Abstract
Butterfly-shaped pigment dystrophy is an eye disease characterized by lesions in the macula that can resemble the wings of a butterfly. Here, we report the identification of heterozygous missense mutations in the α-catenin 1 (CTNNA1) gene in three families with butterfly-shaped pigment dystrophy. In addition, we identified a Ctnna1 missense mutation in a chemically induced mouse mutant, tvrm5. Parallel clinical phenotypes were observed in the retinal pigment epithelium (RPE) of individuals with butterfly-shaped pigment dystrophy and in tvrm5 mice, including pigmentary abnormalities, focal thickening and elevated lesions, and decreased light-activated responses. Morphological studies in tvrm5 mice revealed increased cell shedding and large multinucleated RPE cells, suggesting defects in intercellular adhesion and cytokinesis. This study identifies CTNNA1 gene variants as a cause of macular dystrophy, suggests that CTNNA1 is involved in maintaining RPE integrity, and suggests that other components that participate in intercellular adhesion may be implicated in macular disease.
Butterfly-shaped pigment dystrophy (MIM 608970) belongs to a group of autosomal dominant pattern dystrophies of the retinal pigment epithelium (RPE), first described in a large Dutch family (Family A, Fig. 1a)1-3. The disease is characterized by accumulation of pigmented material in the macula that can resemble the wings of a butterfly3. Affected individuals present from middle age with either normal or slightly diminished best-corrected visual acuity (BCVA) and color vision, and the activity of the RPE measured by electrooculogram (EOG) recordings may be abnormal4-6. Responses of the retina, recorded by full-field electroretinography (ERG), and dark adaptation are generally normal4,7,8. The disease is relatively benign, but can progress to atrophy of the retina and underlying choroid in the macula4,6,8 and to subretinal neovascularization9, both resulting in severe vision loss.
Figure 1.
CTNNA1 mutations in three families with butterfly-shaped pigment dystrophy. (a) Two affected individuals (A-III:7 and A-III:11) of family A were analyzed by whole exome sequencing and the c.953T>C; p.(Leu318Ser) variant in the CTNNA1 gene segregated in all family members. Two additional variants in the CTNNA1 gene were identified in family B and C with complete segregation in both families. (b) The heterozygous mutation c.953T>C; p.[Leu318Ser] (M1) was found in the large Dutch family A, and mutations c.1293T>G; p.[Ile431Met] (M2) and c.919G>A; p.[Glu307Lys] (M3) were found heterozygously in Dutch family B and Belgian family C, respectively. WT, wild type. Arrows indicate the proband of each family.
Mutations in the PRPH2 gene (MIM 179605) have been identified in individuals with butterfly-shaped pigment dystrophy1,4,7,10-15, but in most individuals the genetic cause is unknown. Genetic heterogeneity for butterfly-shaped pigment dystrophy has been demonstrated in a large Dutch family with butterfly-shaped pigment dystrophy (Family A, Fig. 1a), in which the involvement of the PRPH2 gene was excluded8. Subsequently, a novel disease locus on chromosome 5q21.2-q33.2 was identified in this family16.
Here we report the identification of mutations in the CTNNA1 gene (MIM 116805) in the large Dutch family (Family A, Fig. 1a) and in additional families with butterfly-shaped pigment dystrophy. In addition, we describe a Ctnna1 mutation in a chemically induced mouse mutant, tvrm5, and characterize the pathologic events leading to RPE dysmorphology in this model.
Results
CTNNA1 mutations in butterfly-shaped pigment dystrophy
Whole exome sequencing identified 23,783 variants that were shared by individuals A-III:7 and A-III:11 of family A (Fig. 1a). Shared variants located within the linkage interval on 5q21.2-q33.2 (between markers D5S433 and D5S487)16 were filtered for heterozygous (present on ≥20% and ≤80% variant reads), non-synonymous variants, with a frequency of less than 0.5% in the Exome Variant Server database (EVS website), and a high nucleotide conservation (PhyloP score > 2.7). Only one potential causative variant was identified, residing in the CTNNA1 gene [NM_001903]: c.953T>C; p.(Leu318Ser) (PhyloP score 5.1). All affected relatives carried the variant in heterozygous state, while the variant was absent in all unaffected family members. The variant was predicted to be disease-causing by SIFT, affects a residue that is completely conserved among vertebrate species (Supplementary Fig. 1), and was not found in 162 ethnically matched controls nor in the EVS database.
Sequencing of all 17 coding exons of the CTNNA1 gene in 93 unrelated probands with butterfly-shaped pigment dystrophy and other pattern dystrophies identified three additional rare missense variants in the CTNNA1 gene (Supplementary Table 1). Heterozygous variants c.1293T>G; p.(Ile431Met) and c.919G>A; p.(Glu307Lys) were identified in two probands of Dutch and Belgian ancestry, respectively (Fig. 1b), and segregate with the disease in families B and C (Fig. 1a). Both variants were predicted to be disease-causing by Polyphen and SIFT, affect residues that are completely conserved among vertebrate species (Supplementary Fig. 1), and were not identified in 162 ethnically matched controls nor in the EVS database. A third missense variant, c.160C>T; p.(Arg54Cys), was identified in an Italian proband who presented with a small area of RPE atrophy superior to the fovea in the right eye, without classical phenotypical features as seen in pattern dystrophies. The variant was predicted to be disease-causing by Polyphen and SIFT, affects a residue that is completely conserved among vertebrate species (Supplementary Fig. 1), and was not identified in 200 ethnically matched unrelated control individuals nor in the EVS database, but was present in the unaffected 64-year-old father of this individual. Therefore, the pathogenic nature of this variant remains unclear.
The clinical features of the affected individuals of the three families with butterfly-shaped pigment dystrophy are described in Supplementary Table 2 and the results of retinal fundus photography, fluorescence angiography (FA), near-infrared reflectance (NIR) images and ocular coherence tomography (OCT) are provided in Fig. 2.
Figure 2.
Retinal images of individuals with butterfly-shaped pigment dystrophy. (a–b) Retinal imaging of the right eye of individual A-III:11 at age 36. (a) Fundus photography showing a typical butterfly-shaped hyperpigmentation in the macula, surrounded by hypopigmentation. (b) On FA the central pigmented lesions were hypofluorescent radially branching “butterfly-shaped” structures with hyperfluorescence of the surrounding hypopigmented retina. (c–e) Retinal imaging of the left eye of individual B-II:1 at age 62. (c) Fundus photographs showing mild pigmentary changes in the macula. (d) On NIR images the retinal changes are more pronounced. (e) OCT scanning in the plane indicated by black arrow in c shows that the abnormalities on NIR imaging in d are hyperreflective and located at or just anterior to the RPE, and also shows atrophy of the RPE and photoreceptors at the fovea. (f–i) Retinal imaging of the right eye of individual C-II:1 at age 33 (f, g and i) and age 23 (h). (f) Fundus photographs showing pattern dystrophy with a starfish-aspect with thickened fovea and six branches with hyperpigmentation. Accumulation of yellowish macular pigment is seen in the foveal area. (g) Fundus autofluorescence imaging showing hyperautofluorescence of the deposits. (h) FA shows blockage of background fluorescence by pigmented lesions. (i) OCT of the central foveal lesion of the right eye (scan in the plane indicated by black arrowhead in f), showing a dome-shaped sub-RPE deposit in the fovea. ONL, outer nuclear layer; IS/OS, inner and outer segments of the photoreceptors; RPE, retinal pigment epithelium.
Identification of a Ctnna1 mutant mouse model
An indirect ophthalmoscopy screen of mice from a B6J mutagenesis program identified a mutant, tvrm5, that showed widespread fundus mottling and occasional bright spots, often with dark centers (ring spots). Linkage analysis of a tvrm5 × DBA/2J intercross based on the ring spot phenotype revealed a recessive disease locus on chromosome 18 that was narrowed to ∼14.5 Mbp by fine mapping (distal to D18Mit22 and proximal to D18Mit236). Whole exome sequencing identified coding variants in Ctnna1 (NM_009818; c.1307T>C; p.[Leu436Pro]; PhyloP score 2.6) and Pcdhb14 (c.1259C>T; p.[Thr420Ile]; PhyloP score 0.3). The higher PhyloP score, the predicted pathogenicity by SIFT, and the much higher expression of Ctnna1 compared to Pcdhb14 in the retina and RPE (BioGPS website) implicated the Ctnna1 mutation as the causative allele. By Sanger sequencing, this allele was confirmed to be homozygous in 17 affected progeny, heterozygous in two unaffected progeny and absent from eight unaffected progeny as well as from B6J and DBA/2J mice, indicating co-segregation of the mutant allele with the ring spot phenotype. Based on the similarity of human CTNNA1 disease to that of tvrm5 mice, the mutant allele was denoted as Ctnna1tvrm5. The distribution and expression of CTNNA1 protein and the expression of Ctnna1 mRNA in the posterior eye of B6J and heterozygous or homozygous mutant mice was similar (Supplementary Figs. 2 and 3), indicating that the missense mutation does not affect protein stability or expression.
