrophobic or chemically reactive (e.g., epoxy, aldehyde, maleimide) surfaces.8–10 However, this approach can cause denaturation and loss of activity due to immobilization of the protein in a nonproductive orientation or nonspecific binding of the protein to the surface. Approaches to preserve the protein conformation include three-dimensional matrixes such as hydrogels and polyacrylamide, and light-directed biotin–avidin arrays.11,12 Alternatively, one can immobilize antibodies on a DNA array by first modifying the protein of interest with a single-stranded oligonucleotide.13,14 In general, this approach prevents protein denaturation and loss of binding activity associated with printing antibodies on a solid support and potentially allows for greater control of the orientation of the surface bound antibodies.13–17 Not only do these arrays allow for facile and rapid generation of antibody arrays, they have also been shown to have superior binding characteristics when compared to standard antibody arrays. Utilizing DNA directed antibody immobilization on a DNA microarray also allows for concomitant detection of multiple biomolecules, biomarkers, genes or cell types on a single platform.
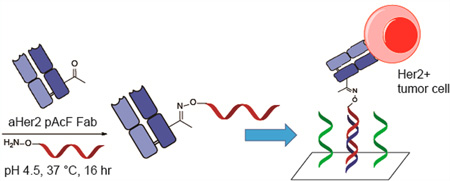
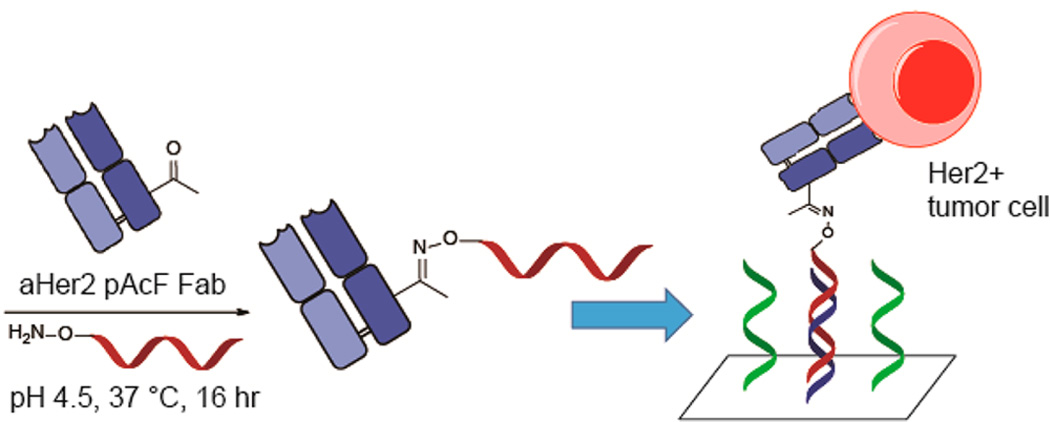
The most common method for conjugating DNA to antibodies is by modification of surface exposed lysine residues. However, coupling to the lysine residues results in a heterogeneous mixture of products which can interrupt antigen binding and cause the antibodies to aggregate.18–20 Random conjugation also prevents control of antibody orientation on the surface, which can lead to loss of activity and specificity. Peluso et al. reported up to a 10-fold increase in analyte binding capacity between a specifically oriented and a randomly oriented antibody using streptavidin-coated surfaces.11 In the context of immuno-PCR, there was a significant difference in signal when comparing site-specific and random DNA conjugation.21 Additionally, site-specific DNA–Fab conjugation has recently been used to develop an extremely sensitive homogeneous immunoassay, detecting PSA at concentrations of 0.27 ng/mL.22 The availability of genetically encoded unnatural amino acids with unique chemical reactivity can provide a solution to these challenges. Previously, we have site-specifically incorporated p-acetylphenylalanine (pAcF) and p-azidophenylalanine (pAzF) into proteins in response to amber nonsense codons in bacteria and mammalian cells.23–27 These amino acids have orthogonal chemical reactivity to the canonical 20 amino acids and can be conjugated efficiently and selectively to alkoxyamine- or alkyne-derived reagents under mild conditions. Herein, we report the development of a DNA-directed antibody microarray technology utilizing site-specific antibody–oligonucleotide conjugates (Figure 1). This versatile platform can be used for cancer cell immobilization, as well as the detection of genes and proteins.
