Abstract
Experimental studies have uncovered a variety of microRNA (miRNA)–target duplex structures that include perfect, imperfect and seedless duplexes. However, non-canonical binding modes from imperfect/seedless duplexes are not well predicted by computational approaches, which rely primarily on sequence and secondary structural features, nor have their tertiary structures been characterized because solved structures to date are limited to near perfect, straight duplexes in Argonautes (Agos). Here, we use structural modeling to examine the role of Ago dynamics in assembling viable eukaryotic miRNA-induced silencing complexes (miRISCs). We show that combinations of low-frequency, global modes of motion of Ago domains are required to accommodate RNA duplexes in model human and C. elegans Ago structures. Models of viable miRISCs imply that Ago adopts variable conformations at distinct target sites that generate distorted, imperfect miRNA-target duplexes. Ago's ability to accommodate a duplex is dependent on the region where structural distortions occur: distortions in solvent-exposed seed and 3′-end regions are less likely to produce steric clashes than those in the central duplex region. Energetic analyses of assembled miRISCs indicate that target recognition is also driven by favorable Ago-duplex interactions. Such structural insights into Ago loading and target recognition mechanisms may provide a more accurate assessment of miRNA function.
INTRODUCTION
Understanding microRNA (miRNA) function in post-transcriptional gene silencing remains challenging due to the diversity of miRNA–mRNA target interactions and their modulation by Argonaute (Ago) and other effector proteins. Target recognition is mediated by the miRNA-induced silencing complex (miRISC) (1–3), comprising Argonaute bound to an individual miRNA ‘guide’ strand capable of recognizing many independent target sites (4–6). While current models of miRNA-target recognition depend heavily on base-pairing in the seed region and predicted secondary structures (7–9), a variety of data challenge the conventional view that RNA-only interactions dominate miRNA target recognition. First, these factors cannot adequately explain known examples of non-canonical target sites (10,11) or recent transcriptome-wide experimental evidence for numerous imperfect seed duplexes and seedless sites (12); a recent assessment of target prediction algorithms indicates that additional structural criteria (size and position of bulges) regarding the seed region are needed to improve prediction algorithms (9). Biophysical and structural studies have shown that miRISC both stabilizes the guide miRNA strand (thereby lowering the entropic cost of seed recognition) (13) and coordinates miRNA-target base pair formation (by releasing the Ago-bound 3′ end of the guide miRNA strand, thus allowing formation of an RNA duplex within Ago's RNA-binding channel) (2,14). Molecular simulations show that both RNA–RNA and RNA–protein interactions contribute to miRNA activity (15), and that Ago domains undergo coordinated motions (16). In addition, Agos are increasingly understood to form complexes with other proteins that regulate translation (e.g. GW182 and Puf family proteins) (1,17–20) or mediate alternative gene silencing pathways (21).
Collectively, these data highlight the need for atomic-resolution models of miRNA-associated complexes to study structural mechanisms and dynamics of these regulatory interactions. Computational analysis using recently solved crystal structures of prokaryotic (T. thermophilus (22,23) and P. furiosus (24)) and eukaryotic (K. polysporous and H. sapiens) Agos, with or without bound RNA (2,25–28), can provide a deeper mechanistic understanding of target recognition by miRISC. For example, studies of C. elegans miRNAs have highlighted the prevalence of imperfect miRNA-target duplexes having a large internal loop and/or nucleotide bulge in the seed region (7,11), such as the C. elegans let-7 miRNA, which binds to its targets to form duplexes having a 5-nt internal loop and a single nucleotide bulge in the seed region. The presence of a large loop or bulge can induce structural distortions, including kink turns and non-canonical base pairs and interactions (29,30). How structurally distorted duplexes are accommodated to form stable ternary complexes, what conformational changes are required for free Ago to fit different RNA duplexes, and the global contribution of these factors to miRNA function are unexplored questions.
Here, we exploit the solved Ago structures to construct models of bacterial and eukaryotic miRISC based on analysis of their global dynamics using elastic network model (ENM) theory (31). We use existing high-resolution miRNA-target datasets, derived from ligation experiments (12) and other biochemical and genetic studies, to generate RNA 3D structure ensembles; fit them with Ago structural models; and then analyze the viability of these predicted miRISC structures. We show that the low-frequency, global modes of motion are conserved between bacterial and eukaryotic Agos, and that the transition from the closed to the open Ago conformation that fits an RNA duplex can be explained by these global modes of motion. This observation allowed us to develop a general algorithm for constructing eukaryotic miRISC structures with variable duplex conformations. Our analysis of how eukaryotic Ago structures accommodate duplexes provides new insights into the structural determinants of miRISC target recognition.
MATERIALS AND METHODS
Argonaute and Argonaute-RNA structures
We performed miRISC dynamics and binding affinity analyses using Ago-RNA crystal structures from bacterial (T. thermophilus (PDB codes 3HJF, 3DLH, 4NCB), A. aeolicus (1YVU)), archaeal (P. furiosus (1U04)), and eukaryotic (K. polysporous (4F1N), H. sapiens (4EI1, 4KRF)) species. These were solved as RNA-free Ago (1YVU,1U04), Ago-guide strand (3DLH, 4F1N, 4EI1, 4KRF) and Ago-duplex (3HJF, 4NCB) structures. RNA-free Ago structures and intermediate Ago-RNA complexes used in analyses were derived from these structures.
C. elegans Argonaute structures from comparative modeling
We built worm Ago structures using Modeller (32,33), a comparative modeling program. Solved human AGO1 (4KRF) (28) and AGO2 (4EI1) (26) structures, having ∼66% sequence identity with ALG-2, were used as templates. No loop refinement was performed since many of the loops are missing in solved human Ago structures; we also removed a long random loop (residues 1–73) from the model ALG-2 structure. The model ALG-2 structure has RMSD values of ∼1.5 Å compared to the template human Ago structures. We also modeled the related ALG-1 structure (sequence identity of ∼85% to ALG-2); it deviates from the ALG-2 and human AGO1/2 template structures by ∼3 Å.
RNA structure modeling
RNA duplex structures were built from input secondary structures using the MC-Sym algorithm (34), as described in our previous work (15). For each secondary structure, we generated a maximum of 1000 3D structures and ranked them using the all-atom AMBER99 force field with atomic interaction and implicit solvation energies.
Computation of RNA–RNA and RNA–protein binding energies
To assess miRISC energetics, we evaluated the RNA–RNA and RNA–protein binding energies separately. As described in our previous work (15), the force field used is a combination of all-atom solute interactions and implicit (water and ions) solvation energies. Briefly, for RNA–RNA interactions, we computed binding free energies with contributions from atomic interaction, solvation and entropic terms. For RNA–protein interactions, we considered the atomic interaction and solvation terms without the entropic term. This modeling approach can predict duplex structures and binding free energies with reasonable accuracy (15).
