Abstract
Hygromycin A (HygA) binds to the large ribosomal subunit and inhibits its peptidyl transferase (PT) activity. The presented structural and biochemical data indicate that HygA does not interfere with the initial binding of aminoacyl-tRNA to the A site, but prevents its subsequent adjustment such that it fails to act as a substrate in the PT reaction. Structurally we demonstrate that HygA binds within the peptidyl transferase center (PTC) and induces a unique conformation. Specifically in its ribosomal binding site HygA would overlap and clash with aminoacyl-A76 ribose moiety and, therefore, its primary mode of action involves sterically restricting access of the incoming aminoacyl-tRNA to the PTC.
INTRODUCTION
Hygromycin A (HygA) is a natural product of Streptomyces hygroscopicus first isolated in 1953 (1,2). It is endowed with promising biological activities and has a unique structure (Supplementary Figure S1) consisting of a furanose, cinnamic acid and aminocyclitol moiety (3). The biosynthetic pathway of HygA has been elucidated (4) and its total chemical synthesis has also been described (5–7). HygA has a relatively broad antimicrobial spectrum, displaying activity against gram-positive bacteria including mycobacteria and actinomycetes (3). In addition this molecule is also active against Serpulina (Treponema) hyodysenteriae (the agent of swine dysentery), leptospira and endomoeba (1,3). The limited activity of HygA against enteric gram-negative bacteria has been attributed to the efficient AcrA/B efflux pump operating in these organisms (8).
The structure and biological activity of HygA are distinct from those of hygromycin B, another antibiotic produced by the same organism, but HygA displays some common features with chloramphenicol (1–3) and orthoformimycin (9). HygA was shown to be a translational inhibitor; or more precisely, HygA was found to bind to the large (50S) ribosomal subunit and to inhibit the peptidyl transferase (PT) activity of the ribosome (10–12). Other translational steps, such as the enzymatic (EF-Tu dependent) binding of aminoacyl-tRNA to the ribosomal A site and the translocation of peptidyl-tRNA from the A to the P site were found to be unaffected by HygA (10). Furthermore, since HygA is a more potent agent than chloramphenicol and inhibits ribosomal binding of chloramphenicol, it was suggested that the binding sites of these two antibiotics are close or partially overlapping (10).
A structural similarity has been observed between HygA, A201A and puromycin (4,13–17). More precisely, the 6′-7′-dihydroxy-α-methylcinnamic acid moiety of HygA (Supplementary Figure S1) and the 7′-hydroxy-α-methylcinnamic acid present in A201A are similar to the tyrosine-derived moiety of puromycin (4).
Similar to HygA, both A201A and puromycin are potent inhibitors of protein synthesis and all three antibiotics prevent peptide bond formation (10,13,17–20). Puromycin, the best characterized of the three antibiotics, binds, as do HygA and chloramphenicol, to the A site of the large subunit where it structurally mimics the aminoacyl-tRNA 3′ terminus and can serve as an acceptor of the polypeptide chain via the 2′ amino group (21,22). Moreover, in its higher affinity form of CC-puromycin, it can induce on the large subunit, the same active conformation of the PTC, as observed on the complete 70S ribosome (22–26). Despite the similarity between the three antibiotics, and in contrast to puromycin, both HygA and A201A do not carry at the 2′ position a reactive amino group ((17) and Supplementary Figure S1) and therefore cannot act as acceptor substrates in peptide bond formation.
In situ chemical probing showed that macrolides with a mycarose containing disaccharide at position 5 of the lactone ring, such as, carbomycin, tylosin and spiramycin, inhibit or compete with HygA for ribosomal binding. On the other hand, HygA can bind to the ribosome concomitantly with macrolides that have only a monosaccharide extension on the lactone ring and do not inhibit the PT reaction (11). Overall, the available data indicate that although HygA binds to the ribosome in a region that overlaps that of other 50S-targeting antibiotics, its binding mode is clearly distinct.
