Abstract
Nuclear export of messenger ribonucleoproteins (mRNPs) through the nuclear pore complex (NPC) can be roughly classified into two forms: bulk and specific export, involving an nuclear RNA export factor 1 (NXF1)-dependent pathway and chromosome region maintenance 1 (CRM1)-dependent pathway, respectively. SUN proteins constitute the inner nuclear envelope component of the linker of nucleoskeleton and cytoskeleton (LINC) complex. Here, we show that mammalian cells require SUN1 for efficient nuclear mRNP export. The results indicate that both SUN1 and SUN2 interact with heterogeneous nuclear ribonucleoprotein (hnRNP) F/H and hnRNP K/J. SUN1 depletion inhibits the mRNP export, with accumulations of both hnRNPs and poly(A)+RNA in the nucleus. Leptomycin B treatment indicates that SUN1 functions in mammalian mRNA export involving the NXF1-dependent pathway. SUN1 mediates mRNA export through its association with mRNP complexes via a direct interaction with NXF1. Additionally, SUN1 associates with the NPC through a direct interaction with Nup153, a nuclear pore component involved in mRNA export. Taken together, our results reveal that the inner nuclear envelope protein SUN1 has additional functions aside from being a central component of the LINC complex and that it is an integral component of the mammalian mRNA export pathway suggesting a model whereby SUN1 recruits NXF1-containing mRNP onto the nuclear envelope and hands it over to Nup153.
INTRODUCTION
In eukaryotic cells, the genome is physically separated from the cytoplasm by the nuclear envelope (NE). The NE contains a double membrane, the inner nuclear membrane (INM) and the outer nuclear membrane (ONM) which dynamically fuse to create pores and the nuclear pore complexes (NPCs) are embedded inside. mRNA transcription takes place in the nucleus, while mRNA translation into functional protein occurs in the cytoplasm. This spatial problem is settled by the efficient mRNA export from the nucleus into the cytoplasm (1–3).
mRNA export is a multi-stage process and can be divided into three different stages: first, pre-mRNA is packaged into an messenger ribonucleoprotein (mRNP) complex after correct transcription and processing; second, the mRNP targets and translocates through the central channel of the NPC; third, cytoplasmic fibrils release the mRNP into the cytoplasm for translation (4). Transcription is the first step of the mRNP formation. To support ongoing transcription many factors such as proteins of the heterogeneous nuclear ribonucleoprotein (hnRNP) family bind to the nascent mRNA transcripts produced by RNA polymerase II (5). In human cells, about 30 different hnRNPs associate with pre-mRNAs in the nucleus. They affect multiple aspects of mRNA metabolism including the packaging of nascent transcripts, alternative splicing, nucleocytoplasmic transport and translational regulation (6) and accompany the mRNA at different stages during mRNP export (7).
A crucial structure involved in mRNA export is the NPC which has functions in selective material transport. In general, the NPC channel is open for small molecules, like water, sugar and ions, however, in case of larger molecules (5–40 nm in diameter) such as proteins, rRNA and mRNP, particular receptors are required for crossing the NPC (3). Until now, there are two major transport receptors known which are involved in two distinct mRNA export pathways: nuclear RNA export factor 1 (NXF1) (also known as TAP) and chromosome region maintenance 1 (CRM1) (also known as exportin 1). Based on this, mRNA export can be roughly classified into two forms: bulk and specific export (8). Most of the constitutively expressed mRNAs are exported by the NXF1-dependent pathway (bulk export). By contrast, special subsets of mRNAs, uridine-rich small nuclear RNAs, rRNAs and signal recognition particle RNA are exported by the CRM1-dependent pathway (specific export) (3).
The nucleus is physically linked to the cytoskeleton by the linker of nucleoskeleton and cytoskeleton (LINC) complex. It is composed of SUN (Sad1 and UNC-84) domain proteins in the INM and KASH (Klarsicht, ANC-1 and Syne homology) domain proteins in the ONM. In the perinuclear space, SUN and KASH domains directly interact with each other to form a linker between the nucleo- and cytoskeleton through the NE. This connection enables the LINC complex to mediate the communication and force transduction between nucleo- and cytoplasm (9,10). SUN1 and SUN2 are the most prominent SUN domain proteins in human. They directly interact with the KASH domain-containing nesprins as well as with lamin A and emerin (11–13). SUN1 proteins form highly immobile oligomeric complexes in the nuclear envelope in interphase cells for which both perinuclear and nucleoplasmic segments of SUN1 are essential (14).
Recently, it was reported that SUN1 and SUN2 can act as disease-modifier genes in individuals with co-segregating mutations in other Emery-Dreifuss muscular dystrophy genes (15,16). So far, they have not been involved in nuclear mRNA export, however, it was shown that SUN1 associates with the NPC, whereby SUN1 but not SUN2 colocalizes with the NPC and displays an importance for NPC assembly in early interphase and for its distribution across the nuclear surface (14,17,18). In this study, we aim to discover novel functions of SUN1 in mammalian cells aside from being a central component of the LINC complex. Thus, the focus of this work is on the roles of SUN1 in mammalian mRNA export.
MATERIALS AND METHODS
Cell culture
Human fibroblast and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin and streptomycin (100 μg/ml), L-glutamine and non-essential amino acids at 37°C with 5% CO2. Human fibroblast cells were transfected by the CLB-Transfection™ System (Lonza) and HeLa cells were transfected by Lipofectamine 2000 (Invitrogen™) according to the manufacturer's instructions. Transfected cells were incubated for 24–48 h post-transfection.
Plasmids and site-directed mutagenesis
We used a plasmid carrying cDNA sequences encoding human SUN1 full length (Green Fluorescent Protein (GFP)-SUN1) (14) to produce the GFP-SUN1-NT (amino acid residues 1–239) and Glutathione S Transferase (GST)-SUN1-NT (residues 1–239) by Polymerase Chain Reaction (PCR). The siRNA resistant (R) plasmid GFP-SUN1R was generated using a site-directed mutagenesis kit (Promega). GFP-SUN2 full length and GST-SUN2-NT (residues 1–138) (19) were generated from SUN2-V5-His (14). All the Nup153 plasmids used in this study were kind gifts from Prof Birthe Fahrenkrog (20). The pGEX-4T-NXF1 and pET-9a-NXT1 plasmids were kind gifts from Prof Stuart A Wilson (21).
Protein–protein interaction studies
Immunoprecipitation
Human fibroblasts were transfected with GFP, GFP-SUN1 or GFP-SUN2 encoding plasmids. The next day cells were scraped into lysis buffer (10 mM Tris/HCl, pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 0.5% NP-40) supplemented with 1 mM DTT, Benzamidine, PMSF and protease inhibitor cocktail (Roche), followed by sonication and centrifugation (16,000 g at 4°C for 10 min). Lysates were precleared with protein A Sepharose (GE Healthcare) for 1 h and then incubated with GFP-Trap beads (Chromotek) for 2 h at 4°C. Beads were pelleted at 1000 g, and then washed several times with lysis buffer minus protease inhibitors. Samples were heated in 5x sodium dodecylsulphate (SDS) sample buffer (95°C, 5 min) and analyzed using 12% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blotting.
GST pulldown
Expression of recombinant GST-SUN2-NT and GST-NXF1: NXT1 polypeptide was in Escherichia coli strain XL1-Blue. GST-SUN1-NT and all GST-Nup153 polypeptides were produced in ArcticExpress RIL. Cells were lysed and the fusion proteins were isolated as described in (22). After several washing steps, the purified GST fusion proteins were incubated with human fibroblast or HeLa total cell lysates (12,000 g, supernatant) for 1.5–3 h at 4°C. GST alone was used as negative control. Beads coupled with protein complexes were washed three times (500 g, 4°C, 1 min) with lysis buffer minus protease inhibitors (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate) and heated in 5x SDS sample buffer (95°C, 5 min). Samples were analyzed using 12% SDS-PAGE followed by western blotting.