Global miRISC dynamic modes from the elastic network model (ENM) theory
To model global dynamics of Ago and miRISC structures, we used the elNemo implementation of ENM (31,35), available at http://www.sciences.univ-nantes.fr/elnemo/. Default parameters were used except for the displacement parameter which was varied between 100 and 800. The nontrivial modes (7 and higher) were relabeled as modes 1, 2, 3, etc. Low-frequency modes reflect global motions, whereas higher-frequency modes tend to sample local dynamics.
Generation of open eukaryotic Argonaute conformations and Argonaute loading
We generated open eukaryotic Ago conformations using an iterative application of the ENM method (Supplementary Figure S1) with a few low-frequency modes. A similar approach was previously used to search for protein conformations that fit X-ray crystallographic data (35). The target or open Ago conformation is the one that accommodates a duplex structure. Starting with a solved Ago structure (RNA removed), we generated conformations using ENM and selected a structure from a low-frequency mode (1, 2 or 3) that improves the fit for a ‘reference’ duplex. We used the DNA–RNA hybrid duplex in bacterial Ago (3HJF) as our reference duplex and placed it within a eukaryotic Ago structure by aligning the eukaryotic and bacterial Ago structures using CE, a protein–protein structure alignment algorithm (36). This procedure was repeated with the new (selected) starting structure. In practice, multiple rounds of ENM application and starting structure selection are needed to produce open Ago structures that are free of major steric clashes with the reference duplex. This approach produces a discrete set of conformations for the transition from a closed to an open conformation.
Ago loading was predicted using the solved T. thermophilus Ago-duplex structure (3HJF) as a reference structure (15). Initially, the guide RNA strand structure was aligned with its corresponding guide DNA structure and then the open Ago structure was aligned with the T. thermophilus Ago. The guide RNA strand alignment was performed for all atoms in the first 11 to 15 bases (from the 5′ end) using TINKER's superpose routine. This procedure was applied to both seed and seedless sites, with the latter alignments starting from the first paired miRNA base. Ago–Ago alignment was performed using CE (36).
The resulting ternary complex was energy minimized with the Amber99 plus GBSA force field as implemented in the TINKER package (37). The conformation of miRNA's unpaired 5′-end base arising from specific interactions with the Mid domain was not modeled as it would require a much longer relaxation time. To account for duplex conformational flexibility, we performed multiple (typically, 5–10) Ago-duplex dockings using low-energy duplexes for a given secondary structure.
A general algorithm for assembling miRISC structures
Many animal miRNA-target duplexes contain multiple base pair mismatches and bulges. These imperfections can introduce significant structural distortions that may require the Ago conformation to change in subtle ways to accommodate the duplex structure. Solving this problem requires simultaneous changes in Ago and duplex conformations to determine optimal complexes. Here, we describe a more general approach for constructing miRISC structures using duplex and Ago conformational ensembles. Our algorithm involves the following steps (Figure 1): First, the duplex and Ago structures ensembles are constructed independently. Since only open Ago conformations can load duplexes without steric clashes, we consider a set of open Ago conformations as determined using the iterative ENM method described above. Our open conformational set is defined by Ago structures generated by a combination of nontrivial ENM modes 1/2 (i.e. mode 1 followed by mode 2), 1/3 and 1/2/3; this set, which includes a range of structure displacement parameters or amplitudes, has ∼100 open Ago structures. Second, Ago-duplex complexes are assembled by docking the duplex to the Ago (described above). For a duplex ensemble with 1000 structures, the maximum number of complexes considered is ∼105; in practice, we used the top 50 energy-ranked duplexes for docking. Third, each complex is scored based the number of atomic overlaps between the Ago and duplex structures (van der Waals radii of atomic types were used to determine atomic overlaps). Structures with a low number of atomic overlaps are then energy minimized or relaxed to remove minor steric clashes. Fourth, predicted complexes are top (∼5) scoring structures that can be energy minimized. The main point of our algorithm is to use independent duplex and Ago structure ensembles to form complexes that are not dynamically correlated; this approach helps avoid being trapped in local energy minima encountered in molecular dynamics simulations. The resulting algorithm is effective for constructing and screening many candidate complexes with varied duplex and Ago conformations.
Figure 1.
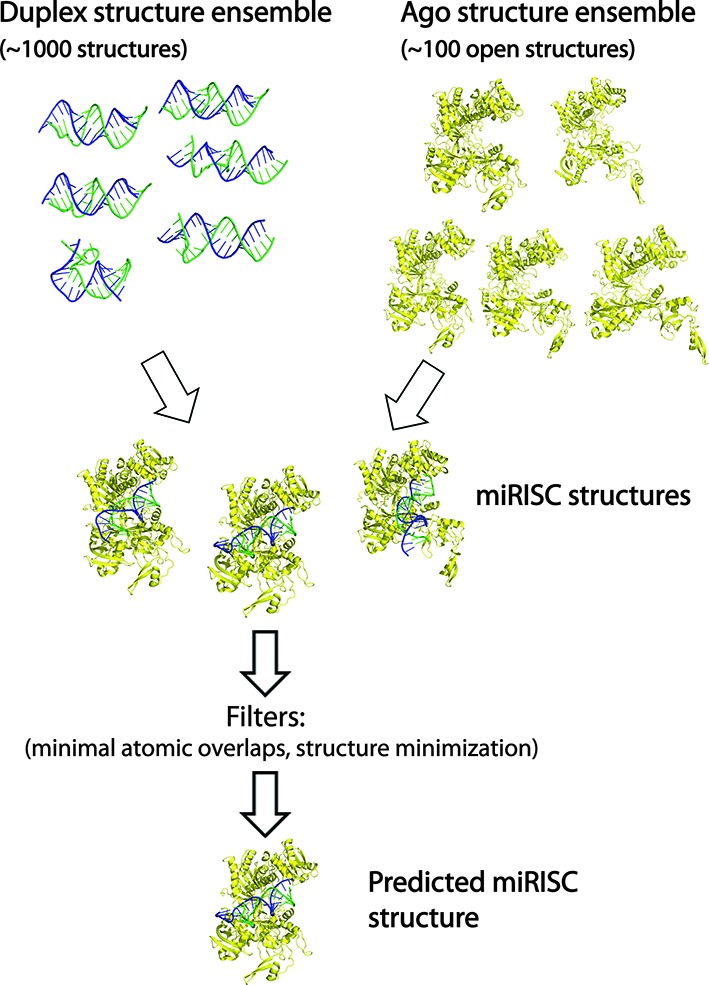
An algorithm for assembling miRISC structures. First, duplex and Ago structure ensembles are independently generated using MC–Sym duplex assembly and ENM methods, respectively. Second, each duplex structure is docked (i.e. guide RNA strand alignment using 3HJF as a reference complex) to each Ago conformation to produce an ensemble of model miRISC structures. Third, the miRISC structures are ranked by the number of atomic overlaps and then the top scoring complexes are energy minimized, which are the predicted structures.