Figure 1.
Synthesis of protein array using an anti-Her2 Fab with site-specific incorporation of pAcF. An excess of synthesized aminooxy-DNA compound (Scheme S1) is added to anti-Her2 pAcF Fab and incubated at 37 °C for 16 h. After purification, the anti-Her2-oligonucleotide conjugate is added to the array and annealed to its complementary strand. Her2 positive tumor cells can then be captured and analyzed.
RESULTS AND DISCUSSION
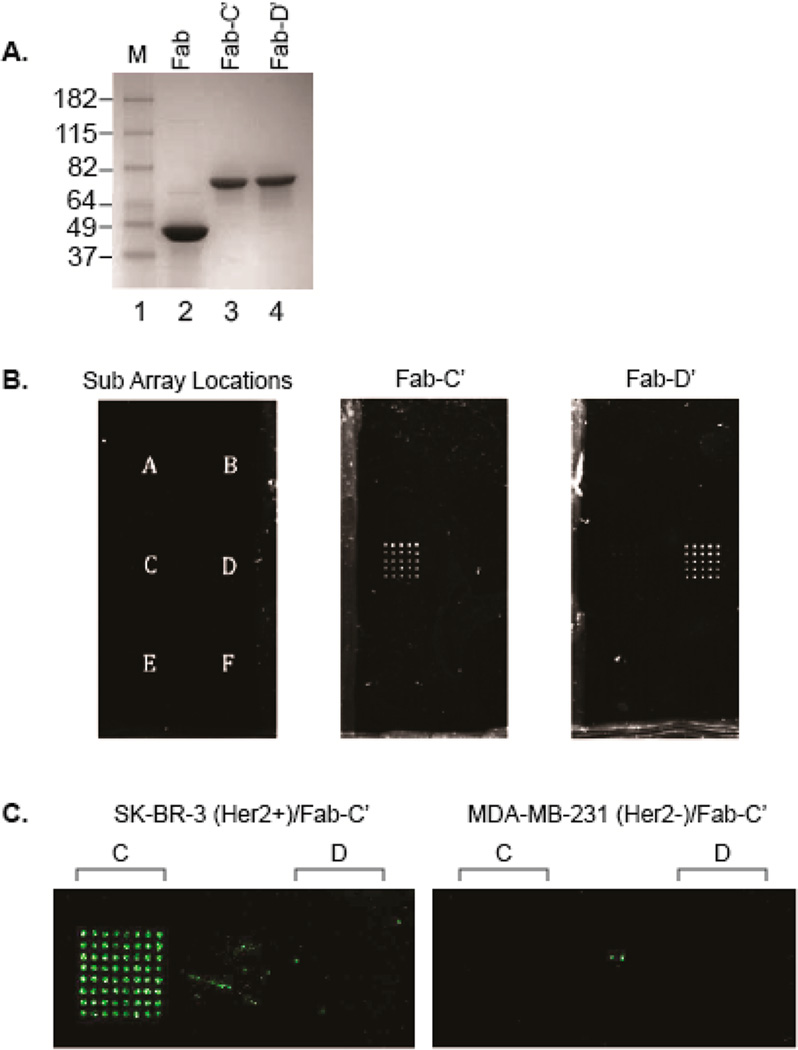
As an initial proof-of-concept, we used this platform for the detection of Her2+ cancer cells. To produce a model DNA-directed antibody microarray, we used trastuzumab (Herceptin), an approved monoclonal antibody for the treatment of Her2 overexpressing breast cancers.28 An anti-Her2 pAcF Fab was expressed using an orthogonal amber suppressor aminoacyl-tRNA synthetase/tRNA pair specific for pAcF. The previously reported pBAD/pEVOL expression system was used to substitute pAcF at surface-exposed site S202 in the light chain. This site was chosen due to its previously reported high expression yields and conjugation efficiency. In addition, modification at this site does not affect antigen binding affinity or specificity.29 Anti-Her2 S202pAcF mutant Fab was expressed in E. coli in good yields (>2 mg/L shake flasks, >400 mg/L fermentation), purified by Protein G, and characterized by SDS-PAGE gel and electrospray-ionization mass spectrometry (ESI-MS) (Expected 47 860 Da; Observed 47 861 Da). As depicted in Figure 2A, lane 2, only one band is observed after Protein G purification, indicating >95% purity. The antibody–oligonucleotide conjugates were then produced via the protocol outlined in Kazane et al., utilizing an aminoxy-functionalized single-stranded oligonucleotide to achieve bio-orthogonal condensation with the ketone moiety and form a stable oxime linkage.21 Anti-Her2 S202pAcF Fab was conjugated to aminooxy-modified oligonucleotide sequences (C′ and D′, Table S1) (100 µM Fab, 3 mM oligonucleotide, 100 mM methoxy aniline, pH 4.5, 37 °C, 16 h), purified by anion exchange chromatography (Mono Q 5/50 GL), and analyzed by SDS-PAGE (Figure 2A and S1). As depicted in Figure 2A, there is only one band (lanes 3 and 4) which correspond to the homogeneous site-specific Fab-oligonucleotide conjugate.
