Abstract
Unusual DNA/RNA structures of the C9orf72 repeat may participate in repeat expansions or pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia. Expanded repeats are CpG methylated with unknown consequences. Typically, quadruplex structures form by G-rich but not complementary C-rich strands. Using CD, UV and electrophoresis, we characterized the structures formed by (GGGGCC)8 and (GGCCCC)8 strands with and without 5-methylcytosine (5mCpG) or 5-hydroxymethylcytosine (5hmCpG) methylation. All strands formed heterogenous mixtures of structures, with features of quadruplexes (at pH 7.5, in K+, Na+ or Li+), but no feature typical of i-motifs. C-rich strands formed quadruplexes, likely stabilized by G•C•G•C-tetrads and C•C•C•C-tetrads. Unlike G•G•G•G-tetrads, some G•C•G•C-tetrad conformations do not require the N7-Guanine position, hence C9orf72 quadruplexes still formed when N7-deazaGuanine replace all Guanines. 5mCpG and 5hmCpG increased and decreased the thermal stability of these structures. hnRNPK, through band-shift analysis, bound C-rich but not G-rich strands, with a binding preference of unmethylated > 5hmCpG > 5mCpG, where methylated DNA-protein complexes were retained in the wells, distinct from unmethylated complexes. Our findings suggest that for C-rich sequences interspersed with G-residues, one must consider quadruplex formation and that methylation of quadruplexes may affect epigenetic processes.
INTRODUCTION
The most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) is a (GGGGCC)n•(GGCCCC)n repeat expansion in the C9orf72 gene (1,2). In unaffected individuals, the most common repeat size is 2–19 units, while the pathogenic repeat size is 250–1600 repeats. ALS-FTD is one of more than 40 neurological, neurodegenerative, and neuromuscular diseases caused by gene-specific repeat expansions, such as myotonic dystrophy type 1 (DM1) and fragile X mental retardation syndrome (FXS). C9orf72-expansions show somatic repeat instability, aberrant CpG methylation, and lead to toxic-RNAs, toxic, RAN-peptides, and loss-of-function, pathological mechanisms, many of which have been reported in DM1 and FXS (3,4).
Epigenetic modifications occur at the C9orf72 gene with increased cytosine DNA methylation at CpG sites both in the promoter and within the repeat of many but not all C9orf72 expansion carriers (5–10). The CpG island and promoter upstream, but not downstream of the GGGGCC repeat is methylated in 36% of expansion carriers. However, methylation within the C9orf72 repeat is present in all individuals with greater than 90 repeats (10). Reports have not distinguished whether the C9orf72 methylation is the canonical 5-methylcytosine (5mCpG) or the more recently recognized 5-hydroxymethylcytosine (5hmCpG). C9orf72 methylation correlated with decreased levels of C9orf72 mRNA, RNA foci and RAN-dipeptide aggregates (6,7,9). The expanded and methylated C9orf72 repeat may differentially bind specific proteins. The expanded C9orf72 repeats, but not non-pathogenic repeats, have been reported to associate with a specific set of modified histones, which coincides with reduced C9orf72 mRNA expression (5). However, no study has assessed both C9orf72 DNA methylation state and protein interaction. Although the role of epigenetics in C9orf72-associated ALD/FTD is not yet understood, characterizing the structural and biological affects of C9orf72 repeat methylation may shed light on the mutagenic and pathogenic nature of the repeat.
Unusual DNA, RNA and R-loop (RNA–DNA) structures have been suggested to be involved in disease-associated repeat instability, or pathogenesis (11–13). Typically, G-rich strands are thought to form four-stranded G-quadruplexes, and C-rich strands have been demonstrated to form intercalated i-motif structures at acidic pH (14,15). Formation of G-quadruplex structures has been reported in the RNA (GGGGCC)n repeat (16–18). The G-rich DNA strand of the C9orf72 repeat can form quadruplexes (13). Several proteins bind preferentially to quadruplex RNA and DNA (19). Numerous proteins that bind quadruplexes have been reported to bind the C9orf72 quadruplex RNAs or RNA foci, including ASF/SF2, nucleolin, and hnRNPK (13,17–20). There is little knowledge of protein interactions with the C9orf72 DNA, and of the structure of the C-rich strand.
Here, we examined the structures formed by the G-rich and C-rich strands of the C9orf72 repeat with various modifications, including 5mCpG, 5hmCpG and N7-deazaG. We show that both the C-rich and G-rich strands of the C9orf72 repeat can form four-stranded structures. We also demonstrate that 5mCpG, 5hmCpG and N7-deazaG modifications affect secondary structure. Furthermore, hnRNPK binds to the C-rich DNA repeat and binding is affected by methylation status.
MATERIALS AND METHODS
DNA oligonucleotide synthesis and Labeling
All the DNA oligodeoxyribonucleotides were purchased as purified from ACGT Corp., Toronto, Canada. The ODNs were dialyzed versus Milli-Q water and lyophilized. The concentrations were determined by recording their absorbance at 260 nm using molar extinction coefficients calculated using the nearest-neighbour method (21). Oligos were then dissolved in the desired buffer to give concentrations ranging from 5 μM to 40 μM depending on the experiment. Oligonucleotides were end-labeled using [γ-32P]ATP. Equal sample amounts, based on Cerenkov counting (5000 cpm/sample), were electrophoresed on 4% or 8% polyacrylamide gels at 15 V/cm for 60 min or 9 V/cm for 120 min, respectively. We also carried out electrophoresis under denaturing conditions. In this case, the samples were heat denatured in formamide, frozen on dry ice until loading and electrophoresed on pre-heated denaturing 6% 8M urea sequencing gels, run at 150 V/cm. Some degree of heterogeneity is evident as both faster and slower migrating species that were resistant to denaturation (Supplementary Figure S1). We, and others have reported heat- and denaturation-resistant structures formed by G- and C-rich sequences that aberrantly migrate faster or slower on denaturing gels (17,18,22–24).