Figure 3.
Live retinal imaging of homozygous Ctnna1tvrm5 mice. (a) Fundus imaging of mice at 12–14 months of age (n=7) revealed bright lesions near the posterior pole and mottling throughout the fundus. (b) OCT analysis of these mice revealed lesions at the same locations. An averaged projection of the OCT volume from the inner nuclear layer to the photoreceptor inner/outer segment junction is shown en face, to guide comparison with the fundus image in a. The lesions identified in a are superimposed as single en face slices (yellow boxes). Scale bar, 100 μm. (c–e) Detail of lesions shown from left to right in a, respectively, obtained from a less saturated image. (f–h) Cropped OCT B-scans corresponding to lesions shown from left to right in b, respectively. Spot lesions (f), show a slight focal thickening of RPE hyperreflectivity (arrowhead) and distortion of the overlying retinal layers. Ring lesions (g and h) include an elevated hyperreflective region surrounding a central hyporeflective area above the RPE. (i) Cropped OCT B-scan showing posterior eye of B6J mice at 12–14 months of age (n=6). The ONL, IS/OS, and RPE are indicated. Scale bars in i are 100 μm and apply to f–i.
RPE lesions and photoreceptor loss in Ctnna1tvrm5 mice
Clinical disease assessment by brightfield fundus imaging of homozygous Ctnna1tvrm5 mice (Fig. 3a) revealed lesions in the central fundus biased toward the posterior pole that were correlated with the spots (Fig. 3c) and ring spot (Fig. 3d, e) phenotypes. Fine pigment mottling was also detected throughout the fundus (Fig. 3a). En face OCT images (Fig. 3b) revealed hyperreflective objects at the same locations as observed by brightfield fundus imaging (Fig. 3b). OCT B-scans of spots (Fig. 3f) showed localized thickening of hyperreflectivity corresponding to the RPE and distortion of the photoreceptor inner/outer segment junction and external limiting membrane toward the vitreous. B-scans of ring spots showed localized thickening surrounding an elevated hyporeflective core (Fig. 3g and h). Notably, the B-scan profiles of lesions in homozygous Ctnna1tvrm5 mice and individuals with butterfly-shaped pigment dystrophy were similar, both for small and large lesions (compare Figs. 2e and 3f, and 2i and 3h, respectively). OCT analysis also revealed a significant decrease in outer nuclear layer (ONL) thickness of ∼9% at 1 and 3 months of age and ∼15% at 12–14 months of age in homozygous Ctnna1tvrm5 compared to wild-type B6J mice (Supplementary Fig. 4a). In heterozygotes, mottling was more apparent with age (Supplementary Fig. 4b), but spots and ring spots were not evident by either fundus imaging or OCT, and ONL thickness was not significantly affected (Supplementary Fig. 4a). Taken together, these findings indicate that pathologic changes occur at the apical surface of the RPE or within the RPE itself and include the outer retina, where progressive photoreceptor loss is observed in homozygous Ctnna1tvrm5 mice (Fig. 3 and Supplementary Fig. 4a).
Decreased light-activated RPE response in Ctnna1tvrm5 mice
To test whether the morphological RPE defects were accompanied by a decrease in RPE function, we examined Ctnna1 mutant mice by direct current electroretinography (DC-ERG). In comparison to wild-type B6J mice, DC-ERG responses were reduced in amplitude at one year of age in both heterozygous and homozygous Ctnna1tvrm5 mice (Fig. 4a). When the main components of the response were quantified, significant differences from B6J were noted for the c-wave, fast oscillation (FO), and off-response in both heterozygous and homozygous Ctnna1tvrm5 mice (Fig. 4b); the amplitude reduction of the light peak (LP) component, which tends to be variable in amplitude in mice, was not interpretable. Differences were not noted in responses between heterozygous and homozygous Ctnna1tvrm5 mice.
Figure 4.
ERG recordings of Ctnna1tvrm5 mice. (a) Mean dc-ERG waveforms of B6J (+/+, n=13), heterozygous (tvrm5/+, n=5), and homozygous Ctnna1tvrm5 (tvrm5/tvrm5, n=9) mice. (b) Dc-ERG amplitude (mean ± SEM) showed significant effect of strain on the c-wave, fast oscillation (FO) and off response (one-way ANOVA F(2,24) = 21.9, 20.2 and 10.6; P < 0.0001, 0.0001 and 0.0005, respectively). Post hoc analysis (Tukey) indicated significantly reduced responses in Ctnna1tvrm5 mice compared to B6J (*P < 0.003; **P < 0.0001), without significant differences between heterozygous and homozygous mutants (P > 0.05). Light peak (LP) variability precluded analysis. (c, d) Strobe flash response functions showing (c) dark-adapted a- and b-wave amplitudes (mean ± SEM) or (d) cone ERG amplitudes (mean ± SEM) for B6J, heterozygous and homozygous Ctnna1tvrm5 mice (n=3, 6 and 10, respectively). A significant effect of strain was observed (two-way repeated measures ANOVA, F(2,16) = 5.7 and 12.3; P = 0.0134 and 0.0006, for dark-adapted a- and b-wave responses, respectively; F(2,16) = 9.1, P = 0.0032 for cone ERG). Multiple comparisons (Tukey) at ≥ 1.38 log cd s/m2 and ≥ –1.22 log cd s/m2 for the a- and b-waves, respectively, revealed significantly decreased responses (P < 0.02) in homozygous as compared to B6J or heterozygous mice, which did not differ significantly (P > 0.05). Multiple comparisons (Tukey) at ≥ 0.9 log cd s/m2 indicated significantly reduced cone responses (P < 0.002) in homozygous Ctnna1tvrm5 as compared to B6J or heterozygous mice, which did not differ significantly (P > 0.05).
As the DC-ERG signal is generated in response to rod photoreceptor activity17, it was essential to determine whether the reduced DC-ERGs of Ctnna1 mutant mice resulted from a decrease in RPE function alone or were accompanied by a decline in rod photoreceptor function. Therefore, we also recorded strobe flash ERGs from B6J and Ctnna1tvrm5 mutant mice. While the overall waveform did not differ across genotypes, the dark-adapted a-wave and b-wave amplitudes were both significantly reduced in homozygous Ctnna1tvrm5 compared to B6J or heterozygous Ctnna1tvrm5 mice (Fig. 4c). Thus, the reduction in the DC-ERG responses in homozygous Ctnna1tvrm5 mice may reflect dysfunction of photoreceptors, RPE cells, or both. By contrast, dark-adapted responses did not differ in heterozygous Ctnna1tvrm5 and B6J mice (Fig. 4c), indicating that the reduced DC-ERG response in heterozygous mice occurs without a loss of photoreceptor function. The cone ERG response was significantly reduced in homozygous Ctnna1tvrm5 mice compared with B6J or heterozygous Ctnna1tvrm5 mice (Fig. 4d). Together with the live imaging results, the ERG observations support a pathogenic mechanism whereby heterozygous Ctnna1tvrm5 mice develop RPE dysfunction without significant photoreceptor damage, while homozygous animals develop a more severe RPE functional deficit causing rod and cone photoreceptor cell loss.
