Abstract
Polyubiquitination, a critical protein post-translational modification, signals for a diverse set of cellular events via the different isopeptide linkages formed between C-terminus of one ubiquitin (Ub) and the ε-amine of K6, K11, K27, K29, K33, K48, or K63 of a second Ub. We assembled di-ubiquitins (Ub2) comprised of every lysine linkage and examined them biochemically and structurally. Of these, K27-Ub2 is unique as it is not cleaved by most deubiquitinases. As this remains the only structurally uncharacterized lysine-linkage, we comprehensively examined the structures and dynamics of K27-Ub2 using NMR, small-angle neutron scattering, and in silico ensemble modeling. Our structural data provide insights into functional properties of K27-Ub2, in particular, that K27-Ub2 may be specifically recognized by K48-selective receptor UBA2 domain from proteasomal shuttle protein hHR23a. Binding studies and mutagenesis confirmed this prediction, further highlighting structural/recognition versatility of polyubiquitins and the potential power of determining function from elucidation of conformational ensembles.
Introduction
Polyubiquitination is undoubtedly one of the most important post-translational modifications of proteins in eukaryotes. Aside from well-known roles in targeting substrates for proteasomal degradation and DNA repair, polyubiquitin (polyUb) chains also signal for critical cellular processes including cell cycle regulation, immunity, mitochondrial protein degradation, and even mRNA stability (Dikic and Dotsch, 2009; Pickart and Fushman, 2004). The incredible diversity of signaling outcomes stems from the ability of ubiquitin (Ub) to form chains via isopeptide linkages between the ε-NH2 group of any of the seven lysines (K6, K11, K27, K29, K33, K48, K63) on Ub and the C-terminus of a second Ub; importantly, all of these linkages are present in the cell at varying levels of abundance(Xu et al., 2009). It has been hypothesized that each of these different linkages imparts unique structural and dynamical properties on a polyUb chain, enabling the chains to be recognized differently by downstream receptor proteins (Pickart and Fushman, 2004). The well-understood functions of polyUb in proteasomal degradation and DNA repair are mediated by chains composed of K48 and K63 linkages, respectively. The cellular functions of the other, so-called non-canonical polyUb chains (linked via K6, K11, K27, K29, K33) are substantially less clear and are the focus of current research efforts.
Ubiquitination with K6-linked chains occurs on the BRCA1-BARD1 Ub ligase and its associated substrates, suggesting a link to DNA repair processes (Wu-Baer et al., 2010). K11-polyUb may interact with the Npl4 adaptor protein during Drosophila development, among other roles associated with the mitotic phase in the cell cycle and ERAD (Meyer and Rape, 2014; Zhang et al., 2013). Very recent findings revealed mostly non-proteolytic roles for K27-, K29-, and K33-polyUbs. Ubiquitination with K33-linked chains regulates T-cell receptor-ζ function by governing its phosphorylation and protein binding profiles (Huang et al., 2010). K33-polyubiquitination also contributes to the stabilization of actin for post-Golgi transport (Yuan et al., 2014). K29-polyUbs participate in growth and development-associated pathways (Wnt/β-catenin signaling), and are also implicated in regulation of mRNA stability via recognition by the adaptor protein UBXD8 (Fei et al., 2013; Zhou et al., 2013). K27-linked chains are observed on mitochondrial trafficking protein Miro1 and slow down its degradation by the proteasome, therefore acting as a marker of mitochondrial damage (Birsa et al., 2014). K27- and K33-polyUb chains are also implicated in regulation of innate immunity (Arimoto et al., 2010; Birsa et al., 2014; Cao et al., 2015; Liu et al., 2014).
Biochemical and structural studies of non-canonical polyUb chains have been hampered by the lack of linkage-specific Ub-conjugating enzymes needed to make the non-canonical linkages. However, recent studies have uncovered linkage-specific enzymes for K6 and K11, and structures have been determined for these chains (Bremm et al., 2010; Castañeda et al., 2013; Hospenthal et al., 2013; Matsumoto et al., 2010). Linkage semi-selective E3s combined with linkage-specific deubiquitinases (DUBs) were utilized to study K29- and K33-linked chains (Kristariyanto et al., 2015; Michel et al., 2015). To our knowledge to date, no structures are yet available for K27-linked polyUb chains. Importantly, the dynamic nature of polyUb chains necessitates that they be characterized in solution (Castañeda et al., 2013).
Significant progress in chemical biology and incorporation of unnatural amino acids has permitted the development of chemical, non-enzymatic assembly methods (reviewed in (Hemantha and Brik, 2013)) including a strategy we developed that utilizes mutually orthogonal removable amine-protecting groups Alloc and Boc (Castañeda et al., 2011a). Using this strategy, here we made Ub2s consisting of every non-canonical Ub linkage (K6, K11, K27, K29, and K33) and examined them by NMR spectroscopy and biochemical assays. We found that of all the chains, K27-Ub2 stood out in that it exhibited the largest spectral perturbations and resisted deubiquitination. We therefore employed NMR, small-angle neutron scattering (SANS), and computational modeling to characterize the conformational ensemble and dynamics of K27-Ub2. We uncovered unique dynamical and functional properties of K27-Ub2 chains which set them apart from all other previously-characterized Ub2s. Furthermore, the structural features of K27-Ub2 suggested unexpected binding preferences of this chain, which were verified experimentally.
Results
Deubiquitination assays reveal uniqueness of K27 among all isopeptide linkages
Fully natural K6-, K11-, K27-, K29-, K33-, and K48-Ub2s with native isopeptide linkages and free of any mutations (Fig S1A) were assembled using nonenzymatic method (Castañeda et al., 2011a). K48-Ub2 (for some measurements) and K63-Ub2 were made enzymatically employing chain-terminating mutations (Varadan et al., 2004; Varadan et al., 2002). To assess whether deubiquitinases (DUBs) are able to disassemble Ub chains of non-canonical linkages (K6, K11, K27, K29, and K33), each Ub2 was screened against six DUBs, representing different DUB families, including Cezanne, OTUB1, AMSH, USP2, USP5 (IsoT), and Ubp6 (Fig 1). As expected, K11-specific Cezanne preferentially cleaved K11-Ub2 chains, while K48-specific OTUB1 and K63-specific AMSH selectively cleaved K48- and K63-Ub2 chains, respectively. Strikingly, linkage non-specific USP2, USP5, and Ubp6 were unable to disassemble K27-Ub2 at all. K27 was the only linkage that resisted cleavage by USP5. USP2 and the yeast-proteasome-associated DUB Ubp6, were also less effective at cleaving K29-Ub2. K27-Ub2 also resisted disassembly by reconstituted proteasome lid core complex containing Rpn11 (Yu et al., 2015). Furthermore, due to its resistance to cleavage by DUBs, K27-Ub2 can act as a competitive inhibitor of DUB activity towards other linkages (Fig S1). These observations that a wide range of DUBs had difficulty in processing K27-Ub2 compared to all other Ub2s, inspired us to investigate the structural and dynamical properties of this chain with the goal of understanding what makes the K27-linkage differ from other linkages.
Figure 1.
Disassembly of Lys-linked Ub2s by various linkage-specific (A) and promiscuous (B) deubiquitinases (DUBs). SDS-PAGE gels showing products after treatment of all Lys-linked Ub2s with indicated DUBs for 24 hours or overnight (O/N) at 30°C. Chain disassembly is evident from appearance of a monoUb band. Note that K27-Ub2 is not reduced to monoUb by any DUB tested here. (See also Fig S1)
K27-Ub2 exhibits no noncovalent interdomain contacts
We employed solution NMR spectroscopy to attain atom-specific information for each Ub2 (Castañeda et al., 2016). 1H-15N NMR spectra were collected separately for each Ub unit (uniformly 15N-enriched) in K6, K11, K27, K29, K33, and K48-Ub2. It turns out that each Ub2 has a distinct NMR spectroscopic signature, and even 1D 1H NMR spectra can be used to distinguish Ub2s of different Lys linkages (Fig S2).