Contact maps of Argonaute–RNA interactions
To display protein–RNA interactions in multiple complexes, we used residue-level resolution contact maps (38). Contact distances were computed using O5’ atoms of RNA strands and Cα atoms of protein residues. Two residues are in contact when their O5’–Cα distance is less than 10 Å. For each Ago-duplex complex, we display its interactions as Ago–miRNA and Ago–mRNA contact maps. Variation of Ago–RNA interactions can be displayed by plotting multiple complexes in a contact map.
RESULTS
Bacterial and eukaryotic Argonaute proteins display similar global conformational dynamics
Since global motions of proteins are determined by their shapes (39), we reasoned that a comparative analysis could reveal the basic modes of conformational dynamics among Ago family members and allow us to infer their relevance for target recognition. To explore global dynamics of Ago, we employ the ENM theory (31), which uses residue-level resolution protein models to compute energetically probable modes of motion; a related all-atom normal mode method was previously used to describe prokaryotic Ago dynamics (40). This approach has been widely used to analyze large-scale motions of biomolecular complexes, including ribosomes and viral coat protein complexes (39,41–42).
Side-by-side comparisons of the three lowest-frequency, ‘nontrivial’ modes of motion for bacterial (T. thermophilus), yeast (K. polysporous), and human Agos reveal similar patterns of relative domain conformational movements (Figure 2). Conformational dynamics are indicated using arrows to highlight the directions of motion between two superimposed end-point structures. For yeast and human Ago structures, the first nontrivial mode is an open/closed, hinge motion between Ago's Mid/PIWI and PAZ domains, which allows Ago's RNA-binding channel to accommodate the width of a straight A-form RNA helix. Comparing crystal T. thermophilus Ago (TtAgo) structures with a guide DNA strand and hybrid DNA–RNA helix shows a similar opening motion to accommodate the hybrid duplex structure (Supplementary Figure S2a).
Figure 2.
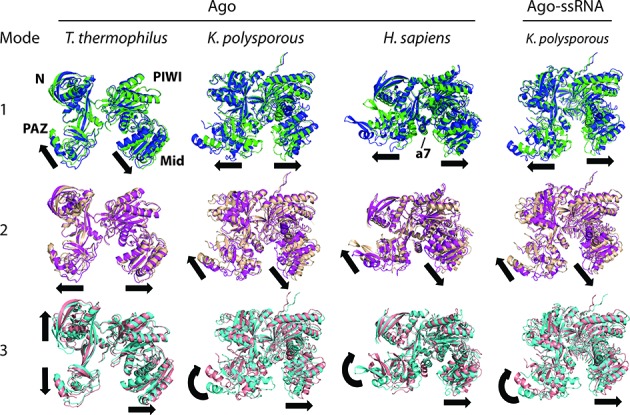
Bacterial and eukaryotic Argonautes have similar conformational dynamics. Conformations of the two end points of motion are superimposed to show the major directions (arrows) of domain movements. A central helical segment in eukaryotic Agos (labeled a7 in the human structure) interacts with the minor groove of the RNA duplex. The first three nontrivial, low-frequency modes of motion were derived from the elastic network model (ENM) theory based on coarse-grained Ago models. For K. polysporous (without and with 9-nt RNA) and H. sapiens Agos, mode 1 is a hinge motion between PAZ and Mid/PIWI domains and mode 2 is a torsional motion between the same domains; for T. thermophilus, the motion types are in reversed mode order. Similarity of the three modes across species suggests they are shared functional motions of Argonaute proteins.
The next mode of motion is a twist or torsional conformational movement between Mid/PIWI and PAZ domains, which allows the placement of alpha-helix 7 (labeled a7 on the human structures and located in the Linker 2 region between PAZ and Mid domains) in the minor groove of the RNA duplex, within or near the seed region (base pairs 6 to 9). Recent crystal structures of TtAgo-DNA duplex also indicate the presence of L2-DNA interactions, although the L2 residues (265–281) are not fully structured (PDB code 4NCA) (23). The third nontrivial mode of motion involves mainly a rotation of the PAZ domain together with the a7 helix. This motion (which is not seen in modes 1 and 2) locks the a7 helix within the minor groove of the seed duplex. Differences in conformational dynamics between yeast and human Ago emerge in higher-order modes of motion (data not shown) and likely reflect local structural differences between the eukaryotic Ago structures.
Eukaryotic and bacterial Ago structures exhibit similarities and subtle differences in global dynamic modes of motion. The bacterial TtAgo also displays dynamic modes 1 and 2, but in reversed order of frequency; its third mode involves a hinge-like motion between the PAZ and N terminal domains, rather than a rotational motion of the PAZ domain. A previous normal mode analysis of the dynamics of prokaryotic (P. furiosus and A. aeolicus) Agos also supports the presence of these lowest-frequency, hinge and torsional motions (40); we have reproduced the low-frequency modes of their Ago structures together with those for a modeled C. elegans ALG-2 (see Materials and Methods, Supplementary Figure S3), and all show global dynamic modes resembling those observed in other bacterial and eukaryotic species (Figure 2). The differences in dynamic modes between bacterial and eukaryotic Agos likely reflect the presence of additional structural elements in eukaryotic Agos (25). Overall, our analysis of six Ago structures from bacterial, archaeal (P. furiosus), and eukaryotic species implies that the conservation of low-frequency conformational dynamics (especially modes 1 and 2) reflects their global structural similarity and their functional requirements for loading RNA or DNA duplexes. A more comprehensive picture of Ago's global dynamics will emerge as additional structures for distinct Ago family members are solved. Phylogenetic analyses of Ago genes from plants, animals and fungi suggest the existence of diverse Ago family proteins that participate in gene silencing and defense against mobile genetic elements, as well as other functions (43,44).
Argonaute loading requires a closed to open transition involving multiple modes of conformational movements
Yeast and human Ago structures complexed with a guide RNA strand (25–28) appear to be in the closed conformation, as attempts to load straight RNA duplexes lead to steric clashes, especially with Ago's a7 helix in the L2 region (Supplementary Figure S2b), as noted previously (3). The idea that Ago undergoes a transition from closed to open conformation to accommodate the RNA duplex is supported by experimental studies: Dicer-mediated loading of an RNA duplex requires an open Ago conformation maintained by the chaperone HSP90 (45–47). However, such studies have not shed light on the precise conformational nature of the open state.