Ectopic cells and inclusions in RPE of Ctnna1tvrm5 mice
To assess cellular changes that might provide clues to the underlying cause of RPE lesions and functional defects in Ctnna1tvrm5 mice, we examined fixed retinal tissues by histology. Compared to the B6J retina at one month of age (Fig. 5a), the retina in homozygous Ctnna1tvrm5 mice (Fig. 5b) was grossly normal apart from slight ONL thinning. However, in the homozygous mutants, outer retinal distortion was occasionally noted in areas where unusual features were found in the underlying RPE. First, ectopic pigmented cells were present on the RPE apical surface in focal lesions appearing as a slightly thickened RPE layer contiguous with the epithelium (Fig. 5c) or as a double layer (Fig. 5d). Single pigmented cells were also observed (Fig. 5e), which may be shed RPE cells. The observation of nuclei with no evidence of surrounding pigment (Fig. 5f) suggested the presence of additional cell types in the subretinal space. Second, eosinophilic inclusions were found frequently within the RPE cell layer (Fig. 5g), often with an internal laminar appearance. Serial sections revealed nuclei on the edges of these inclusions and sparse melanin pigment enclosed within a well-defined boundary, compatible with an intracellular location. Finally, pigmented cells on the RPE apical surface and eosinophilic inclusions were both present in larger lesions (Fig. 5h–k), suggesting a common pathogenic basis of these features. In agreement with live imaging results, these unusual features were observed at one month and at 12–14 months in homozygous Ctnna1tvrm5 mice but were not detected in heterozygous and B6J animals.
Figure 5.
Light micrographs of hematoxylin and eosin stained paraffin-embedded ocular sections from B6J (n=3) and Ctnna1tvrm5 mice (n=6) at one month of age. (a) B6J retina. (b) Homozygous Ctnna1tvrm5 retina showing normal layered organization with outer retinal distortion in an area of abnormal RPE. (c–k) Detail of RPE lesions in eyes of homozygous Ctnna1tvrm5 mice. (c) Slight RPE thickening. (d) A double layer of pigmented cells, possibly RPE. (e) A single pigmented, nucleated cell on the RPE apical surface. (f) A nucleus without surrounding pigment (arrowhead) on the RPE apical surface and a small eosinophilc inclusion (asterisk). (g) An eosinophilic inclusion bounded by melanin pigment. (h–k) Sections 3, 5, 8 and 13 from a series of 14 sections of a large RPE lesion containing both pigmented cells on the RPE surface and an eosinophilic inclusion. Nuclei (asterisks) in i may be in the same cell as the eosinophilic inclusion. Scale bars in panels b, g and k are 100, 20 and 20 μm, respectively, and apply to each row of images. ONL, outer nuclear layer; RPE, retinal pigment epithelium.
These findings provide a histological correlate of the live imaging results. The pigmented cells accumulating on the apical surface of the RPE (Fig. 5c and d) corresponded to the bright spots observed by fundus imaging and en face OCT (Fig. 3a and b), and to the localized thickening of the hyperreflective RPE layer in OCT B-scans (Fig. 3f–h). Accumulation of eosinophilic material in the epithelial layer (Fig. 5g) may also contribute to localized thickening observed by OCT. Lesions containing large eosinophilic inclusions flanked by double layers of pigmented cells corresponded to the ring spots observed by live imaging (Fig. 5h–k).
RPE dysmorphology and multinucleate cells
As an additional test for RPE structural defects, we stained RPE/choroid/sclera flatmounts at one month of age to detect nuclei and the F-actin cytoskeleton at RPE cell boundaries. Flatmounts from B6J mice revealed mono- and binucleate polygonal RPE cells with vertices shared by three cells (Fig. 6a). In contrast, homozygous Ctnna1tvrm5 mice exhibited unusual RPE morphology comprising four or more cells surrounding an F-actin circle of variable size (Fig. 6b–d) or a shared vertex (Fig. 6e). Orthogonal views of B6J image stacks indicated that RPE nuclei are normally located below F-actin structures at the RPE apical surface (Fig. 6f, left), presumably microvilli18, and below the circumferential F-actin band that defines the polygonal cell boundary (Fig. 6f, right). In homozygous Ctnna1tvrm5 mice, nuclei were occasionally observed directly above the circular F-actin motif (Fig. 6g), consistent with apical shedding of an RPE cell. However, in most of these structures, nuclei were not apparent or were positioned basally to the circular F-actin motif, possibly indicating that basal shedding also occurs.
Figure 6.
RPE cell dysmorphology in RPE/choroid/sclera flatmounts of Ctnna1tvrm5 mice stained with rhodamine phalloidin (red) and DAPI (blue) to reveal F-actin and nuclei, respectively. (a) Merged projections of a confocal image stack show normal mono- and binucleate polygonal cells in B6J mice at one month of age (n=5 mice). (b–e) Circular F-actin structures (b–d) or vertices (e) shared by four or more cells were frequent in homozygous Ctnna1tvrm5 mice (n=5 mice). (f) Orthogonal displays at two z-positions of cells in panel a show that RPE nuclei in B6J mice lie below F-actin on the apical surface (left) and in the circumferential band (right). (g) Orthogonal display of cells from panel c showing a flattened nucleus between apical F-actin and the circular F-actin structure in homozygous Ctnna1tvrm5 mice. (h) RPE cells B6J mice at 12–14 months of age remained mostly mono- and binucleate (n=5 mice). (i) Heterozygous Ctnna1tvrm5 mice at 12–14 months of age showed large multinucleate RPE cells (n=5 mice). (j) Homozygous Ctnna1tvrm5 mice exhibited multinucleate cells as well bright F-actin tangles and rounded cells with bright F-actin boundaries (n=6 mice). Scale bars in panels e and j are 25 and 50 μm, respectively, and apply to the corresponding rows of images.
RPE dysmorphology was also evident at 12–14 months of age. Compared to normal polygonal cells in B6J mice (Fig. 6h), the RPE cells in heterozygous (Fig. 6i) and homozygous Ctnna1tvrm5 mice (Fig. 6j) were highly variable in size and included extremely large, irregularly-shaped multinucleate cells. In addition to the absence of apical F-actin boundaries in the interior of these abnormal RPE cells, the disordered clustering of nuclei suggested a lack of intercellular barriers. Bright F-actin structures, including tangles and intense staining at the boundaries of rounded cells, were more pronounced in homozygous than in heterozygous Ctnna1tvrm5 mice. In summary, a primary consequence of the Ctnna1tvrm5 mutation appears to be significant dysmorphology in the form of RPE cell shedding and the accumulation of large multinucleate RPE cells.
Structural and functional implications of CTNNA1 variants
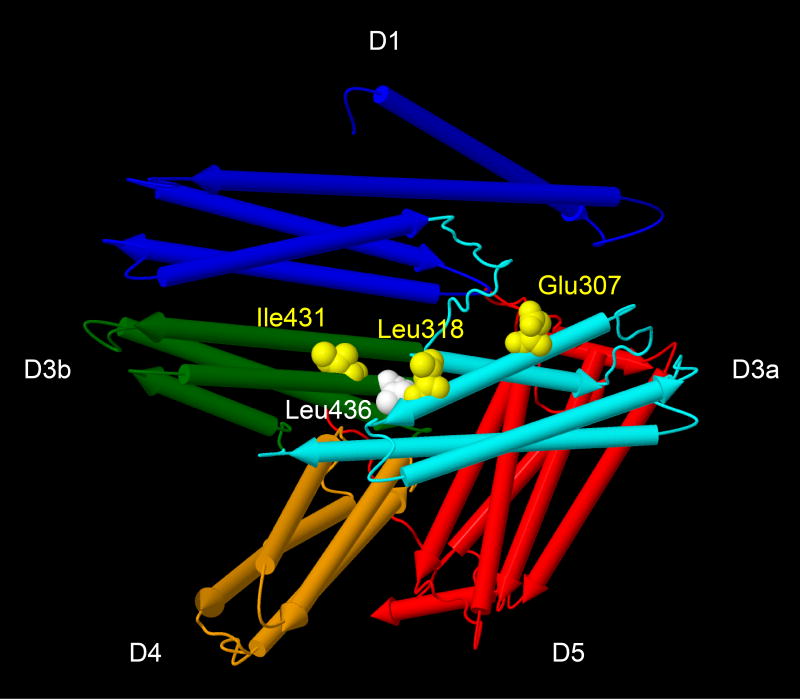
CTNNA1 encodes α-catenin 1 (also known as αE-catenin or CTNNA1), which stabilizes intercellular adherens junctions as a force-sensing adaptor between cell surface cadherins and the actin cytoskeleton19,20. CTNNA1 responds to increased intercellular tension by unfurling binding sites for actin-associated proteins, such as VCL (vinculin), afadin, α-actinin, and formin19,20. Given the importance of adherens junctions for RPE integrity21, we explored possible structural and functional consequences of the CTNNA1 variants. The CTNNA1 variants detected in the three families with butterfly-shaped pigment dystrophy (p.[Glu307Lys], p.[Leu318Ser], and p.[Ile431Met]) and the mouse Ctnna1tvrm5 variant (p.[Leu436Pro]) are predicted to alter residues in CTNNA1 that are completely conserved among vertebrate species (Supplementary Fig. 1). As shown in an x-ray crystallographic structural model of the human protein22 (Fig. 7), four of the five variants affect amino acid residues (p.Glu307, p.Leu318, p.Ile431, p.Leu436) clustered in the middle of protein. The variants map to the proposed force-sensing module (domains D3–D4, amino acid residues 260–630) and to protein binding domains within this module that may be sensitive to tension (D3a, residues 260–400; D3b, residues 400–507)20. The four variants map in or near the region reported to bind VCL20,23-25. The fifth variant, which is of uncertain pathogenic significance, affects Arg54 within the N-terminal domain that binds β-catenin, a direct binding partner of cadherin. To test for an effect of the variants on VCL binding, the wild-type and mutant CTNNA1 proteins were heterologously co-expressed with VCL. Although co-immunoprecipitations confirmed interaction of CTNNA1 with VCL, the mutations did not affect the interaction of these proteins (Supplementary Fig. 5).