To distinguish between the two Ub units in Ub2, the Ub whose C-terminus participates in the isopeptide linkage is termed ‘distal’, while Ub that contributes the lysine side chain to the isopeptide bond is termed ‘proximal’ (Fig S2). Differences in NMR spectra between the distal or proximal Ub and monoUb were quantified as amide chemical shift perturbations (CSPs) (Fig 2A). In all Ub2 chains, the largest CSPs were observed for C-terminal residues 74-76 of the distal Ub, reflecting chemical modifications that accompany formation of the isopeptide bond. The other residues in the distal Ub are not directly affected by the chemical bonding to the proximal Ub. Therefore, we used CSPs in the distal Ub as indicators of noncovalent interactions between the Ub units in each Ub2. Our results suggest that, except for K6- and K48-Ub2, the noncovalent interdomain contacts in the rest of Ub2s are weak or transient, but nevertheless involve the hydrophobic surface patch (residues L8, I44, V70 (Beal et al., 1996)) of the distal Ub (Castañeda et al., 2016). Notably, the distal Ub in K27-Ub2 exhibited the smallest CSPs of all the Ub2s studied.
Figure 2.
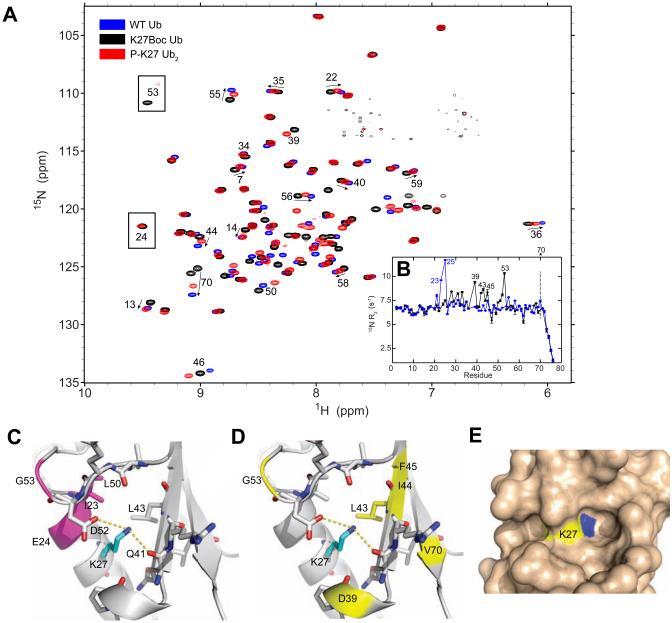
NMR characterization of the Ub2 chains. (A) CSPs (Δδ) for the distal and proximal Ubs in each Ub2 versus monoUb as a function of residue number. For each Ub2, the isopeptide linkage is indicated by arrows connecting the C-terminus of the distal Ub with the target lysine of the proximal Ub. In the third column, CSPs represent spectral differences between the corresponding Lys(Boc) variant and WT monoUb. The structure of Lys(Boc) is shown on the top. (B) CSPs for Lys(Boc) vs. WT Ub are mapped on the solution structure of monoUb (PDB ID 1D3Z); CSPs > 0.04 ppm and > 0.10 ppm are shown in orange and red, respectively. The modified lysine is shown in blue. The structure of Lys(Boc) is shown at the top; the part that mimics the isopeptide bond is circled in red. (See also Fig S2)
In contrast to the distal Ub, the proximal Ub of K27-Ub2 showed strong CSPs (Fig 2A), which were the largest and most widespread CSPs among all Ub2s. The presence of spectral perturbations in only one of the two Ub units within a chain was puzzling and triggered additional examination. Generally, CSPs in the proximal Ub could be caused by: (1) noncovalent interactions with the distal Ub, (2) alteration in the electronic microenvironment arising from chemical modification of the isopeptide-linked lysine, and/or (3) changes to the 3-D structure of the proximal Ub. We found that for most of the chains (except for K6- and K48-Ub2) the proximal Ub CSPs could be replicated in a monoUb variant containing Lys(Boc), a lysine derivative whose bond chemistry and charges at the Nζ mimic the isopeptide bond. This suggests that the large CSPs in the proximal Ub of these chains result primarily from the covalent (isopeptide) bonding and not from noncovalent interdomain interactions.
Structural basis for the observed effects of K27 modifications
Despite the large spectral differences between K27(Boc) Ub or the proximal Ub of K27-Ub2 and WT Ub (Fig 3A), chemical shift index analyses indicate that the secondary structure of Ub was unaffected by the K27 modifications (Fig S3).
Figure 3.
The effects of K27 modification. (A) Superimposition of 1H-15N TROSY-HSQC spectra of the proximal Ub of K27-Ub2 (red), K27(Boc) Ub (black), and WT Ub (blue). Practically the same residues show perturbations in the proximal Ub of K27-Ub2 and in K27(Boc) Ub. (B) Comparison of 15N R2 rates for WT Ub (blue) and K27(Boc) Ub (black). The R2 of V70 in K27(Boc) Ub could not be accurately quantified (dotted line) because its signal was severely exchange-broadened beyond the first time point in the experiment. (C, D) Structural microenvironment of K27 in Ub (PDB ID 1D3Z). Residues that either exhibit elevated 15N R2s or are exchange-broadened beyond detection are highlighted in (C) for WT Ub, colored magenta, and in (D) for K27(Boc) Ub (and the proximal Ub of K27-Ub2), colored yellow. Putative hydrogen bonds between K27 and the side chains of Q41 and D52 are marked with yellow lines. (E) Surface representation highlighting the low solvent accessibility of the K27 side chain in Ub. The Nζ atom is colored blue. (See also Fig S3)
We were surprised to observe the amide signals of E24 and G53 in the NMR spectra of K27(Boc) Ub and the proximal Ub of K27-Ub2, as these signals are typically not present in Ub spectra at these conditions (Fig 3A). Given this observation, we inspected backbone dynamics first in the K27(Boc) Ub variant by measuring 15N transverse relaxation rates (R2) for all backbone amides (Fig 3B). In WT Ub, only residues I23 and N25 exhibit elevated 15N R2s, with signals of E24 and G53 exchange-broadened beyond detection. These residues are located either on the adjacent (to K27) turn of the α helix or in close spatial proximity (G53). The backbone dynamics in K27(Boc) Ub are drastically different (Fig 3B,D), with residues 23-25 exhibiting “normal” 15N R2s, while amides in the C-terminal part of the α helix, as well as in residues 39, 43-45, 51-53, and 70 all exhibiting elevated 15N R2s and large CSPs. These observations point to changes in the local dynamics on the μs-ms timescale in the vicinity of K27 in K27(Boc) Ub. As shown in the next section, these observations extend to the proximal Ub of K27-Ub2.
What is the structural basis for the strong effect of K27 modifications? A detailed inspection of the microenvironment and contacts of K27 in WT Ub (Fig 3C,D,E) shows that in stark contrast with other lysines in Ub, which are solvent exposed, K27 is almost entirely buried (its solvent accessibility is < 5% of that for Lys in a Gly-Lys-Gly tripeptide). Of all lysines in Ub, the ε-amine of K27 participates in the most hydrogen bonds (with Q41 and E52) and is the least mobile (the ε–NH3+ has the highest S2axis order parameter, 0.71) (Esadze et al., 2011; Huang et al., 2014). The CH2-rich component of the K27 side chain contributes significantly to Ub's hydrophobic core, through contacts with I23, P38, Q41, and L43. We surmise that, given K27's microenvironment, the neutralization of the positive charge of its ε-amine stemming from either the K27(Boc) substitution or isopeptide bond formation has significant impact on the chemical shifts of nearby residues. Furthermore, the concomitant perturbations to the interactions involving K27 side chain would account for the changes in dynamics observed for K27(Boc) Ub and the proximal Ub of K27-Ub2.