To elaborate essential Ago dynamic modes for the closed/open conformational transition, we iteratively applied ENM to load a 16 base-pair duplex into the yeast and human Ago structures, using the low-frequency modes of motion shown in Figure 2. Iterative ENM generates new conformations from an input (closed or partially open) structure to simulate Ago's conformational trajectory toward accommodation of the growing duplex during target recognition (Methods and Supplementary Figure S1). For human Ago, all three lowest-frequency modes are needed to fit the duplex. Relative to the crystal structure (4EI1)—which is in the closed state with no duplex—the open conformation has an RMSD of 3.1 Å, and shows a7 helix placed in the minor groove of the RNA duplex near the seed region, which has been proposed as a sensor for the arrival of target RNA (3). In addition, the minor groove at the other end of the duplex is held in place by the loops between helices in the Mid/PIWI domain. For yeast Ago, only two dynamic modes are needed to fit the duplex, producing a similar degree of tight fitting as seen for the open human Ago structure; the open conformation has an RMSD of 2.7 Å when superimposed with the solved structure (4F1N).
These two examples illustrate that open Ago conformations can be generated from low-frequency Ago dynamic modes. The fact that only low-frequency modes are needed to achieve open conformations suggests that they are the essential conformational moves for making the transition from the closed state (represented by crystal structures without a duplex). Although the global conformational move sequences selected (e.g., native→2→1, native→2→1→3) may not represent the exact dynamic steps in miRISC-target recognition in vivo, it is a reasonable way to build final miRISC models.
Stability of intermediate Argonaute–RNA complexes suggests that both duplex and Ago-duplex binding affinities are drivers of target recognition
To better understand the various steps in target recognition (seed annealing to 3′-end base pairing and associated Ago movements), we analyzed binding energies of intermediate complexes using duplexes of 8 to 16 base-pairs (Figure 3A). As expected, the duplex binding free energy becomes more favorable as the number of base pairs increases, for both bacterial and eukaryotic miRISC structures (Figure 3B, left). Notably, a similar trend is also found for the dependence of the Ago-duplex interaction energy on the length of the duplex or base-pair number (Figure 3B, right). Considering duplex-Ago interactions, the average binding energy (without entropic contributions) is similar for Agos from all three species. Clearly, these results suggest that base-pair formation and Ago-duplex interactions together drive the intermediate complexes toward the completion of the target recognition process.
Figure 3.
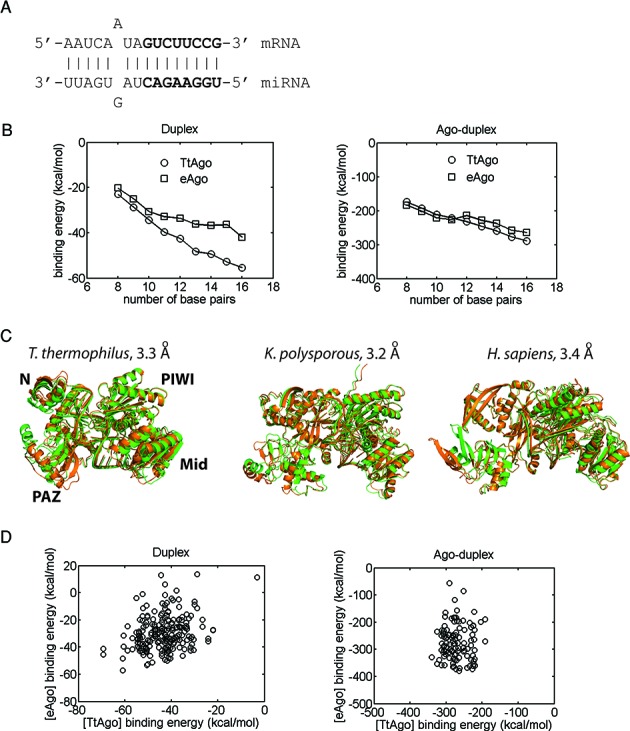
Comparison of the binding affinity trends and dynamics for various bacterial and eukaryotic Ago–RNA complexes. (A) Secondary structure of the full duplex in complexes; seed region in bold letters. (B) Duplex binding affinities (left) and Ago-duplex binding affinities with no entropic contributions (right) of bacterial (T. thermophilus, TtAgo) and eukaryotic (eAgo, averaged K. polysporous (Kp) and human) miRISCs as a function of the duplex length or the number of base pairs. Shorter duplexes were derived from the full duplex by truncation from the 3′ end of the miRNA. Each data point represents an average for 10 complexes with different low-energy duplex conformations. (C) Comparison of the dynamics (mode 1) of Tt, Kp, and human miRISCs with the full duplex. Superimposed structures of the two end points of motion are shown (RMSD values indicated). (D) Scatterplots of bacterial (TtAgo) and eukaryotic (eAgo) miRISC binding affinities for 11 full duplexes, each differing by a single base-pair mismatch (with 10 constructed miRISCs per duplex): (left) duplex binding energies and (right) Ago-duplex binding energies with no entropic contributions within miRISCs.
More specific information about miRISCs can be gained by analyzing the relation between structure and dynamics of pre-target recognition, intermediate and final complexes. For the complex between yeast Ago and the guide RNA strand, the first three modes of motions are similar to those for the RNA-free Ago, i.e. closed/open (hinge) and twist (torsional) motions (Figure 2, right column), except for some damping of the closed/open motion due to the presence of the guide strand. Similar dynamic features are also observed in human miRISC with a guide RNA (data not shown). For T. thermophilus miRISC with a guide DNA (Supplementary Figure S4), dynamic modes 1 and 2 are similar to modes 2 and 1 for yeast miRISC with a guide RNA, respectively. Overall, the similarity of low-frequency modes for RNA-free Ago and Ago-guide RNA structures indicates that pre-target recognition complexes exhibit essentially the same functional motions. In contrast, the lowest-frequency mode of the Ago-seed duplex complex shows markedly reduced closed/open motion because the seed duplex (8 base-pairs) forms extensive contacts with the Mid/PIWI domain and L2 (data not shown). For bacterial, human and yeast miRISCs with the full 16-base pair duplex, the key observation is that the Mid/PIWI domain now forms a rigid block with the duplex, such that the clearly observable mobility is associated with the PAZ domain (mode 1 in Figure 3C and Supplementary Figure S4). Our analysis suggests that miRISC's conformational flexibility becomes progressively diminished from the mobile pre-recognition complex (Ago-guide RNA) to the seed-target and intermediate recognition complexes and then to the final, stable miRISC with base-paired miRNA and target strands. Thus, by identifying the interactions that affect miRISC dynamics, we gain information complementary to the existing static structures of RNA-free Ago, Ago-guide RNA and Ago-duplex from crystallographic studies.