Figure 7.
Structural model of human CTNNA122 showing the location of amino acid residues altered by CTNNA1 mutations identified in three families with butterfly-shaped pigment dystrophy and in Ctnna1tvrm5 mice. The model (Protein DataBank entry 4IGG, polypeptide chain A) includes residues 82–635 and 666–861. The α-helices are depicted as rockets and colored according to domain classification (D1, N-terminal domain; D3a, D3b and D4, middle domains also known as MI, MII and MIII, respectively; D5, C-terminal domain). Affected residues are shown in space-filling representation (yellow, human variants; white, mouse variant). The sequences of human CTNNA1 and mouse Ctnna1 are identical at the residues shown.
Discussion
This work implicates mutations in the CTNNA1 gene as a cause of butterfly-shaped pigment dystrophy. Previously, this condition has been associated with mutations in peripherin 2, a protein involved in outer segment disc biogenesis of retinal photoreceptor cells1. The finding that the disease is also caused by variants in CTNNA1, a central component of adherens junctions, broadens our perspective on its cellular and molecular basis. Intriguingly, a juvenile form of macular dystrophy (MIM 601553) that results in RPE atrophy and pigmentary changes has been linked to variants in CDH3, which encodes P-cadherin26,27. Together, these findings support the idea that other components of the cadherin-based intercellular adhesion machinery may be implicated in macular degenerative disease.
The parallel ocular phenotypes in affected individuals carrying a CTNNA1 variant allele and in homozygous Ctnna1tvrm5 mice suggest that similar mechanisms underlie pathogenesis in both species and that Ctnna1tvrm5 mice accurately model the human disease. Importantly, the dysmorphology of RPE cells in Ctnna1tvrm5 mice raises the possibility that RPE defects are a precipitating event in the disease. Although disease phenotypes were stronger in homozygous Ctnna1trmv5 mice, RPE defects were also observed in heterozygous mutants, implying a gene-dosage effect. This likely arises from an altered function of the variant protein, as we found similar Ctnna1 mRNA and CTNNA1 expression levels in the posterior eye of wild-type and heterozygous or homozygous Ctnna1tvrm5 mutant mice. Although our work and previous studies28-31 (BioGPS.org) indicated that the mRNA and protein are expressed in both the RPE and the retina, the disease phenotypes may be due to perturbation of CTNNA1 function specifically in the RPE. The human disease mainly affects the RPE1-8, while the mouse Ctnna1tvrm5 mutation has only slight effects on retinal integrity and is less disruptive than an embryonic lethal gene-trap allele32, or a conditional knockout of Ctnna1 in retinal progenitor cells, which causes retinal developmental defects29. Thus, our results support a causative role of RPE defects in butterfly-shaped pigment dystrophy due to CTNNA1 variants.
Eosinophilic inclusions observed in homozygous Ctnna1tvrm5 RPE appear similar to hard drusen, small dome-shaped eosinophilic masses between the RPE and Bruch's membrane that are associated with macular degeneration when abundant33-36. The laminar substructure of these inclusions is sometimes evident in hard drusen37, and resembles the “onion-skin” characteristic of drusen associated with Doyne honeycomb retinal dystrophy/Malattia leventinese38. As these inclusions correspond to elevated lesions in OCT B-scans , we suggest that similar lesions detected by OCT in patients with butterfly-shaped pigment dystrophy derive from eosinophilic inclusions in the RPE.
Pigmented cells on the RPE apical surface and aberrant F-actin structures in Ctnna1tvrm5 mice were similar to shedding structures in chick embryonic RPE39 and in mouse models of RPE injury40,41. Shed RPE cells may survive to accumulate and/or proliferate locally at lesions, creating a double layer of pigmented cells as observed in our study, in mouse models that target damage to the RPE40,42 and in geographic atrophy43 . Alternatively, shed RPE cells may die and be phagocytosed by intact RPE or immune cells, yielding subretinal macrophages containing melanin pigment granules as found in mouse models of retinal degeneration44,45. We propose that RPE shedding is an initial event leading to focal RPE thickening and changes in melanin pigment distribution in CTNNA1-associated butterfly-shaped pigment dystrophy. RPE shedding may prove to be a common attribute of macular dystrophic disease, as subretinal pigmented cells and RPE cytoskeletal derangements similar to those observed in Ctnna1tvrm5 mice have also been associated with human age-related macular degeneration.46,47
Abnormally large multinucleate RPE cells in Ctnna1tvrm5 mutant mice may arise from RPE hyperproliferation and a cytokinesis defect due to the mutation, through repeated nuclear divisions without cell division. Alternatively, altered RPE cell adhesion due to the Ctnna1tvrm5 mutation may lead to cell fusion and syncytia formation. For example, large multinucleate cells observed upon conditional deletion of Ranbp2 in the mouse RPE were interpreted previously as syncytia48.
The positional clustering of pathogenic human and mouse variants within CTNNA1 suggests a shared effect on the activity of this protein. The variants may disrupt adherens junctions, which are critical for maintaining RPE integrity21. As the affinity of CTNNA1 variants for VCL was normal, the mutations may disrupt binding to other actin-associated proteins or alter the conformational response to mechanical stress. Recent studies demonstrate that CTNNA1 participates in molecular networks independent of cell adhesion, such as the NF-KB, Ras-MAPK, Hedgehog and Hippo signaling pathways. Consequently, the variants may perturb CTNNA1 function in cellular processes regulated by these pathways, including cytokinesis, cell differentiation or growth49-51.
Future studies may focus on other RPE cell adhesion components that contribute to macular disease, and may uncover the mechanisms by which CTNNA1 dysfunction affects RPE turnover, proliferation, and the pathogenesis of pattern dystrophy.
Methods
Clinical Evaluations of Human Subjects
Fourteen affected and 12 unaffected individuals from three butterfly-shaped pigment dystrophy families were included in this study. Participants received the diagnosis of butterfly-shaped pigment dystrophy based on the aspect of the lesions on ophthalmoscopy sometimes combined with fluorescein angiography (FA). The medical histories, age at onset, (best-corrected) Snellen visual acuity (BCVA) and fundus appearance, including slit-lamp examination, fundus examination by indirect ophthalmoscopy and fundus photography were retrieved from medical records of all included subjects. Additionally, FA was available in 11 affected subjects, fundus autofluorescence and NIR images were obtained in 6 and spectral-domain optical coherence tomography (OCT) in 4 affected participants. Eleven affected subjects underwent electro-oculography (EOG) and electroretinography (ERG) and color vision testing were available in 6 and 7 affected subjects, respectively. ERG and EOG were recorded and interpreted as previously described.52 In 6 patients, EOG was performed according to the International Society for Clinical Electrophysiology of Vision (ISCEV) protocol. The study was approved by the local medical ethics committees (Commissie Mensgebonden Onderzoek regio Arnhem-Nijmegen, Ethics Committee of Ghent University Hospital, and Ethical Committee of the Second University of Naples) and adhered to the tenets of the Declaration of Helsinki. All participants provided written informed consent prior to participation in the study.