CD spectroscopy
CD spectra were recorded at room temperature using an Aviv model 62 DS spectropolarimeter using 5- and 20-μM samples dissolved in the indicated buffers. The pH was adjusted to 7.5 with HCl. All the samples were heated to 95ºC and allowed to cool to room temperature overnight. Three CD spectra, over the wavelength range of 320–220 nm were recorded in a 1-mm cuvette and averaged.
UV spectroscopy
Melting experiments were carried out on a Cary model 300 Bio spectrophotometer. Depending on the concentration, the samples were assessed in a 1-mm or 1-cm pathlength cuvette. Oligos were dissolved in 10 mM Tris containing either 100 mM NaCl, KCl or LiCl, pH 7.5. Melting was monitored at 280 nm (for all oligos but unmodified and G-rich 5mCpG in KCl) or 290 nm. The heating rate was 1ºC/min. All experiments were done in triplicate.
Electrophoretic mobility shift assays
Reactions were carried out in a 20-μl volume containing ∼80 fmol of [γ-32P]ATP end-labeled DNA with the indicated amounts of hnRNPK (Abnova). Binding reactions contained a final buffer concentration of 10 mM Tris (pH 7.4), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.05% non-idet P40. DNA was preincubated with the buffer described above for 30 min prior to the addition of protein and poly(dI-dC). Reactions were incubated at room temperature for 20 min, loaded onto 4% native polyacrylamide gels, electrophoresed at 200 V for 1 h in 1× Tris borate-–EDTA (0.089M Tris base, 0.089 M boric acid, 0.002 M EDTA), dried and autoradiographed. The extent of binding was empirically assessed, using ImageQuant software, by comparing the relative intensity of the band corresponding to the bound complex with that corresponding to the unbound material.
RESULTS
Quadruplex formation by both C9orf72 repeat strands and the effect of CpG methylation
We determined the structural effect of three different modifications, 5mCpG, 5hmCpG and N7-deazaG, on both the G-rich and C-rich strands of the C9orf72 repeat by CD spectroscopy and UV melting, in the presence of various ions (Figures 1 and 2). In KCl, the unmodified G-rich strand exhibits a CD spectrum with an intense positive peak at 290 nm, a negative peak around 260 nm a positive peak around 240 nm (Figure 1B). This spectrum is typical of antiparallel G-quadruplexes (25). In LiCl, the intensity at 290 nm decreases and the negative peak at 260 nm turns into a positive shoulder with a small negative peak around 240 nm. In LiCl, the population is shifted into a higher percentage of parallel quadruplex structures reflected by the 260-nm shoulder (Figure 1A). Methylating the cytosines at CpG sites of the G-rich strand results in a more intense 260 nm peak. In KCl the spectra of the unmodified and 5mCpG G-rich strand are similar. When cytosines of CpG sites are 5-hydroxymethylated, in all salt conditions, the CD spectra exhibits a positive peak at 285 nm, a shoulder around 260 nm with a negative peak around 240 nm. Positive peaks at 285 nm suggest duplex base stacking (25). The stability of the cyclic structure of G•G•G•G-tetrads, a critical component of G-quadruplexes, depends upon the ability of the N7 of guanine to accept a hydrogen bond (26). We directly tested the importance of this by replacing guanine residues with N7-deazaguanine, where the N7 position of guanine has been replaced by CH, which is not a hydrogen bond acceptor. Previous use of N7-deazaG revealed a strong dependence of G4-quadruplex (26). Here, modifying the N7 position of all guanines in the G-rich strands, in all salt conditions, results in a broad positive peak around 270 nm and negative peak around 250 nm showing complete structural transition. This spectrum has been associated with G•C•G•C-tetrad formation (27). These results support the suggestion that the peaks at 290 and 260 nm observed for the unmodified and 5mCpG G-rich strands arise from quadruplex formation. All G-rich sequences were also studied in the presence of LiCl or NaCl (Supplementary Figures S2 and S3). Although LiCl is generally considered too small to stabilize G-quadruplexes, interestingly the CD spectrum of the C9orf72 oligos in the presence of LiCl is very similar to that in NaCl (Supplementary Figures S2 and S3). Quadruplex formation in the presence of LiCl has previously been reported for the FRAXA and telomeric repeats (28,29).
Figure 1.
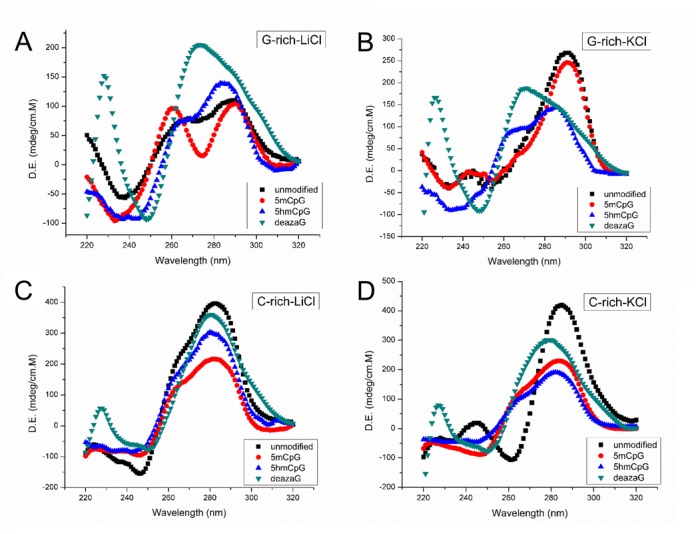
CD spectra of indicated C9orf72 DNAs. All experiments were done at 5 μM strand concentration, in 10 μM Tris (pH 7.5), and 100 μM of the indicated salt.