In light of these data and the frequent occurrence of resistance and cross-resistance phenotypes acquired by pathogenic bacteria to 50S-targeting antibiotics, the use of HygA as a pharmacophore for the development of new anti-infectives capable of overcoming existing resistance mechanisms, requires a precise knowledge of the relationship between the binding site of HygA and that of chloramphenicol and other 50S inhibitors, the macrolides in particular. In this study we used a combination of X-ray crystallography and biochemical approaches to address the ribosomal localization of HygA and describe its relationship to other anti-infectives. Taken together the structural and biochemical data presented indicate that HygA binds within the PTC such that it would clash with the aminoacyl-ribosyl moiety at the 3′end of the A-site tRNA. Therefore its primary mode of action involves blocking the accommodation of the A-site tRNA within the PTC. At the same time HygA induces conformational changes in functionally important residues of the PTC, which perturb the active site conformation and may have a secondary role in HygA's inhibitory mechanism.
MATERIALS AND METHODS
Biochemical assays
In vitro mRNA translation (driven by 027IF2Cp(A) mRNA) and tests of individual translational steps (e.g. Phe-tRNA binding to the ribosomal A site, fMet-puromycin formation and dipeptide (fMet-Phe) formation) were carried out as described (27,28). In situ probing of the 23S rRNA by hydroxyl radical cleavage was carried out as described (29). EF-Tu-dependent A-site binding of proflavine-labeled Phe-tRNA to MFmRNA-programmed 70S ribosomes carrying P-site bound fMet-tRNA (70S IC), was carried out in the absence or presence of 20 μM HygA as described (30).
X-ray crystallography
Crystals of the 50S ribosomal subunit from Deinococcus radiodurans were prepared as previously described (31,32), soaked overnight in a stabilizing solution with or without 20 μM HygA and flash-frozen in liquid nitrogen. X-ray diffraction data were collected with a PILATUS detector at the X06SA beamline at the Swiss Light Source (Villigen, Switzerland), and processed to 2.9 Å and 3.0 Å for the complex and apo structures, respectively, using the XDS (33) and CCP4 (34) program packages. A previously reported structure of the D. radiodurans 50S subunit (PDB accession code: 2ZJR) (35) was fully refined against the newly collected data set for the native 50S subunit, and the resulting model was used to phase the 50S–HygA complex with Phenix (36). The same set (∼5%) of the reflections data was omitted during refinement for the free R factor calculation. The binding position of the drug was determined based on σA-weighted difference maps (37). Electron density indicating the presence of HygA and alterations in nearby nucleotides was unambiguously observed in the A site of the PTC in the initial unbiased electron density maps obtained with G2061, A2451-C2452, A2503-C2507, A2572-C2573, G2583-U2585 and A2602 excluded from calculation (Supplementary Figure S2). Although the positions of the phenol-, alkene- and peptide moieties within the HygA structure suggest a conjugated system, with a preferential continuous planarity in solution along the C1′–C4′ backbone of HygA, during model refinement three separate planarity restraints for each of these moieties were created. This resulted in small violations (<10°) in the planarity of the system, possibly due to the specific chemical environment of the ribosome, which can affect the mesomerism of the conjugated system (i.e. along the C1′–C4′ atoms, Supplementary Figure S1). Concerning the aminocyclitol moiety, during refinement either no torsion restraints for this moiety were active or torsion restraints were present to support either a chair or twisted boat conformation; the aminocyclitol moiety of HygA has been reported to acquire a twisted boat conformation in solution (38). Independently from the applied setup the models always converged to the same final model with a chair like conformation. Compared to the twisted boat conformations, the chair like conformation also fits best the density of the unbiased map or the 2Fo–Fc map obtained imposing the twisted boat conformation through out the refinement. Modeling and refinement were carried out using Coot (39) and Phenix (36). rRNA nucleotide suites were thoroughly corrected using ERRASER (40) and RCrane (41). Chemical structures of HygA and relevant compounds were depicted using MarvinSketch (ChemAxon: http://www.chemaxon.com). Coordinates and restraints of the drug suitable for refinement were obtained using the GRADE server (42) and eLBOW (43). rRNA residues were numbered according to the Escherichia coli scheme and helices indicated using the standard nomenclature.