Whole Exome Sequencing and Mutation Analysis in Human Subjects
To identify the genetic defect in a large Dutch family with nine individuals affected by butterfly-shaped pigment dystrophy (family A; Fig. 1A), the DNA of two affected individuals (A-III:7 and A-III:11) was analyzed using whole exome sequencing. The exomes were enriched using the SureSelect Human All Exon v.2 Kit (50Mb), which targets the exonic sequences of approximately 21,000 genes, according to the manufacturer's protocol (Agilent). Sequencing was performed on a 5500xl SOLiD sequencing platform (Life Technologies). BioScope software v.1.3 (Life Technologies) was used to map color space reads along the hg19 reference genome assembly. The high-stringency calling DiBayes algorithm was used for single-nucleotide variant calling, and small insertions and deletions were detected using the small Indel Tool.
All coding exons and intron-exon boundaries of the CTNNA1 gene were screened for mutations in 93 unrelated probands with butterfly-shaped pigment dystrophy (N=19) or other pattern dystrophies (N=74) by Sanger sequencing. Primers for PCR amplification and sequencing were designed with Primer3 software (Supplementary Table 3). Missense variants detected in the CTNNA1 gene were analyzed in ethnically matched control individuals by restriction enzyme analysis (TaqI for c.953T>C; and BccI for c.1293T>G;), allele-specific PCR (c.919G>A), or Sanger sequencing (c.160C>T).
Mouse Maintenance and Mutagenesis
All procedures used in animal experiments for this study were approved by the Institutional Animal Care and Use Committees of the Cleveland Clinic and The Jackson Laboratory and were in accordance with the “Guide for the Care and Use of Experimental Animals” established by the National Institutes of Health (1996, Revised 2011) and the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Mice were bred and maintained on a 12 hour light-dark cycle in the Research Animal Facility vivarium of The Jackson Laboratory (JAX). Pasteurized JL Mouse and Rat Auto 4F/No Hysil 5K54 diet (fat content, 4%, PMI Nutrition International) and HCl-acidified water (pH 2.8-3.2) were provided ad libitum.
The tvrm5 mutant (JAX stock number 021610) was identified among G3 mice from an ethylnitrosourea mutagenesis screen of C57BL/6J (B6J; JAX stock number 000664) mice generated by the Center for New Mouse Models of Heart, Lung, Blood and Sleep Disorders at JAX.53 Phenotypic screening by indirect ophthalmoscopy at ∼21 weeks of age was performed by the Translational Vision Research Models (TVRM) program.54 Once heritability was established, tvrm5 mice were backcrossed to B6J for seven generations and maintained in homozygous state by intercrossing. B6J mice were used as wild-type controls and in segregating crosses to generate heterozygous mice from the inbred mutant stock.
tvrm5 Mapping and Sequence Analysis
To identify the causative locus, a mutant tvrm5 male was outcrossed with a DBA/2J (+/+) female, the resulting F1 progeny were intercrossed, and 31 F2 generation mice were analyzed for linkage using a pool scan with genome-wide simple sequence length polymorphic (SSLP) markers. Map location was confirmed in individual F2 mice from the pool and additional SSLP markers were used to refine the map position. For whole exome sequencing, genomic DNA (1 μg) was fragmented to a peak size of 300 bp by sonicating on low power (30 s power on-off cycle) for 10 min using a Diagenode Bioruptor UCD-200TM-EX (Denville). The pre-capture paired end library was constructed using the TruSeq DNA Sample Preparation Kit (FC-121-100, Illumina) without size selection step and 18 cycles of PCR. The pre-capture library was hybridized to the NimbleGen Mouse Exome capture probe set (#9999042611, Roche Nimblegen) according to the manufacturer's instructions. The sequencing library was quantified by qPCR, pooled with two similar libraries, and sequenced on a single lane of a HiSeq 2000 (Illumina) using a 2×100 bases (paired end) sequencing protocol. Genotypes were confirmed by sequencing the PCR product amplified with primers Ctnna1F and Ctnna1R (Supplementary Table 3) at 97°C, 3 min; 45 cycles of 95°C, 10 s and 50°C, 30 s; 72°C, 30 s; 72°C, 3 min.
qRT-PCR and Western Analysis
For molecular analysis, eyes of mice asphyxiated with carbon dioxide were removedand dissected immediately in ice-cold phosphate-buffered saline (PBS) to yield an RPE/choroid/sclera complex or placed in RNAlater (Thermo Scientific) at room temperature and punctured at the limbus with a needle. To recover RNA, the RPE/choroid/sclera complex was treated with RNAprotect (Qiagen) immediately following dissection and processed to yield a pigmented pellet enriched for RPE RNA55. Alternatively, enucleated eyes in RNAlater were dissected to recover the combined retina and RPE, which adhered tightly as evidenced by RPE nuclei and a polygonal melanin pattern on the retinal surface. Excess RNAlater was removed bymicropipetting. The RPE-enriched pellet (two eyes per animal) or the combined retina and RPE (one eye per animal) were processed with the RNeasy Micro Kit (Qiagen) as recommended by the manufacturer, including on-column DNase I treatment. Purified RNA was quantified using a Nanodrop-1000 spectrophotometer (Thermo Scientific). RNA was reverse-transcribed with the RETROscript Reverse Transcription Kit (Thermo Scientific) according to the manufacturer's instructions using random priming. Quantitative real-time PCR (qRT-PCR) was performed with the iTaq Universal SYBR Green SuperMix (Bio-Rad) and Ctnna1rtF2 and Ctnna1rtR2 primers (Supplementary Table 3), using the CFX96 Real-Time PCR Detection System (Bio-Rad). The comparative CT method (ΔΔCT) was applied with Actb as an internal calibrator and the relative fold change in Ctnna1 transcript was calculated as 2-ΔΔCT. Melting curve analyses were performed to validate target gene amplification. Error bars represent the additive propagation of error calculated as described previously56.
For protein analysis, combined retina and RPE samples were resuspended and sonicated for 10 s in Laemmli sample buffer (Bio-Rad) containing Protease Inhibitor Cocktail Set III, EDTA-free (EMD Millipore) and 50 mM D,L-dithiothreitol. An equal volume of each lysate and a volume of a BJ6 lysate greater than and less than this volume were heated at 95°C for 5 min and electrophoresed on Any kD Mini-PROTEAN TGX gels (Bio-Rad). Precision Plus Dual Color Proteins were included to estimate protein molecular weights. Electrophoresed proteins were transferred to nitrocellulose with a Trans-Blot Turbo system (Bio-Rad) using the High MW standard protocol. Blots were stained with Ponceau S for 1 h, washed twice with distilled water and imaged with visible light on a G:Box Chemi XRQ gel imaging system (Syngene). After blocking for 2-12 h at 4°C in 5% nonfat dry milk in Tris-buffered saline containing 0.05% w/v Tween-20 (TBST), blots were incubated overnight with a 1:1,000 dilution of rabbit monoclonal anti-CTNNA1 antibody (EP1793Y, Abcam) in blocking buffer. Following three 15-min washes with TBST, blots were incubated for 1.5 h at room temperature with a 1:1,000 dilution of goat anti-rabbit IgG conjugated with horseradish peroxidase (7074; Cell Signaling Technology), washed three times for 15 min with TBST, developed with ECL Prime Western Blotting reagent (GE Healthcare Bio-Sciences) and imaged forchemiluminescence with the G:Box Chemi XRQ system. To determine relative expression levels, TIF-format images were quantified in Fiji57. Total protein was determined from Ponceau S images by first adjusting background intensity, subtracting a constant value over the entire image. The line tool was then adjusted to the width and full length of each lane and the integrated density of the selected region was measured. A linear fit of total integrated density to load volume was determined in Excel (Microsoft) from the integrated intensities of B6J samples loaded at varying volumes, and used to calculate the volume equivalent of each sample. A similar approach yielded volume equivalents for CTNNA1 staining from chemiluminiscence images without background subtraction. The volume equivalent of CTNNA1 was divided by that of total protein and the resulting values were tested statistically. Values from heterozygous and homozygous samples were averaged and divided by the corresponding B6J value to yield the relative expression level, which was graphed with the relative propagating error calculated from the square root of the sum of the squares of the relative error for each group.
