A subgroup of patients infected with nontyphoidal Salmonella develops a persistent infection. Most of these are presented as symptomatic relapse gastroenteritis, illuminating a previously overlooked manifestation. During persistence, Salmonella gains genetic and phenotypic changes, affecting antibiotic resistance, and host-pathogen interactions.
Keywords: Salmonella enterica, salmonellosis, persistent infection, WGS, epidemiology
Abstract
Background. Although chronic infections by typhoidal Salmonella are well-known, prolonged human infections by nontyphoidal Salmonella (NTS) are poorly characterized.
Methods. We retrospectively analyzed 48 345 culture-confirmed NTS infections that occurred in Israel 1995–2012. A case-control study was performed to identify risk factors associated with persistent infections. Whole-genome-sequencing, pulsed-field gel electrophoresis (PFGE), and a mouse infection model were used to study genetic and phenotypic differences between same-patient persistent, recurring isolates.
Results. In total, 1047 cases of persistent NTS infections, comprising 2.2% of all reported cases of salmonellosis, were identified. The persistence periods ranged between 30 days to 8.3 years. The majority (93%) of the persistently infected patients were immunocompetent, and 65% were symptomatic with relapsing diarrhea, indicating a distinct clinical manifestation from the asymptomatic carriage of typhoidal Salmonella. Four NTS serovars (Mbandaka, Bredeney, Infantis and Virchow) were found to be significantly more frequently associated with persistence than others. Comparative genomics between early and later isolates obtained from the same patients confirmed clonal infection and showed 0 to 10 SNPs between persistent isolates. A different composition of mobile genetic elements (plasmids and phages) or amino acid substitutions in global regulators was identified in multiple cases. These changes resulted in differences in phenotype and virulence between early and later same-patient isolates.
Conclusions. These results illuminate the overlooked clinical manifestation of persistent salmonellosis that can serve as a human reservoir for NTS infections. Additionally, we demonstrate mechanisms of in-host microevolution and exhibit their potential to shape Salmonella pathogenicity, antimicrobial resistance and host-pathogen interactions.
Salmonella enterica is a Gram-negative, facultatively intracellular human and animal pathogen posing a major public health concern worldwide [1]. The species S. enterica includes more than 2600 serovars, which are taxonomically classified into 6 subspecies, sharing high sequence similarity [2, 3]. Subspecies I serovars are divided into 2 clinically relevant groups according to the disease they cause. Infections with the human-restricted S. Typhi, S. Paratyphi A and B, and S. Sendai elicit an invasive, life-threatening systemic disease referred to as typhoid or enteric fever [4]. On the other hand, nontyphoidal serovars (NTS) normally cause self-limited gastroenteritis, associated with intestinal inflammation and diarrhea that lasts for 5–7 days, in immunocompetent individuals [5].
Salmonella are excreted in the feces of infected animals and people during the course of the disease but may also endure during convalescence. In sum, 1%–4% of individuals infected with S. Typhi become asymptomatic chronic carriers that continue to excrete the pathogen for more than 12 months [4]. Long-term carriage of S. Paratyphi is less characterized than S. Typhi, but a recent study in Nepal suggests a similar prevalence of persistence for serovars Typhi and Paratyphi A in endemic regions [6].
In contrast to the well-documented cases of prolonged infections by S. Typhi, persistent NTS infections are far less studied and the prevalence of long-term NTS carriers in the population is not known. Here, we integrated epidemiological, microbiological and genomics approaches to demonstrate that at least 2.2% of all confirmed NTS infections in Israel result in prolonged infection. Moreover, we show genetic and phenotypic changes between related isolates obtained from the same patients over the course of the infection. We further show that these differences can alter virulence-associated phenotypes in vitro and Salmonella pathogenicity in vivo, using the colitis mouse model.
MATERIALS AND METHODS
Study Population
Salmonellosis is a notifiable disease in Israel by law. All microbiology laboratories countrywide are required to submit Salmonella isolates from all sources to the national Salmonella reference center (NSRC), where serological identification is performed according to the Kauffmann–White-Le-Minor scheme [7]. All the serotyped isolates, their source, sending laboratory, date of isolation and patient identification number (ID) are routinely documented [8]. In this study we analyzed 48 345 cases of NTS infections reported to the NSRC between 1995 and 2012. The minimal duration of infection for each case was calculated, based on the number of days between the first and the last submitted culture, if the same-patient samples were of the same serovar.
A Case-control Study
The case-control study included telephone interviews with 103 NTS persistent patients (cases, from which 2 or more Salmonella samples of the same serovar had been submitted with a minimal interval of 30 days) and 135 randomly chosen nonmatched controls from 2011 to 2012, that submitted a single stool sample (see Supplementary Text). All interviewees answered a standardized questionnaire addressing demographic details, health condition and background diseases, the course of the disease and the treatment they received (including antibiotic treatment before or during the infection), and whether other pathogens were isolated as well.