Figure 2.
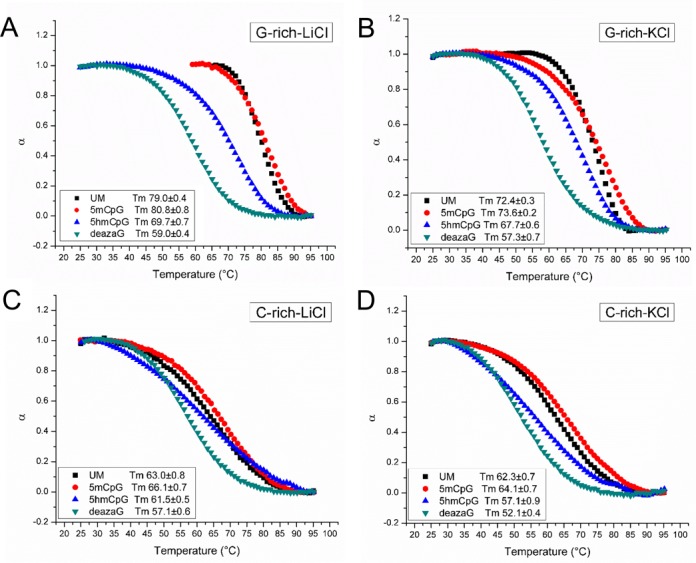
UV melting curves of indicated C9orf72 DNAs. All experiments were done at 5 μM strand concentration, in 10 μM Tris (pH 7.5), and 100 μM of the indicated salt. Melting temperature units are °C.
The CD spectra of the C-rich strands are shown in Figure 1C and D. Spectra for the unmodified, 5mCpG, and 5hmCpG C-rich strands were qualitatively similar in LiCl, with a positive peak at 280 nm, a negative peak at 250 nm and positive shoulder at 260 nm. Also in LiCl, an isodichroic point is seen at 260 nm. A positive 260-nm shoulder was not evident for the unmodified strand in KCl, rather a negative 260-nm peak is evident which may due to the stabilizing effect of KCl, turning the shoulder into a more prominent peak. Methylation or hydroxymethylation of CpGs do not alter the CD spectra significantly. Both 5mCpG, and 5hmCpG of the C-rich strands decreased the intensity of the 280-nm peak, but retained the overall shape of the spectra. The CD spectrum of the N7-deazaG C-rich strands lost the 260-nm shoulder in LiCl, and retained the 280-nm and 250-nm peaks. This suggests that the 260-nm peak arises from quadruplex formation even in the C-rich strands, which is probably due to the high fraction of guanines present. Tetrabutylammonium (TBA) is too large to stabilize G-quartet formation (30). Interestingly, in the presence of TBA, while the 280-nm peak of the unmodified, 5mCpG, 5hmCpG C-rich strands was retained, the 260-nm shoulder disappeared. For N7-deazaG C-rich strands, the CD spectra were the same in TBA, KCl and NaCl. Similar to the G-rich strands, all spectra were similar in LiCl and NaCl, suggesting that the conformation does not depend strongly on the nature of the cation (Supplementary Figures S2 and S3).
We performed UV melting on the same samples, under identical conditions as above. The unmodified and 5mCpG G-rich strands exhibit the highest thermal stability in all three ions (Figure 2A, Supplementary Figure S2). Interestingly, the unmodified and 5mCpG, G-rich strands exhibited lower stability in KCl than in NaCl or LiCl. This is likely due to the fact that in in KCl there is a single folding unit of antiparallel quadruplexes and the melting curve reflects these species (measured at 290 nm), while in NaCl and LiCl the CD spectra suggests multiple folding forms and the melting profiles may reflect denaturation of other folding units (recorded at 280 nm). The 5hmCpG G-rich strands all have lower stability than their unmodified forms, but higher than the N7-deazaG strands. For the C-rich strands the melting temperatures are lower than for the G-rich strands (Figure 2C and D). The unmodified and 5mCpG oligos all show concentration-dependent melting temperatures, suggesting possible multimolecular structure formation. Both the G-rich and the C-rich N7-deazaG-substituted oligos are more stable in LiCl than in than NaCl or KCl.
Methylation status of the C9orf72 repeat DNA affects its electrophoretic migration
To further evaluate the structural heterogeneity and complexity of the various forms of the (GGGGCC)8 and (GGCCCC)8 oligonucleotides, we assessed their electrophoretic migration on native polyacrylamide gels following pre-incubation in various salt solutions. An unusual pattern of electrophoretic species was evident for each, with slight variations in migration existing between the different salts (Figure 3). In general, each oligo migrates as a series of closely-spaced bands, with the C-rich repeats migrating slightly slower than their G-rich counterparts. A slower migrating, but minor species, was evident for the G-rich strand in all of its forms, with the exception of the N7-deazaG variant. This second band migrated similarly for the samples incubated with LiCl and NaCl, which was faster than the samples that were in the KCl or no salt condition. This slower migrating band disappears in denaturing gel conditions (Supplementary Figure S1). The 5hmCpG and N7-deazaG oligos migrate much slower than the unmethylated and 5mCpG forms. The greatest heterogeneity is displayed by the 5′-hydroxymethylated CpG forms of the G- and C-rich repeats, as evidenced by the broad smear of electrophoretic species. Curiously, unlike the 5mCpG, which migrated faster than the unmethylated forms, the 5hmCpG migrated the slowest. Furthermore, the 5hmCpG oligos are distinctly different from the unmethylated and 5mCpG methylated oligos in terms of their migration. The 5hmCpG oligos migrate much slower in comparison, suggesting they form different structures than the unmethylated and canonical 5mCpG modification. The N7-deazaG modified DNA, which is unable to form G-quadruplex structures, exhibits much more structural heterogeneity than its unmethylated counterparts, as evidenced by the broad smear.