Live Imaging
Live fundus images were obtained with a Micron III Retinal Imaging System (Phoenix Research Laboratories) and an R2200 ultrahigh resolution spectral domain OCT system (Bioptigen). Image acquisition and processing were performed as described previously.58 ONL thickness in one eye of each mouse was determined from OCT volume datasets imported into Fiji at four positions, each ∼250 μm from the optic nerve head in the superior, nasal, temporal and temporal fundus. Thickness was determined from 50-pixel wide lines drawn vertically across the ONL at each position. To identify the ONL boundaries reproducibly, the Plot Profile tool was used to plot image intensity by distance, and the length (in pixels) of a horizontal line between the midpoints of the intensity peaks on either side of the ONL was determined. Values from each eye were averaged and converted to ONL thickness by an axial factor of 1.53 μm/pixel determined from the OCT acquisition program.
Histology of Mouse Eyes
For conventional histology, eyes of mice asphyxiated with carbon dioxide were removed and treated with a 3:1:4 mixture of methanol:acetic acid:PBS with subsequent paraffin embedding and staining with hematoxylin and eosin as described.59 Brightfield images were acquired using a Leica DMLB light microscope with 5× (HC PL FLUOTAR, 40× (HCX PL APO), and 100× (N PLAN) objectives. Images were collected using a QImaging Retiga 2000R camera and RGB Slider in color mode with QCapture software. Processing of exported image files in Fiji included gamma adjustment for color balance, brightness and contrast adjustment, and unsharp mask.. For immunohistochemistry, enucleated eyes were placed in Tissue-Tek optimal cutting temperature compound (Sakura Finetek) and snap frozen in N2(l)-chilled 2-methylbutane. Cryosections (10 μm) were air-dried on ColorFrost Plus glass microscope slides (Thermo Scientific), fixed for 10 min on ice with 4% w/v paraformaldehyde in PBS and rinsed in PBS. Sections were blocked in PBS containing 0.3% v/v Triton X-100 and 2% w/v normal horse serum for 30 min at room temperature, followed by overnight incubation at 4°C with rabbit monoclonal anti-CTNNA1 antibody (EP1793Y; Abcam) diluted 1:200 in blocking solution. Following three washes in PBS, sections were incubated for 1 hour at room temperature in Cy3-conjugated F(ab′) donkey anti-rabbit IgG secondary antibody (711-166-152; Jackson ImmunoResearch) diluted 1:200 in blocking solution. Samples were washed three times in PBS and mounted on slides with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). Stained slides were imaged with an LD Plan-Neofluar 20×/0.4 Korr Ph2 M27 or a Plan-Apochromat 100×/1.40 Oil M27 objective on a ZeissAxio Observer Z.1 microscope with filter sets 43 or 49 using Zen 2012 software and a Hamamatsu ORCA-ER-1394 camera.. Illumination intensity and exposure times were constant for all samples, and control sections that were treated identically except for the omission of primary antibody showed little to no fluorescence under the imaging conditions selected. At both magnifications, image brightness and contrast adjustments were applied uniformly across all samples in Fiji.
For flatmounts, enucleated eyes were fixed, dissected, and stained with DAPI and rhodamine phalloidin as described for retina,58 except that the RPE/choroid/sclera complex was retained and examined. Confocal image stacks were obtained using a Leica SP5 confocal microscope, HCX PL APO CS 63.0× 1.30 GLYC objective, and Leica Acquisition Suite software. Conventional fluorescence images were obtained at 10× (Plan-Apochromat 10×/0.45 M27) using the Zeiss filter sets and microscope system described above. Confocal images were processed in Fiji by dividing the blue by the green channel after denoising to reduce the contribution of autofluorescence to the DAPI signal, and images were oriented with the Interactive Stack Rotation plugin. An image stack subset was maximally projected to visualize the F-actin circumferential boundary with minimal overlap from apical F-actin. Conventional fluorescence image stacks were oriented with the Interactive Stack Rotation plugin to optimize the display of F-actin at RPE cell boundaries, and brightness and contrast were adjusted in Fiji.
Mouse Visual Electroretinography
Mice were studied using ERG stimulation and recording protocols to evaluate outer retinal and RPE function. Mice were dark-adapted overnight, anesthetized with ketamine (80 mg/kg) and xylazine (16 mg/kg), and placed on a temperature-regulated heating pad throughout the testing sessions. Pupils were dilated with eye drops (2.5% phenylephrine HCl, 1% cyclopentolate, 1% tropicamide). The corneal surface was anesthetized with 1% proparacaine HCl.
Outer retina function was examined using a conventional strobe-flash ERG protocol.60 ERGs were recorded using a stainless-steel wire active electrode referenced to a needle electrode placed in the cheek. A second needle electrode placed in the tail served as ground lead. Responses were differentially amplified (0.3–1,500 Hz), averaged, and stored using a UTAS E-3000 signal averaging system (LKC Technologies). White light strobe flashes were initially presented in darkness within a Ganzfeld bowl with increasing luminance, from -3.6–2.1 log cd s/m2. Cone ERGs were isolated by superimposing stimuli (-0.8–1.9 log cd s/m2) upon a steady adapting field (20 cd/m2). The a-wave amplitude was measured 8 ms after flash onset from the prestimulus baseline. The b-wave amplitude was measured from the a-wave trough to the peak of the b-wave or, if no a-wave was present, from the prestimulus baseline.
RPE ERG components were examined using a dc-ERG protocol.60 Responses were obtained from the corneal surface of the left eye using a capillary tube with filament (BF100-50-10; Sutter Instrument), filled with Hank's buffered salt solution to contact an Ag/AgCl wire electrode. A similar electrode contacting the right eye served as the reference. Responses were differentially amplified (dc-100 Hz), digitized at 20 Hz, and stored using LabScribe Data Recording Software (iWorx). White light stimuli were derived from an optical channel using a Leica microscope illuminator as the light source and delivered to the test eye with a 1-cm-diameter fiber-optic bundle. Stimulus luminance was 2.4 log cd/m2. Stimulus timing and duration was controlled at 7 min by a shutter system (Uniblitz). The c-wave amplitude was measured from the prestimulus baseline to the c-wave peak. The fast oscillation (FO) amplitude was measured from the c-wave peak to the trough of the FO. The light peak (LP) amplitude was measured from the FO trough to the asymptotic value. The off-response amplitude was measured from the LP value just prior to stimulus light offset to the peak of the initial component.
Statistical Analysis
In mouse studies, which were exploratory, n≥3 age-matched animals were analyzed. The number of female and male mice used in each experiment is given in Supplementary Table 4. Sample size pre-estimation, blinding and randomization were not implemented. All mice selected for quantitative studies were included in the statistical analysis. ONL thickness data at each age, dc-ERG component amplitudes for each stimulus condition, replicate ΔCT (Ctnna1-Actb) values from qRT-PCR analysis, and the ratios of CTNNA1 to total protein volume equivalents drom western blotting were analyzed by one-way ANOVA. Statistically significant comparisons (p < 0.05) were further characterized by Tukey HSD post hoc analysis. Response functions obtained for ERG a- and b-wave amplitudes were compared across genotypes using a two-way repeated measures ANOVA, with multiple comparisons by Tukey post hoc analysis at each flash intensity. Assumptions for ANOVA modeling were met, as a quantile-quantile plot of ANOVA residuals against a normal distribution approximated the diagonal, and variances in each analysis except for the dc-ERG LP response were found to be equal (Levene test). Statistical tests were performed with JMP (SAS) and Prism (GraphPad Software).
Co-immunoprecipitations
Co-immunoprecipitation analyses were performed to test the interaction of wild-type and mutant CTNNA1 with VCL (vinculin, [MIM 193065]). Expression constructs to produce 3×HA-CTNNA1 (wild-type and variants c.160C>T; p.[Arg54Cys], c.919G>A; p.[Glu307Lys], c.953T>C; p.[Leu318Ser], c.1293T>G; p.[Ile431Met], and c.1307T>C; p.[Leu436Pro]) and 3×FLAG-VCL proteins were co-transfected in human embryonic kidney 293T (HEK293T) cells. As a negative control, production of the functionally unrelated p63 protein (MIM 603273) was used. As positive controls, the previously described interactions between NPHP4 (nephrocystin-4, MIM 607215) and RPGRIP1 (MIM 605446) were used.61 Following transfection, cells were cultured for 24 hr and subsequently lysed on ice in lysis buffer (50 mM Tris-HCL [pH 7.5], 150 mM NaCl, and 0.5% Triton X-100) supplemented with complete protease inhibitor cocktail (Roche). Lysates were incubated with anti-FLAG M2 agarose from mouse (Sigma-Aldrich) for 2 hr at 4°C. After incubation, beads with bound protein complexes were washed in lysis buffer, resuspended in 4× NuPAGE Sample Buffer and heated for 10 min at 70°C. Beads were precipitated by centrifugation, and supernatant was run on a NuPAGE Novex 4%–12% Bis-Tris SDS-PAGE gel. The interaction between 3×HA-CTNNA1 and 3×FLAG-VCL was assessed by immunoblotting, followed by staining with polyclonal rabbit anti-HA (1:1,000; Sigma-Aldrich) or monoclonal mouse anti-FLAG (1:1,000; Sigma-Aldrich) as a primary antibody, and goat anti-rabbit IRDye800 (1:20,000; Li-Cor) or goat anti-mouse Alexa Fluor 680 (1:20,000; Molecular Probes) as a secondary antibody. Fluorescence was analyzed on a Li-Cor Odyssey 2.1 infrared scanner. HEK293 cells were obtained from the cell culture facility of the Department of Human Genetics at Radboud University Medical Center and were tested for mycoplasma.