Statistical Analysis
Statistical analyses were performed with R (v 3.0.3). Multinomial logistic regression was performed for the multivariable analysis (see Supplementary Text).
Pulsed-field Gel Electrophoresis (PFGE)
PFGE was performed according to the Pulsenet International Standardized Protocol [9] using XbaI endonuclease.
Whole Genome Sequencing (WGS) Bioinformatics and Phylogeny
Whole genome shotgun assemblies for 11 pairs of persistent S. Typhimurium isolates were generated at Expression Analysis (Durham, NC) using 50 cycles of Illumina's paired end chemistry to a depth of approximately 175-fold coverage and assembled using Velvet [10]. Maximum likelihood tree was constructed using RAxML [11] with 100 replications. See Supplementary Text for bioinformatical workflow. All the sequences were deposited into the NCBI Sequence Read Archive (SRA) under accession numbers: SRS1190306, SRS1190305, SRS1190304, SRS1190303, SRS1190300, SRS1190301, SRS1190299 SRS1190298, SRS1190297, SRS1190296, SRS1190295, SRS1190294, SRS1190293, SRS1190290, SRS1190289, SRS1190284, SRS1190283, SRS1190282, SRS1190281, SRS1190280, SRS1190279, and SRS1190278.
Virulence-associated Phenotypes Comparison
Intra-macrophage survival, motility, biofilm formation, and growth assays were performed as described in the Supplementary Text.
Mouse Infection
Mouse competitive index experiments were conducted using Female C3H/HeNHsd mice infected at an age of 7–8 weeks. Streptomycin (20 mg per mouse) was given by oral gavage 24 hours prior to the infection. Early and later isolates obtained from the same patients harboring pWSK129 (Kmr) and pWSK29 (Ampr), respectively, were grown in Luria broth (LB) with the appropriate antibiotics for 16 hours and diluted in saline. 0.2 mL containing equal numbers (5 × 106–1 × 107 colony-forming units [CFU]) of each isolate was administrated orally. At day 4 post infection, mice were sacrificed, and the bacterial load was enumerated. The competitive index (C.I.) was calculated as [later isolate (Ampr) /early isolate (Kmr)] output / [later isolate /early isolate] input.
Ethics
The study was approved by the Institutional Review Board (IRB) of the Sheba Medical Center, approval numbers 7072-09-SMC and 8142-10-SMC. Informed consents were obtained and documented on each interview sheet (See Supplementary Text). Mouse experiments were conducted in line with national ethical guidelines and approved by the IRB (Approval # 933/14).
RESULTS
The Prevalence of Persistent NTS Infections in Israel, 1995–2012
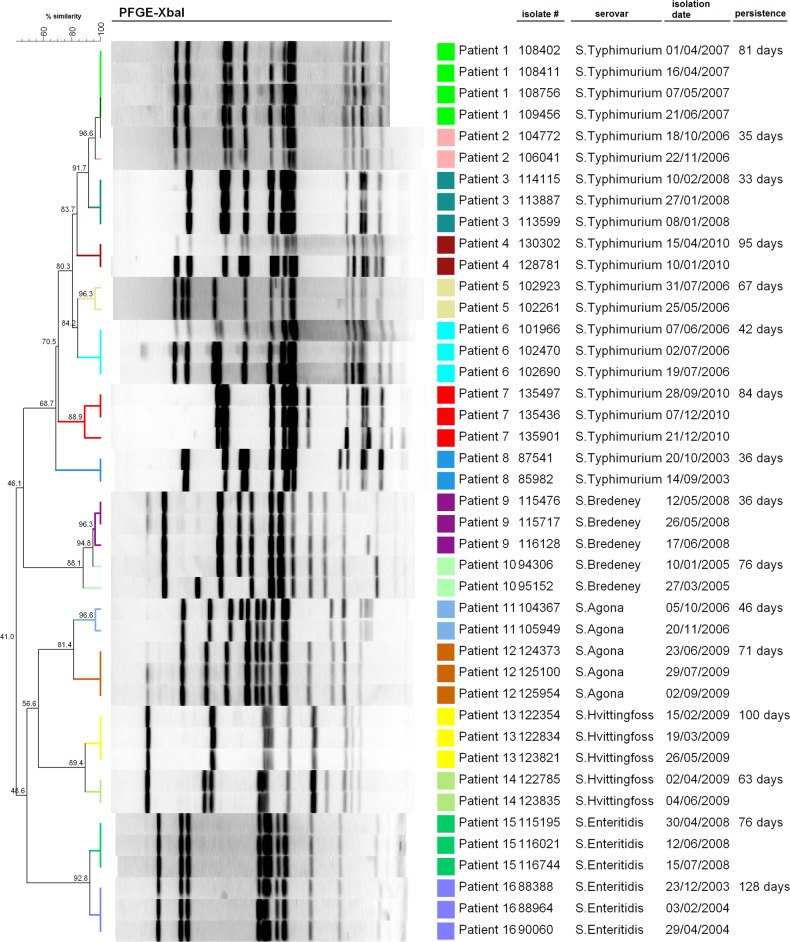
Analysis of 48 345 documented salmonellosis cases revealed 1047 independent cases of persistent infections, based on the submission of two or more isolates of the same serovar, separated by 30 days or more (Table 1). Setting aside the less likely possibility of reinfection with the same strain over an extended period of time (see below), this analysis indicated that at least 2.2% (95% confidence interval [CI], 2.0–2.3) of all reported cases of salmonellosis in Israel are long-term infections that persist 30 days or more. The median minimal persistent period of all cases was 55 (IQR 39–91) days, and the maximal period of NTS persistence found was 8.3 years caused by an S. Newport infection of a 75-year-old male. Because the minimal infection duration was inferred from the time intervals between the isolation of first and the last Salmonella isolates, the actual period of the infection is expected to be longer. In the majority (94.2%) of the persistent cases, persistent Salmonella was isolated only from stool samples, in 2.2% of cases the pathogen was