Figure 2.
(A) SDS-page gel analysis of anti-Her2 S202pAcF Fab after conjugation to oligonucleotide-C′ and oligonucleotide-D′. Conjugates were purified by anion exchange chromatography prior to gel analysis. Lanes 3 and 4 correspond to oligonucleotide site-specifically coupled to anti-Her2 pAcF mutant Fab. (B) Antibody localization on the DNA array. The 5 × 5 subarrays each consist of a single-stranded oligonucleotide sequences A–F (left). Anti-Her2 S202pAcF-C′ (middle) or anti-Her2 S202pAcF-D′ (right) (2 mM) was incubated on the slide for 15 min followed by addition of anti-κ-R-PE (1:1000 dilution). The slide was then imaged with 488 nm excitation. White dots indicate localization of the anti-Her2–oligonucletoide conjugate. (C) Cell immobilization. SK-BR-3 (Her2+) or MDA-MB-231 (Her2−) were stained with CellTracker Orange and added to the slides preannealed with anti-Her2 S202pAcF-oligonucleotide-C′. An evanescence field reader was used to image the slides at a 555 nm excitation. Green dots indicate cells immobilized on the DNA array.
Immobilization experiments with anti-Her2–oligonucleotide conjugates were performed on slides containing six 5 × 5 sub arrays (Figure 2B, left) with each sub array comprising different DNA sequences (DNA sequences A–F, Table S1). Thus, the subarray C would be expected to hybridize to its complementary oligonucleotide C′, for example. These arrays were printed onto NHS-ester coated glass slides, which were blocked for 1 h with 50 mM ethanolamine in 0.01% PBST. The antibody conjugate was added onto the DNA arrays via the direct addition of 100 µL of 2 µM (100 µg/mL) anti-Her2 Fab–oligonucleotide conjugate and hybridized for 15 min at room temperature (rt). SYBR green (which has a much higher sensitivity for dsDNA than for ssDNA) was used to verify that the DNA on the array had hybridized to its complement and therefore bound the Fab–oligonucleotide conjugate (data not shown). Additionally, anti-κ R-Phycoerythrin (R-PE) was used to confirm the presence of the antibody at the desired positions, and to show this method’s ability to localize and image labeled proteins. As shown in Figure 2B, anti-Her2 S202pAcF-C′ (middle) and anti-Her2 S202pAcF-D′ (right) both hybridized to the correct subarray with minimal nonspecific binding to the other subarrays (Figure S2).
Next, we analyzed the binding and capture of Her2 positive tumor cell lines. For the cell immobilization experiments, an evanescence field (EF) reader was utilized to image wet slides containing the immobilized live cells. Experiments were performed on slides containing 8 × 8 subarrays of DNA sequences C and D. The initial conditions used were as follows: hybridization of 10 µg (100 µL of a 100 µg/mL (2 µM) solution) of the antibody conjugate (15 min rt), followed by the addition of 106 cells/mL of SK-BR-3 (Her2+) or MDA-MB-231 (Her2−) cells (30 min rt). The cells were stained with CellTracker Orange (Life Technologies) so that live cells could be imaged using a 555 nm excitation. Her2 positive cells (SK-BR-3) bound to the protein array, with >1000-fold difference in signal compared to Her2 negative cells (MDA-MB-231) (Figure 2C and Figure S3). To ensure that the cells were selectively bound to the array, we performed the experiment in the absence of the Fab–oligonucleotide conjugate: no binding was observed for either of the cell lines (Figure S4). For further verification that the duplex DNA formed correctly when the conjugates were hybridized to the array, restriction sites were engineered into the DNA sequences. Following the previously described protocol for cell localization, the slide was imaged before and after treatment with the requisite restriction enzyme for 2 h at 37 °C. The anti-Her2–oligonucleotide/cell complexes were cleaved from the arrays treated with the restriction enzyme as indicated by the loss of signal at the 555 nm excitation wavelength. The arrays in the absence of the restriction enzyme retained the cells (Figure S5).