Location of Variant Residues in CTNNA1
The three-dimensional structure of CTNNA1 was drawn from an x-ray crystallographic structural model (Protein DataBank entry 4IGG) using Jmol, an open-source Java viewer for chemical structures in three dimensions (Jmol website).
Supplementary Material
Acknowledgments
We thank S. Kohl and C. Hamel for providing DNA samples of individuals with pattern dystrophies, J. Hansen for help with animal care, and JAX Scientific Services, including Genome Technologies, Histopathology Sciences and Imaging Sciences. This research was supported by Foundation Fighting Blindness Center Grant C-GE-0811-0548-RAD04 to the Radboud University Nijmegen Medical Center, Netherlands Organization for Scientific Research Vidi Innovational Research Award 016.096.309 to A.I.d.H., Nederlandse Oogonderzoek Stichting and Diana Hermens Stichting awards to C.B.H., Research Foundation – Flanders grant to E.D.B. and B.P.L., FWO Flanders grant 3G079711 to E.D.B., Belgian Science Policy Office Interuniversity Attraction Poles programme P7/43 award to E.D.B. and B.P.L., Netherlands Organization for Scientific Research Vici Innovational Research Award 865.12.005 to R.R., the Foundation Fighting Blindness (C-CMM-0811-0546-RAD02) to R.R., a U.S. Veterans Administration Medical Research Service grant to N.S.P., a Foundation Fighting Blindness Center Grant to the Cole Eye Institute, Cleveland Clinic, an unrestricted award from Research to Prevent Blindness to the Department of Ophthalmology, Cleveland Clinic Lerner College of Medicine, U.S. National Institutes of Health (NIH) and National Eye Institute grant EY016501 to P.M.N., and NIH National Cancer Institute award P30CA034196 to JAX.
Footnotes
URLS: BioGPS website: www.BioGPS.org
EVS website: http://evs.gs.washington.edu/EVS/
Jmol website: http://www.jmol.org
Accession codes: Information about ENU-induced Ctnna1-mutant mice can be accessed at https://www.jax.org/strain/021610. Whole-exome sequencing data have not been deposited into a public database as patients did not provide their consent for these data sets to be publicly released.
Author Contributions: N.T.M.S., M.P.K., P.M.N. and A.I.d.H. wrote the manuscript.
N.T.M.S., S.A.-L., E.D.B, S.W., S.B., F.S., F.P.M.C., C.J.F.B., B.P.L. and C.B.H. performed clinical examinations in patients and families and/or provided patient samples.
M.P.K., W.H., L.S., L.R., G.B.C. and J.R.C. performed genetic studies, live imaging, morphological studies and expression analysis in Ctnna1tvrm5 mice.
F.E.S.-K. and T.W.v.M. performed CTNNA1 mutation analysis in patients and families.
M.Y. and N.S.P. performed electrophysiology in Ctnna1tvrm5 mice.
S.J.L. and R.R. performed co-immunoprecipitations of CTNNA1 with VCL.
K.N. provided bioinformatic support of whole exome sequencing experiments in family A.
M.P.K., C.B.H., P.M.N. and A.I.d.H. supervised the work.
Conflict of interest: The authors have no proprietary or commercial interest in any materials discussed in this article.
References
- 1.Boon CJ, et al. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Retin Eye Res. 2008;27:213–235. doi: 10.1016/j.preteyeres.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A. Gass' Atlas of Macular diseases. 5th. Elsevier; 2012. pp. 254–266. [Google Scholar]
- 3.Deutman AF, van Blommestein JD, Henkes HE, Waardenburg PJ, Solleveld-van Driest E. Butterfly-shaped pigment dystrophy of the fovea. Arch Ophthalmol. 1970;83:558–569. doi: 10.1001/archopht.1970.00990030558006. [DOI] [PubMed] [Google Scholar]
- 4.Fossarello M, et al. Deletion in the peripherin/RDS gene in two unrelated Sardinian families with autosomal dominant butterfly-shaped macular dystrophy. Arch Ophthalmol. 1996;114:448–456. doi: 10.1001/archopht.1996.01100130444016. [DOI] [PubMed] [Google Scholar]
- 5.Pinckers A. Patterned dystrophies of the retinal pigment epithelium. A review Ophthalmic Paediatr Genet. 1988;9:77–114. doi: 10.3109/13816818809031483. [DOI] [PubMed] [Google Scholar]
- 6.Prensky JG, Bresnick GH. Butterfly-shaped macular dystrophy in four generations. Arch Ophthalmol. 1983;101:1198–1203. doi: 10.1001/archopht.1983.01040020200005. [DOI] [PubMed] [Google Scholar]
- 7.Nichols BE, et al. Butterfly-shaped pigment dystrophy of the fovea caused by a point mutation in codon 167 of the RDS gene. Nat Genet. 1993;3:202–207. doi: 10.1038/ng0393-202. [DOI] [PubMed] [Google Scholar]
- 8.van Lith-Verhoeven JJ, Cremers FP, van den Helm B, Hoyng CB, Deutman AF. Genetic heterogeneity of butterfly-shaped pigment dystrophy of the fovea. Mol Vis. 2003;9:138–143. [PubMed] [Google Scholar]
- 9.Marano F, Deutman AF, Aandekerk AL. Butterfly-shaped pigment dystrophy of the fovea associated with subretinal neovascularization. Graefes Arch Clin Exp Ophthalmol. 1996;234:270–274. doi: 10.1007/BF00430421. [DOI] [PubMed] [Google Scholar]
- 10.Grover S, Fishman GA, Stone EM. Atypical presentation of pattern dystrophy in two families with peripherin/RDS mutations. Ophthalmology. 2002;109:1110–1117. doi: 10.1016/s0161-6420(02)01029-1. [DOI] [PubMed] [Google Scholar]
- 11.Nichols BE, et al. A 2 base pair deletion in the RDS gene associated with butterfly-shaped pigment dystrophy of the fovea. Hum Mol Genet. 1993;2:1347. doi: 10.1093/hmg/2.8.1347-a. [DOI] [PubMed] [Google Scholar]
- 12.Richards SC, Creel DJ. Pattern dystrophy and retinitis pigmentosa caused by a peripherin/RDS mutation. Retina. 1995;15:68–72. doi: 10.1097/00006982-199515010-00013. [DOI] [PubMed] [Google Scholar]
- 13.Vaclavik V, Tran HV, Gaillard MC, Schorderet DF, Munier FL. Pattern dystrophy with high intrafamilial variability associated with Y141C mutation in the peripherin/RDS gene and successful treatment of subfoveal CNV related to multifocal pattern type with anti-VEGF (ranibizumab) intravitreal injections. Retina. 2012;32:1942–1949. doi: 10.1097/IAE.0b013e31824b32e4. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, et al. A novel RDS/peripherin gene mutation associated with diverse macular phenotypes. Ophthalmic Genet. 2004;25:133–145. doi: 10.1080/13816810490514388. [DOI] [PubMed] [Google Scholar]
- 15.Zhang K, Garibaldi DC, Li Y, Green WR, Zack DJ. Butterfly-shaped pattern dystrophy: a genetic, clinical, and histopathological report. Arch Ophthalmol. 2002;120:485–490. doi: 10.1001/archopht.120.4.485. [DOI] [PubMed] [Google Scholar]
- 16.den Hollander AI, et al. Identification of novel locus for autosomal dominant butterfly shaped macular dystrophy on 5q21.2-q33.2. J Med Genet. 2004;41:699–702. doi: 10.1136/jmg.2004.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Peachey NS, Marmorstein AD. Light-evoked responses of the mouse retinal pigment epithelium. J Neurophysiol. 2004;91:1134–1142. doi: 10.1152/jn.00958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaughan DK, Fisher SK. The distribution of F-actin in cells isolated from vertebrate retinas. Exp Eye Res. 1987;44:393–406. doi: 10.1016/s0014-4835(87)80173-2. [DOI] [PubMed] [Google Scholar]
- 19.Kobielak A, Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 21.Sandig M, Kalnins VI. Subunits in zonulae adhaerentes and striations in the associated circumferential microfilament bundles in chicken retinal pigment epithelial cells in situ. Exp Cell Res. 1988;175:1–14. doi: 10.1016/0014-4827(88)90250-9. [DOI] [PubMed] [Google Scholar]
- 22.Rangarajan ES, Izard T. Dimer asymmetry defines alpha-catenin interactions. Nat Struct Mol Biol. 2013;20:188–193. doi: 10.1038/nsmb.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provost E, Rimm DL. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr Opin Cell Biol. 1999;11:567–572. doi: 10.1016/s0955-0674(99)00015-0. [DOI] [PubMed] [Google Scholar]