Spin-relaxation measurements reveal unique dynamical properties of K27-Ub2
The 15N R1 and R2 rates and 1H-15N steady-state hetNOEs for both Ubs in K27-Ub2, are generally comparable with those for K11-Ub2 and K48-Ub2 (Fig 4). The average longitudinal relaxation time T1 (=1/R1) for secondary structure residues is 716 ± 50 ms for both Ubs, consistent with the expected T1 for a molecular species of 17-20 kDa (Varadan et al., 2005) and suggesting that the two Ub units in K27-Ub2 tumble together as a single entity rather than independent ‘beads on a flexible string’.
Figure 4.
Comparison of backbone dynamics in K11-, K27-, and K48-Ub2 chains. (A) 15N relaxation rates, R1 and R2, {1H}-15N hetNOEs, and squared order parameters (S2) for each residue in the distal (left) and proximal (right) Ub unit in K11-Ub2 (green), K27-Ub2 (black), and K48-Ub2 (magenta). Relaxation data for K11-Ub2 and K48-Ub2 chains have been reported in (Castañeda et al., 2013) and (Varadan et al., 2002), respectively. Panels B-F highlight unique features of K27-Ub2 relaxation data. Note near-‘normal’ 15N R2 values for residues 24-25 (panel C) and increased 15N R2 for residues 39, 43-45, 53 and 70 in the proximal Ub of K27-Ub2 (panel D), and elevated hetNOE values for the C-terminus of the distal Ub in K27-Ub2 (panel E). Notably, residues 53 and 70 in the proximal Ub exhibit drastically increased 15N R2 values of 26 s−1 and 21 s−1, respectively. (Contributions to 15N R2 from conformational exchange are shown in Fig S4.)
As in other Ub2s, high order parameters (S2 > 0.8) revealed that most residues in both Ubs of K27-Ub2 are well structured, while near-zero or negative hetNOE values and low order parameters (S2~0) were only observed in the C-terminal residues of the proximal Ub, consistent with high flexibility of Ub’s free C-terminus (Fushman et al., 2004). These data suggest that the K27-linkage did not result in new large-amplitude backbone motions on the ps-ns timescale.
In general, compared to the proximal Ub, the C-terminus of the distal Ub in K11-, K27-, and K48-Ub2 is considerably rigidified (hetNOE > 0.35) as a result of its tethering to the proximal Ub. In contrast to other Ub2s, C-terminal residues of the distal Ub in K27-Ub2 exhibit 15N R2 rates similar to those in well-structured regions, and G75 and G76 have elevated (~0.55) hetNOE values (Fig 4B, E). These data indicate that the C-terminus of the distal Ub in K27-Ub2 is more ordered than in the other Ub2s. Given the confined environment and almost negligible solvent accessibility of K27 (see above) and elevated order parameters for the distal Ub’s C-terminus, the interdomain mobility of K27-Ub2 is likely significantly restricted compared to K11- and K48-Ub2.
The 15N R2 pattern in the proximal Ub was also distinct from K11- and K48-Ub2 or WT monoUb but very similar to that seen for the K27(Boc) monoUb variant (compare Fig 4C,D with Fig 3B). As in K27(Boc) Ub, residues 23-25 showed R2 values similar to the average R2 (Fig 4C), while elevated R2 values were observed for residues 39, 43-45, and especially for 53 and 70 (Fig 4D), due to significant contributions from conformational exchange (Fig S4). These data imply increased rigidity for the helical turn immediately preceding K27 in the proximal Ub of K27-Ub2 and increased μs-ms motions in the β-strand opposite K27.
Each Ub unit retains its three-dimensional structure in K27-Ub2
In light of the substantial amide CSPs and conformational exchange in the proximal Ub, we assessed the structure of each Ub unit in K27-Ub2 by measuring 15N-1H residual dipolar couplings (RDCs) (Fig S5). Using the NMR structure of monoUb (PDB ID 1D3Z) and residues belonging to structured regions, we determined the alignment tensor for each Ub unit in K27-Ub2 (Table S1). For both Ubs there was excellent agreement between experimental RDCs and those back-calculated from the monoUb structure using the derived alignment tensor (Fig 5A). Importantly, these results indicate that the 3-D structure of the Ub units is unaffected by the isopeptide linkage at K27.
Figure 5.
Agreement with experimental data. (A) Agreement between experimental and back-calculated values of RDCs (A) or relaxation rates ratio ρ (B) for secondary structure residues for the distal (left panel) or proximal (right panel) Ubs in K27-Ub2, analyzed separately. Solution structure of monoUb (PDB ID 1D3Z) was used for each Ub. (C, D) Agreement for both Ub units analyzed together, between experimental values of RDCs (C) or ρ (D) and those back-calculated from the angle-optimized single-structure representations of K27-Ub2 (see Fig 6A,B). Data for distal and proximal Ubs are colored blue and red, respectively. Dashed line corresponds to absolute agreement. Pearson’s correlation coefficient (r) and quality factors (Q) are indicated. Lower Q means better agreement. (E) L-curve analysis of the ensemble size for RDC data of K27-Ub2 and K48-Ub2 chains using SES. Red squares denote the minimum number of conformers needed to reproduce experimental RDC data. The dashed line represents the relative error for the best possible ensemble solution of size > 0. (F) Agreement between experimental RDCs and RDCs predicted for 1-, 2-, and 3-conformer ensembles. (See also Fig S5)
Average “single-snapshot" structures of K27-Ub2 from RDCs and 15N relaxation data
The similarity between the distal and the proximal Ubs in the overall range of RDCs (Fig S5) and in the alignment tensors (Table S1) suggests that the two Ubs units in K27-Ub2 orient together essentially as a single entity. We therefore used RDCs and 15N relaxation data to determine single-structure representations of K27-Ub2 (Fushman et al., 2004). The RDC-derived structure that best reproduces the RDC data for both Ub units taken together (Fig 6A) provides a strong agreement between the experimental and back-calculated RDCs (Fig 5C), with the r (0.99) and Q (0.10) values nearly as good as for the individual Ub units in K27-Ub2 (Fig 5A, Table 1). This suggests that, despite the lack of major interfacial Ub/Ub contacts, a single-structure representation is capable of capturing some major RDC-relevant features of interdomain orientations in K27-Ub2. This is in stark contrast with K11-Ub2 and K48-Ub2, where a single RDC-derived structure could not reproduce well the experimental RDCs for both Ubs taken together (Berlin et al., 2013; Castañeda et al., 2013). Combined with the elevated hetNOEs and order parameters of the distal Ub C-terminus, these results imply that K27-Ub2 is the least flexible of the Ub2s studied thus far.
Figure 6.
Structures of K27-Ub2. (A,B) Best single-structure representations from RDC and 15N-relaxation data. Distal Ub is on the left, proximal is on the right. The backbone of Ub is shown as ribbon, the yellow spheres represent side chains of the hydrophobic patch residues L8, I44, V70. For each Ub, red, blue, and yellow sticks represent x, y, and z axes, respectively, of the alignment (A) or diffusion (B) tensor of Ub2 (Table S1). The structures were determined using PATIDOCK (Berlin et al., 2010) (A) or ELMDOCK (Berlin et al., 2011) (B). Two structures are shown for each (differing by 180° rotation of the proximal Ub) due to orientational degeneracy of RDCs and relaxation rates (Fushman et al., 2004). (C) Single-conformer structures identified by SES from the SASSIE ensemble that are in best agreement with RDC data. (D) Three color-coded clusters of population-weighted two-conformer ensembles of K27-Ub2 in agreement with experimental RDC data. Up to four ensembles (superimposed by the distal Ub) are shown per cluster to illustrate the convergence of solutions. The numbers indicate the relative weights of the conformers. (E) Agreement between experimental SANS data (black dots) and calculated (lines) using Xtal2sas (Curtis et al., 2012) for the conformational ensembles shown in D. Calculated data are for one representative ensemble from each cluster, color-coded according to the corresponding cluster. (F) Previously-determined complexes of K48- and K63-Ub2 with their linkage-specific binding partners, UBA2 and Rap80, respectively. (See also Fig S6)
Table 1.