isolated from extra-intestinal sites and in 3.6% of cases Salmonella was found both in stool and extra-intestinal sites during persistence. To explain the nature of recurrent NTS isolates and to exclude reinfection of the same patient with different strains of the same serovar, we analyzed the genetic similarity of 42 recurrent isolates, obtained from 16 different patients. PFGE showed that different isolates originated from the same person are highly similar on the genetic level, but distinct from other same-serovar strains isolated from different patients (Figure 1). As reinfection of the same host with the exact same clone separated by a long period of time is unlikely and potentially diminished even further by immunity, these results together with whole-genome sequencing (see below) support persistent infection by these NTS serovars.
Table 1.
Distribution of Nontyphoidal Salmonella Isolates Associated With 1047 Persistent Infection Cases Between 1995 and 2012 in Israel
| Number of Submitted Isolates | Cases | % of Cases | Median Duration of Infection in Days (Interquartile Range) | Minimum Infection Period in Days | Maximum Infection Period in Days |
|---|---|---|---|---|---|
| 2 | 529 | 50.5 | 49 (37–82) | 30 | 2751 (7.5 y) |
| 3 | 316 | 30.2 | 54.5 (40–86) | 30 | 3037 (8.3 y) |
| 4 | 129 | 12.3 | 70 (46–104) | 30 | 1687 (4.6 y) |
| 5 | 39 | 3.7 | 93 (71–134) | 32 | 417 (1.1 y) |
| 6 | 20 | 1.9 | 100 (67–135) | 47 | 211 |
| 7 | 11 | 1.1 | 234 (188–277) | 152 | 1745 (4.7 y) |
| 9 | 2 | 0.2 | 1112 (667.5–1556) | 223 | 2001 (5.4 y) |
| 10 | 1 | 0.1 | 492 (492–492) | 492 (1.3 y) | 492 (1.3 y) |
Figure 1.
Recurrent NTS isolates obtained from the same patients are genetically related. 42 recurrent NTS isolates from 16 different patients were analyzed by PFGE using XbaI endonuclease. Isolate number, NTS serovar, isolation date and the minimal duration of persistence are indicated on the right. A dendogram showing the percentage of similarity between isolates is shown on the left. Abbreviations: NTS, nontyphoidal Salmonella; PFGE, pulsed-field gel electrophoresis.
Host and Pathogen Factors Associated With Persistent Infections
To identify host factors that may contribute to a persistent infection, a retrospective case-control study, which included 103 NTS persistent patients (cases) and 135 randomly selected controls was performed. The median minimal infection duration of the persistent cases was 66 days (IQR 40.5–111.5) and the median number of submitted isolates was four. This analysis showed that persistent NTS infections were not associated with an immunocompromised status, as only 6.8% of the cases reported poor general health (all cases and controls were human immunodeficiency virus (HIV) negative, Supplementary Table 1). Notably, 62 out of 96 (64.6%) patients that recalled the disease reported prolonged symptomatic illness with relapsing diarrhea (Table 2). To evaluate factors associated with symptomatic and asymptomatic persistent infections in comparison to random nonpersistent controls we performed multinomial logistic regression analysis (Table 2). Symptomatic, but not asymptomatic, persistent infections were significantly associated with younger age at infection (0.96 odds ratio [OR] 95% CI, .93–.99), receiving antibiotic treatment against Salmonella (OR 3.11 95% CI, 1.22–7.97), hospitalization due to salmonellosis (OR 3.07 95% CI, 1.1–8.55), and coinfection with other enteric pathogens (OR 11.37 95% CI, 3.07–42.08). Anemia and receiving probiotics were associated both with symptomatic and asymptomatic persistence as compared to the nonpersistent controls.
Table 2.