RESULTS
HygA binds in the A site of the peptidyl transferase center and alters its conformation
To elucidate the molecular mechanism of action of HygA we first determined its ribosomal binding site using X-ray crystallography. The initial unbiased map clearly showed extra electron density in the A site of the peptidyl transferase center (PTC; Supplementary Figure S2). Its elongated shape with prominent densities at either end allowed unambiguous assignment of the terminal furanose and aminocyclitol groups and the subsequent placement of the central cinnamic acid moiety. Moreover, by comparison with the native 50S structure conformational changes in 23S rRNA residues were also detected in the proximity of HygA (Supplementary Figure S2). The altered nucleotides, including U2506, U2585 and A2602, were remodeled and the resulting structure refined to convergence with a final resolution of 3.0 Å (Table 1; final 2Fo–Fc map seen in Figure 1C and D). As seen in Figure 1, HygA binds in a pocket delimited by A2503, the G2061-A2451 and C2452-U2504 base pairs, and the G2505-U2506 segment. The rRNA residues comprising this binding pocket are universally conserved (>98% conservation) and, therefore, presumably similar across species. Toward the bottom of the pocket (i.e. near the ribosomal tunnel) the two hydroxyl groups of the furanosyl moiety potentially hydrogen bond with nucleotides A2503 and G2505, apparently serving as anchoring points for this moiety. On the contrary, the acetyl group on the furanose ring protrudes into the lumen of the ribosomal tunnel and, therefore, makes no significant interactions with the ribosome (Figures 1C–D and 2C). The central cinnamic acid moiety stacks against the walls of the pocket, namely the G2061-A2451 and C2452-U2504 base pairs, with the C6′ hydroxyl group of HygA within hydrogen-bonding distance of the O2′ and O3′ of G2505 (Figures 1C-D and 2A–C). In addition, the C1′ carboxyl oxygen, in the cinnamic acid moiety, is within hydrogen bonding distance of both the 2′-OH of A2451 and the N6 of A2602 (Figure 2A and C). The aminocyclitol moiety is positioned at the entrance of the binding pocket. In this moiety, the oxygen atoms at positions 5 and 6 potentially hydrogen bond with the N4 of C2573, while its amide nitrogen at position 2 and hydroxyl oxygen at position 3 could form hydrogen bonds with the O2 of U2506 and O4 of U2585, respectively (Figure 2A and C).
Table 1. Data Collection and refinement Statistics.
| Complex | 50S apo | 50S–HygA |
|---|---|---|
| PDB accession code | 5DM6 | 5DM7 |
| Data collection statistics | ||
| Space group | I222 | I222 |
| Cell dimensions, Å (a, b, c) | 169.90, 410.76, 696.12 | 169.82, 411.54, 695.65 |
| Resolution range, Å | 59.3–2.90 (3.06–2.90) | 59.3–3.00 (3.16–3.00) |
| Resolution where I/σI = 2, Å | 3.2 | 3.7 |
| No. of observed reflections | 11 664 835 (1 754 909) | 2 197 039 (315 299) |
| No. of unique reflections | 531 662 (78 466) | 479 454 (68 449) |
| Multiplicity | 21.9 (22.3) | 4.58 (4.60) |
| Completeness,% | 99.8 (99.6) | 99.5 (99.1) |
| Rmerge,% | 24.1 (786.2) | 21.5 (562.3) |
| I/σI | 10.38 (0.71) | 6.28 (0.33) |
| CC(1/2),% | 99.9 (47.8) | 99.8 (18.5) |
| Wilson B-factor, Å2 | 78.94 | 66.56 |
| Refinement statistics | ||
| Rwork/Rfree,% | 24.1/28.2 | 26.5/31.5 |
| No. of atoms | ||
| RNA | 62 274 | 62 274 |
| Protein | 27 162 | 27 162 |
| Ligand | 0 | 36 |
| Ions | 198 | 197 |
| RMSD | ||
| Bond lengths, Å | 0.014 | 0.027 |
| Bond angles, ° | 2.323 | 2.407 |
Values in parentheses refer to the highest resolution bin.