T. thermophilus, yeast and human miRISCs have similar binding affinity trends
Since different species employ distinct sets of Agos (21,43), it is interesting to probe possible Ago-dependent effects on miRNA function by analyzing how differences in T. thermophilus, yeast and human Ago structures affect miRISC binding affinities. We compared duplex and Ago-duplex binding free energies to load a set of eleven 16 base-pair duplexes (based on the duplex in Figure 3A) that differ by a single base-pair mismatch; the in vivo activities of these duplexes were previously measured in D. melanogaster (10).
The binding free energies of the 16-base-pair duplexes within T. thermophilus and eukaryotic (yeast and human) Ago structures show rough linear correlations (R2 = 0.21) over a range of 60 kcal/mol (Figure 3D, left). The more favorable duplex affinities for T. thermophilus could reflect either that it is a solved crystal structure (whereas the open eukaryotic Ago structures were predicted) or, more likely, that the additional structural elements in eukaryotic Agos (25) may restrict the duplex within the RNA-binding channel. Ago-duplex binding affinities (without entropic contributions), which provide a direct measure of the energies of RNA–protein interactions, fall within the same range (−200 kcal/mol to −350 kcal/mol) for both bacterial and eukaryotic miRISC with no apparent correlations between them (Figure 3D, right), implying that the single base-pair mismatches have little influence on Ago-duplex interactions. This similarity of binding affinity trends and values is remarkable given the fact that eukaryotic Ago structures are much larger (904 versus 665 residues for solved yeast and bacterial Ago structures, respectively) and contain multiple structural elements not found in the T. thermophilus Ago structure.
Models of human miRISC suggest that Argonaute conformation adapts to accommodate distinct target site structures
To determine how distorted duplex structures are accommodated within the Ago structure, we build miRISC models using a general algorithm that accounts for conformational adaptation in the Ago loading process (see Materials and Methods and Figure 1). We consider building human miRISC structures based on recent high-resolution datasets from chimeric miRNA–mRNA reads derived from ligation of human miRNA and target sequences bound to AGO1 (12). Structures of human miRNA-target duplexes complexed with Ago (Figure 4 and Supplementary Figure S5) show considerable conformational variability. We observe that minor duplex imperfections (e.g. single base pair mismatches) retain straight conformations, whereas major imperfections (e.g. internal loops with 4 or more unpaired bases) lead to helix bending, as seen in duplexes let-7a::BIRC6 (site 2), let-7b::SLC25A13, let-7a::HMGA2, miR-15a::CCD2 and miR-17*::VIM (site 2).
Figure 4.
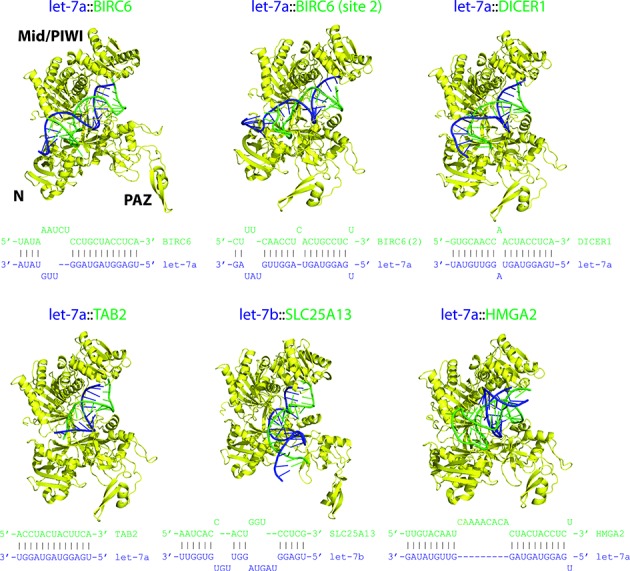
Models of human miRISC structures. All viable structures (free of steric clashes) were assembled using the method outlined in Figure 1; each displayed structure is ranked in the top 5. Complexes were built from human AGO1 and a set of experimentally determined miRNA-target duplexes (secondary structures shown) which are supported by at least five ligated miRNA-target sequence reads. Imperfect duplexes lead to diverse duplex conformations which are accommodated by variable Ago structures. Color scheme: Ago, yellow; guide RNA strand, blue; and mRNA strand, green.
The 13 human miRNA-target duplexes displayed can be fitted into open AGO1 (4KRF) structures (Figure 4 and Supplementary Figures S5 and S6). All duplexes, except for the shorter 13-base-pair duplex let-7a::TAB2, extend beyond the RNA-binding channel of AGO1 without steric clashes. This result confirms a previously proposed model of duplex docking based on the crystal yeast Ago structure, which is in a closed conformational state (25). We then quantified structural deviations between the (open) model and crystal Ago structures to gain insight into conformational adaptation associated with Ago's binding to different targets. Combining all 13 human miRISC structures (top 5 miRISC models/target), the distribution of RMSD values for superimposed model and crystal (4KRF) AGO1 structures peaks at ∼2 Å, with most cases <3 Å (Figure 5A, left). The RMSD values between Ago conformations from distinct target sites have a wider range (5 Å), with peaks at ∼2 Å and ∼4 Å (Figure 5C, left). In contrast, superimposed Ago structures from the same target sites (using 5 independent miRISC models) have a much narrower RMSD distribution with a strong peak at ∼0.5 Å (Figure 5B, left) and a mean of 1.4 Å, indicating that the open Ago conformation for any given target of miRISC is stable with only minor structural changes. Together, these results suggest that the open Ago state adapts to the miRNA recognition site determined by the miRNA–target duplex structure. Focusing on the human let-7a/b miRNA family (Figure 4), which has six distinct targets in our dataset, the average RMSD value for superimposed Ago structures at distinct target sites is 2.5 Å versus 1.5 Å for Ago structures at the same targets (Figure 5D, left). For all human miRNA families considered, the superimposed Ago structures from distinct sites also have an average RMSD value of ∼2.5 Å, suggesting that Ago structural changes associated with the let-7 miRNA family/targets reflect Ago's behavior with other miRNA families.
Figure 5.