Bivariate Analysis of Cases and the Controls Characteristics and a Multinomial Logistic Regression Analysis of Factors Correlating With Asymptomatic and Symptomatic Persistent Infection as Compared to the Nonpersistent Controls
| Bivariate Analysis |
Multinomial Logistic Regressiona,b |
||||||
|---|---|---|---|---|---|---|---|
| Controls | Persistent Infection Cases | P Value | Persistent Asymptomatic vs. Controls OR (95% CI) | P Value | Persistent Symptomatic vs. Controls OR (95% CI) | P Value | |
| Number | 135 | 103 | |||||
| Age at infection, years (median (IQR)) | 2.9 (1.29–20.6) | 0.65 (0.36–1.25) | .002 | 0.98 (.95–1) | .108 | 0.96 (.93–.99) | .003 |
| Female Sex (%) | 62 (45.9) | 47 (45.6) | 1 | 2.05 (.83–5.06) | .121 | 1.19 (.49–2.88) | .697 |
| General poor health (%) | 16 (11.9) | 7 (6.8) | .277 | 0.29 (.03–3.03) | .301 | 1.06 (.19–5.93) | .951 |
| N of household members (median (IQR)) | 4 (3–5) | 4 (3–5) | .204 | 0.83 (.6–1.16) | .274 | 0.87 (.63–1.2) | .408 |
| Reason for submission of multiple samples (%): | |||||||
| Do not remember | … | 7 ( 6.8) | |||||
| Symptomatic | … | 62 (60.2) | |||||
| To ensure clearance | … | 34 (33.0) | |||||
| Antibiotics against Salmonella infection (%)a | 69 (54.8) | 74 (74.0) | .004 | 1.07 (.43–2.69) | .886 | 3.11 (1.22–7.97) | .018 |
| Anemia (%) | 22 (16.3) | 29 (28.2) | .04 | 3.7 (1.3–10.52) | .014 | 4.66 (1.69–12.85) | .003 |
| Probiotics (%) | 7 (5.2) | 24 (23.3) | <.001 | 8.4 (2.1–33.58) | .003 | 18.52 (5.1–67.28) | <.001 |
| Hospitalization/ER visit (%) | 21 (15.6) | 30 (29.1) | .018 | 2.35 (.75–7.32) | .141 | 3.07 (1.1–8.55) | .032 |
| Coinfection with other enteric pathogens (%)a | 8 (6.2) | 18 (17.6) | .011 | 2.26 (.39–13.01) | .360 | 11.37 (3.07–42.08) | <.001 |
| Infection year (median, range) | 2011 (2011–2012) | 2010 (2000–2012) | <.001 | 0.51 (.35–.75) | .001 | 0.48 (.33–.7) | <.001 |
Abbreviations: CI, confidence interval; ER, emergency room; IQR, interquartile range; OR, odds ratio.
a Complete-case analysis.
b Adjusted for: Age at infection, Sex (Female vs Male), Poor self-reported health (yes/no), Number of household members, Antibiotics against Salmonella infection (yes/no), Anemia (yes/no), Probiotics (yes/no), Hospitalization/ER visit (yes/no), Coinfection with other enteric pathogens (yes/no), Infection year. The full model was based on 122 controls, 33 persistent asymptomatic cases and 60 persistent symptomatic cases.
Next, we asked whether some NTS serovars are more frequently associated with human persistent infections than others, by comparing the prevalence of the different serovars among persistent (N = 1047) vs sporadic (N = 47 298) cases. The occurrence of the ubiquitous serovars Enteritidis and Typhimurium was found to be significantly lower among persistent infections than expected by their general prevalence. On the other hand, the frequency of 4 serovars (Mbandaka, Bredeney, Infantis, and Virchow) appeared to be significantly higher among persistent infections than in sporadic cases (Table 3). These results suggest that serovar-specific genetic or ecological factors are expected to contribute to the establishment of persistent infection in humans.
Table 3.
The Leading Nontyphoidal Salmonella Serovars and Their Association With Sporadic and Persistent Infections in Humans in Israel
| Rank | Serotype | Number of Sporadic Cases | % of Sporadic Cases | Number of Persistent Cases | % of Persistent Cases | Persistence Indexa | P Value |
|---|---|---|---|---|---|---|---|
| 1 | Enteritidis | 11069 | 23.4 | 129 | 12.3 | 0.5 | .001 |
| 2 | Virchow | 5887 | 12.4 | 174 | 16.6 | 1.3 | .001 |
| 3 | Typhimurium | 5644 | 11.9 | 89 | 8.5 | 0.7 | .001 |
| 4 | Infantis | 5214 | 11.0 | 156 | 14.9 | 1.4 | .001 |
| 5 | Hadar | 4000 | 8.5 | 84 | 8.0 | 0.9 | Ns |
| 6 | Bredeney | 1254 | 2.7 | 53 | 5.1 | 1.9 | .001 |
| 7 | Montevideo | 1025 | 2.2 | 27 | 2.6 | 1.2 | Ns |
| 8 | Agona | 987 | 2.1 | 18 | 1.7 | 0.8 | Ns |
| 9 | 9,12:l,v:- | 891 | 1.9 | 28 | 2.7 | 1.4 | .082 |
| 10 | Blockley | 777 | 1.6 | 21 | 2.0 | 1.2 | Ns |
| 11 | Heidelberg | 771 | 1.6 | 16 | 1.5 | 0.9 | Ns |
| 12 | Newport | 763 | 1.6 | 22 | 2.1 | 1.3 | Ns |
| 13 | Muenchen | 683 | 1.4 | 23 | 2.2 | 1.5 | .061 |
| 14 | Kentucky | 662 | 1.4 | 16 | 1.5 | 1.1 | Ns |
| 15 | Java | 656 | 1.4 | 13 | 1.2 | 0.9 | Ns |
| 16 | Mbandaka | 469 | 1.0 | 22 | 2.1 | 2.1 | .001 |
| 17 | Anatum | 438 | 0.9 | 9 | 0.9 | 0.9 | Ns |
| 18 | Tennessee | 437 | 0.9 | 14 | 1.3 | 1.4 | Ns |
| Other | 5671 | 12.0 | 133 | 12.7 | 1.1 | ||
| Total | 47298 | 100 | 1047 | 100 |
a Persistence index [% of persistent cases]/ [% of sporadic cases]; P-values are based on a χ2 test; ns, not significant.