Each dataset was obtained from a single crystal.
Figure 1.
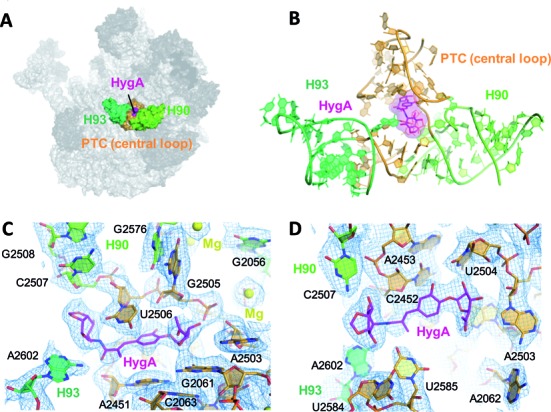
The HygA binding site. (A) An overview showing the location of HygA (colored in magenta) on the Deinococcus radiodurans 50S ribosomal subunit. (B) The HygA binding site is formed by 23S rRNA helices H90 and H93 (light and dark green, respectively) and the PTC central loop (orange). (C and D) Final 2Fo-Fc electron density map sharpened, contoured at 1 sigma and shown from two different perspectives.
Figure 2.
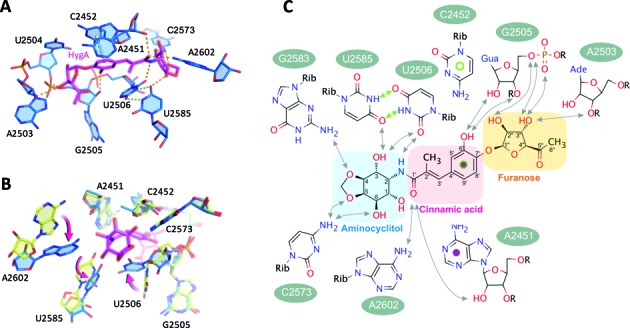
HygA-dependent perturbations in the PTC. (A) The structure of the PTC's A-site in the presence of HygA (50S–HygA). In the 50S–HygA complex nucleotides critical for tRNA binding acquire a unique conformation. Potential hydrogen bonds between HygA and the 23S rRNA are indicated in orange and those newly formed solely within the 23S rRNA are green. (B) An overlay of the 50S–HygA (blue) and apo-50S (yellow) structures with the significant displacements of U2506, U2585 and A2602 indicated with arrows. The position of the A2602 base in the apo-50S structure is only indicative as this residue is very dynamic in the absence of the drug while it acquires a more stable conformation in the presence of HygA. (C) A schematic representation of the potential interactions formed between HygA and 23S rRNA residues that are predicted on the basis of geometric constrains. Potential hydrogen bonds between HygA and the 23S rRNA are indicated with black arrows and HygA dependent hydrogen bonds formed within the 23S rRNA are indicated with green arrows. The stacking interaction between the C2452, the cinnamic acid moiety and A2451 are indicated with green and purple circles. Note the orientation of the drug in D is mirrored relative to panels A–C.
Comparing the structures of the PTC's A site in the presence and absence of HygA, reveals that HygA binding induces conformational changes in functionally important PTC nucleotides (Figure 2B). Namely, in the presence of HygA, U2506 and U2585 are shifted and interact, forming a non-canonical Watson–Crick base pair with their N3 and O4 positions alternately hydrogen-bonded (Figure 2A and C, green dotted lines; Supplementary Figure S2C and D). The formation of the U2506 and U2585 base pair in the presence of HygA is in good agreement with chemical probing experiments showing that in the presence of HygA the N3 of both U2585 and U2506 are protected from modification (11). A similar base pair is observed when homoharringtonine, a eukaryotic specific anti-tumorigenic compound targeting the A-site, is bound to the Haloarcula marismortui 50S subunit (44). Furthermore, A2602 is disordered in the native structure (50S apo) but its density becomes more evident in the presence of HygA (Figure 1C and D, Supplementary Figure S2C and D) which likely reflects that it is more stably positioned. Accordingly, our model places the extracyclic amine of A2602 within hydrogen bonding distance of the carboxyl oxygen of the cinnamic acid moiety (Figure 2A–C). Taken together, the X-ray crystallographic analysis indicates that upon binding to the A-site of the PTC, HygA influences the conformation of several nucleotides (i.e. A2602, U2585, U2506) that play critical roles in various stages of translation including positioning the A and P-site tRNAs during peptide bond formation and translocation (25,26,45).