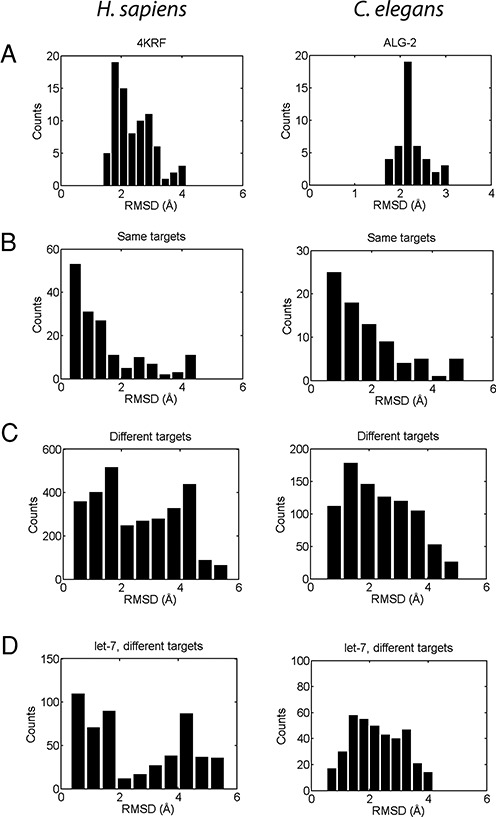
Quantification of conformational changes in human and worm Ago structures associated with target recognition. Ago–Ago RMSD values for: (A) Agos at all targets superimposed with a reference structure (AGO1 (4KRF) for human and modeled ALG-2 for worm). (B) Agos binding the same targets. (C) Agos binding different targets. (D) Agos associated with a let-7 guide miRNA binding to different targets. In all cases, 5 model miRISC structures were assessed for each target.
Accommodation of duplex conformations by human AGO1 is possible partly because of the presence of pockets within Ago's RNA-binding channel and partly because duplexes can protrude out of the channel to solvent-exposed regions in order to avoid steric clashes (Figure 4 and Supplementary Figures S5 and S6). The former scenario accounts for accommodation of most duplexes with a moderate degree of duplex distortions or distortions occurring near Ago's pockets. For example, the let-7a::BIRC6 duplex has a bulge, with bases A5 and A6 of the mRNA strand flipped out, but still accommodated by a pocket between the N-terminal group and linker L1 of AGO1 (Supplementary Figure S6a). Similarly, for the second target site on the BIRC6 mRNA (site 2), the flipped out base U16 of the miRNA strand is accommodated by a pocket in the N terminal group (Supplementary Figure S6b).
Several duplexes with strong structural distortions are also accommodated by the AGO1 structure. The 3′ end of the let-7b::SLC25A13 duplex is drastically bent from the helical axis and protrudes out of the AGO1's RNA-binding channel, which helps avoid steric clashes with Ago; this duplex also has a large internal loop (8 nt covering seed and mid-duplex regions) where most bases are flipped out but avoid steric clashes with the N terminal, L2 and PIWI domains (Supplementary Figure S6c). Another example is the let-7a::HMGA2 duplex with a 9-nt internal loop that produces a sharp bend enabling it to avoid steric clashes with the N-terminal and PIWI domains (Figure 4). The duplex miR-15a::CCND2 has a 5-nt internal loop (bases 12–16) on the miRNA strand, which generates a distorted loop that protrudes into the solvent-accessible side of the RNA-binding channel and interacts with the PAZ domain (Supplementary Figure S6d). Finally, the seedless duplex miR-17*::VIM (site 2) has three flipped out mRNA bases U3, C4 and U5 causing a severe bend but avoids steric clashes with the AGO1 structure; the U3 base is solvent exposed, whereas the C4 and U5 bases are positioned between the linker L2 and the PAZ domain (Supplementary Figure S6e). These cases illustrate the myriad ways in which significant duplex structural distortions can be accommodated by the pockets and solvent-exposed surfaces of the Ago structure.
Beyond these cases, mapping the general character of Argonaute-RNA interactions would provide some guidance for experimental work. To perform this analysis, we used the same dataset of chimeric miRNA–mRNA reads which are supported by other experiments (targets in Supplementary Table S3 in (12)). We constructed 78 AGO1-duplex complexes with diverse seed and seedless duplex structures (number of unpaired bases range from 0 to 18); we considered only the best fitting complexes with the lowest number of atomic overlaps. To analyze the effects of imperfect duplexes on AGO1-RNA interactions, we divided the duplexes into structure classes 1 to 4 by the number of unpaired bases (0, 1–4, 5–8, 9–12) and used residue-resolution contact maps to represent protein-RNA interactions (see Methods). For straight, perfect duplexes or class 1 structures, their miRNA strands exhibit localized interactions with only the L1, L2, Mid and PIWI domains (Supplementary Figure S7, left). In contrast, class 2 duplexes (1–4 unpaired bases) display significant interactions with the N terminal domain in addition to the other domains. Further, more interaction residues are involved (larger dotted regions) compared with those in the class 1 plot. This outcome is also observed for structure classes 3 and 4, indicating that structural imperfections in duplexes cause significant changes in AGO1-miRNA interactions not observed in class 1 duplexes. A similar general observation also holds for AGO1-mRNA interactions (Supplementary Figure S7, right). The mRNA strand of class 1 duplexes interacts strictly with the L1, L2, Mid and PIWI domains of AGO1. In contrast, the mRNA strand of duplex structure classes 2–4 interacts with AGO1's N terminal group and PAZ domain. In particular, as the number of unpaired bases in the duplex increases (progressing from class 1 to class 4 duplexes), the contact maps show a corresponding increase in the number of mRNA bases interacting with the L1, L2 and PIWI domains. This is clearly due to the size of the internal loops and bulges of these duplexes, allowing multiple diverse opportunities to interact with AGO1 domains.
Models of C. elegans miRISC structures indicate that the complex's quality depends on the duplex region where structural distortions occur
To further assess the impact of structurally distorted duplexes on miRISC's viability, we now examine a set of genetically and biochemically verified C. elegans miRNA-target duplexes (6,48). Since experimental C. elegans Ago structures are not available, we used comparative modeling to build C. elegans ALG-2 structures based on homologous human AGO1 and AGO2 structures (∼66% sequence identity with ALG-2; see Methods). The optimal C. elegans miRISC structures show that there is a varying quality of fit within the open ALG-2 structures (Figure 6 and Supplementary Figure S8): six duplexes can be fully accommodated (let-7::daf-12, let-7::hbl-1, let-7::lin-41, lin-4::lin-14, lin-4::lin-28, miR-61::vav-1), whereas the other six duplexes can only be partially accommodated (let-7::nhr-25, let-7::pha-4, let-7::lss-4, miR-273::die-1, lsy-6::cog-1, miR-84::let-60). In all miRISC structures the solvent-exposed seed duplexes are unhindered. Steric clashes tend to occur in the central region of the duplex (base pair positions 10–14) where it meets the PIWI domain. The 3′ end region of the duplex (positions 14 and beyond) is largely free of steric clashes because the N-terminal region is mostly solvent-exposed. Similar factors explain how the seed and 3′-end duplex regions avoid steric clashes with human AGO1. These observations suggest miRNA-target duplex regions that can tolerate significant structural distortions induced by bulges and internal loops.