Genetic Changes Between Early and Later Same-patient Isolates
Although PFGE of the persistent isolates has demonstrated high genetic similarity between isolates from the same patients, a few cases (patients 2, 5, 7, 9, and 10) showed subtle differences in the PFGE profile between recurrent isolates. To further characterize possible genetic changes in a higher resolution we sequenced the entire genome of 22 S. Typhimurium isolates obtained from 11 persistent infection patients at 2 different time points during the infection. We chose to focus on serovar Typhimurium because of its high global prevalence and the availability of an established mouse model, which allows phenotypic and virulence comparison between isolates. The minimal time interval between isolation dates was 33 days, and hereafter these time points are referred to as “early” and “later.” Phylogenetic analysis showed that in all 11 cases, the isolates from the same person always clustered together in discrete nodes with high support values, that were separate from isolates submitted by other persistent cases, and 164 S. Typhimurium reference strains that were included as controls to reduce random clustering (Supplementary Figure 1). These results reinforced the results obtained using the PFGE and confirmed that these recurring cases are continuous persistent infections caused by clonal lineages.
In 5 out of the 11 persistent cases we found different compositions of mobile genetic elements (MGEs) including prophages and plasmids between same-patient related isolates, explaining the nonidentical PFGE pattern found in a few examples. S. Typhimurium isolates obtained at early and later time points from patients B, D, I, and K presented gain and/or loss of various plasmids. Similarly, the later persistent isolate from patient E lacked a 40 Kb Gifsy-1-like prophage that was present in its early corresponding isolate (Supplementary Table 2).
To further confirm changes in plasmid composition between recurrent isolates of persistent cases we applied an S1 nuclease digest followed by PFGE of the 9 isolates obtained from patients B, D, I, and K. This analysis confirmed the WGS data and showed clear differences in plasmid presence between related isolates from the same patients (Figure 2). Interestingly, all 3 isolates from patient K did not harbor the 95 kb pSLT plasmid of S. Typhimurium, indicating that the pSLT virulence plasmid is not required to maintain a persistent infection in humans. The number of core-genome single nucleotide polymorphisms (SNPs) between early and later isolates obtained from the same patient ranged from 0 to 10 per patient. Overall, 15 synonymous changes, 10 nonsynonymous changes, 1 premature stop-codon, and 2 changes in noncoding regions were identified within pairs of strains (pooled-average of 1 SNP/ 24 days, Supplementary Tables 2 and 3). Despite the high genetic similarity of the core genome, multiple SNPs were found to be nonsynonymous substitutions or nonsense mutations in global virulence regulatory genes including dksA [12, 13] in patient A; rpoS [14] in patient C; hilD [15], melR [16] and a nonsynonymous mutation in rfc [17], in patient D; and barA [18] in patient F. In addition, 10 SNPs were mapped to shdA that was previously linked to prolonged fecal shedding [19], in patient I.
Figure 2.
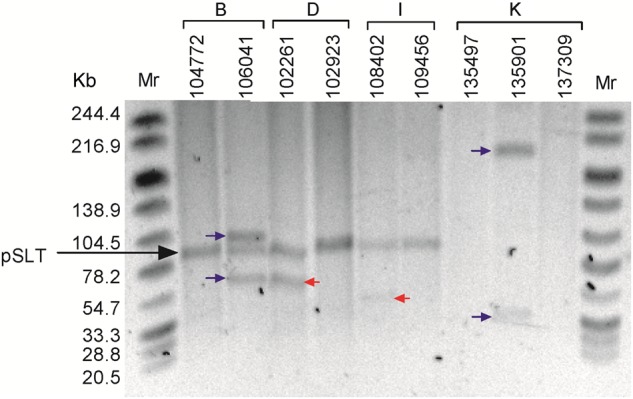
Plasmids gained and lost in related persistent isolates. Nine S. Typhimurium persistent isolates obtained from patients B, D, I, and K were subjected to an S1 nuclease digest followed by PFGE. Plasmids that were found in the later isolates, but not in the early isolates are indicated by a blue arrow. Plasmids that were found in the early, but not the later, isolates are indicated by a red arrow. The prevalent approximately 95 kilobase (Kb) virulence S. Typhimurium plasmid, pSLT, is indicated by a black arrow. Abbreviation: PFGE, pulsed-field gel electrophoresis.