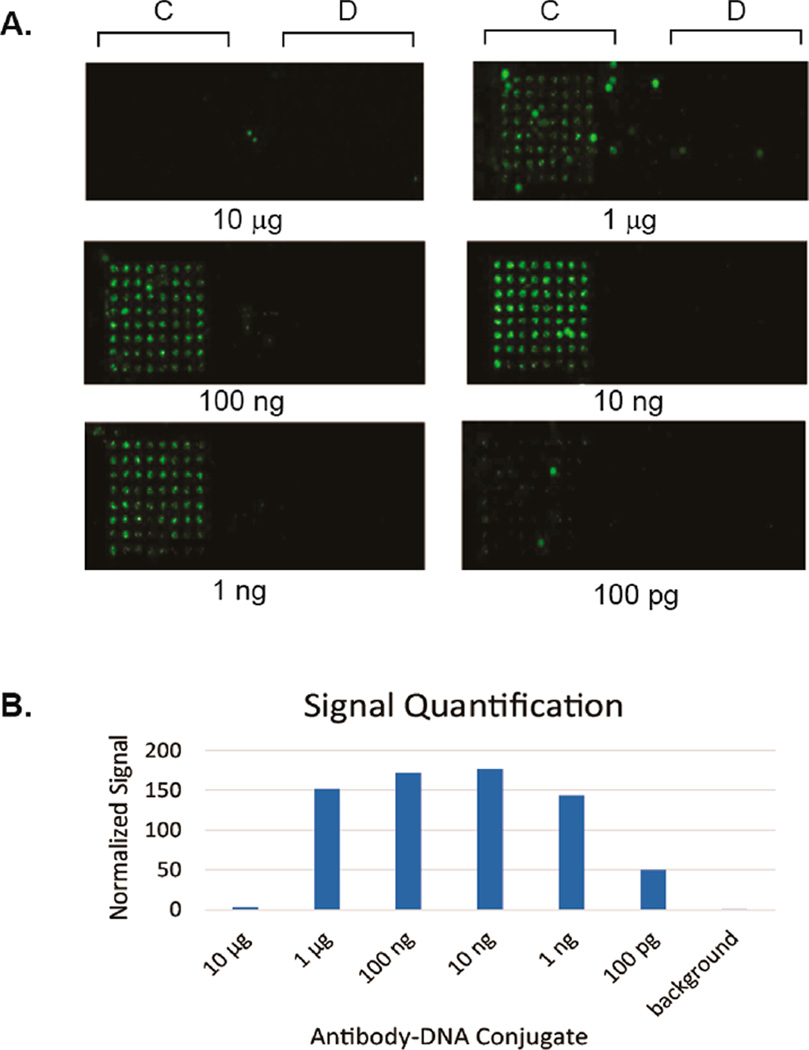
We next tested the sensitivity of this method via a titration of the concentration of cells. Anti-Her2 S202pAcF-C′ was hybridized to the array and different concentrations of SK-BR-3 cells (2 × 106 to 1 × 104 cells/mL) were added to each well of the slide (100 µL/array) and an evanescence field reader was used to image the slides at 555 nm excitation. Signal can be easily detected at the lowest concentration (~1000 cells/array), whereas comparable methods (Herceptin localized via printed anti-human Fc antibody, and Herceptin directly printed on the slide) had significantly worse detection limits (Figure S6). For further confirmation, we imaged the cells with both an evanescence field reader and bright field microscopy to confirm the presence of cells on the array (Figure S7). We then investigated if precomplexing the antibody conjugate with the cells would impact the array binding. A series of dilutions of the antibody conjugate were prepared and combined with aliquots of SK-BR-3 cells (106 cells/mL) before being added to the DNA array (Figure 3). Using the previously described conditions (10 µg Fab-oligonucleotide and 106 cells/mL), almost no binding was observed. This is most likely due to excess antibody–oligonucleotide conjugate hybridizing to the array and competing with the cell/antibody complex. However, we found that precomplexing the antibody conjugate with the cells allows for the use of orders of magnitude less antibody, with a visible signal all the way down to 20 pM (100 pg/well) of antibody conjugate (with a maximum signal at 2 nM (10 ng/well) of conjugate).