Agreement between K27-Ub2 structures and NMR data
| Structure or Conformational Ensemble | Data a | rb | Qc |
|---|---|---|---|
| RDC, aligned tensors d | RDC-SVD | 0.98 | 0.11 |
| RDC, angle-optimized e | RDC-SVD | 0.99 | 0.10 |
| 15N Relaxation, angle-optimized e | RDC-SVD | 0.95 | 0.22 |
| 15N Relaxation, aligned tensors d | Relaxation | 0.90 | 0.31 |
| 15N Relaxation, angle-optimized e | Relaxation | 0.91 | 0.29 |
| SES, 1-conformer ensemble | RDC-SVD | 0.98 | 0.12 |
| SES, 1-conformer ensemble | Pred-RDC | 0.97 | 0.16 |
| SES, 2-conformer ensemble | Pred-RDC | 0.99 | 0.07 |
For RDC-SVD, the agreement is calculated between experimental RDCs and RDCs back-calculated from NH vectors using SVD. For Pred-RDC, the agreement is calculated between experimental RDCs and RDCs predicted directly from structure using PATI(Berlin et al., 2009).
Pearson’s correlation coefficient.
Quality factor for RDCs (Clore and Garrett, 1999) or 15N relaxation data (Ghose et al., 2001). Lower Q means better agreement.
Structures labeled ‘aligned tensors’ were obtained by orienting the two Ubs such that the corresponding axes of the alignment tensors (RDC) or diffusion tensors (15N relaxation) reported by each Ub analyzed separately were parallel to each other (Fushman et al., 2004).
Structures labeled ‘angle-optimized’ were generated using ELMDOCK (Berlin et al., 2011) (for 15N relaxation data) or PATIDOCK (Berlin et al., 2010) (for RDC data) to obtain the best agreement between the experimental and predicted relaxation or RDC data for both Ubs taken together.
The single-structure representation of K27-Ub2 derived from 15N relaxation data (Fig 6B) also provides a good agreement between experimental and back-calculated data (ratio of relaxation rates, ρ) (Fig 5D, Table 1). Remarkably, essentially the same Euler angles (α, β) were obtained for the alignment and diffusion tensors (Table S1), indicating similar average interdomain orientations sensed by the RDCs and the 15N relaxation data. Indeed, the K27-Ub2 structures constructed from these two sets of data are very similar (Fig 6A,B) except for rotations about the γ angle (Table S1). As further evidence of the similarity between the two sets of structures, the RDC values back-calculated using the relaxation-derived structure are in good agreement with experimental RDCs (Table 1). The close agreement between the measurements of physically different phenomena (alignment vs. diffusion) provides strong support for the structural data obtained here.
Perhaps the most intriguing feature of the derived structures of K27-Ub2 (Fig 6A,B) is the positioning of the hydrophobic patches of the two Ub units, which could allow simultaneous interaction of both Ubs with a receptor. Curiously, some of these derived structures resemble the open conformation of K48-Ub2 in complex with the UBA2 domain of hHR23a (compare Figs 6B and 6F). It is surprising that the two chains can adopt similar conformations despite the substantial difference in the location of the conjugation site on the proximal Ub, namely, the β-sheet face in K48-Ub2 versus the α helix in K27-Ub2.
Conformational ensembles of K27-Ub2
Despite reduced interdomain mobility compared to other Ub2s, K27-Ub2 is not fully rigid. Given the transient nature of noncovalent interdomain contacts it is natural to anticipate some degree of interdomain mobility in this chain. Indeed, the backbone flexibility of the Ub-Ub linker (residues 72-76 of the distal Ub) is comparable to that in flexible loops (e.g. residues 7-11) (Fig 4A). In addition, although both Ubs reported similar τc values (Table S1), the actual range of the observed ρ values for the proximal Ub was noticeably narrower than for the distal Ub (Fig 5B). These observations point to averaging by interdomain motions on a time scale comparable to or faster than the overall tumbling.
We therefore asked whether considering multiple conformations of K27-Ub2 instead of a single structure could further improve agreement with our experimental data. We used the sparse ensemble selection (SES) method (Berlin et al., 2013; Castañeda et al., 2016), to determine representative conformational ensembles for K27-Ub2 from a starting set of 23000 sterically-allowed structures of the chain, generated in silico using SASSIE (Curtis et al., 2012) (Fig S6).
The SES analysis revealed that consideration of two conformers improves significantly the agreement between experimental and predicted RDCs for K27-Ub2 (Fig 5E). Only marginal improvement was seen beyond two conformers, therefore, we did not consider larger ensembles (Fig 5F). When a single-conformer ensemble is considered, the best agreement between experimental and predicted RDCs gives r = 0.97 with Q = 0.16 (see also Supplemental Information).
For the 2-conformer ensembles, the agreement between experimental and predicted RDC data for both distal and proximal Ubs taken together is as good as when Ubs are considered individually (compare Fig 5F with Fig 5A). Our analysis revealed at least three ensemble clusters, with the major conformer in each ensemble accounting for 63-70% of the total population (Fig 6D). Notably, the major conformer in all of these ensembles has interdomain orientation similar to the RDC-derived and 15N-relaxation derived structures, as well as the 1-conformer SES ensembles (Fig 6A,B). In all major conformers the two Ub units are oriented with their hydrophobic patches toward each other and at least 10 Å apart (indicative of the absence of a Ub/Ub interface, in line with almost negligible distal-Ub CSPs). These major conformers are related to each other by a 180° rotation of the proximal Ub about the horizontal axis. Similarly, the minor states of these ensembles are related by a 180° rotation about the vertical axis. Both cases are likely a consequence of the orientational degeneracy inherent in RDCs, i.e. the inability to distinguish directionality of the alignment tensor axes (z vs. −z, etc.) (Fushman et al., 2004). Therefore, we conclude that all three sets of conformational ensembles are indeed related to each other.
To further test the conformational ensembles determined by SES, we compared the population-weighted predicted SANS profiles for these ensembles with the experimental data for K27-Ub2. The overall agreement was good already for the 1-conformer solutions, and improved slightly for the 2-conformer ensembles (Figs 6E, S6). Thus we conclude that SANS data generally validate the conformational ensembles determined here.
Can K27-Ub2 recognize UBA2 in a manner similar to K48-Ub2?
Together, our structural and conformational analyses suggest that K27-Ub2 is capable of adopting a conformation where the hydrophobic patches of both Ubs face each other, for at least part of the time (Fig 6A-D). Interestingly, this arrangement is reminiscent of the conformation that K48-Ub2 adopts when it forms a ‘sandwich-like’ complex with the UBA2 domain of hHR23a (Fig 6F), in which the UBA2 interacts with the hydrophobic patches of both Ubs simultaneously (Varadan et al., 2005). Therefore we hypothesized that K27-Ub2 might interact with the UBA2 domain in a similar manner. To test this hypothesis we conducted NMR titration studies of K27-Ub2 binding to UBA2.
To our surprise, strong signal attenuations, indicative of tight binding, were observed in many residues (7, 8, 43, 46, 47, 48, 50, 68) in the proximal domain of K27-Ub2 early in the course of its titration with UBA2 (Fig 7A). This behavior is reminiscent of UBA2 binding to K48-Ub2 (Varadan et al., 2005). A comparison of UBA2-induced changes in K27-Ub2 spectra with those of K11-Ub2 and K48-Ub2 under nearly identical titration conditions revealed that the K27-Ub2 signals indeed behave more similar to K48-Ub2 than to K11-Ub2 signals. Just as in K27-Ub2, signals of residues 7, 8, 46, and 47 of the K48-Ub2's proximal Ub attenuated strongly during the course of the titration. Note in this regard that no signal attenuations were observed for K11-Ub2 or K63-Ub2 which showed weaker affinity for UBA2 (Castañeda et al., 2013; Varadan et al., 2004).
Figure 7.