Figure 3.
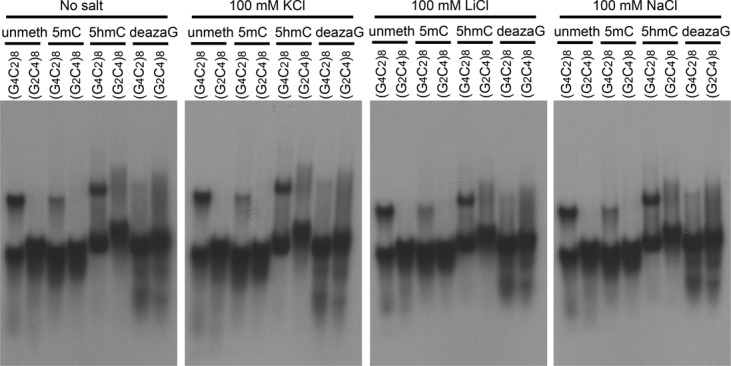
Methylation type and salt influence electrophoretic migration patterns. Migration of [γ-32P] ATP end-labeled DNA (frozen and thawed at room temperature) in native 8% polyacrylamide gel. [γ-32P] ATP end-labeled (GGGGCC)8 and (CCCCGG)8 containing the indicated modifications were dissolved in 50 mM Tris–HCl (pH 7.4) with the indicated salt, incubated for 20 min and then electrophoresed for 2 h at 120 V (9 V/cm).
The (GGCCCC)8 strand is preferentially bound by hnRNPK and affected by methylation
Toward addressing a biological effect of C9orf72 repeat modifications, we assessed the binding of hnRNPK. To date, there is no evidence on the effect of DNA methylation at the C9orf72 repeat and protein interactions. Using recombinant hnRNPK, we observed preferential binding of the protein to the C-rich repeat DNA (Figure 4). Unmethylated C-rich DNA was bound with complexes resolved as two distinct slower migrating species. The other modifications gave rise to broad smears of species with increasing amounts of hnRNPK, with most of the DNA-protein complex unable to enter the gel, likely due to increased structural complexity. Notably, the 5hmCpG form was preferentially bound (38%) over the 5mCpG form (14%), while the unmethylated form most efficiently bound (68%). Previously, protein interactions with r(GGGGCC)n and other repeat sequences reported the retention of the nucleic acid-protein complex in the well (18,31,32). Treatment with proteinase K and SDS broke down these complexes, allowing the DNA to migrate similarly to the control without protein. The unmethylated G-rich DNA was also able to bind, although very minimally, to the protein, creating a complex that migrated faster than the DNA-protein complexes of C-rich DNA. We did not observe binding to the modified G-rich sequences. These results imply that methylation status and type can alter the biological recognition of the C9orf72 repeats.
Figure 4.
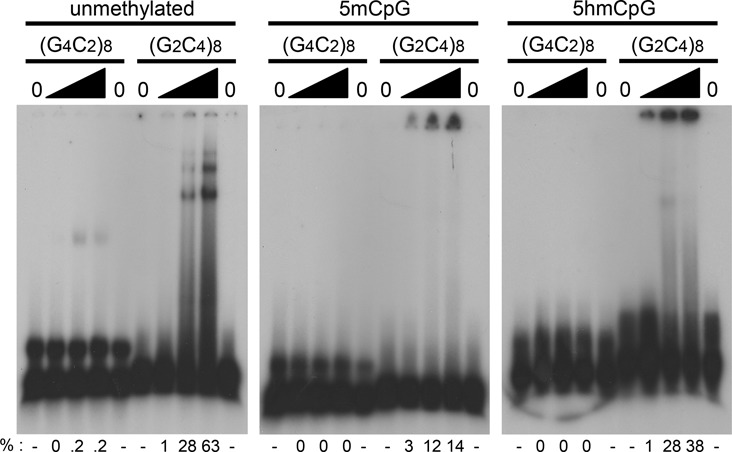
Methylation type influences DNA-protein interactions. Human recombinant hnRNPK band-shift with the [γ-32P] ATP end-labeled d(GGGGCC)8 or d(GGCCCC)8 DNAs (∼80 fmol) and 0, 50, 200 or 400 ng of purified recombinant hnRNPK. DNAs are either not CpG methylated, or contain 5-methylcytosine or 5-hydroxymethylcytosine bases at each CpG within the sequence. Samples were pre-incubated with the protein in a buffer solution containing 10 mM Tris (pH 7.4), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, and 0.05% Non-idet P40. Samples were electrophoresed on a 4% native polyacrylamide gel for 1 hour at 200 volts (15V/cm). Lanes were densimetrically assessed using ImageQuant software to quantify the percent bound (%).
DISCUSSION
We demonstrate that both the C-rich and G-rich DNA strands of the C9orf72 repeat can form a heterogenous population of structures, including those with features of four-stranded structures. It is accepted that a given G-rich and C-rich sequences can form numerous structural conformations (33). Previously, it was reported for shorter G-rich C9orf72 repeats (≤4 repeat units) that these form high levels of structural variants (34) – our findings are consistent with their the suggestion that such structural variation may be reflected in vivo and may be considerable for longer repeat tracts. That the C-rich strand can assume a quadruplex is distinct from many other studies, most of which have focused upon the G-rich strand, with little attention to the complementary C-rich strand (telomere repeats is one example). In cases where the complementary C-rich strand has been studied, it has often been reported that these form i-motifs under non-physiological pH. We propose that the four-stranded species formed by C-rich C9orf72 strands are stabilized by G•C•G•C-tetrads and C•G•C•G-tetrads, which would sandwich C•C•C•C-tetrads (a predicted structure is shown in Figure 5A). Our findings suggest that for C-rich strands, which also contain G-residues, one must consider four-stranded quadruplex formation.