Figure 6.
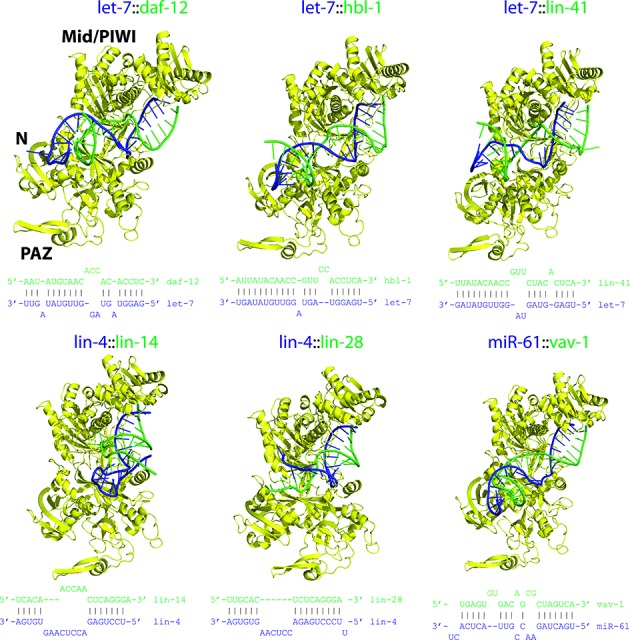
Models of C. elegans miRISC structures. All viable structures were assembled using the method outlined in Figure 1; each displayed structure is ranked in the top 5. Complexes were derived from ALG-2 and a set of miRNA-target duplexes (secondary structures shown) from experimental literature; the initial ALG-2 structure (without the unstructured residues 1–73) was modeled using human AGO1 and AGO2 templates via comparative modeling. Color scheme: Ago, yellow; guide RNA strand, blue; and mRNA strand, green.
The worm Ago conformation shows a similar pattern of changes in response to different duplex structures as observed for human AGO1. The RMSD distribution for models of open ALG-2 structures superimposed with the closed structure has a sharp peak at ∼2 Å (Figure 5A, right). For superimposed ALG-2 structures from distinct targets, the RMSD distribution is again broad (range of ∼5 Å and mean of 2.4 Å; Figure 5C, right). For superimposed ALG-2 structures from the same target sites, the RMSD values are small (distribution peak at ∼1.0 Å with a mean of 1.8 Å; Figure 5B right), indicating only minor variations in the Ago structure at a particular binding site. Note that since some duplexes cannot be completely accommodated within ALG-2, the results represent the optimal Ago conformations for the duplexes. Similarly, the results for the let-7 miRNA family reflect ALG-2 changes in broader miRNA families: average RMSD of 2.3 Å for distinct targets (Figure 5D, right) and 1.7 Å for the same targets.
DISCUSSION
Assembly of miRISC structures
Computational assembly of miRISC structures exploiting experimentally-determined Ago structures and miRNA–target duplexes can provide in-depth analysis of the key components of miRNA-mediated gene silencing machinery. Our miRISC assembly algorithm is based on several main ingredients (Figure 1): treat miRISC assembly as loading of preformed duplexes into Ago structures; derive open Ago structures capable of fitting duplexes by using an iterative ENM method to sample Ago's conformational space; and assemble candidate miRISCs from duplex and Ago structure ensembles to simulate the effects of Ago–RNA conformational dynamics. The complexity of miRISC assembly was made tractable by using multi-resolution (atomic, residue-level, and implicit solvent) models. This general algorithm allows Ago loading with straight duplexes and distorted duplexes arising from imperfect base pairings, which frequently occur in animals (7,11). Although further refinements of the algorithm are possible, we expect our Ago-RNA models can be used to probe their interactions with other effector proteins (e.g. chaperone HSP90 (45–47) and GW proteins (18)) and serve as starting points for detailed, all-atom simulation studies (e.g., effects of divalent ions in target cleavage (2)).
The miRISC assembly algorithm can be improved in several ways. Consideration of higher-frequency modes (nontrivial modes >3) is expected to refine miRISC structures by incorporating local structural changes. The assembled structures can also be refined using molecular dynamics simulations to equilibrate local interactions and sample fluctuations near the optimal conformation. Our algorithm uses preformed duplexes as targets for the Ago structure to describe the final, equilibrated states of target recognition. Describing the complex dynamics of target recognition would involve computing Ago modulation of duplex formation (2), which is beyond the scope of this work. Finally, our entropy computations are still limited to small structures (<1500 atoms); overcoming this limitation will be important for estimating Ago-RNA binding free energies that can be compared with experiments.
Eukaryotic miRISC structures
Solved bacterial miRISC structures have provided many insights into the structural mechanisms of miRNA-mediated gene silencing (2,23). Since eukaryotic Agos load RNA duplexes rather than DNA duplexes found in bacteria (23,49), lack of solved eukaryotic miRISC structures has hampered our ability to adequately analyze their interactions (3). Our assembly of eukaryotic miRISC structures, including human and C. elegans miRISC structures with known duplexes, helps fill current gaps in solved structures and enables comparative analysis of their interactions and dynamics. Our key findings can be summarized as follows: RNA-free eukaryotic (yeast and human) Agos have similar dynamic modes of motion but display subtle differences from bacterial Agos (Figure 2); the binding affinity trends for T. thermophilus, yeast and human miRISCs (RNA loaded only) are similar (Figure 3); the majority of duplexes can be accommodated by open Ago conformations, especially the duplexes derived from miRNA-target ligation experiments (Figure 4 and Supplementary Figure S5); and modeled human and C. elegans miRISC structures suggest that Ago adapts it conformation at distinct targets to fit diverse miRNA–target duplex structures (Figure 5).
The observed similarities of low-frequency dynamic modes and affinity trends in bacterial and eukaryotic Ago and miRISC structures reflect their global structural similarity. Some aspects of such Ago dynamics in prokaryotes have been observed in crystal structures (22) and inferred from computational analysis (40). Our complementary RNA–RNA and Ago–RNA affinity analyses reinforce the view that Ago's DNA/RNA-binding surfaces play the same basic function of stabilizing the duplex and Ago–duplex interactions. The main differences in structural elements between these Agos lie mainly outside of the nucleic acid binding surfaces and may be more relevant for understanding their interactions with other proteins.