UBA2 recognizes K27-Ub2. (A) Overlay of 1H-15N NMR spectra of the proximal Ub at three titration points (blue=start, yellow=middle, red=endpoint), for K11-Ub2, K27-Ub2, and K48-Ub2. (B) CSPs in the distal or proximal Ubs of K27-Ub2 at the endpoint of titration with unlabeled UBA2. Significant attenuations are denoted with gray bars. Note that none of the residues that attenuate in the proximal Ub exhibit conformational exchange in the ligand-free Ub2 (except for L43). CSPs were mapped (right) onto a SASSIE-generated K27-Ub2 conformation. (C) Titration curves for select residues in either the proximal Ub of K27-Ub2 or UBA2 as a function of total ligand concentration ([Lt]). Lines represent fits to 1:1 binding model. Initial analyte concentrations were 100 μM (K27-Ub2) and 30 μM (UBA2), respectively. (D) CSPs in UBA2 upon titration with unlabeled K27-Ub2, monoUb, K48-Ub2, or K11-Ub2. Residues uniquely perturbed by binding to K27-Ub2 are outlined in red. (E) CSPs were mapped onto the surface of UBA2 (PDB ID 1DVO). (See also Fig S7)
Spectral perturbations in K27-Ub2 upon titration with UBA2 are consistent with the involvement of the hydrophobic patch residues from both Ubs in the interactions with UBA2 (Fig 7B,C). Of importance are also large CSPs and signal attenuations in the C-terminus of the distal Ub, suggesting that the Ub-Ub linker participates in UBA2 binding (Fig S7H,I). Significant perturbations were also detected in several residues in the α helix and strands β3-β5 of the proximal Ub that were affected by formation of the K27 isopeptide linkage (Fig 2); these CSPs likely reflect a rearrangement around K27 and/or changes in local dynamics upon UBA2 binding.
On the UBA2 side (Fig 7D), a number of significant CSPs and signal attenuations were detected, supporting strong binding between UBA2 and K27-Ub2. Uniquely, UBA2 resonances for residues 344, 346, and 358 were perturbed in a manner similar to that when UBA2 was titrated with K48-Ub2 (Table S3). Importantly, the observed perturbations map to both faces of the UBA2 domain (Fig 7E), further suggesting that a single UBA2 molecule interacts with both Ub units of K27-Ub2 simultaneously and in a bidentate binding mode, akin to that for K48-Ub2.
The dissociation constant (Kd) was determined from the titration curves for both Ubs of K27-Ub2 and for UBA2 on a per-residue basis (Fig 7C). Assuming a 1:1 stoichiometry of binding, the Kd was 42 μM ± 8 μM, for the proximal Ub (Table S4), and 63 μM ± 17 μM for the distal Ub. A 2:1 binding model was also tested, but the fit was poor and the residuals showed systematic deviations. For residues in UBA2 the estimated Kd was 6 μM ± 6 μM; this value should be treated as approximate as we did not attain saturation by the end of the titration (see Fig 7 legend). In support of the 1:1 stoichiometry, the average T1 at titration endpoint for residues in secondary structure was 941 ms ± 67 ms, consistent with the expected size of such complex (Varadan et al., 2005). Overall, these data indicate that UBA2 binds K27-Ub2 preferentially over K11-Ub2 (150-200 μM) (Castañeda et al., 2013), K63-Ub2 (180-280 μM) (Varadan et al., 2004), and monoUb (~300-600 μM) (Mueller et al., 2004; Raasi et al., 2005) and comparably to or somewhat weaker than K48-Ub2 (8-20 μM) (Varadan et al., 2005).
Encouraged by these findings we performed paramagnetic spin-labeling experiments to further map the interactions between UBA2 and K27-Ub2. A nitroxide spin label (MTSL) was attached to C344 of UBA2, and its effects on K27-Ub2 were examined using NMR. Paramagnetic relaxation enhancement (PRE) was detected in both Ubs, indicative of close distances between K27-Ub2 and UBA2. Particularly strong PREs (signals wiped out) were observed in and around the hydrophobic patch residues. Notably, the PRE effects on the distal Ub were different from those for the proximal Ub, particularly for residues 25-42 (Fig 8A). From the PRE data we reconstructed the position of the spin label’s unpaired electron relative to each Ub unit separately. The results show that the spin label is located somewhat closer to the distal than to the proximal Ub. The back-calculated PRE profiles are in excellent agreement with experimental data (Fig 8A). However, we found that the resonances for K6, T7, L8, I44, and V70 of the proximal Ub and G75 and G76 of the distal Ub shifted by a maximum of 0.1 ppm in the 1H dimension, whereas signals of L8, V70, and L71 of the distal Ub did not re-attain full intensity after the spin label was reduced. After MTSL was cleaved from C344 of UBA2 (using TCEP), the Ub signals returned to their positions as seen previously. The overall similarity of the CSPs and signal attenuations in the proximal Ub before and after MTSL cleavage from UBA2 suggests that binding equilibrium and structure may have been affected slightly, but not dramatically by the presence of MTSL (Fig S7G). In fact, a titration using UBA2 with reduced MTSL showed a slight decrease in binding affinity (Kd ~ 77 μM ± 21 μM) for residues in the proximal Ub. These observations implicate C344 as a residue near the binding interface between UBA2 and K27-Ub2.
Figure 8.
Structural model of K27-Ub2 bound to UBA2. (A) PREs in the distal and proximal Ubs of K27-Ub2 caused by MTSL attached to C344 of UBA2. Gray bars depict experimental PREs for each Ub, red lines are back-calculated PREs from the reconstructed position of the spin label. Structural cartoons on the right show spin label’s location (yellow sphere) relative to each Ub (ribbon); Ub residues are colored according to I/I0 magnitude, ranging from red (I/I0 = 0) to blue (I/I0 = 1). (B) Agreement between the experimental PREs (gray bars) and back-calculated PREs (red line) for the HADDOCK model of the UBA2:K27-Ub2 complex, with UBA2 in green and Ubs colored as in A. Also shown are the reconstructed location of the spin label (yellow sphere) and the side chain of residue 344 in UBA2 (yellow sticks). (C) Top cluster result from HADDOCK (superposition of four complex structures). (D) Comparison of the UBA2 interface with monoUb (left) and proximal Ub of K27-Ub2 (right). (E) PREs in UBA2 and distal Ub caused by MTSL attached to C77 of the proximal Ub are mapped onto the model of the UBA2:K27-Ub2 complex. (F) The effect of L8A+I44A mutations in the distal Ub on the proximal Ub and UBA2. Residues that exhibit significant differences in titration trajectories and CSPs as a result of the mutations are colored red. Sites of L8A and I44A substitutions are represented as yellow spheres. (See also Figs S8,S9,S10)
Given the PRE pattern for K27-Ub2, we asked whether any conformer in the 23000-member SASSIE ensemble of K27-Ub2 was consistent with the PRE data, assuming a single spin-label position. The conformer that agreed best with the PRE data is shown in Figure S7A, S7B. The back-calculated PRE profile for both Ubs taken together is in excellent agreement with experimental PRE data; it is nearly indistinguishable from the agreements for each individual Ub analyzed separately. Importantly, this conformer supports a binding model where the hydrophobic patches of both Ubs interact with UBA2 in a sandwich-like mode, similar to the K48-Ub2:UBA2 interaction (Varadan et al., 2005). However, the structure of K48-Ub2:UBA2 complex is inconsistent with our experimental PRE data for K27-Ub2 (Fig S7C,D). Given these observations, we set out to determine a structural model of the K27-Ub2:UBA2 complex.
Structural model of K27-Ub2 complex with UBA2
We used the biomolecular docking program HADDOCK (de Vries et al., 2010) to obtain models of the K27-Ub2:UBA2 complex using the CSP and PRE data accrued here as intermolecular NMR constraints (Table S5). The highest-scoring HADDOCK cluster is shown in Figure 8C; its agreement with experiment is depicted in Figures 8B and S7E,F. The back-calculated PREs are in strong agreement with the experimental data (almost as good as for individual Ubs treated separately), and the reconstructed location of the spin label is near the site of spin label’s attachment to UBA2 (C344). The docked structures of K27-Ub2 in complex with UBA2 (Fig 8C,E) are similar to the one (Fig S7B) extracted from the K27-Ub2 SASSIE ensemble.