Figure 5.

Proposed quadruplex structure in the C9orf72 8-repeat sequence and biological implications regarding R-loop formation. (A) Model of quadruplex structure forming in the (GGGGCC)8 and (GGCCCC)8 strands with various tetrad assemblies. Green colored boxes represent methylatable cytosines within the repeat sequence. (B) Intramolecular quadruplexes can form by either the G-rich or C-rich DNA strands of the C9orf72 repeat and may stabilized R-loops. They can also reflect the ‘pearls’ in a ‘pearls-on-a-string’ model of slipped-quadruplexes. See text.
We propose that both the C-rich and G-rich C9orf72 quadruplexes involve G•C•G•C-tetrads and C•G•C•G-tetrads (Figure 5A). Mixed G•C•G•C-tetrads are structurally distinct from G•G•G•G-tetrads. We propose that the distinct CD peak or a shoulder at 260 nm that we, and others observed for both C9orf72 DNA strands (13) reflects the presence of G•C•G•C-tetrads. Notably, G•C•G•C-tetrads can assume at least four different inter-base interactions (27,35–46). Unlike G•G•G•G-tetrads, which are stabilized by hydrogen bonding with the N7 position of the guanine residues (26), some, but not all G•C•G•C-tetrads form independent of the N7-G, with hydrogen bonding of the cytosine amino protons either with the O6 oxygen of guanine (35,46). Other G•C•G•C-tetrad conformations do utilize hydrogen bonding with the N7 nitrogen of guanine (35,46). The absence of an absolute requirement of the N7-G may explain the ability of the C-rich C9orf72 strand to form four-stranded structures at physiological pH. The limited effect of N7-deazaG on the CD spectrum of the C-rich strand, which is not predicted to involve G•G•G•G-tetrads, may be due to some of the GCGC-tetrads formed by the C-rich strand involve the N7 of G residues and hence are affected by the N7-deazaG. Moreover, unlike G•G•G•G-tetrads, which require cation coordination for stability, G•C•G•C-tetrads do not require cations for stability (35–44,46), which may explain the insensitivity of C9orf72 quadruplex formation to LiCl or TBA. It is notable that some G4-quadruplexes, including that formed by the FRAXA CGG repeat, are extremely stable even in the presence of lithium ions (28,29). A recent study of an oligo with a repeat of (GGGGCC)3GG(Br)GG with an 8-bromoguanine substitution, did not detect G•C•G•C-tetrads, which may be due to the preferential adoption of a syn-glycosidic conformation by 8-bromoguanines (47). Similarly, an NMR study of short tracts of (GGGGCC)n with either 1, 2 or 4 repeats, did not reveal G•C•G•C-tetrads (34). It is possible that the involvement of G•C•G•C-tetrads may require longer tracts of at least eight C9orf72 repeat units.
Our data imply that the C-rich C9orf72 quadruplex can involve C•C•C•C-tetrads (Figure 5A). These may be stabilized via stacking interactions that arise from sandwiching them between sets of G•C•G•C-tetrads or G•G•G•G-tetrads. Sandwiched C•C•C•C-tetrads in the FRAXA (CGG)n repeat were proposed to be stabilized by adjacent G•G•G•G-tetrads (48) and elsewhere (49).
Most quadruplexes reported involve G•G•G•G-tetrads. However, some G-rich oligos with limited numbers of C residues can form quadruplexes involving G•C•G•C-tetrads and C•C•C•C-tetrads (27,35,36,42,46,50,51). For example, the disease-associated fragile X (CGG)n repeat can form quadruplexes involving G•C•G•C-tetrads and C•C•C•C-tetrads (35,36,42,48). Notably, the FRAXA CD spectra also revealed a 260 nm peak/shoulder (48). It is widely accepted that quadruplex structures will involve G4 tetrads composed of four interacting guanine residues. Such an expectation has led to the prediction that any sequence conforming to the sequence consensus G≥3N1–7G≥3N1–7G≥3N1–7G≥3 will form G4 quadruplexes. However, quadruplexes may also be stabilized by G•C•G•C-tetrads; thus, C-rich sequences with interspersed G residues can also form stable, four-stranded structures (35–45). Our finding here that the C-rich C9orf72 strand presents as a quadruplex-forming sequence does not conform to the expected consensus. Thus, while it is reasonable to expect that any sequence that conforms to the G≥3N1–7G≥3N1–7G≥3N1–7G≥3 consensus will form a quadruplex, the reverse is not necessarily true, in that all quadruplex-forming sequences may not conform to the consensus. Interestingly, G•C•G•C-tetrad-containing quadruplexes have long been proposed to act in various biological processes including the junction of replication forks (38) and homologous pairing of DNAs by the intercoiling of two Watson-Crick paired helices, which may facilitate recombination (52).