Immunofluorescence analysis and antibodies used
Fixation of cultured cells grown on coverslips was with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min followed by permeabilization with 0.5% Triton X-100 for 5 min for primary antibody staining. Alternatively, cells were fixed in cold methanol (−20°C) for 10 min. The following antibodies were used: rabbit monoclonal anti-SUN1 (Abcam), rabbit monoclonal anti-SUN2 (Abcam), mouse monoclonal anti-hnRNP F/H (ImmuQuest), mouse monoclonal anti-hnRNP K/J (ImmuQuest), rabbit polyclonal anti-Lamin B1 (Abcam), mouse monoclonal anti-NXF1 (Abcam), mouse monoclonal anti-RUVBL1 (Abcam), mouse monoclonal anti-Nuclear Pore Complex Proteins (mAb414) (Abcam), mouse monoclonal anti-Nup153 [QE5] (Abcam), rabbit polyclonal anti-GANP (Bethyl Laboratories) and mouse monoclonal anti-GAPDH−POD (Sigma). The secondary antibodies used were conjugated with Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes, Sigma). The samples were counterstained with DAPI (Sigma) for DNA and mounted in gelvatol. Samples were analyzed using confocal laser scanning microscopy (TCS-SP5, Leica) or Leica Structured Illumination (integration of the OptiGrid® module, Leica research microscopes and Leica MM AF software).
RNAi
RNAi was performed as described (23). Allstars negative control siRNA and SUN1 siRNA (5′-TTACCAGGTGCCTTCGAAA-3′) were purchased from QIAGEN and Microsynth, respectively. 5×103 HeLa cells were seeded in 24-well plate for immunofluorescence analysis and 5×104 HeLa cells were seeded in 6-well plates for western blot analysis. RNAi transfection was performed at a final siRNA concentration of 20 nM per target gene using transfection reagent (INTERFERin®, Polyplus) according to the manufacturer's protocol. Cells were either fixed or harvested 72 h after transfection. For rescue experiments, HeLa cells were transfected with GFP-SUN1R expression vector by Lipofectamine® 2000 (Invitrogen) after 24 h siRNA treatment. Cells were processed for detection of poly(A)+RNA by in situ hybridization after 72-h siRNA treatment.
Subcellular fractionation
After RNAi transfection, HeLa cells were processed for subcellular fractionation as described (24). Low concentrations of NP-40 (0.1%) disrupt cytoplasmic, but not nuclear membranes and short centrifugation times allow intact nuclei to come down leaving soluble cytosolic proteins in the supernatant. Cells grown in 6-well plates were washed, trypsinized and centrifuged in 1.5 ml centrifuge tubes (1000 rpm, 5 min at 4°C). After removing the supernatants, the cell pellets were resuspended in fractionation buffer (0.1% NP-40 in PBS) and triturated five times using a p1000 micropipette. Afterward, part of the lysate was removed and kept as the ‘whole cell lysate.’ The rest samples were centrifuged with a table top microfuge (∼10 s), and then the supernatants were kept as the ‘cytosolic fraction.’ Next the pellets were washed with fractionation buffer once and centrifuged as above. After removing the supernatants, the pellets (referred to as nuclei) were resuspended in fractionation buffer and designated as the ‘nuclear fraction.’ Finally, the ‘whole cell lysates’ and the ‘nuclear fractions’ were sonicated and heated along with the ‘cytosolic fraction’ at 95°C for 5 min. Samples were resolved by SDS-PAGE and analyzed by western blotting with appropriate antibodies.
In situ hybridization and cell proliferation assay
HeLa cells were cultured and processed as described (25). To localize poly(A)+RNA in HeLa cells, 5×103 cells were seeded in 24-well plates and fixed after 72-h RNAi transfection. For leptomycin B (LMB) treatment, HeLa cells were treated with LMB at a final concentration of 7 ng/ml (37°C, 2 h) after 72-h RNAi transfection. After fixation with 4% paraformaldehyde, the cells were permeabilized by 0.2% Triton X-100 in PBS. Then prehybridization was performed by incubating the cells with prehybridization buffer (125 μg/ml tRNA, 0.5 mg/ml ssDNA, 1 mg/ml BSA, 50% formamide, 1 mM vanadyl ribonucleoside complexes and 0.1 g/ml dextran sulfate in 1x SSC) at 37 °C for 1 h (in a tissue culture incubator). Then the poly(A)+RNA was localized by hybridization with 1 ng/μl of Cy3-conjugated Oligo d(T)50 probe (Gene Link, Inc.) in prehybridization buffer at 37°C for 2 h. After washing with SSC and PBS, the samples were stained with DAPI and mounted with gelvatol. Images were acquired using confocal laser scanning microscopy (TCS-SP5, Leica) and processed with LAS AF Lite software (Leica). The mean Cy3 intensity was determined for the nuclear and cytoplasmic distribution of 500 individual cells at each condition. Nuclear/cytoplasmic (N/C) ratios were calculated and plotted on line graphs for the indicated conditions. For cell proliferation assays, HeLa cells were counted using a TC10™ Automated Cell Counter (Bio-Rad) for viable cell number at each indicated time point following transfection with control or SUN1 siRNA.
Poly(A) + RNA isolation
1.5×107 HeLa cells were harvested for nuclear poly(A)+RNA immunoprecipitation essentially as described (26), except Dynabeads® Oligo (dT)25 were used instead of oligo(dT) cellulose. Briefly, the harvested HeLa cell pellet was washed in 1 ml ice cold PBS and centrifuged at 1000 rpm for 2 min. The pellet was suspended in 1 ml PBS homogenization buffer (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH 7.2, 0.2% NP-40 and 100 μg/ml PMSF) and homogenized using a glass homogenizer (tight pestle). The nuclear fraction was isolated after 10 min centrifugation (2000 g, 4°C). After removing the supernatant (cytoplasmic fraction), the pellet was lysed in 450 μl PBS lysis buffer (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH 7.2, 100 μg/ml PMSF, 5 mM vanadyl ribonucleoside complex and 0.2 U/μl of RNasin from Promega) with three times (5 s each) sonication and 10 min centrifugation (2000 g, 4°C) afterward. The resulting supernatant was the soluble nuclear extract. Then the Dynabeads® Oligo (dT)25 and 0.05% NP-40 were added and incubated with nuclear extract for 45 min at 4°C with gentle rotation. The poly(A)+RNA was isolated after washing the beads thoroughly for three times (500 μl each) with PBS lysis buffer plus 0.1% NP-40 by placing on the magnet at room temperature. Samples were resolved by SDS-PAGE and analyzed by western blotting with appropriate antibodies.
RESULTS
SUN1 and SUN2 interact with hnRNP F/H and hnRNP K/J
The largest member of the SUN domain family, SUN1, contains 812 amino acids corresponding to a molecular weight of ∼90 kD, while SUN2 contains 717 amino acids with a molecular weight of ∼80 kD (27). The SUN domain proteins consist of an N-terminus, transmembrane, coiled- coil and a SUN domain at the C-terminus located in the lumen of the NE (Figure 1A). The N-terminus of SUN domain protein associates with the nucleoskeleton by interactions with A-type lamins, chromatin-binding proteins and other nuclear proteins.
Figure 1.