Phenotypic Changes Between Early and Later Same-patient Isolates
Gain or loss of large genetic elements as well as SNPs in key regulatory genes may confer substantial changes affecting host-pathogen interactions. Therefore, we next screened for phenotypic differences, between early and later isolates obtained from 11 patients with persistent S. Typhimurium. Comparison of these virulence-relevant phenotypes included (i) growth in minimal media (Supplementary Figure 2); (ii) biofilm formation (Supplementary Figure 3A); (iii) motility (Supplementary Figure 3B); and (iv) replication inside macrophages (Supplementary Figure 4). These assays showed, in independent cases, significant differences between early and later isolates obtained from the same patients over the course of the infection, suggesting that diversity in virulence-associated phenotypes can be maintained or acquired during persistence. In addition, antibiogram profiling demonstrated that the later isolate from patient K (135901) had a multidrug resistance (MDR) to piperacillin, ceftriaxone, and trimethoprim/sulfamethoxazole. This phenotype was consistent with the acquisition of a large plasmid, conferring extended-spectrum beta-lactamase activity, demonstrating that the resistance profile of persistent Salmonella may change during the course of the infection, in a manner that could affect clinical treatment.
Variation in the Virulence of Persistent Isolates In-vivo
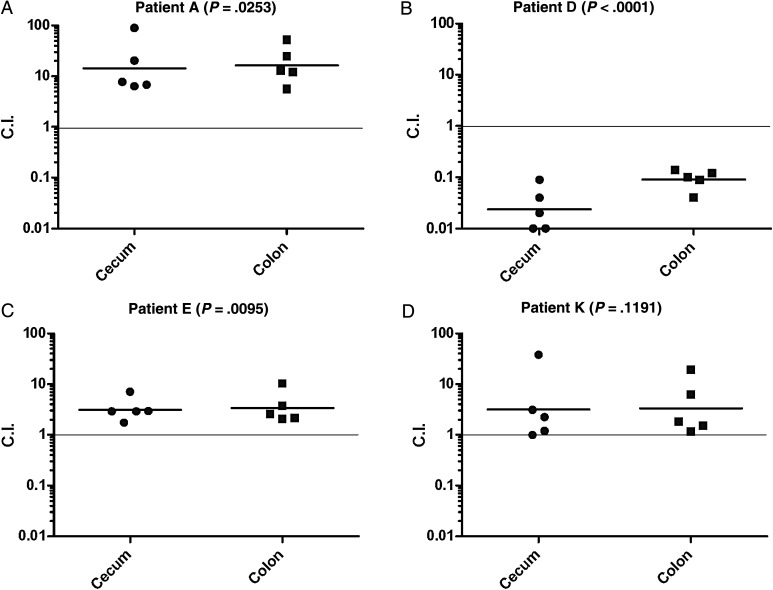
Finally, to explore whether these changes can affect Salmonella pathogenicity in vivo, we chose 4 pairs of isolates from patients A, D, E, and K that showed genetic and phenotypic differences in vitro and performed competitive index infections using the streptomycin pretreated mouse colitis model, which resembles a gastrointestinal disease. In 3/4 cases, these experiments demonstrated significant differences in intestinal infection between the early and the later isolates that were obtained from the same patient (Figure 3). These results show that the demonstrated genetic differences between related persistent NTS isolates can confer a significant and quantifiable change in the fitness and the pathogenicity of these variants in vivo.
Figure 3.
Related persistent isolates differ in their pathogenicity in vivo. Groups of 5 female C3H/HeNHsd mice were treated with streptomycin and 24 hours later were infected orally with a 1:1 mixed inoculum of early and later S. Typhimurium isolates (5 × 106–1 × 107 CFU from each isolate) obtained from patients A (isolate 85982 and 87541; panel A), D (102261 and 102923; panel B), E (128781 and 130302; panel C), and K (135497 and 137309; panel D). Four days post infection mice were sacrificed, and the colonization of the later isolates relative to the early isolate was determined in the cecum and colon as explained in “Materials and Methods” section. Each point shows the competitive index (C.I.) value in a single mouse, whereas the geometrical mean of each group is indicated by the horizontal line. Two-tailed 1-sample t-test against a theoretical mean of 1.00 was used to determine statistical significance and is shown in brackets. Abbreviation: CFU, colony-forming unit.