Methylation of DNA is an important epigenetic mark associated with altered gene expression, replication and genetic instability. Both methylcytosine and hydroxymethylcytosine at CpG sites of each repeat unit could alter the structures of both C9orf72 strands. The increased and decreased melting temperatures of the 5mCpG and 5hmCpG forms may serve a regulatory role when DNA strands must separate, as during DNA transcription, replication, repair, or recombination. Increased and decreased thermal stability have been reported for 5mCpG and 5hmCpG sites at non-repeated sequences (53–57). The altered thermal stability of the methylated repeats may affect the propensity of the C9orf72 repeats to form unusual DNA structures. The faster and slower electrophoretic migration of the 5mCpG and 5hmCpG forms we observed (Figure 3) may reflect increased and decreased compaction of the DNA. The methylation state of non-repetitive sequences has been shown to enhance or impair the ability to form unwound DNA, bent-DNA, cruciforms, Z-DNA, quadruplexes and triplex–DNA (58–60). We propose that 5mCpG of the C9orf72 repeats stabilizes the quadruplex, similar to its effect upon FRAXA repeats (29), while 5hmCpG destabilizes the quadruplexes. Unusual structure formation by the C9orf72 repeats may contribute to pathogenesis and their genetic length instability, as demonstrated for R-loops (13,17). In fragile X patients, an absence of CpG methylation of the CGG/CCG repeats has been linked to enhanced somatic contractions of the expanded repeats in patient tissues (61–63). It will be interesting to assess the possible link of methylation to C9orf72 repeat instability. Our findings suggest that methylation state, as well as the form of methylation, may alter C9orf72 repeat structure. Distinguishing the kind of CpG methylation at the C9orf72 promoter and repeat, in specific tissues may reveal functional roles of this epigenetic mark.
The expanded and methylated C9orf72 repeat may differentially bind specific proteins. Specific histone modifications have been reported at the expanded C9orf72 repeats, but not non-pathogenic repeats (5,6); however, the DNA methylation status in these studies was not assessed. We show CpG methylation-sensitive binding of hnRNPK to C9orf72 repeat DNAs. hnRNPK bound preferentially to the C-rich DNA strand, consistent with previous studies showing an association of hnRNPK with the C-rich RNA foci in cells with C9orf72 expansions (13,64). However, direct binding of hnRNPK to the C-rich C9orf72 RNA was not observed (64). Our finding that hnRNPK bound to the C-rich DNA strand, is consistent with the reported preference of hnRNPK to bind DNA over RNA (65). Our data shows that hnRNPK binding is affected by CpG methylation of the C9orf72 repeats. Protein complexes with the r(GGGGCC)n and r(CUG)n repeat sequences have been reported to be retained in the wells of electrophoresis gels (18,31). We postulate that the methylated C9orf72 DNA–protein complexes are retained in the wells because they form multimeric DNA-protein structures incapable of entering the gels as opposed to a structural alteration of the DNA. This is supported by the observation that treatment of the complexes with proteinase K permits the protein-free DNA to co-migrate with the starting DNA. The implications of this differential binding is unknown, however, the observation that DNA methylation can alter both the biophysical traits and the interaction of proteins supports the concept that this epigenetic modification may be mediated by structural and protein selective avenues. hnRNPK functions in many genetic processes, including transcription, translation, chromatin remodeling and DNA repair (66).
Considerable evidence supports the formation of unusual DNA, RNA or RNA-DNA structures by disease associated repeats, structures that participate in mutagenic and pathogenic processes. It is very likely that our preparations contain mixtures of different folded forms, antiparallel, parallel quadruplexes along with hairpins, as previously suggested (13). The intramolecular structures formed by the G-rich and C-rich C9orf72 DNA strands that we report here, could reflect the displaced DNA strand in the RNA-DNA hybrids (R-loop) (Figure 5B), previously shown to form on either of the C9orf72 strands (13,17). R-loops can be biophysically stabilized by quadruplex formation of the displaced G-rich DNA strand at non-repetitive sequences (67). Notably, our observation herein, that the C-rich C9orf72 DNA can also assume a quadruplex structure suggests that it may stabilize R-loops having RNAs hybridized to the G-rich strand. Thus, R-loop formation on either DNA strand of the C9orf72 repeats could be stabilized by quadruplex formation by either of the displaced DNA strands. Similarly, the intramolecular quadruplexes formed by the G-rich and C-rich C9orf72 DNA strands that we report here, could reflect the ‘pearls’ in a ‘pearls-on-a-string’ type model of slipped-quadruplexes (Figure 5B). Alternately, a ‘bubble’ of separated strands, where each forms a series of quadruplexes may stabilize strand separation for some function, possibly the initiation of DNA replication, as several studies reveal that metazoan replication initiation sites coincide with quadruplex-forming sequences (68–72). Since transcription could potentially occur in the 64% of C9orf72 expansion carriers that do not have adjacent promoter CpG methylation but have a methylated repeat, the kind of methylation could modulate the formation of the above structures. Although the role of epigenetics in C9orf72-associated ALD/FTD is not understood, characterizing the structural and biological affects of C9orf72 repeat methylation may shed light on the mutagenic and pathogenic nature of the repeat.
Supplementary Material
Acknowledgments
CEP conceived, designed, and coordinated the study. BZ, MM, RBM and CEP wrote the manuscript. BZ performed and analyzed the experiments in Figures 1 and 2. RBM designed and interpreted the results of Figure 1. MM performed and analyzed the experiments in Figures 3 and 4. KB provided intellectual input regarding hnRNPK. All authors reviewed the results and approved the final version of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Weston Brain Institute (to C.E.P.); Amyotrophic Lateral Sclerosis Association of America (to C.E.P.); Natural Science and Engineering Research Council of Canada (to C.E.P., R.B.M.); National Institutes of Health [R01 DK083310, R21 GM111439, R33 CA191135 to K.B.]. Funding for open access charge: Weston Brain Institute Grant to Christopher E. Pearson.
Conflict of interest statement. None declared.