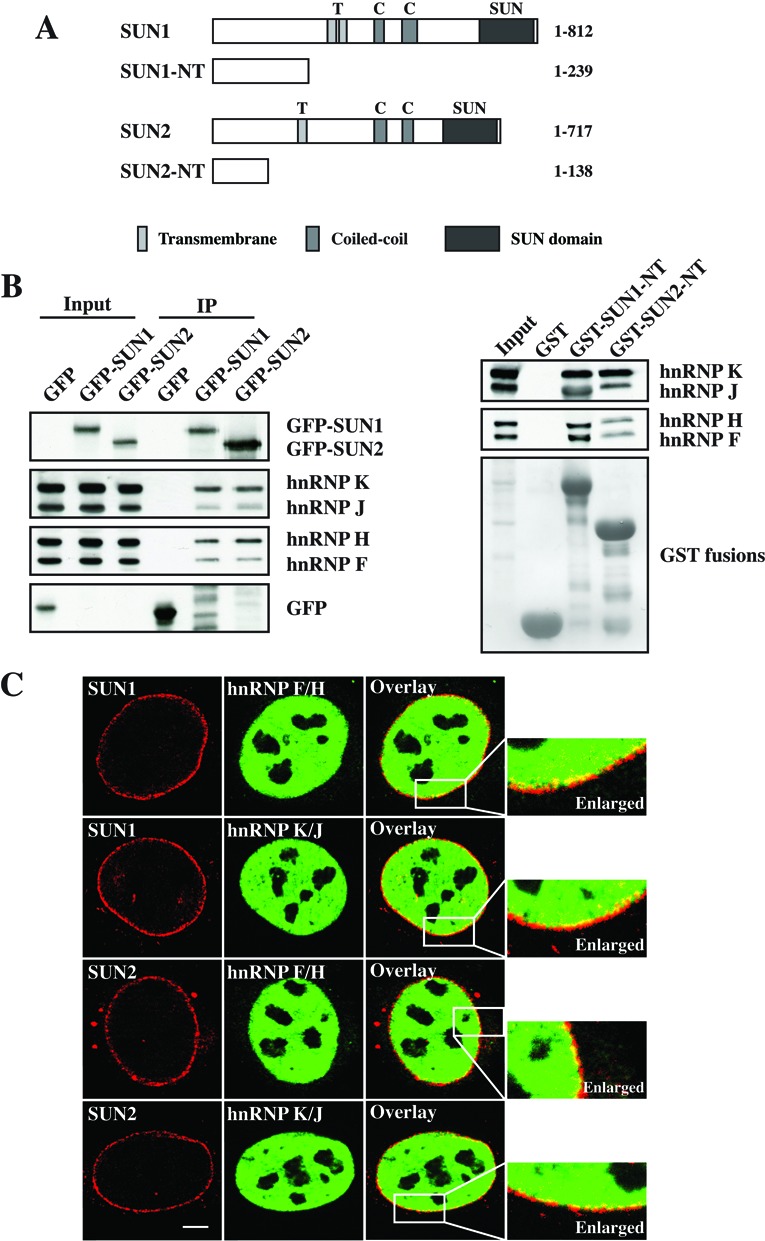
Interaction and colocalization of SUN1 and SUN2 with hnRNPs. (A) Schematic of the full length and the N-termini of SUN1 and SUN2 used in this study. Transmembrane, coiled-coil and SUN domain are shown. (B) Western blot analysis of Co-IP and pulldown between SUN1 and SUN2 with hnRNPs. For Co-IP, lysates from human fibroblasts expressing GFP alone, GFP-SUN1 or GFP-SUN2 were immunoprecipitated by using GFP-Trap beads and detected by GFP monoclonal antibodies. For pulldown experiment, GST, GST-SUN1-NT and GST-SUN2-NT were used and visualized by Ponceau S staining. The proteins were separated by SDS-PAGE (12% acrylamide) and the interactions were detected by hnRNP F/H and hnRNP K/J antibodies. (C) Colocalization of endogenous SUN1 and SUN2 with hnRNP F/H or hnRNP K/J. HeLa cells were stained with appropriate antibodies and analyzed by confocal microscopy. Bar: 5 μm.
In a previous study, in order to further understand the role of SUN proteins, we screened for SUN interacting proteins by applying pulldown experiment and LC–MS analysis (19). Among the proteins identified were hnRNPs, like hnRNP F/H and hnRNP K/J. The hnRNP F/H are well-known for their functions in the regulation of alternative splicing, and the hnRNP K, one of the shuttling hnRNPs, plays an important role in many aspects of mRNA metabolism, including mRNA silencing, transcription, splicing, regulation of mRNA stability and translation (6,7).
To confirm the interactions between SUN1/2 and hnRNPs, we performed co-immunoprecipitation (Co-IP) and pulldown experiments. For the Co-IP studies, wild-type fibroblasts were transfected with GFP, GFP-SUN1 or GFP-SUN2 and immunoprecipitated by GFP-Trap beads. For pulldown assays, GST tagged N-termini of SUN1 (residues 1–239) and SUN2 (residues 1–138) (Figure 1A) were directly incubated with the cell lysates and GST alone was used as negative control. All samples were subjected to western blot analysis and probed with appropriate antibodies. Strong signals of hnRNP F/H and K/J were detected in both experiments confirming that SUN1 and SUN2 interacted with hnRNP F/H and hnRNP K/J, suggesting that the N-termini of SUN proteins might be involved in mRNA biogenesis according to the functions of hnRNP F/H and hnRNP K/J and that SUN1 and SUN2 might interact with hnRNP–mRNA complexes in vivo (Figure 1B). We further examined the localization of the endogenous proteins by immunofluorescence. Consistent with previous studies, hnRNP F/H and hnRNP K/J were shown mainly in the nucleus and colocalized with both SUN1 and SUN2 along the inner nuclear envelope, suggesting that SUN1 and SUN2 associate with hnRNPs on the NE and that they might play a role in mRNA biogenesis at a late stage, such as mRNA export (Figure 1C).
hnRNP F/H and hnRNP K accumulate in the nucleus upon SUN1 depletion
The hnRNP proteins accompany mRNA at different stages during mRNA export (7). Before an mRNP (mature mRNA) translocates through the NPC, the nuclear-restricted hnRNPs containing nuclear-retention signals start to get released from the mRNP and stay in the nucleus, whereas the shuttling hnRNPs will accompany the mRNA through the NPC channel and into the cytoplasm (4,7). Therefore, the shuttling hnRNPs become an important marker reflecting the efficiency of mRNA export. One such well-known shuttling hnRNP involved in our study is hnRNP K (hnRNP F/H were unknown until now), which interacts with both SUN1 and SUN2.
To confirm our hypothesis that SUN proteins are involved in mRNA export, we performed siRNA-mediated knockdown studies of SUN1. Immunofluorescence and western blot analysis confirmed the efficient knockdown of SUN1 in HeLa cells (Figure 2A and B). Immunofluorescence analysis by confocal microscopy of HeLa cells showed strong NE staining for SUN1 in control siRNA cells and very weak staining in SUN1 siRNA cells (Figure 2A). Western blot analysis showed that SUN1 was absent from HeLa lysates after siRNA depletion (Figure 2B). To test whether SUN1 depletion influences the distribution of hnRNPs, we performed subcellular fractionation in control and SUN1 knockdown cells (Figure 2C). The results showed that SUN1 was only observed in the whole cell lysate and the nuclear fraction of control cells, but was not detectable in SUN1 knockdown cells. Lamin B1 and GAPDH were used as nuclear and cytosolic marker, respectively. Interestingly, based on the levels of lamin B1 and GAPDH, both hnRNP F/H and K appeared decreased in the cytosolic fraction of SUN1 knockdown cells and increased in the nuclear fraction compared to control knockdown cells (Figure 2C). These results indicate that, in the absence of SUN1, the hnRNP F/H and hnRNP K accumulate in the nucleus suggesting SUN1 depletion affects the nucleocytoplasmic transport of the shuttling hnRNP protein hnRNP K, supposed to be a marker of mRNA export.
Figure 2.