Figure 3.
(A) Titration of anti-Her2 Fab–oligonucleotide conjugates. Different concentrations of anti-Her2 S202pAcF-C′ was added to SK-BR-3 cells (106 cell/mL, stained with CellTracker Orange). The mixture was incubated on the array for 30 min before washing. An evanescence field reader was used to image the slides at a 555 nm excitation. Green dots indicate cells immobilized on the DNA array. (B) Quantification was performed using Image Studio Lite software. The values shown are the total signal (sum of the red, green, and blue channel signal values) normalized to background signal.
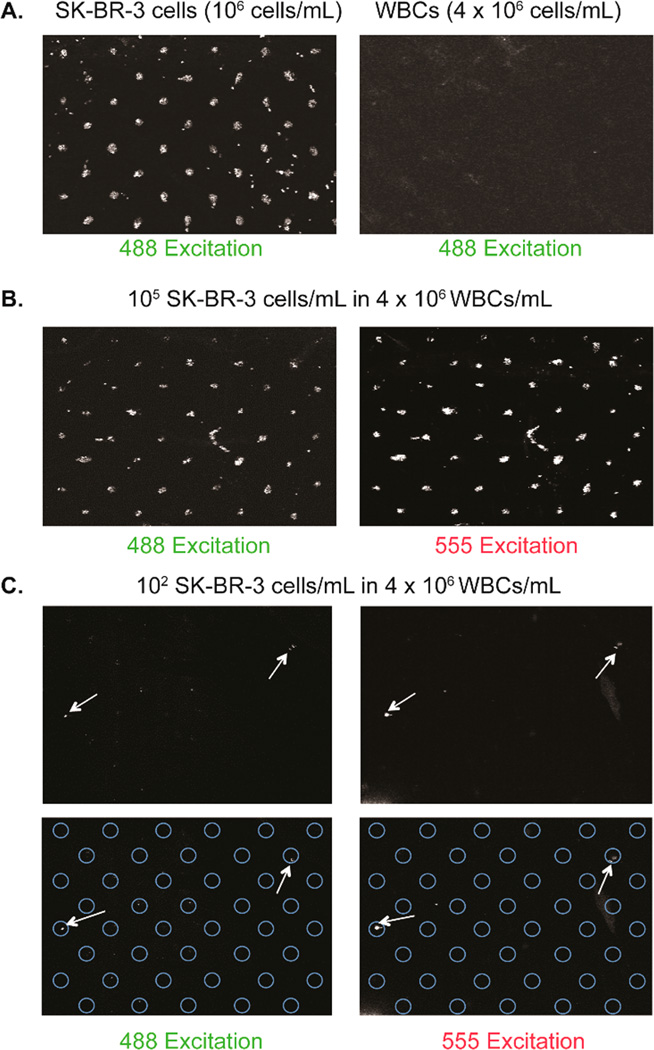
In order to use this application as a diagnostic tool, the DNA-encoded antibody microarray must be able to bind specific cells in a complex mixture. In this regard, we performed a mixing experiment with white blood cells and tumor cells. Red blood cells (RBCs) from a healthy donor were lysed (150 mM NH4Cl, 10 mM KHCO3, 130 nM EDTA), pelleted, and the RBCs in the supernatant were aspirated. The remaining white blood cells (WBCs) were resuspended in PBS (pH 7.4). Varying concentrations of SK-BR-3 cells (stained with CellTracker Orange) were spiked into 4 × 106 WBCs/mL. One microgram of anti-Her2 S202pAcF-C′ conjugate was hybridized to the slide and the spiked sample was added followed by the addition of an anti-Her2-AlexaFluor 488 conjugate (made through site-specific conjugation with anti-Her2 S202pAcF Fab and aminooxy-Alexa Fluor 488 from Life Technologies). The inclusion of the additional anti-Her2–Alexa Fluor 488 conjugate allows for imaging on both the 555 and 488 nm excitation channels and enables confirmation that the SK-BR-3 cells are bound to the array. As depicted in Figure 4, 105 SK-BR-3 cells/mL spiked into 4 × 106 WBCs/mL bound to anti-Her2 S202pAcF-C′ with excellent selectivity (compared to the negative control of only 4 × 106 WBCs/mL). Additionally, cells were detected down to a concentration of 102 SK-BR-3 cells/mL in 4 × 106 WBCs/mL, which is equivalent to ~10 SK-BR-3 cells per array (100 µL of 102 SKB3 cells/mL in 4 × 106 WBCs/mL) (Figures 4C and S8).