REFERENCES
- 1.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gendron T.F., Belzil V.V., Zhang Y.J., Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson C.E., Nichol Edamura K., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 5.Belzil V.V., Bauer P.O., Prudencio M., Gendron T.F., Stetler C.T., Yan I.K., Pregent L., Daughrity L., Baker M.C., Rademakers R., et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belzil V.V., Bauer P.O., Gendron T.F., Murray M.E., Dickson D., Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res. 2014;1584:15–21. doi: 10.1016/j.brainres.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu E.Y., Russ J., Wu K., Neal D., Suh E., McNally A.G., Irwin D.J., Van Deerlin V.M., Lee E.B. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russ J., Liu E.Y., Wu K., Neal D., Suh E., Irwin D.J., McMillan C.T., Harms M.B., Cairns N.J., Wood E.M., et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015;129:39–52. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xi Z., Zinman L., Moreno D., Schymick J., Liang Y., Sato C., Zheng Y., Ghani M., Dib S., Keith J., et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am. J. Hum. Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi Z., Zhang M., Bruni A.C., Maletta R.G., Colao R., Fratta P., Polke J.M., Sweeney M.G., Mudanohwo E., Nacmias B., et al. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol. 2015;129:715–727. doi: 10.1007/s00401-015-1401-8. [DOI] [PubMed] [Google Scholar]
- 11.Reddy K., Schmidt M.H., Geist J.M., Thakkar N.P., Panigrahi G.B., Wang Y.H., Pearson C.E. Processing of double-R-loops in (CAG).(CTG) and C9orf72 (GGGGCC).(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014;42:10473–10487. doi: 10.1093/nar/gku658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axford M.M., Wang Y.H., Nakamori M., Zannis-Hadjopoulos M., Thornton C.A., Pearson C.E. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLoS Genet. 2013;9:e1003866. doi: 10.1371/journal.pgen.1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan A.T., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks T.A., Kendrick S., Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B. Jr, Pearson C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamiri B., Reddy K., Macgregor R.B. Jr, Pearson C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014;289:4653–4659. doi: 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon-Ortiz A.M., Svendsen J., Boulton S.J. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair (Amst.) 2014;19:152–162. doi: 10.1016/j.dnarep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S., et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 22.Shida T., Yokoyama K., Tamai S., Sekiguchi J. Self-association of telomeric short oligodeoxyribonucleotides containing a dG cluster. Chem. Pharm. Bull. (Tokyo) 1991;39:2207–2211. doi: 10.1248/cpb.39.2207. [DOI] [PubMed] [Google Scholar]
- 23.Hirao I., Naraoka T., Kanamori S., Nakamura M., Miura K. Synthetic oligodeoxyribonucleotides showing abnormal mobilities on polyacrylamide gel electrophoresis. Biochem. Int. 1988;16:157–162. [PubMed] [Google Scholar]
- 24.Hirao I., Nishimura Y., Naraoka T., Watanabe K., Arata Y., Miura K. Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: d(GCGAAAGC) Nucleic Acids Res. 1989;17:2223–2231. doi: 10.1093/nar/17.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorlickova M., Kejnovska I., Bednarova K., Renciuk D., Kypr J. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality. 2012;24:691–698. doi: 10.1002/chir.22064. [DOI] [PubMed] [Google Scholar]
- 26.Murchie A.I., Lilley D.M. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viladoms J., Escaja N., Frieden M., Gomez-Pinto I., Pedroso E., Gonzalez C. Self-association of short DNA loops through minor groove C:G:G:C tetrads. Nucleic Acids Res. 2009;37:3264–3275. doi: 10.1093/nar/gkp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J.Y., Yoon J., Kihm H.W., Kim D.S. Structural diversity and extreme stability of unimolecular Oxytricha nova telomeric G-quadruplex. Biochemistry. 2008;47:3389–3396. doi: 10.1021/bi702013d. [DOI] [PubMed] [Google Scholar]
- 29.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan H.Y., Shek Y.L., Amiri A., Dubins D.N., Heerklotz H., Macgregor R.B. Jr, Chalikian T.V. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011;133:4518–4526. doi: 10.1021/ja110495c. [DOI] [PubMed] [Google Scholar]
- 31.Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warf M.B., Berglund J.A. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Largy E., Mergny J.L. Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014;42:e149. doi: 10.1093/nar/gku751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sket P., Pohleven J., Kovanda A., Stalekar M., Zupunski V., Zalar M., Plavec J., Rogelj B. Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging. 2015;36:1091–1096. doi: 10.1016/j.neurobiolaging.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Zhang N., Gorin A., Majumdar A., Kettani A., Chernichenko N., Skripkin E., Patel D.J. Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs. J. Mol. Biol. 2001;312:1073–1088. doi: 10.1006/jmbi.2001.5002. [DOI] [PubMed] [Google Scholar]
- 36.Kettani A., Kumar R.A., Patel D.J. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 37.Kettani A., Bouaziz S., Gorin A., Zhao H., Jones R.A., Patel D.J. Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 38.Lowdin P.O. Some aspects of the hydrogen bond in molecular biology. Ann. N. Y. Acad. Sci. 1969;158:86–95. doi: 10.1111/j.1749-6632.1969.tb56215.x. [DOI] [PubMed] [Google Scholar]
- 39.Kubitschek H.E., Henderson T.R. DNA rplication. Proc. Natl. Acad. Sci. U.S.A. 1966;55:512–519. doi: 10.1073/pnas.55.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGavin S. Models of specifically paired like (homologous) nucleic acid structures. J. Mol. Biol. 1971;55:293–298. doi: 10.1016/0022-2836(71)90201-4. [DOI] [PubMed] [Google Scholar]