Our NMR-based model of the K27-Ub2:UBA2 complex provides a molecular basis for understanding UBA2's stronger affinity for K27-Ub2 over monoUb and K11-, K63-Ub2. In the UBA2-bound conformation of K27-Ub2, the hydrophobic surface patches on both Ubs form an extended hydrophobic surface shaped as a pocket that accommodates UBA2. This enables a number of polar and hydrophobic contacts between UBA2 and K27-Ub2. UBA2 is positioned such that its helix α3 sits in the hydrophobic pocket where it contacts both Ubs. Noteworthy, UBA2’s contacts with the proximal Ub resemble those in the monoUb:UBA2 complex (Mueller et al., 2004) in that UBA2’s helices α1 and α3 and the α1/α2 loop face the proximal Ub (Fig 8D). Importantly, UBA2’s residues 344-346, and 358, which exhibit unique CSPs upon addition of K27-Ub2, reside at the interface between UBA2 and the distal Ub (Fig 8E).
To test the validity of this model, we engineered a K27-Ub2 variant where we introduced mutations (L8A+I44A) in the hydrophobic patch of the distal Ub. Via NMR titration experiments, we observed effects on UBA2 and the proximal Ub simultaneously (Figs 8F, S8, S9). As expected, the binding affinity was reduced compared to WT K27-Ub2 (Kd of 70 μM ± 7 μM for residues in the proximal Ub (Fig S8)). Weaker binding is also supported by the absence of strong signal attenuations in almost all residues of the proximal Ub. Importantly, the largest perturbations localized to the parts of the complex next to L8 and I44 of the distal Ub. In UBA2, residues 344-349 exhibited the largest reduction in CSPs in comparison with WT K27-Ub2; these residues face I44 in our model. Residues 344, 346, 351, and 359 all titrated with different trajectories; each of these face either L8 or I44 of the distal Ub in the complex (Figs 8F, S9). Notably, residues 44-49 in the proximal Ub were substantially affected by the distal-Ub substitutions (Figs S8, S9), consistent with the location of these residues in a loop that directly abuts L8 of the distal Ub. For example, the amide resonance of I44 does not move at all in WT K27-Ub2, but shifts substantially in the L8A+I44A variant (Fig S9). Together, these mutagenesis data are consistent with our model of the UBA2 complex with K27-Ub2.
To further validate our model of the complex, MTSL was positioned on a Cys (C77) introduced at the C terminus of the proximal Ub of WT K27-Ub2 via native chemical ligation. The observed PREs localized to one surface of helices α2 and α3 in UBA2, and to the hydrophobic patch of the distal Ub facing these helices (Figs 8E, S10), as well as to the C terminus of the distal Ub, all located in the vicinity of the C terminus of the proximal Ub in the complex. Thus, by and large, the mutagenesis and spin-labeling data support the proposed model of the K27-Ub2:UBA2 complex. However, given that the structures obtained here were derived from limited experimental data they should be treated as low-resolution models of the complex.
Although K27 and K48 are located on opposite sides of Ub, the UBA2-bound conformation of K27-Ub2 is very similar to that of K48-Ub2 in complex with UBA2 (Fig 9). Similarly to K48-Ub2:UBA2 complex, UBA2’s helix α3 is positioned in the hydrophobic pocket, thus contacting both Ubs. However the actual orientation of UBA2 differs by almost 180°: in the case of K48-Ub2 helices α1 and α2 face the distal Ub while helix α2 on the back side of UBA2 faces the proximal Ub.
Figure 9.
Comparison of the UBA2 complexes with (A) K27-Ub2 and (B) K48-Ub2 (PDB ID 1ZO6). UBA2 is colored green, linkage-forming G76 and K27 (or K48) are shown as red sticks. (C) Conformational differences between the free (yellow, from Fig 6B) and UBA2-bound (gray) K27-Ub2; the arrow indicates the rotation of the proximal Ub upon complex formation.
Discussion
The K27 linkage remains the only (as yet) structurally uncharacterized linkage found in polyUb chains. In general, little is known about K27-linked polyUb chains and the biological signals they elicit in the cell. In this work we have addressed the dearth of structural data for K27-linked polyUb chains, particularly in the context of all Ub2 chains, and placed the implications of these observations in a functional perspective.
Of all seven lysines in Ub, K27 is the least solvent-accessible, is the most ordered, and is involved in several hydrophobic contacts and hydrogen bonds. These features, combined with perturbations to K27's microenvironment upon isopeptide bond formation, may have major implications for the signaling mechanisms involving K27-linkage, including polyUb formation, molecular recognition of this chain type, and cleavage by DUBs. In fact, position 27 in Ub is sensitive to mutations: point mutations of K27 in Ub can cause growth defects in yeast, and only a small number of amino acid substitutions are tolerated at this position (Roscoe et al., 2013). Here we demonstrated that disruption to K27's microenvironment by the isopeptide linkage produces large changes in NMR spectra of the proximal Ub in K27-Ub2. Importantly, the overall structure of the proximal Ub is intact, but backbone dynamics are altered, primarily for residues around K27. Our conclusion is also corroborated by a recent structural study of the so-called K0 Ub variant commonly used in biological assays (Huang et al., 2014). In that variant, all lysines (including K27) are replaced with arginines, resulting in large NMR spectral changes, but largely unaffecting the overall structure and dynamics of Ub.
Interestingly, K27 plays important roles beyond chain linkage, in other proteins similar to Ub. Nedd8, a Ub-like modifier protein whose conjugation to target proteins elicits unique biochemical signals in the cell, mirrors Ub structurally and contains many of the same lysines (K6, K11, K27, K33, K48). Just as in Ub, K27 is the least solvent-accessible lysine in Nedd8. Although K27 in Nedd8 is not known to participate in chain conjugation, it plays an important role in the mechanism of protein neddylation (Sui et al., 2015): with a K27R substitution, Nedd8 can no longer be conjugated to its target substrates.
We found that the Ub-Ub linker of K27-Ub2 confers unique properties to this polyUb chain. The C-terminal residues G75-G76 in the distal Ub are more ordered than in other Ub2 chains. Combined with K27's low solvent accessibility, this makes the Ub-Ub linker the least mobile and least accessible of all polyUb chains. This may explain why K27-Ub2 was not cleaved by any of the DUBs assayed here, especially USP2 and USP5 which were capable of cleaving all other lysine linkages (Fig 1). These results are consistent with prior observations with USP2 treatment of the ubiquitinated proteome (Kim et al., 2011). Only two DUBs (OTUD2 and OTUD6A) are known to cleave K27-Ub2 chains, however these DUBs are not K27-specific; they also cleave K29- and K33-Ub2s (Mevissen et al., 2013).
To date, no binding partners are known to specifically interact with K27-linked polyUb. Therefore, the mechanisms by which these chains interact with receptor proteins are unknown. Based on our structural/conformational analyses, we hypothesized that K27-Ub2 can utilize hydrophobic patches on both Ubs to interact with a target receptor in a bidentate manner, analogous to how K48-Ub2 binds the UBA2 domain from the proteasomal shuttle protein hHR23a (Varadan et al., 2005). Our binding studies confirmed this prediction. With an affinity for UBA2 near that of K48-Ub2, K27-Ub2 stands apart from monoUb or K11-Ub2 and K63-Ub2. Our model of the K27-Ub2:UBA2 complex (Fig 8) suggests that UBA2's increased affinity for K27-Ub2 could arise from the interactions it makes with both Ubs simultaneously, due to the sandwich-like arrangement of the binding partners. The NMR titration data also revealed that the Ub-Ub linker in K27-Ub2 is affected by UBA2 binding, and the CSPs are significantly stronger than in K11-Ub2 (Fig S7H,I) or K63-Ub2. This could be a result of direct interaction between the linker and UBA2 or a consequence of domain rearrangement upon complex formation. Note however that the K27-Ub2:UBA2 complex is distinct from the K48-Ub2:UBA2 complex (Fig 9). Not all contacts are similar, particularly for helices α1 and α3 and residues on the back side of UBA2 (C344, K346). The extended hydrophobic pocket accommodating UBA2 in K27-Ub2 appears less deep than in K48-Ub2, resulting in a lesser buried surface area (1450 Å2 vs. 2600 Å2). These differences might explain the differences in the affinity of the two chains for UBA2. It should be pointed out here that the differences in the UBA2 binding contacts within the sandwich-like complexes with K27-Ub2 and K48-Ub2 exemplify the versatility of possible polyUb:receptor interactions depending on or shaped by the linkage type.