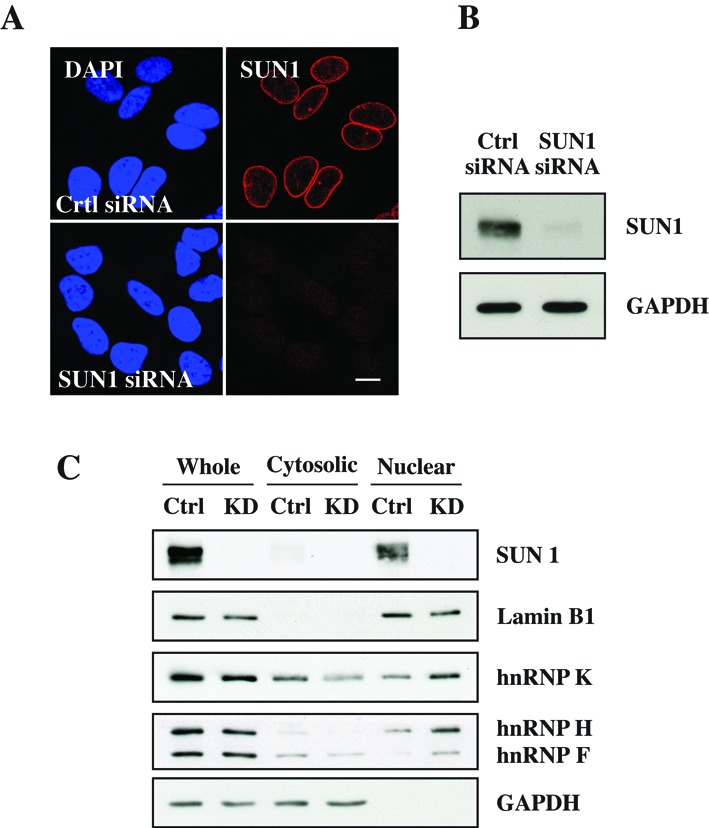
SUN1 depletion results in nuclear accumulation of hnRNPs. (A) Immunofluorescence analysis of HeLa cells treated with control and SUN1 siRNA and stained with SUN1 antibodies. Nuclei are indicated by DAPI staining. Bar: 10 μm. (B) Western blot analysis of SUN1 expression levels in control and SUN1 siRNA cells. GAPDH was used as loading control. (C) Separation of nuclear and cytoplasmic proteins by subcellular fractionation in control and SUN1 knockdown cells. Lamin B1 and GAPDH were used as nuclear and cytosolic markers, respectively.
RNA fluorescence in situ hybridization reveals nuclear accumulation of poly(A)+RNA in SUN1 depleted cells
Our observation that hnRNP F/H and hnRNP K accumulates in the nucleus after SUN1 depletion provides a clue that SUN1 might participate in hnRNP-involved mRNA export. To investigate this further, we examined the cellular distribution of poly(A)+RNA in control and SUN1 siRNA cells via RNA fluorescence in situ hybridization (FISH) with an oligo (dT) probe. In control cells, most poly(A)+RNA was cytoplasmic, except for a few discrete foci in the nucleus. In contrast, the SUN1 siRNA cells showed strong nuclear accumulation of poly(A)+RNA (Figure 3A). These results indicate that the poly(A)+RNA distribution is altered in SUN1 depleted cells, whereby poly(A)+RNA strongly accumulated in SUN1 depleted nuclei compared to control. Further, rescue of the mRNA export defect was performed by expressing a GFP-tagged siRNA-resistant (R) SUN1 cDNA (GFP-SUN1R) under siRNA treatment condition. We found that the expression of GFP-SUN1R with SUN1 siRNA partially rescued the mRNA export defect confirming that the SUN1 knockdown phenotype was not due to an off-target effect (Figure 3A). Importantly, the nuclear export of the shuttling proteins BRCA1 (28) and c-Fos (29) took place in the absence of SUN1 (Supplementary Figure S1), demonstrating that NPCs were functional for bidirectional transport of protein–cargo complexes in SUN1-depleted cells. Consistent with these results, the SUN1 depletion also caused a defect in cell proliferation. At 24- and 48-h post-transfection, the knockdown cells showed no defect in cell proliferation and were comparable with control siRNA transfected cells. However, significant defects were seen at 72 and 96 h after SUN1 depletion with a reduction to 86 and 83%, respectively (Figure 3B). Our results indicate that SUN1 depletion results in a defect of mRNA export and cell proliferation concluding that SUN1 contributes to the efficient nuclear export of mRNA.
Figure 3.
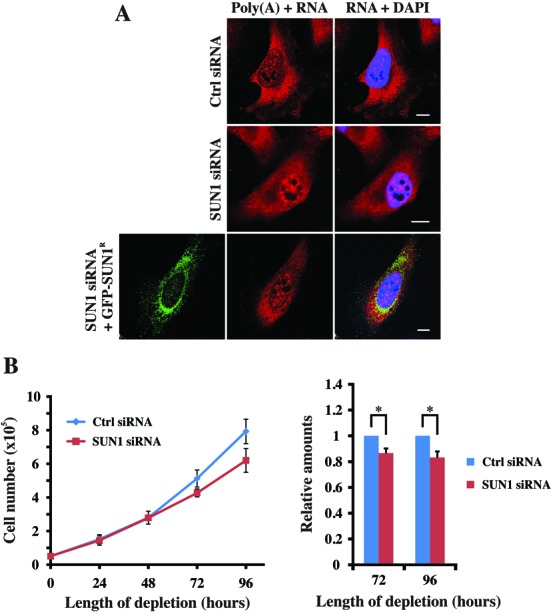
(A) SUN1 depletion results in nuclear accumulation of poly(A)+RNA. RNA FISH showed poly(A)+RNA accumulation in the nucleus of SUN1 depleted cells after 72 h of siRNA transfection (upper two panels). For rescue experiment, HeLa cells were transfected with a GFP-SUN1R expression vector and processed for poly(A)+RNA detection by FISH after 24- and 72-h siRNA treatment, respectively (last panel). Bar: 10 μm. (B) Time courses of cell proliferation defects in cells depleted for SUN1. Viable cell number was determined in SUN1-depleted samples 24–96-h post-transfection and represent the mean of independent readings from three independent depletion experiments ±SD. P-values are shown from three independent experiments (*P < 0.05).
SUN1 functions in the NXF1-dependent mRNA export
Recently two pathways, the NXF1-dependent pathway and CRM1-dependent pathway, were characterized in mammalian mRNA export (8). Next we asked which mRNAs require SUN1 for export and in which export pathway SUN1 is involved. To answer these questions, we used LMB to block the CRM1-dependent export under RNAi condition and compared the distribution of poly(A)+RNA in both LMB untreated and treated cells (Figure 4A). For each trial, the N/C ratio of the poly(A)+RNA distribution was determined by measuring fluorescence intensity. Without LMB treatment, the N/C ratios of the control cells are centered at the range (0.9–1.3) with a mean value of ∼1.15 which is similar as previously reported in HeLa control cells (25). Interestingly, the N/C ratios of most SUN1 siRNA cells are centered at the range of (1.3–1.7) with a mean value of ∼1.63 reflecting higher nuclear accumulation of poly(A)+RNA (Figure 4B and C). After treatment with 7 ng/ml of LMB, the N/C ratios of both control- and SUN1 siRNA-transfected cells were significantly shifted to (1.3–1.7) and (1.7–2.1) with an increased mean value up to ∼1.56 and ∼1.84, respectively (Figure 4B and C). In control cells, the increase of the N/C ratio after LMB treatment revealed that the drug treatment worked under our experimental condition. Most strikingly, in SUN1 siRNA cells, the shift of the N/C ratio range and the increase of the mean value indicates that LMB treatment can cause additional nuclear accumulation of poly(A)+RNA above that caused by SUN1 depletion. Form these results we conclude that in SUN1 knockdown cells there exists a CRM1-dependent and a CRM1-independent pathway which in turn suggests that SUN1 might participate in the NXF1-dependent pathway.