Figure 4.
Cell spiking experiments. (A) Standard cell immobilization experiment with the addition of anti-Her2-Alexa Fluor 488 (left). Negative control using only the 4 × 106 WBCs/mL and anti-Her2-Alexa Fluor 488 (right). (B) 1 mg of anti-Her2 S202pAcF-C′ conjugate was hybridized to the slide and 105 SK-BR-3 cells/mL in 4 × 106 WBCs/mL were added. Anti-Her2–Alexa Fluor 488 was added for detection in the 488 channel. Cells were stained with CellTracker Orange for detection in the 555 channel. White dots indicate cells immobilized on the array. (C) 102 SK-BR-3 cells/mL in 4 × 106 WBCs/mL (10 cells/array). White arrows indicate cells visible in both channels.
The number of CTCs per unit volume of blood can vary widely depending on the type and stage of cancer. Screens of patients with a variety of cancers have shown CTC counts between 1 and 103 cells/mL of whole blood.7,30,31 Assays utilizing either conventionally produced antibody microarrays or DNA encoded antibody microarrays generated using randomly labeled antibodies have shown cell detection as low as 103 cells/mL; however, usually require cell concentrations on the order of 106–107 cells/mL.13,32–36 The detection of 100 cells/mL (10 cells/well) in 4 × 106 WBCs/mL is within the reported range for CTC counts, though further experiments using clinical samples (and detection of multiple biomarkers) will be necessary to confirm the utility of this assay as a method for the detection of CTCs. Additionally, because these arrays are printed on coated microscope slides, any immobilized cells could undergo immunohistochemistry (IHC) or immunofluorescence (IF) for detection and analysis of other CTC markers.
CONCLUSION
In summary, by utilizing the site-specific incorporation of the pAcF unnatural amino acid into a Fab, we developed a DNA directed antibody microarray system that allows for the facile generation of antibody microarrays while circumventing many of the drawbacks of conventionally produced antibody arrays (e.g., denaturation of the antibody during production, and poor shelf life).13,14 Additionally, because this microarray is generated on a DNA array platform, it allows for concomitant analysis of genes, proteins, and/or cell types. This microarray has been shown to immobilize cells at cell concentrations significantly lower than that of comparable methods, making it potentially useful as a method for detecting circulating tumor cells.
Supplementary Material
Acknowledgments
We would like to thank the DNA core facility at the Scripps Research Institute for use of their equipment and expertise, and Omar Bazirgan at Sevion Therapeutics for recombinant trastuzumab. This work was supported by grants 1RCIE-BO10745 (NIH) and RSG-09-1601 (American Cancer Society) to V.S.