The finding that a UBA domain of hHR23a, a protein also involved in DNA damage recognition, has binding preference for K27-Ub2 might suggest a role for K27-linked chains in DNA repair. Although no interaction has been reported yet between K27-polyUb and hHR23a or its yeast homologue Rad23, recent evidence implicates K27-linked Ub chains in the DNA repair pathway (Gatti et al., 2015). RNF168, an E3 ligase known to participate in chromatin ubiquitination, builds K27-linked chains, and these chains directly interact with proteins in the DNA double-strand repair pathway, particularly the tandem UIMs of Rap80, the UDR domain of 53BP1, and RNF169, which also bind K63-linked chains (Gatti et al., 2015; Sims and Cohen, 2009). Intriguingly, some of the less-populated conformers of K27-Ub2 have the hydrophobic patches of both Ubs arranged in an extended manner resembling the structure of K63-Ub2 in the Rap80 bound state (Fig 6D, F). We can speculate that, similar to K63-chains, such an extended conformation might enable avid interaction of K27-chains with tandem UIMs of Rap80.
A comparison of the structures of free and UBA2-bound K27-Ub2 (Fig 9C) illustrates the role of conformational flexibility of this chain and suggests that it might be capable of binding/accommodating other UBAs (or small compact proteins) in a similar manner. Despite scarcity of data on functional roles of K27-linked chains and their recognition by receptors, we found a few recent examples in the literature where such interactions might be relevant. For example, NEMO, a modulator of NF-κB activation, is modified by K27-linked Ub chains at different lysines, each affording a different biological outcome (Arimoto et al., 2010; Liu et al., 2014). A recent study revealed recognition of NEMO's K27-linked ubiquitination by a UBA domain of Rhbdd3 (Liu et al., 2014), a rhomboid membrane protein serine protease. This UBA interacts with K27-linked Ub chains on NEMO, ultimately leading to inhibition of the TLR-triggered activation of NF-κB and also inhibition of interleukin-6 (IL-6) production. There is only 21% sequence identity (29% sequence similarity) between hHR23a UBA2 and Rhbdd3 UBA. However, the Rhbdd3 UBA equivalents of hHR23a UBA2 residues 327 and 330-332, known to interact with Ub (Fig 8, and (Mueller et al., 2004)), are conserved. It is tempting to speculate that Rhbdd3's UBA domain may interact with K27-linked Ub chains in a similar manner as found here for hHR23a UBA2.
The structural and dynamic properties of K27-Ub2 set it apart from other Ub chains. Much remains to be determined concerning the mechanisms by which this chain is built, recognized by receptor proteins, and disassembled into Ub monomers. Our results demonstrate that although K27-Ub2 exists in open conformations in solution with no defined Ub-Ub interface (in contrast to K48-Ub2 or K6-Ub2) the flexibility of the linker allows this chain to adopt conformational states that enable concerted interactions of both Ubs with a ligand. We find it remarkable that the conformational ensembles derived from NMR data allowed us to predict an unanticipated binding mode and binding partner, which were then confirmed experimentally. From our studies, it appears that K27-linked Ub2 chains are versatile in their ability to be recognized by various downstream receptor proteins. We anticipate that these findings will lead to further studies into the mechanisms of how K27-linked polyUb chains are recognized by other proteins.
EXPERIMENTAL PROCEDURES
Preparation of Proteins
Ub variants containing Lys(Boc) substitutions were expressed and purified as described(Castañeda et al., 2011b). Ub2 constructs were assembled as detailed elsewhere (Castañeda et al., 2011a; Varadan et al., 2004; Varadan et al., 2002). See Supplemental Information for further details.
DUB Assays
Deubiquitinase assays were performed using 25 μM Ub2 in PBS pH 7.4 buffer at 30 °C, except for the Ubp6 assay which used 50 μM Ub2. DUB concentrations were 1 μM (IsoT/USP5, OTUB1, USP2), 250 nM GST-Cezanne, 5 μM GST-AMSH, or 8 μM Ubp6. For each assay, aliquots were taken at 1-hr, 2-hr, 4-hr, and 20-hr, quenched with 5× loading dye, and frozen at −20 °C until ready to be run on a 15% SDS-PAGE gel. DUB inhibition assays were performed as detailed in Figure S1.
NMR Experiments
NMR experiments were performed at 23°C on 600 MHz or 800 MHz spectrometers equipped with cryoprobes. Proteins were prepared in 20 mM sodium phosphate buffer (pH 6.8) containing 0.02% NaN3 and 5% D2O. Unless indicated otherwise, Ub2 constructs used for NMR studies had a single Ub unit (either distal or proximal) enriched with 15N. CSPs were quantified as Δδ = [(ΔδH)2 + (ΔδN/5)2]1/2 where ΔδH and ΔδN are the differences in 1H and 15N chemical shifts for the same residue between Ub2 and monoUb. 15N-1H RDCs were measured and analyzed as detailed in Supplemental Information.
Longitudinal (R1) and transverse (R2) 15N relaxation rates, and {1H}-15N steady-state heteronuclear Overhauser enhancement (hetNOE) were measured for Ub2 samples (125 μM – 200 μM) using established protocols (Hall and Fushman, 2003). The ratio ρ of relaxation rates was determined for each residue as ρ = (2R2'/R1' – 1)−1, where R1' and R2' are modified longitudinal (R1) and transverse (R2) 15N relaxation rates with the high-frequency contributions subtracted (Fushman et al., 2004).
Site-directed spin labeling was achieved by attaching the nitroxide paramagnetic spin label, 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanesulfonate (MTSL), to C344 of hHR23a UBA2 or to C77 introduced in the proximal Ub of K27-Ub2. PRE effects were quantitated as the ratio (I/I0) of the signal intensities in the 1H-15N HSQC spectra recorded with MTSL in the oxidized (I) and reduced (I0) states.
Conformational Ensemble Analysis
We employed SASSIE (Curtis et al., 2012) to generate structural ensembles for K27-Ub2. 30000 trial structures were generated for K27-Ub2. Monte Carlo moves about the φ/ψ backbone torsion angles were permitted only for residues 72-76 of the distal Ub. Trial structures were rejected if there were Cα-atom steric clashes within 3 Å. This yielded ca. 23000 sterically-allowed structures. For analyses of experimental data using these structures, the solution structure of monoUb (PDB ID: 1D3Z) was superimposed on the distal and proximal Ubs using residues 1-71 and 1-72, respectively. The conformation of the linker was the same as in the conformer’s structure. The implementation of Sparse Ensemble Selection (SES) for RDC-based analysis of these structures is detailed in Supplemental Information.
Further details on the experimental procedures and analyses are in Supplemental Information.
Supplementary Material
Highlights.