Figure 4.
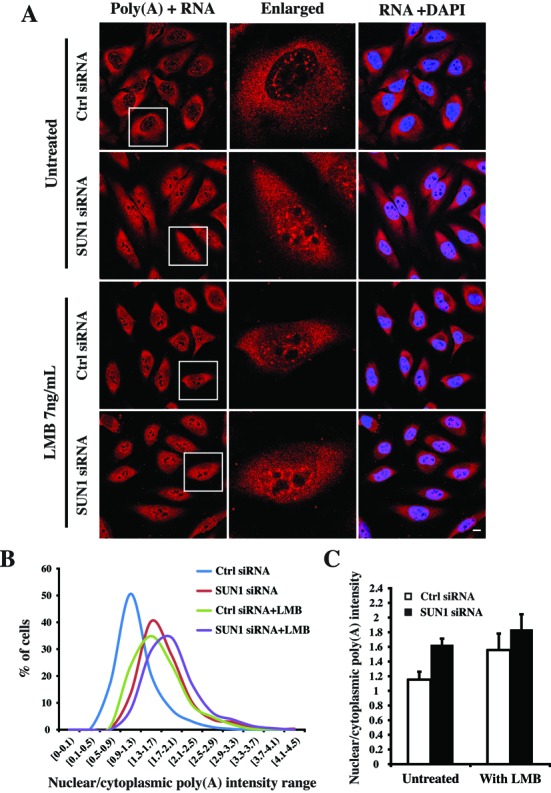
Comparison of the nuclear accumulation of poly(A)+RNA caused by SUN1 depletion and LMB treatment. (A) RNA FISH showed the distribution of poly(A)+RNA in both LMB untreated and treated cells under siRNA condition. The single cells (boxed) are enlarged. The treatment was performed by adding 7 ng/ml of LMB for 2 h after 72-h siRNA transfection. Bar: 10 μm. (B) The nuclear/cytoplasmic (N/C) ratio of the poly(A)+RNA distribution was determined by measuring fluorescence intensity, and the percentages of cells in each N/C ratio intensity range are shown. For each condition ∼500 cells were analyzed. (C) The mean nuclear/cytoplasmic (N/C) ratio of LMB untreated and treated cells under siRNA condition. The values represent the mean ±SD of three independent experiments.
SUN1 associates with nuclear mRNP through a direct interaction with NXF1, a general mRNA export factor in mammals
In higher eukaryotes, NXF proteins exist as a protein family due to the amplification of Nxf genes. The best known NXF protein is NXF1 (Mex67p in yeast) which directly interacts with mRNA (30) and has an essential role in bulk mRNA export as a heterodimer with NXT1 (31,32). In eukaryotic cells, the formation of the mRNP complex is the first step of mRNA export (4). To test whether SUN1 is associated with mRNP, we harvested the nuclear poly(A)+RNPs from mammalian nuclei with Dynabeads® Oligo (dT)25. Endogenous SUN1 specifically copurified with the poly(A) fraction as did NXF1 (Figure 5A). Moreover, hnRNP K, hnRNP H and a reduced amount of hnRNP J and hnRNP F were also observed in the poly(A) fraction, but not RUVBL1 which we used for control (Figure 5A). RUVBL1 is a AAA+ protease linked to a wide range of cellular processes and present in cellular complexes like the histone acetyltransferase Tip60 complex, chromatin remodeling complexes Ino80 and SWR-C, and the telomerase complex. Taken together, our results indicate that SUN1 associates with NXF1-containing nuclear mRNPs, either directly or indirectly through other proteins.
Figure 5.
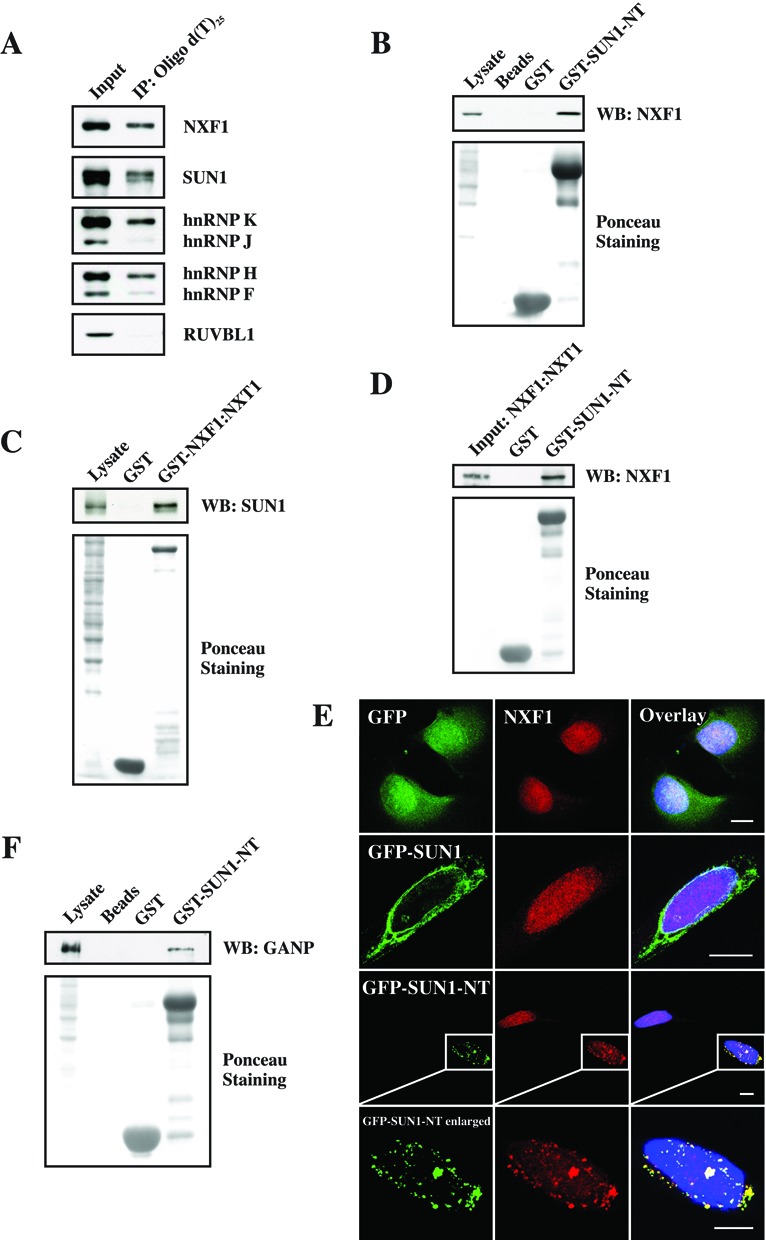
SUN1 associates with mRNP, NXF1 and GANP. (A) SUN1 interacts with nuclear poly(A)+RNPs. Nuclear poly(A)+RNPs purified from soluble nuclear extract of HeLa cells was analyzed by western blotting for associated proteins. (B) The N-terminus of SUN1 interacts with NXF1. Endogenous NXF1 was pulled down from HeLa cell lysates by GST-tagged N-terminus of SUN1. (C) NXF1 interacts with SUN1. The proteins encoded by plasmids pGEX-4T-NXF1 and pET-9a-NXT1 were coexpressed in Escherichia coli strain XL1-Blue and purified by Glutathione Sepharose 4B. Endogenous SUN1 was precipitated by GST-tagged full-length NXF1:NXT1. (D) The N-terminus of SUN1 directly interacts with NXF1:NXT1 released from the GST part after thrombin cleavage. (E) Immunofluorescence showed the altered localization of NXF1 in GFP-, GFP-SUN1- and GFP-SUN1-NT-overexpressing cells. Bar: 10 μm. (F) The N-terminus of SUN1 interacts with GANP. Endogenous GANP was pulled down from HeLa cell lysates by GST tagged N-terminus of SUN1. For all pulldown experiments, proteins were separated by SDS-PAGE (12% acrylamide) and the GST fusion proteins were visualized by Ponceau S staining. GST was used as negative control.