Footnotes
ASSOCIATED CONTENT
Experimental procedures, oligonucleotide sequences, characterization data, and supplementary array data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev. Proteomics. 2011;8:61–79. doi: 10.1586/epr.10.99. [DOI] [PubMed] [Google Scholar]
- 2.Brennan DJ, O’Connor DP, Rexhepaj E, Ponten F, Gallagher WM. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer. 2010;10:605–617. doi: 10.1038/nrc2902. [DOI] [PubMed] [Google Scholar]
- 3.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingvarsson J, Wingren C, Carlsson A, Ellmark P, Wahren B, Engstrom G, Harmenberg U, Krogh M, Peterson C, Borrebaeck CA. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8:2211–2219. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- 5.Carlsson A, Wingren C, Ingvarsson J, Ellmark P, Baldertorp B, Ferno M, Olsson H, Borrebaeck CA. Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. Eur. J. Cancer. 2008;44:472–480. doi: 10.1016/j.ejca.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Chen GY, Yao SQ. Recent advances in microarray technologies for proteomics. Chem. Biol. 2013;20:685–699. doi: 10.1016/j.chembiol.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Thirumalapura NR, Ramachandran A, Morton RJ, Malayer JR. Bacterial cell microarrays for the detection and characterization of antibodies against surface antigens. J. Immunol. Methods. 2006;309:48–54. doi: 10.1016/j.jim.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 10.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 11.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 12.Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A. Protein microchips: use for immunoassay and enzymatic reactions. Anal. Biochem. 2000;278:123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- 13.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins. J. Am. Chem. Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn AL, Gomez J, Bailey RC. DNA-encoding to improve performance and allow parallel evaluation of the binding characteristics of multiple antibodies in a surface-bound immunoassay format. Anal. Chem. 2011;83:3572–3580. doi: 10.1021/ac200317z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boozer C, Ladd J, Chen S, Jiang S. DNA-directed protein immobilization for simultaneous detection of multiple analytes by surface plasmon resonance biosensor. Anal. Chem. 2006;78:1515–1519. doi: 10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- 16.Boozer C, Ladd J, Chen S, Yu Q, Homola J, Jiang S. DNA directed protein immobilization on mixed ssDNA/oligo(ethylene glycol) self-assembled monolayers for sensitive biosensors. Anal. Chem. 2004;76:6967–6972. doi: 10.1021/ac048908l. [DOI] [PubMed] [Google Scholar]
- 17.Ladd J, Boozer C, Yu Q, Chen S, Homola J, Jiang S. DNA-directed protein immobilization on mixed self-assembled monolayers via a streptavidin bridge. Langmuir. 2004;20:8090–8095. doi: 10.1021/la049867r. [DOI] [PubMed] [Google Scholar]
- 18.Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, Kissler KM, Bernhardt SX, Kopcha AK, Zabinski RF, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin. Cancer Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 19.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Amphlett G, Blattler WA, Lambert JM, Zhang W. Structural characterization of the maytansinoid-monoclonal antibody immunoconjugate, huN901-DM1, by mass spectrometry. Protein Sci. 2005;14:2436–2446. doi: 10.1110/ps.051478705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazane SA, Sok D, Cho EH, Uson ML, Kuhn P, Schultz PG, Smider VV. Site-specific DNA-antibody conjugates for specific and sensitive immuno-PCR. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3731–3736. doi: 10.1073/pnas.1120682109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pakkila H, Peltomaa R, Lamminmaki U, Soukka T. Precise construction of oligonucleotide-Fab fragment conjugate for homogeneous immunoassay using HaloTag technology. Anal. Biochem. 2015;472:37–44. doi: 10.1016/j.ab.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Alfonta L, Tian F, Bursulaya B, Uryu S, King DS, Schultz PG. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudis CA. Trastuzumab–mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 29.Hutchins BM, Kazane SA, Staflin K, Forsyth JS, Felding-Habermann B, Schultz PG, Smider VV. Site-specific coupling and sterically controlled formation of multimeric antibody fab fragments with unnatural amino acids. J. Mol. Biol. 2011;406:595–603. doi: 10.1016/j.jmb.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belov L, de la Vega O, dos Remedios CG, Mulligan SP, Christopherson RI. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res. 2001;61:4483–4489. [PubMed] [Google Scholar]
- 33.Ellmark P, Belov L, Huang P, Lee CS, Solomon MJ, Morgan DK, Christopherson RI. Multiplex detection of surface molecules on colorectal cancers. Proteomics. 2006;6:1791–1802. doi: 10.1002/pmic.200500468. [DOI] [PubMed] [Google Scholar]
- 34.Haab BB. Applications of antibody array platforms. Curr. Opin. Biotechnol. 2006;17:415–421. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Peelen D, Kodoyianni V, Lee J, Zheng T, Shortreed MR, Smith LM. Specific capture of mammalian cells by cell surface receptor binding to ligand immobilized on gold thin films. J. Proteome Res. 2006;5:1580–1585. doi: 10.1021/pr050467e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang CX, Liu HP, Tang ZM, He NY, Lu ZH. Cell detection based on protein array using modified glass slides. Electrophoresis. 2003;24:3279–3283. doi: 10.1002/elps.200305424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.