DISCUSSION
Altogether, this study provides important clinical and public health insights, indicating that a small fraction (at least 2.2%) of NTS-infected patients continue to shed Salmonella for an extended period of time (months and even years). These individuals possibly serve as a human reservoir for NTS transmission. Noteworthy, about 65% of the persistent patients experienced a symptomatic disease with relapsing diarrhea, indicating that NTS persistent infection is a clinically distinct manifestation from the known asymptomatic carriage of typhoidal Salmonella.
A case-control study was implemented to identify host factors that correlate with persistent infections. In line with previous reports [20, 21] our study suggests that antibiotic treatment and young age are associated with prolonged symptomatic infection. We also report that hospitalization due to Salmonella, probiotics administration (in line with a recent double-blind, placebo-controlled clinical trial [22]) and coinfection with other enteric pathogens, independently correlate with symptomatic persistence. Moreover, the case-control study suggests a high rate of antibiotic prescription even among the nonpersistent controls (55%), despite the fact that antibiotic therapy is not recommended for an uncomplicated nontyphoid Salmonella infection [21, 23].
To characterize the intra-host genomic plasticity of Salmonella during persistent infection, we applied whole-genome sequencing of recurrent same-patient isolates. In 5 out of the 11 sequenced pairs of isolates we identified differences in MGEs, including prophages and plasmids, between isolates obtained from the same person. In at least 2 cases, these variable elements may affect virulence: loss of gogB, a type 3 secretion system effector gene [24] that is encoded on Gifsy-1 prophage in patient E and acquisition of an MDR plasmid in patient K. Besides differences in MGEs presence, we observed a low occurrence of SNPs, which ranged between 0 and 10 per patient. These results are consistent with the observed number of SNPs that were found in a few recent studies analyzing sequential isolates obtained from patients who were persistently infected with Burkholderia pseudomallaei [25], E. coli associated with asymptomatic bacteriuria [26], and invasive S. Typhimurium pathovar ST313 in sub-Saharan Africa [27]. Recently, Octavia et al also reported a similarly low SNPs rate in S. Typhimurium associated with short and long-term carriage [28].
Noteworthy, we did not observe a clear directionality of neither genetic nor phenotypic changes between early and later same-patient isolates. Mobile genetic elements were both lost and acquired in the later isolates leading to either improved or reduced phenotypes in comparison with the early isolates. Nevertheless, the possibility that this pathogen can alter during the persistence within the human host is intriguing. Multiple nonsynonymous SNPs were found in key virulence regulators including DksA, RpoS, HilD, MelR, and BarA. Mutations in these regulators have the potential to affect virulence and host-pathogen interactions [29–32]. It is therefore possible that these regulatory mutations increase Salmonella in-host fitness and are being subjected to a positive selection leading to the emergence of specific pathoadaptive variants [33]. Phenotypic assays and infection experiments in the colitis mouse model showed, in independent cases, significant differences in the virulence of related NTS persistent isolates, indicating that such genetic variation can alter host-pathogen interactions and affect Salmonella virulence. We speculate that the observed gain or loss of MGEs and the SNPs in global regulators provide the pathogen with effective mechanisms to generate genetic and phenotypic heterogeneity, facilitating clonal competition and microevolution of pathoadaptive variants within the host during persistence.
Due to the retrospective study design, one cannot determine the temporal relationship between persistent infection and the identified correlating factors, which may be either the cause or the consequence of NTS persistence. Misclassification between cases and controls also cannot be ruled out as a small fraction of the controls might have had a disease for more than 30 days without seeking medical care. However, this possibility is unlikely, considering the low incidence of persistent NTS infection. An additional limitation is the possibility of underrepresentation of nonsymptomatic persistent patients due to their possible reduced motivation toward sample submission, which may lead to an underestimation of the asymptomatic persistent infection prevalence and overrepresentation of the symptomatic persistent patients.
Despite the abovementioned limitations, we were able to demonstrate that at least 2% of the reported NTS infections in Israel result in a persistent infection with the same strain for months or even years, that is often manifested as a symptomatic relapse, illuminating a previously overlooked manifestation. Furthermore, we identified the contribution of both human and pathogen-related risk factors to this manifestation and exhibited the gain of genetic and phenotypic differences between related isolates and their potential to affect Salmonella pathogenicity in vivo.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank all the patients who have dedicated their time to participate in the study. We also thank Miri Bartal for the help with the interviews.