NXF1 is a general metazoan mRNA export factor to transport the mRNP through the NPC (33). In order to find out how SUN1 associates with the mRNP and functions in the NXF1-dependent pathway, we performed several studies. Endogenous NXF1 was pulled down from HeLa cell lysates by GST-SUN1-NT (Figure 1A and 5B). Consistently, endogenous SUN1 was precipitated by GST-tagged full-length NXF1:NXT1 as well (Figure 5C). GST was used as negative control. A direct interaction study between NXF1:NXT1 and GST-SUN1-NT was performed by pulldown of NXF1:NXT1 which was released from GST by thrombin. NXF1 was detected by NXF1 antibodies in the GST-SUN1-NT precipitate (Figure 5D). These results indicate that SUN1 interacts directly with NXF1 in vitro and the N-terminus of SUN1 is responsible for this interaction. Further, immunofluorescence studies showed that NXF1 uniformly localized in the nucleus of GFP- and GFP-SUN1-overexpressing cells (Figure 5E; panel 1, 2). GFP-SUN1 localized along the NE as previously reported (15). By contrast, overexpressed GFP-SUN1-NT was not localized at the NE; instead, it formed aggregate-like structures within and outside the nucleus which were also positive for NXF1 (Figure 5E; panel 3, 4). The nuclear staining of NXF1 was reduced and only faint staining was seen in the nucleus as compared to untransfected cells (Figure 5E; panel 3). The same experiment was performed for hnRNP F/H and hnRNP K/J as well, however, the GFP-SUN1-NT distribution did not affect the localization of these hnRNPs (data not shown). These results indicate that SUN1 presumably associates with mRNP cargo through a direct interaction with NXF1 which is recruited to the mRNP cargo in the nucleus prior to export, and that the N-terminus of SUN1 (residues 1–239) is essential for this connection.
Moreover, transcription-export (TREX) complex and TREX-2 have important roles in the early stages of nuclear mRNA export (34). One component of TREX-2 is the germinal centre-associated nuclear protein (GANP) which acts as a courier to collect and deliver the NXF1-containing mRNPs to the NPC (26). In pull down experiments we found that GST-SUN1-NT interacted with GANP (Figure 5F), indicating that the mRNP cargo presumably is delivered to SUN1 by GANP and then handed over to the NPC.
SUN1 associates with the NPC through a direct interaction with Nup153, a nuclear FG nucleoporin at the NPC basket involved in mRNA export
The second step in mRNA export is the mRNP targeting and translocation through the central channel of the NPC. Our observation that SUN1 interacts directly with NXF1, which occurs at the NE, suggests a role for SUN1 in mammalian mRNA export whereby SUN1 recruits NXF1-containing mRNP onto the NE and hands it over to the NPC. One precondition of this hypothesis is that SUN1 should colocalize with NPCs. This was evidenced by immunofluorescence microscopy using antibodies against the endogenous proteins (Figure 6A).
Figure 6.
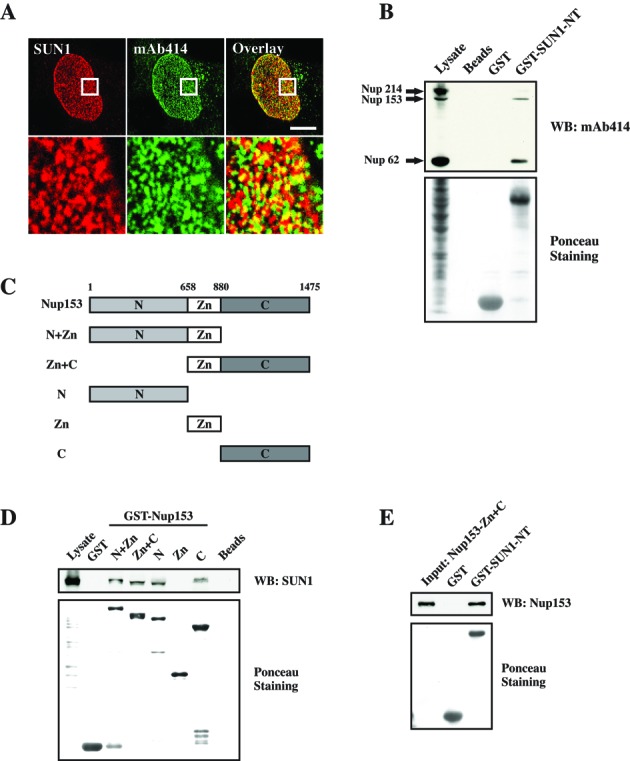
SUN1 colocalization with NPC and interactions with Nup153. (A) Images of the HeLa nuclear surface reveal SUN1 colocalization with NPC labled by mAb414 antibodies. The boxed areas in the top panels are magnified in the bottom panels. Bar: 10 μm. (B) The N-ternimus of SUN1 interacts with Nup62, Nup153 and Nup214. GST-SUN1-NT was used in pulldown study, and NPC proteins were detected with mAb414 antibodies. (C) The predicted domain structure and a schematic of several Nup153 polypeptides which contain N-terminus, Zinc finger and/or C-terminus. (D) Both the N- and the C-terminus of Nup153 interact with SUN1. (E) The N-terminus of SUN1 directly interacts with Nup153-Zn+C polypeptide which was obtained by PreScission protease cleavage and detected by Nup153 antibodies. In all pulldown experiments, proteins were separated by SDS-PAGE (12% acrylamide) and the GST fusion proteins were visualized by Ponceau S staining. GST was used as negative control.
Furthermore, previous reports highlighting the association of SUN1 and NPC revealed that SUN1 is important for NPC assembly in early steps in interphase and for its distribution across the nuclear surface (17,18), but there is no evidence related to SUN1 and NPC-mediated mRNA export. To further confirm the association between SUN1 and NPC, we performed pulldown experiments. Western blot analysis showed that endogenous Nup62, Nup153 and a faint band of Nup214 were observed in GST-SUN1-NT precipitates as revealed by using mAb414 antibodies (Figure 6B). This result indicates that SUN1 associates with the NPC via either a direct or indirect interaction with a subset of nucleoporins, such as Nup62, Nup153 and Nup214.
To date, apart from vertebrate pore proteins Nup98, Nup133, Nup160, Tpr and CAN/Nup214, Nup153 has been involved in the export of mRNA (35–38). Moreover, compared to Nup62 and Nup214, GST-SUN1-NT showed a preference for Nup153, given that there was much lower amount of Nup153 present in the lysates (Figure 6B). Thus, we extended our interaction studies to SUN1 and Nup153. According to the domain structure of Nup153, we used a series of plasmids which coded for the N-terminus, Zinc finger and C-terminus of Nup153 for pulldown study (Figure 6C). Endogenous SUN1 was precipitated by all GST fusion proteins which contain either N-terminal or C-terminal sequences, but not by the Zinc finger alone (Figure 6D). This result indicates that Nup153 interacts with SUN1, and that both the N- and the C-terminus are important for this binding. Further, a direct interaction study between Nup153 and GST-SUN1-NT was performed by pulldown using PreScission protease cleaved purified Nup153-Zn+C fragment. A strong signal of Nup153-Zn+C was detected by Nup153 antibodies in the GST-SUN1-NT precipitates (Figure 6E). GST alone was used as negative control. Therefore, SUN1 associates with the NPC through Nup153, which is a nuclear FG nucleoporin at the NPC basket and plays an important role in mRNA export.