Di-Ubs of all possible lysine linkages were made and studied by NMR and DUB assays
K27-Ub2 has unique NMR characteristics, dynamics, and resistance to DUB cleavage
K27-Ub2 adopts open conformations in solution capable of bidentate binding to receptors
K27-Ub2 binds UBA2 domain of hHR23A through bidentate interactions similar to K48-Ub2
Acknowledgements
This work was funded by a NSF postdoctoral award to C.A.C. and NIH R01 grants GM065334 and GM021248 to D.F. and GM084396 to T.A.C., utilized NMR instrumentation supported in part by NSF grant DBI1040158 and NIH shared instrumentation grant 1S10OD012254 and neutron-scattering facilities supported in part by the NSF under Agreement No. DMR-0944772, and benefitted from CCP-SAS software developed through a joint EPSRC (EP/K039121/1) and NSF (CHE-1265821) grant. We thank Konstantin Berlin for helpful discussions regarding SES. Some mass spectrometry data were collected on a Shimadzu 8040 MS, supported in part by a Shimadzu instrumentation award to C.A.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.A.C., E.D., A.C., M.A.N., M.R.R., T.A.C. made the proteins; C.A.C., E.D., A.C., M.A.N., D.F., S.K. conducted the experiments; C.A.C., D.F., O.W. analyzed the data; J.E.C. developed code for generating ensembles; C.A.C. and D.F. conceived the study and wrote the paper.
References
- Arimoto K-I, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci USA. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, Castañeda CA, Schneidman-Duhovny D, Sali A, Nava-Tudela A, Fushman D. Recovering a representative conformational ensemble from underdetermined macromolecular structural data. J Am Chem Soc. 2013;135:16595–16609. doi: 10.1021/ja4083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, O'Leary DP, Fushman D. Improvement and analysis of computational methods for prediction of residual dipolar couplings. J Mag Reson. 2009;201:25–33. doi: 10.1016/j.jmr.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, O'Leary DP, Fushman D. Structural assembly of molecular complexes based on residual dipolar couplings. J Am Chem Soc. 2010;132:8961–8972. doi: 10.1021/ja100447p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, O'Leary DP, Fushman D. Fast approximations of the rotational diffusion tensor and their application to structural assembly of molecular complexes. Proteins. 2011;79:2268–2281. doi: 10.1002/prot.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsa N, Norkett R, Wauer T, Mevissen TET, Wu H-C, Foltynie T, Bhatia K, Hirst WD, Komander D, Plun-Favreau H, Kittler JT. K27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Freund SMV, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, Nedelsky NB, Huang H, Ng A, Gardet A, et al. Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity. 2015;43:715–726. doi: 10.1016/j.immuni.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Chaturvedi A, Camara CM, Curtis JE, Krueger S, Fushman D. Linkage-specific conformational ensembles of non-canonical polyubiquitin chains. Phys Chem Chem Phys. 2016;18:5771–5788. doi: 10.1039/c5cp04601g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Kashyap TR, Nakasone MA, Krueger S, Fushman D. Unique structural, dynamical, and functional properties of K11-linked polyubiquitin chains. Structure. 2013;21:1168–1181. doi: 10.1016/j.str.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. Non-enzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J Am Chem Soc. 2011a;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Liu J, Kashyap TR, Singh RK, Fushman D, Cropp TA. Controlled enzymatic synthesis of natural-linkage, defined-length polyubiquitin chains using lysines with removable protecting groups. Chem Comm (Cambridge) 2011b;47:2026–2028. doi: 10.1039/c0cc04868b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Garrett DS. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 1999;121:9008–9012. [Google Scholar]
- Curtis JE, Raghunandan S, Nanda H, Krueger S. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun. 2012;183:382–389. [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nature Protocols. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Dikic I, Dotsch V. Ubiquitin linkages make a difference. Nat Struct Mol Biol. 2009;16:1209–1210. doi: 10.1038/nsmb1209-1209. [DOI] [PubMed] [Google Scholar]
- Esadze A, Li D-W, Wang T, Bruschweiler R, Iwahara J. Dynamics of lysine side-chain amino groups in a protein studied by heteronuclear 1H-15N NMR spectroscopy. J Am Chem Soc. 2011;133:909–919. doi: 10.1021/ja107847d. [DOI] [PubMed] [Google Scholar]
- Fei C, Li Z, Li C, Chen Y, Chen Z, He X, Mao L, Wang X, Zeng R, Li L. Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/β-Catenin signaling. Mol Cell Biol. 2013;33:4095–4105. doi: 10.1128/MCB.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Varadan R, Assfalg M, Walker O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Prog. Nucl. Magn. Reson. Spectrosc. 2004;44:189–214. [Google Scholar]
- Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Reports. 2015;10:226–238. doi: 10.1016/j.celrep.2014.12.021. [DOI] [PubMed] [Google Scholar]
- Ghose R, Fushman D, Cowburn D. Determination of the rotational diffusion tensor of macromolecules in solution from NMR relaxation data with a combination of exact and approximate methods - Application to the determination of interdomain orientation in multidomain proteins. J Mag Reson. 2001;149:204–217. doi: 10.1006/jmre.2001.2295. [DOI] [PubMed] [Google Scholar]
- Hall JB, Fushman D. Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J Biomol NMR. 2003;27:261–275. doi: 10.1023/a:1025467918856. [DOI] [PubMed] [Google Scholar]
- Hemantha HP, Brik A. Non-enzymatic synthesis of ubiquitin chains: where chemistry makes a difference. Bioorg Med Chem. 2013;21:3411–3420. doi: 10.1016/j.bmc.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Hospenthal MK, Freund SMV, Komander D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat Struct Mol Biol. 2013;20:555–565. doi: 10.1038/nsmb.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jeon M.-s., Liao L, Yang C, Elly C, Yates Iii JR, Liu Y-C. K33-Linked Polyubiquitination of T Cell Receptor-ζ Regulates Proteolysis-Independent T Cell Signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Li J, Byrd RA. Solution structure of lysine-free (K0) ubiquitin. Protein Science. 2014;23:662–667. doi: 10.1002/pro.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett Eric J., Huttlin Edward L., Guo A, Li J, Possemato A, Sowa Mathew E., Rad R, Rush J, Comb Michael J., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristariyanto YA, Abdul Rehman SA, Campbell DG, Morrice NA, Johnson C, Toth R, Kulathu Y. K29-Selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of K29 polyubiquitin. Mol Cell. 2015;58:83–94. doi: 10.1016/j.molcel.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Han C, Xie B, Wu Y, Liu S, Chen K, Xia M, Zhang Y, Song L, Li Z, et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat Immunol. 2014;15:612–622. doi: 10.1038/ni.2898. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Mevissen Tycho E.T., Hospenthal Manuela K., Geurink Paul P., Elliott Paul R., Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H-J, Rape M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel MA, Elliott PR, Swatek KN, Simicek M, Pruneda JN, Wagstaff JL, Freund SMV, Komander D. Assembly and specific recognition of K29- and K33-linked polyubiquitin. Mol Cell. 2015;58:95–109. doi: 10.1016/j.molcel.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DNA. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of Rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y, Liu Y, Xu G. A lysine-to-arginine mutation on NEDD8 markedly reduces the activity of cullin RING E3 ligase through the impairment of neddylation cascades. Biochem. Biophys. Res. Commun. 2015;461:653–658. doi: 10.1016/j.bbrc.2015.04.085. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg N, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys(63)-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Ludwig T, Baer R. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol. Cell. Biol. 2010;30:2787–2798. doi: 10.1128/MCB.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Livnat-Levanon N, Kleifeld O, Mansour W, Nakasone MA, Castaneda CA, Dixon EK, Fushman D, Reis N, Pick E, Glickman MH. Base-CP proteasome can serve as a platform for stepwise lid formation. Biosci Rep. 2015;35:e00194. doi: 10.1042/BSR20140173. art. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W-C, Lee Y-R, Lin S-Y, Chang L-Y, Tan Yen P., Hung C-C, Kuo J-C, Liu C-H, Lin M-Y, Xu M, et al. K33-Linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lv X, Yin W.-c., Zhang X, Feng J, Wu W, Hui C.-c., Zhang L, Zhao Y. Ter94 ATPase complex targets K11-linked ubiquitinated Ci to proteasomes for partial degradation. Dev Cell. 2013;25:636–644. doi: 10.1016/j.devcel.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Zhou H-L, Geng C, Luo G, Lou H. The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 2013;27:1046–1058. doi: 10.1101/gad.215681.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.