- 24.Peng X, Maiers JL, Choudhury D, Craig SW, DeMali KA. alpha-Catenin uses a novel mechanism to activate vinculin. J Biol Chem. 2012;287:7728–7737. doi: 10.1074/jbc.M111.297481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huveneers S, et al. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J Cell Biol. 2012;196:641–652. doi: 10.1083/jcb.201108120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sprecher E, et al. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nat Genet. 2001;29:134–136. doi: 10.1038/ng716. [DOI] [PubMed] [Google Scholar]
- 27.Mason JO, 3rd, Patel SA. A case of hypotrichosis with juvenile macular dystrophy. Retin Cases Brief Rep. 2015;9:164–167. doi: 10.1097/ICB.0000000000000127. [DOI] [PubMed] [Google Scholar]
- 28.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Lewis B, Moran A, Xie T. Cadherin-mediated cell adhesion is critical for the closing of the mouse optic fissure. PLoS One. 2012;7:e51705. doi: 10.1371/journal.pone.0051705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell M, et al. Aberrant retinal tight junction and adherens junction protein expression in an animal model of autosomal dominant Retinitis pigmentosa: the Rho(-/-) mouse. Exp Eye Res. 2006;83:484–492. doi: 10.1016/j.exer.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 32.Torres M, et al. An alpha-E-catenin gene trap mutation defines its function in preimplantation development. Proc Natl Acad Sci USA. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams MA, Craig D, Passmore P, Silvestri G. Retinal drusen: harbingers of age, safe havens for trouble. Age Ageing. 2009;38:648–654. doi: 10.1093/ageing/afp136. [DOI] [PubMed] [Google Scholar]
- 34.Hammond CJ, et al. Genetic influence on early age-related maculopathy: a twin study. Ophthalmology. 2002;109:730–736. doi: 10.1016/s0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 35.Klein R, et al. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114:253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 36.Klein R, Klein BE, Tomany SC, Meuer SM, Huang GH. Ten-year incidence and progression of age-related maculopathy: The Beaver Dam eye study. Ophthalmology. 2002;109:1767–1779. doi: 10.1016/s0161-6420(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 37.Rudolf M, et al. Prevalence and morphology of druse types in the macula and periphery of eyes with age-related maculopathy. Invest Ophthalmol Vis Sci. 2008;49:1200–1209. doi: 10.1167/iovs.07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohn EH, et al. Comparison of drusen and modifying genes in autosomal dominant radial drusen and age-related macular degeneration. Retina. 2015;35:48–57. doi: 10.1097/IAE.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai H, Kalnins VI. Normally occurring loss of single cells and repair of resulting defects in retinal pigment epithelium in situ. Exp Eye Res. 1996;62:55–61. doi: 10.1006/exer.1996.0007. [DOI] [PubMed] [Google Scholar]
- 40.Longbottom R, et al. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci USA. 2009;106:18728–18733. doi: 10.1073/pnas.0902593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia H, Krebs MP, Kaushal S, Scott EW. Enhanced retinal pigment epithelium regeneration after injury in MRL/MpJ mice. Exp Eye Res. 2011;93:862–872. doi: 10.1016/j.exer.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakoczy PE, et al. Progressive age-related changes similar to age-related macular degeneration in a transgenic mouse model. Am J Pathol. 2002;161:1515–1524. doi: 10.1016/S0002-9440(10)64427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarks JP, Sarks SH, Killingsworth MC. Evolution of geographic atrophy of the retinal pigment epithelium. Eye (Lond) 1988;2(Pt 5):552–577. doi: 10.1038/eye.1988.106. [DOI] [PubMed] [Google Scholar]
- 44.Hawes NL, et al. Retinal degeneration 6 (rd6): a new mouse model for human retinitis punctata albescens. Invest Ophthalmol Vis Sci. 2000;41:3149–3157. [PubMed] [Google Scholar]
- 45.Fogerty J, Besharse JC. 174delG mutation in mouse MFRP causes photoreceptor degeneration and RPE atrophy. Invest Ophthalmol Vis Sci. 2011;52:7256–7266. doi: 10.1167/iovs.11-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ach T, et al. Lipofuscin redistribution and loss accompanied by cytoskeletal stress in retinal pigment epithelium of eyes with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:3242–3252. doi: 10.1167/iovs.14-16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zanzottera EC, Messinger JD, Ach T, Smith RT, Curcio CA. Subducted and melanotic cells in advanced age-related macular degeneration are derived from retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2015;56:3269–3278. doi: 10.1167/iovs.15-16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patil H, et al. Selective impairment of a subset of Ran-GTP-binding domains of ran-binding protein 2 (Ranbp2) suffices to recapitulate the degeneration of the retinal pigment epithelium (RPE) triggered by Ranbp2 ablation. J Biol Chem. 2014;289:29767–29789. doi: 10.1074/jbc.M114.586834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, et al. Alpha-catenins control cardiomyocyte proliferation by regulating yap activity. Circ Res. 2015;116:70–79. doi: 10.1161/CIRCRESAHA.116.304472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlegelmilch K, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saksens NT, et al. Dominant cystoid macular dystrophy. Ophthalmology. 2015;122:180–191. doi: 10.1016/j.ophtha.2014.07.053. [DOI] [PubMed] [Google Scholar]
- 53.Svenson KL, Bogue MA, Peters LL. Invited review: Identifying new mouse models of cardiovascular disease: a review of high-throughput screens of mutagenized and inbred strains. J Appl Physiol 1985. 2003;94:1650–1659. doi: 10.1152/japplphysiol.01029.2003. discussion 1673. [DOI] [PubMed] [Google Scholar]
- 54.Won J, et al. Translational vision research models program. Adv Exp Med Biol. 2012;723:391–397. doi: 10.1007/978-1-4614-0631-0_50. [DOI] [PubMed] [Google Scholar]
- 55.Xin-Zhao Wang C, Zhang K, Aredo B, Lu H, Ufret-Vincenty RL. Novel method for the rapid isolation of RPE cells specifically for RNA extraction and analysis. Exp Eye Res. 2012;102:1–9. doi: 10.1016/j.exer.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nordgard O, Kvaloy JT, Farmen RK, Heikkila R. Error propagation in relative real-time reverse transcription polymerase chain reaction quantification models: the balance between accuracy and precision. Anal Biochem. 2006;356:182–193. doi: 10.1016/j.ab.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Low BE, et al. Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci. 2014;55:387–395. doi: 10.1167/iovs.13-13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakamoto K, McCluskey M, Wensel TG, Naggert JK, Nishina PM. New mouse models for recessive retinitis pigmentosa caused by mutations in the Pde6a gene. Hum Mol Genet. 2009;18:178–192. doi: 10.1093/hmg/ddn327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu M, et al. Age-related changes in visual function in cystathionine-beta-synthase mutant mice, a model of hyperhomocysteinemia. Exp Eye Res. 2012;96:124–131. doi: 10.1016/j.exer.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roepman R, et al. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc Natl Acad Sci U S A. 2005;102:18520–18525. doi: 10.1073/pnas.0505774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.