DISCUSSION
It is well-known that SUN1 is the INM component of the LINC complex and has important roles related to the LINC complex. In this study, we show that SUN1 has a novel function in mammalian mRNA export aside from its role in the LINC complex. Our results showed that the SUN proteins interact and colocalize with hnRNP F/H and hnRNP K/J, and SUN1 depletion results in the nuclear accumulation of these hnRNPs (Figures 1 and 2C). This interaction occurs at the N termini of SUN1 and SUN2 (Figure 1). Based on the altered nucleocytoplasmic distributions of hnRNP F/H and hnRNP K in SUN1-depleted cells, we propose that SUN1 contributes to the nucleocytoplasmic shuttling of hnRNP K which we now suggest to be a marker of mRNA export. Besides, lamin A/C, an interaction partner of SUN1, was reported to interact with hnRNP E1 (39), another well-known shuttling hnRNP protein (40,41), suggesting that SUN1 might play a role in hnRNP-involved mRNA export via either direct or indirect interactions.
The TREX complex, an important component of the mRNA export machinery, is formed by the association of the THO complex with six subunits (UAP56, CIP29, CHTOP, PDIP3, ZC11A and UIF) (34) and the depletion of some TREX component, such as ALY (also known as THOC4) (42) and UIF (21), results in the nuclear accumulation of mRNA. Similarly, the deletion of any TREX-2 subunits (Sac3, Thp1, Cdc31, Sus1 and Sem1) in yeast results in mRNA export defects (43–47) and the depletion of most TREX-2 subunits (GANP, PCID2, CETN2, ENY2 and DSS1) in human results in nuclear accumulation of mRNA as well (26,48). Our FISH results reveal that SUN1 depletion results in the nuclear accumulation of poly(A)+RNA indicating that SUN1 might be a so far unknown part of the mRNA export machinery (Figure 3A). Although Sun1−/− mice showed no apparent pathologies expect for infertility (49), cell proliferation analysis showed a reduced cell growth of SUN1 knockdown HeLa cells (Figure 3B). One explanation could be that there are other proteins (such as SUN2) or export pathways (such as membrane budding) (50) compensating for the SUN1-involved mRNA export pathway in the mice.
LMB is a well-known inhibitor of CRM1 which can directly block the CRM1-dependent pathway (51,52). LMB-treated SUN1 depleted cells showed an additive accumulation of nuclear poly(A)+RNA (Figure 4), indicating that SUN1 functions in mRNA export required by the NXF1-dependent pathway. It was known that the TREX component UIF (21) and the TREX-2 component GANP (26) both interact with NXF1. In our study we also reported that SUN1 is associated with nuclear mRNPs through a direct interaction with NXF1 and that the N terminal of SUN1 is essential for this interaction (Figure 5A-D). This strongly supports our hypothesis that SUN1 contributes to the NXF1-dependent pathway. Compared to GFP-SUN1, overexpressed GFP-SUN1-NT accumulates NXF1 within and outside the nucleus (Figure 5E). Whereas overexpressed GFP-SUN1 still localizes at the NE and can transfer the NXF1-containing mRNPs to the NPC for export, the overexpressed GFP-SUN1-NT does not localize at the NE. Instead it forms aggregate-like structures within and outside the nucleus. Since these structures are far away from the NPC, the NXF1 (or NXF1-containing mRNPs) cannot be transferred to the NPC for export, leading to the accumulation of NXF1 along with GFP-SUN1-NT. TREX components are involved in the release of spliced mRNA from nuclear speckle domains, and the depletion of some export factors, such as ALY, results in poly(A)+RNA accumulation in nuclear speckle domains (42). By contrast, the depletion of other export factors, such as GANP, causes the poly(A)+RNA accumulation in more punctuate foci (26,34). In our study, the poly(A)+RNA accumulation due to SUN1 depletion is more close to punctuate foci similar to the effect of GANP depletion, suggesting that the function of SUN1 in mRNA export might be related to GANP. Indeed, this relation of SUN1 with GANP exists as the N terminus of SUN1 interacts with GANP in vitro (Figure 5F).
Consistent with our previous study, SUN1 showed puncta staining and colocalized with the NPC at the nuclear surface which provides the possibility of SUN1 functions in mRNA export (Figure 6A) (14). The association of TREX-2 with the NPC requires the basket nucleoporins Nup153 and Tpr (53). Our pulldown study showed that the N terminus of SUN1 interacts with Nup62, Nup153 and Nup214, and displays a preference for Nup153 (Figure 6B). Similarly, GANP interacts with Nup153 and Nup358 but not Nup62 (26), suggesting that Nup153 might be the crucial connection between mRNP and the NPC at a late stage of mRNA export. Thus, there should be some relations between mRNP and Nup153. Indeed, these relations exist. Nup153 is able to recognize two parts of the mRNP, a single-stranded stretch of mRNA and the mRNA export receptor NXF1 which interacts with the C-terminal region of Nup153 (40). Importantly, we also revealed that the N terminus of SUN1 directly interacts with Nup153 (Figure 6D and E), further confirming our hypothesis that SUN1 hands over the NXF1-containing mRNPs to the NPC component Nup153.
Taken together, mRNA is transcribed by RNA polymerase II, packaged into pre-mRNP by hnRNPs and other RNA-binding proteins, then spliced and processed to form mature mRNA (mRNP). The mRNP is labeled by NXF1 in the nucleus and recruited onto the NE by the INM protein SUN1. Prior to the export from the NPC, the nuclear restricted hnRNPs and some RNA binding proteins are released from the mRNP complex and stay in the nucleus, whereas the shuttling hnRNPs accompany the mRNA into the cytoplasm. Finally, SUN1 hands over the mRNP to the NPC component Nup153, which most likely translocates the mRNP cargo through the NPC either directly by itself or indirectly together with other proteins. This is consistent with a model for SUN1 functions in mammalian mRNA export whereby SUN1, acting as a docking point, recruits NXF1-containing mRNP onto the NE, and hands it over to the NPC component Nup153, which enables the nuclear mRNA export to be performed systematically (Figure 7).
Figure 7.
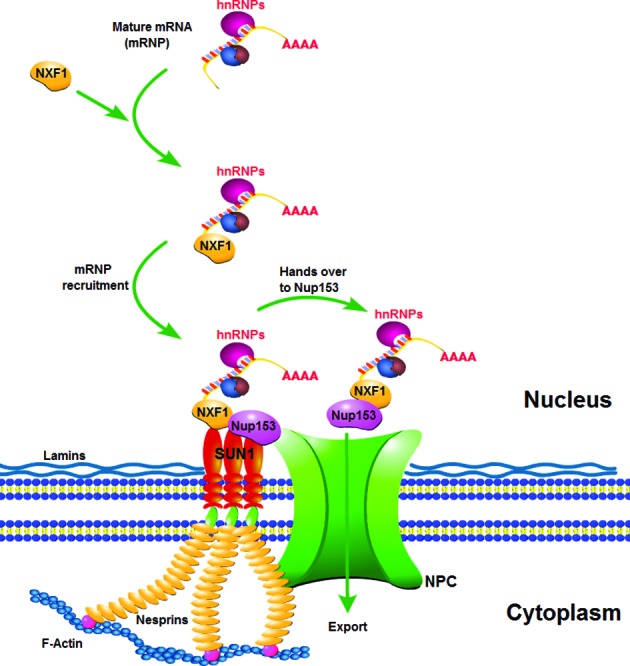
A model of SUN1 functions in mammalian mRNA export. We propose that SUN1 acts as a docking point to recruit NXF1-containing mRNP onto the NE and hands it over to Nup153.