Financial support. This work was supported by a grant number 1096.39.11/2010 from the German-Israeli Foundation for Scientific Research and Development (GIF); and by a grant number 999/14 from the Israel Science Foundation (ISF) awarded to Ohad Gal-Mor. Michael McClelland acknowledges support from the National Institutes of Health (NIH) subcontract HHSN272200900040C, AI039557, AI052237, AI073971, AI075093, AI077645, AI083646, US Department of Agriculture (USDA) 2009-03579 and 2011-67017-30127, the Binational Agricultural Research and Development Fund and a grant from the Center for Produce Safety.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Galanis E, Lo Fo Wong DM, Patrick ME et al. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis 2006; 12:381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai PT, Porwollik S, Long F et al. Evolutionary genomics of the Salmonella enterica subspecies. mBio 2013; 4:e00579–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards RA, Olsen GJ, Maloy SR. Comparative genomics of closely related salmonellae. Trends Microbiol 2002; 10:94–9. [DOI] [PubMed] [Google Scholar]
- 4.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med 2002; 347:1770–82. [DOI] [PubMed] [Google Scholar]
- 5.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol 2006; 85:112–8. [DOI] [PubMed] [Google Scholar]
- 6.Dongol S, Thompson CN, Clare S et al. The microbiological and clinical characteristics of invasive Salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PloS one 2012; 7:e47342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guibourdenche M, Roggentin P, Mikoleit M et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 2010; 161:26–9. [DOI] [PubMed] [Google Scholar]
- 8.Marzel A, Desai PT, Nissan I et al. Integrative analysis of salmonellosis in Israel, 1995–2012 reveals association of serovar 9, 12: l, v:-with extraintestinal infections, dissemination of endemic S. typhimurium DT104 biotypes and a severe under-reporting of outbreaks. J Clin Microbiol 2014. JCM. 00399-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribot EM, Fair MA, Gautom R et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006; 3:59–67. [DOI] [PubMed] [Google Scholar]
- 10.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688–90. [DOI] [PubMed] [Google Scholar]
- 12.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R III, Foster JW. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol 1999; 34:112–23. [DOI] [PubMed] [Google Scholar]
- 13.Azriel S, Goren A, Rahav G, Gal-Mor O. The stringent response regulator DksA is required for Salmonella typhimurium growth in minimal media, motility, biofilm formation and intestinal colonization. Infect Immun 2016; 375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee IS, Lin J, Hall HK, Bearson B, Foster JW. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol 1995; 17:155–67. [DOI] [PubMed] [Google Scholar]
- 15.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 2007; 10:24–9. [DOI] [PubMed] [Google Scholar]
- 16.Webster C, Gardner L, Busby S. The Escherichia coli melR gene encodes a DNA-binding protein with affinity for specific sequences located in the melibiose-operon regulatory region. Gene 1989; 83:207–13. [DOI] [PubMed] [Google Scholar]
- 17.Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R 3rd. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun 2011; 79:4227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teplitski M, Goodier RI, Ahmer BM. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol 2003; 185:7257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsley RA, van Amsterdam K, Kramer N, Baumler AJ. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun 2000; 68:2720–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchwald DS, Blaser MJ. A review of human salmonellosis: II. Duration of excretion following infection with nontyphi Salmonella. Rev Infect Dis 1984; 6:345–56. [DOI] [PubMed] [Google Scholar]
- 21.Sirinavin S, Garner P. Antibiotics for treating salmonella gut infections. Cochrane Database Syst Rev 1999; doi:10.1002/14651858.CD001167. [DOI] [PubMed] [Google Scholar]
- 22.Lonnermark E, Lappas G, Friman V, Wold AE, Backhaus E, Adlerberth I. Effects of probiotic intake and gender on nontyphoid Salmonella infection. J Clin Gastroenterol 2015; 49:116–23. [DOI] [PubMed] [Google Scholar]
- 23.Hohmann EL. Approach to the patient with nontyphoidal Salmonella in a stool culture. Available at: http://www.uptodate.com/contents/approach-to-the-patient-with-nontyphoidal-salmonella-in-a-stool-culture Accessed 27 April 2015.
- 24.Coombes BK, Wickham ME, Brown NF et al. Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. J Mol Biol 2005; 348:817–30. [DOI] [PubMed] [Google Scholar]
- 25.Hayden HS, Lim R, Brittnacher MJ et al. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PloS One 2012; 7:e36507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zdziarski J, Brzuszkiewicz E, Wullt B et al. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog 2010; 6:e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okoro CK, Kingsley RA, Quail MA et al. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin Infect Dis 2012; 54:955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Octavia S, Wang Q, Tanaka MM, Sintchenko V, Lan R. Genomic variability of serial human isolates of Salmonella enterica serovar Typhimurium associated with prolonged carriage. J Clin Microbiol 2015; 53:3507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 2005; 57:691–705. [DOI] [PubMed] [Google Scholar]
- 30.Martinez LC, Yakhnin H, Camacho MI et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 2011; 80:1637–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice CJ, Ramachandran VK, Shearer N, Thompson A. Transcriptional and Post-Transcriptional Modulation of SPI1 and SPI2 Expression by ppGpp, RpoS and DksA in Salmonella enterica sv Typhimurium. PloS One 2015; 10:e0127523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rychlik I, Barrow PA. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev 2005; 29:1021–40. [DOI] [PubMed] [Google Scholar]
- 33.Koskiniemi S, Gibbons HS, Sandegren L et al. Pathoadaptive mutations in Salmonella enterica isolated after serial passage in mice. PloS One 2013; 8:e70147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.