- 41.Mitas M., Yu A., Dill J., Kamp T.J., Chambers E.J., Haworth I.S. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitas M., Yu A., Dill J., Haworth I.S. The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn. Ganti base pairs. Biochemistry. 1995;34:12803–12811. doi: 10.1021/bi00039a041. [DOI] [PubMed] [Google Scholar]
- 43.Leonard G.A., Zhang S., Peterson M.R., Harrop S.J., Helliwell J.R., Cruse W.B., d'Estaintot B.L., Kennard O., Brown T., Hunter W.N. Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution. Structure. 1995;3:335–340. doi: 10.1016/s0969-2126(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 44.Salisbury S.A., Wilson S.E., Powell H.R., Kennard O., Lubini P., Sheldrick G.M., Escaja N., Alazzouzi E., Grandas A., Pedroso E. The bi-loop, a new general four-stranded DNA motif. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5515–5518. doi: 10.1073/pnas.94.11.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim K.W., Alberti P., Guedin A., Lacroix L., Riou J.F., Royle N.J., Mergny J.L., Phan A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Escaja N., Gomez-Pinto I., Pedroso E., Gonzalez C. Four-stranded DNA structures can be stabilized by two different types of minor groove G:C:G:C tetrads. J. Am. Chem. Soc. 2007;129:2004–2014. doi: 10.1021/ja066172z. [DOI] [PubMed] [Google Scholar]
- 47.Brcic J., Plavec J. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015;43:8590–8600. doi: 10.1093/nar/gkv815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen F.M. Acid-facilitated supramolecular assembly of G-quadruplexes in d(CGG)4. J. Biol. Chem. 1995;270:23090–23096. doi: 10.1074/jbc.270.39.23090. [DOI] [PubMed] [Google Scholar]
- 49.Meyer M., Hocquet A., Suhnel J. Interaction of sodium and potassium ions with sandwiched cytosine-, guanine-, thymine-, and uracil-base tetrads. J. Comput. Chem. 2005;26:352–364. doi: 10.1002/jcc.20176. [DOI] [PubMed] [Google Scholar]
- 50.Patel P.K., Bhavesh N.S., Hosur R.V. NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG. Biochem. Biophys. Res. Commun. 2000;270:967–971. doi: 10.1006/bbrc.2000.2479. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D., Huang T., Lukeman P.S., Paukstelis P.J. Crystal structure of a DNA/Ba2 +G-quadruplex containing a water-mediated C-tetrad. Nucleic Acids Res. 2014;42:13422–13429. doi: 10.1093/nar/gku1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson J.H. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc. Natl. Acad. Sci. U.S.A. 1979;76:3641–3645. doi: 10.1073/pnas.76.8.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renciuk D., Blacque O., Vorlickova M., Spingler B. Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine. Nucleic Acids Res. 2013;41:9891–9900. doi: 10.1093/nar/gkt738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thalhammer A., Hansen A.S., El-Sagheer A.H., Brown T., Schofield C.J. Hydroxylation of methylated CpG dinucleotides reverses stabilisation of DNA duplexes by cytosine 5-methylation. Chem. Commun. (Camb.) 2011;47:5325–5327. doi: 10.1039/c0cc05671e. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez Lopez C.M., Guzman Asenjo B., Lloyd A.J., Wilkinson M.J. Direct detection and quantification of methylation in nucleic acid sequences using high-resolution melting analysis. Anal. Chem. 2010;82:9100–9108. doi: 10.1021/ac1024057. [DOI] [PubMed] [Google Scholar]
- 56.Severin P.M., Zou X., Schulten K., Gaub H.E. Effects of cytosine hydroxymethylation on DNA strand separation. Biophys. J. 2013;104:208–215. doi: 10.1016/j.bpj.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hardin C.C., Corregan M., Brown B.A. 2nd, Frederick L.N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 58.Diekmann S. DNA methylation can enhance or induce DNA curvature. EMBO J. 1987;6:4213–4217. doi: 10.1002/j.1460-2075.1987.tb02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hodges-Garcia Y., Hagerman P.J. Cytosine methylation can induce local distortions in the structure of duplex DNA. Biochemistry. 1992;31:7595–7599. doi: 10.1021/bi00148a022. [DOI] [PubMed] [Google Scholar]
- 60.Zacharias W. Methylation of cytosine influences the DNA structure. EXS. 1993;64:27–38. doi: 10.1007/978-3-0348-9118-9_3. [DOI] [PubMed] [Google Scholar]
- 61.Nichol Edamura K., Leonard M.R., Pearson C.E. Role of replication and CpG methylation in fragile X syndrome CGG deletions in primate cells. Am. J. Hum. Genet. 2005;76:302–311. doi: 10.1086/427928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wohrle D., Salat U., Glaser D., Mucke J., Meisel-Stosiek M., Schindler D., Vogel W., Steinbach P. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J. Med. Genet. 1998;35:103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor A.K., Tassone F., Dyer P.N., Hersch S.M., Harris J.B., Greenough W.T., Hagerman R.J. Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am. J. Med. Genet. 1999;84:233–239. [PubMed] [Google Scholar]
- 64.Cooper-Knock J., Higginbottom A., Stopford M.J., Highley J.R., Ince P.G., Wharton S.B., Pickering-Brown S., Kirby J., Hautbergue G.M., Shaw P.J. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. 2015;130:63–75. doi: 10.1007/s00401-015-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomonaga T., Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 66.Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 67.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintome C., Riou J.F., Prioleau M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshina S., Yura K., Teranishi H., Kiyasu N., Tominaga A., Kadoma H., Nakatsuka A., Kunichika T., Obuse C., Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J. Biol. Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cayrou C., Coulombe P., Puy A., Rialle S., Kaplan N., Segal E., Mechali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lombrana R., Almeida R., Alvarez A., Gomez M. R-loops and initiation of DNA replication in human cells: a missing link? Front. Genet. 2015;6:158. doi: 10.3389/fgene.2015.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartholdy B., Mukhopadhyay R., Lajugie J., Aladjem M.I., Bouhassira E.E. Allele-specific analysis of DNA replication origins in mammalian cells. Nat. Commun. 2015;6:7051. doi: 10.1038/ncomms8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