Our data suggest that the functional significance of the interactions between SUN1 and the mRNA export machinery lies in the recognition of mRNPs and their concentration onto the NE for efficient mRNA export. Additionally, the LINC complex plays an important role in mechanosignaling and a crosstalk between both sides of the NE (9,10,54). Nesprins-1 and -2, the ONM components of the LINC complex, contain an actin-binding domain at the N-terminus, enabling them to interact with cytoplasmic actin filaments for mediating the association with cytoskeleton (55,56). This opens new avenues for investigations since it leads to the intriguing possibility that the assembly and disassembly of actin filaments might cause a movement of the LINC complex which in turn enhances SUN1 handing over the mRNP to Nup153.
Supplementary Material
Acknowledgments
P.L. is the recipient of a scholarship from the China Scholarship Council. This work was supported by a grant from CECAD. Prof Birthe Fahrenkrog (Institute for Molecular Biology and Medicine, Université Libre de Bruxelles, Charleroi, Belgium) and Prof Stuart A. Wilson (Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, UK) are acknowledged for providing the Nup153 and NXF1:NXT1 plasmids, respectively. Prof Susan R. Wente (Department of Cell and Developmental Biology, Vanderbilt University School of Medicine, Nashville, TN) is acknowledged for sharing the standard protocol of in situ hybridization. Dr Siu-yuen Chan (Department of Paediatrics & Adolescent Medicine, LKS Faculty of Medicine, University of Hong Kong, Hong Kong) is also acknowledged for providing reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: This work was supported by a grant from Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hocine S., Singer R.H., Grunwald D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Natalizio B.J., Wente S.R. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 2013;23:365–373. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmody S.R., Wente S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matunis E.L., Matunis M.J., Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 7.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 8.Culjkovic-Kraljacic B., Borden K.L. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013;23:328–335. doi: 10.1016/j.tcb.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang W., Worman H.J., Gundersen G.G. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothballer A., Kutay U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 2013;122:415–429. doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 12.Ostlund C., Folker E.S., Choi J.C., Gomes E.R., Gundersen G.G., Worman H.J. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque F., Mazzeo D., Patel J.T., Smallwood D.T., Ellis J.A., Shanahan C.M., Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W., Gotzmann J., Sironi L., Jaeger V.M., Schneider M., Luke Y., Uhlen M., Szigyarto C.A., Brachner A., Ellenberg J., et al. Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim. Biophys. Acta. 2008;1783:2415–2426. doi: 10.1016/j.bbamcr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Li P., Meinke P., Huong le T.T., Wehnert M., Noegel A.A. Contribution of SUN1 mutations to the pathomechanism in muscular dystrophies. Hum. Mutat. 2014;35:452–461. doi: 10.1002/humu.22504. [DOI] [PubMed] [Google Scholar]
- 16.Meinke P., Mattioli E., Haque F., Antoku S., Columbaro M., Straatman K.R., Worman H.J., Gundersen G.G., Lattanzi G., Wehnert M., et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014;10:e1004605. doi: 10.1371/journal.pgen.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talamas J.A., Hetzer M.W. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J. Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q., Pante N., Misteli T., Elsagga M., Crisp M., Hodzic D., Burke B., Roux K.J. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taranum S., Vaylann E., Meinke P., Abraham S., Yang L., Neumann S., Karakesisoglou I., Wehnert M., Noegel A.A. LINC complex alterations in DMD and EDMD/CMT fibroblasts. Eur. J. Cell Biol. 2012;91:614–628. doi: 10.1016/j.ejcb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duheron V., Chatel G., Sauder U., Oliveri V., Fahrenkrog B. Structural characterization of altered nucleoporin Nup153 expression in human cells by thin-section electron microscopy. Nucleus. 2014;5:601–612. doi: 10.4161/19491034.2014.990853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hautbergue G.M., Hung M.L., Walsh M.J., Snijders A.P., Chang C.T., Jones R., Ponting C.P., Dickman M.J., Wilson S.A. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libotte T., Zaim H., Abraham S., Padmakumar V.C., Schneider M., Lu W., Munck M., Hutchison C., Wehnert M., Fahrenkrog B., et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turgay Y., Champion L., Balazs C., Held M., Toso A., Gerlich D.W., Meraldi P., Kutay U. SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J. Cell Biol. 2014;204:1099–1109. doi: 10.1083/jcb.201310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki K., Bose P., Leong-Quong R.Y., Fujita D.J., Riabowol K. REAP: A two minute cell fractionation method. BMC Res. Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folkmann A.W., Collier S.E., Zhan X., Aditi, Ohi M.D., Wente S.R. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell. 2013;155:582–593. doi: 10.1016/j.cell.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickramasinghe V.O., McMurtrie P.I., Mills A.D., Takei Y., Penrhyn-Lowe S., Amagase Y., Main S., Marr J., Stewart M., Laskey R.A. mRNA export from mammalian cell nuclei is dependent on GANP. Curr. Biol. 2010;20:25–31. doi: 10.1016/j.cub.2009.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez J.A., Henderson B.R. Identification of a functional nuclear export sequence in BRCA1. J. Biol. Chem. 2000;275:38589–38596. doi: 10.1074/jbc.M003851200. [DOI] [PubMed] [Google Scholar]
- 29.Malnou C.E., Salem T., Brockly F., Wodrich H., Piechaczyk M., Jariel-Encontre I. Heterodimerization with Jun family members regulates c-Fos nucleocytoplasmic traffic. J. Biol. Chem. 2007;282:31046–31059. doi: 10.1074/jbc.M702833200. [DOI] [PubMed] [Google Scholar]
- 30.Hautbergue G.M., Hung M.L., Golovanov A.P., Lian L.Y., Wilson S.A. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izaurralde E. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell Biol. 2002;81:577–584. doi: 10.1078/0171-9335-00273. [DOI] [PubMed] [Google Scholar]
- 32.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 33.Gruter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B.K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 34.Wickramasinghe V.O., Laskey R.A. Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- 35.Dimaano C., Ullman K.S. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 37.Bastos R., Lin A., Enarson M., Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullman K.S., Shah S., Powers M.A., Forbes D.J. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong N., Radu G., Ju W., Brown W.T. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem. Biophys. Res. Commun. 2005;338:855–861. doi: 10.1016/j.bbrc.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Michael W.M., Siomi H., Choi M., Pinol-Roma S., Nakielny S., Liu Q., Dreyfuss G. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 1995;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- 41.Pinol-Roma S. HnRNP proteins and the nuclear export of mRNA. Semin. Cell Dev. Biol. 1997;8:57–63. doi: 10.1006/scdb.1996.0122. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 43.Fischer T., Strasser K., Racz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer T., Rodriguez-Navarro S., Pereira G., Racz A., Schiebel E., Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Navarro S., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 46.Wilmes G.M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., Cagney G., Collins S.R., Whitworth G.B., Kress T.L., Weissman J.S., et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faza M.B., Kemmler S., Jimeno S., Gonzalez-Aguilera C., Aguilera A., Hurt E., Panse V.G. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jani D., Lutz S., Hurt E., Laskey R.A., Stewart M., Wickramasinghe V.O. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012;40:4562–4573. doi: 10.1093/nar/gks059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi Y.H., Cheng L.I., Myers T., Ward J.M., Williams E., Su Q., Faucette L., Wang J.Y., Jeang K.T. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speese S.D., Ashley J., Jokhi V., Nunnari J., Barria R., Li Y., Ataman B., Koon A., Chang Y.T., Li Q., et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herold A., Teixeira L., Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. The EMBO journal. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kudo N., Wolff B., Sekimoto T., Schreiner E.P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 53.Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 2013;126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- 54.Osmanagic-Myers S., Dechat T., Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhen Y.Y., Libotte T., Munck M., Noegel A.A., Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 56.Padmakumar V.C., Abraham S., Braune S., Noegel A.A., Tunggal B., Karakesisoglou I., Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