Our miRISC models indicate that Ago undergoes a closed/open conformational transition to accommodate the duplex structure, as suggested by experimental Ago loading studies (21). Notably, our models imply that this transition is predominantly mediated by a few low-frequency dynamic modes. Furthermore, Ago's open state allows for structural variations to accommodate diverse duplex structures formed by distinct miRNA families and target sites. The verification of these aspects of target recognition awaits future experimental studies of miRISC structures with distorted duplexes and the dynamics of intermediate Ago-RNA complexes.
Our eukaryotic miRISC models also suggest that the quality of duplex accommodation within Ago structures depends on the nature of duplex structure distortions. Structural distortions in the seed (nt positions 2–8) and 3′ end (positions 14–21) regions are less likely to cause steric clashes with the Ago structure than those in the central duplex region (positions 10–14). This implies that the Ago structure imposes a constraint on duplex structure formation and therefore plays a role in target selection: targets leading to greater miRISC stability are preferred relative to the lower affinity complexes. Thus, a more complete analysis of the targets of miRNAs should incorporate the role of Ago structure. The set of eukaryotic miRISC structures examined here provides a rough guide to the relationship between duplex structure and miRISC stability; specifically, the contact map analysis suggests that duplex imperfections alter the character of duplex interactions with the Ago domains. To further characterize the structural factors influencing miRNA function, larger-scale miRISC modeling studies using high-resolution datasets (such as from chimeric reads (12)) are needed to map this relationship more accurately.
Energy landscapes of the target recognition process
Our analysis of binding energies of these complexes suggests that target recognition steps (increasing number of base pairs) are accompanied by progressive stabilization of miRISC (Figure 3B). This trend is driven by both duplex and Ago-duplex binding energies, and is observed in T. thermophilus, yeast and human complexes. The relationship between binding energy and base pair number describes the trajectory or energy landscape of the target recognition process. From this viewpoint, imperfect duplexes are expected to have more rugged landscapes compared with near perfect duplexes since accommodation of structural distortions in such duplexes requires significant adaptation of Ago conformation. Indeed, our analysis indicates that some C. elegans duplexes may encounter energy barriers that prevent them from being fully loaded into the Ago structure. Energy landscape analysis could thus provide information about the target recognition process complementary to that from structural data.
Implications of miRISC structure modeling
Assembling eukaryotic miRISC complexes highlights the complexity of protein-RNA interactions and requires a careful consideration of the complex's dynamics and energetics. We have identified structural and energetic determinants of Ago loading (e.g., Ago's conformational state, structural distortions in duplex regions, binding affinity trends) complementary to those offered by existing primary and secondary structure-based approaches (7). Although genome-wide target analysis using tertiary structure modeling still poses a considerable challenge due to computing requirements of miRISC structures, these structure-derived features will allow more rigorous future assessment of many known and potential miRNA target sites. Further, we expect development in miRISC modeling could help elucidate the mechanism of seedless sites, possible cooperative effects of adjacent miRNA binding sites (19), interactions of miRISC with other regulatory proteins to modulate translation (17,18), and effects of miRNA target site polymorphisms (50).
Supplementary Material
Acknowledgments
We thank Suse Broyde, Glenn Butterfoss and Sean West for useful discussions on modeling issues and comments on the manuscript. The computations were performed on the High Performance Computing resources at New York University Abu Dhabi.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [U01 HG004276, U01 HG004276-S, RC2 HG005639 to K.C.G.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn C.D., Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013;38:263–271. doi: 10.1016/j.tibs.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Fang Z., Rajewsky N. The impact of miRNA target sites in coding sequences and in 3′ UTRs. PLoS.ONE. 2011;6:e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 6.Zisoulis D.G., Lovci M.T., Wilbert M.L., Hutt K.R., Liang T.Y., Pasquinelli A.E., Yeo G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 9.Cao S., Chen S.J. Predicting kissing interactions in microRNA-target complex and assessment of microRNA activity. Nucleic Acids Res. 2012;40:4681–4690. doi: 10.1093/nar/gks052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan H.H., Gunsalus K.C. Tertiary structure-based analysis of microRNA-target interactions. RNA. 2013;19:539–551. doi: 10.1261/rna.035691.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Li Y., Ma Z., Yang W., Ai C. Mechanism of microRNA-target interaction: molecular dynamics simulations and thermodynamics analysis. PLoS Comput. Biol. 2010;6:e1000866. doi: 10.1371/journal.pcbi.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friend K., Campbell Z.T., Cooke A., Kroll-Conner P., Wickens M.P., Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broderick J.A., Salomon W.E., Ryder S.P., Aronin N., Zamore P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cevec M., Thibaudeau C., Plavec J. Solution structure of a let-7 miRNA:lin-41 mRNA complex from C. elegans. Nucleic Acids Res. 2008;36:2330–2337. doi: 10.1093/nar/gkn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leontis N.B., Lescoute A., Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suhre K., Sanejouand Y.H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti-Renom M.A., Stuart A.C., Fiser A., Sanchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 35.Suhre K., Sanejouand Y.H. On the potential of normal-mode analysis for solving difficult molecular-replacement problems. Acta Crystallogr. Section D, Biol. Crystallogr. 2004;60:796–799. doi: 10.1107/S0907444904001982. [DOI] [PubMed] [Google Scholar]
- 36.Shindyalov I.N., Bourne P.E. A database and tools for 3-D protein structure comparison and alignment using the Combinatorial Extension (CE) algorithm. Nucleic Acids Res. 2001;29:228–229. doi: 10.1093/nar/29.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pappu R.V., Hart R.K., Ponder J.W. Analysis and application of potential energy smoothing and search methods for global optimization. J. Phys. Chem. B. 1998;102:9725–9742. [Google Scholar]
- 38.Pietal M.J., Szostak N., Rother K.M., Bujnicki J.M. RNAmap2D - calculation, visualization and analysis of contact and distance maps for RNA and protein-RNA complex structures. BMC Bioinformatics. 2012;13:333. doi: 10.1186/1471-2105-13-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tama F., Wriggers W., Brooks C.L. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 40.Ming D., Wall M.E., Sanbonmatsu K.Y. Domain motions of Argonaute, the catalytic engine of RNA interference. BMC Bioinformatics. 2007;8:470. doi: 10.1186/1471-2105-8-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tama F., Valle M., Frank J., Brooks C.L. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahar I., Lezon T.R., Yang L.W., Eyal E. Global dynamics of proteins: bridging between structure and function. Annu. Rev. Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Johnston M., Geoffroy M.C., Sobala A., Hay R., Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J., et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sethupathy P., Collins F.S. MicroRNA target site polymorphisms and human disease. Trends Genet. : TIG. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