A structural alignment of the PTC residues in the 50S–HygA structure with those from structures containing a bound A-site aminoacyl tRNA substrate (25) clearly show that the aminocyclitol and cinnamic acid groups of HygA would overlap and sterically clash with the A76 ribose and aminoacyl moiety, respectively, of the A-site substrate (Figure 3A). This structural alignment also makes it evident that despite the presence in HygA of the aminocyclitol and furanose moiety, which are absent in the A76-aminoacyl tRNA, the portion of HygA that shares a structural similarity with puromycin ((4); highlighted atoms in Figure 3C and F) and thereby also with the A76-aminacyl moiety of the A site tRNA, is similarly located in the two complexes (Figure 3A and B). More specifically, the chemical backbone of HygA, comprised by the three hydroxyl, C2 position and two amino of the aminocyclitol group along with the cinnamic acid moiety of HygA, are similarly located and structurally resemble on the 50S–HygA complex, the 2′ hydroxyl and 3′ amino moiety of the A-site tRNA terminal ribose (A76) and the phenylalanine amino acid residue, of the A-site tRNA on the 70S complex (Figure 3A and B). Moreover, these two related moieties, present in HygA and the A76-aminoacyl tRNA, also establish a network of interactions with the same surrounding ribosomal elements i.e. residues A2451, A2602, U2585 and U2506 (Figure 3C–F). For example, the hydroxyl oxygen at position 3 of the aminocyclitol moiety potentially forms a hydrogen bond with O4 of U2585 (Figure 3E and F) similar to that made by the 2′-OH of the A76 of the A-tRNA (Figure 3C and D). Likewise the carboxyl oxygen in the cinnamic acid moiety is located in H bonding distance to both the 2′ OH of A2451 and the N6-amine of A2602 (Figure 3E and F), similar to the carboxyl oxygen of the A-site bound aminoacyl moiety (Figure 3C and D).
Figure 3.
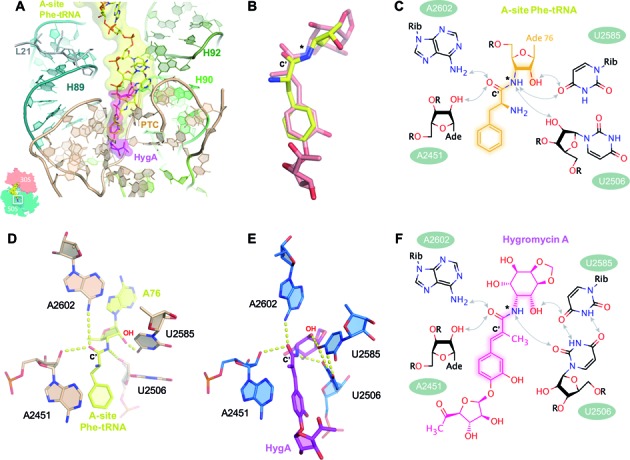
The binding site of HygA on the ribosome overlaps with that of the aminoacyl-A76 ribosyl moiety of the A-site tRNA. (A) The 50S–HygA HygA–5 complex was aligned to a 70S ribosome with an accommodated A-site (PDB accession code: 4QCP; (25)) using 23S rRNA residues 2063–2092, 2227–2258, 2435–2457 and 2494–2608 showing that the aminoacyl-A76 ribosyl moiety at the acceptor end of the A-site tRNA (yellow) overlaps with the binding site of HygA (magenta). In (B) only HygA and the aminoacyl-A76 ribosyl moiety from the aligned structures (panel A) are shown to highlight their similarity. When aligned using PTC residues (as shown in A) the 15 atoms spanning from the 3-hydroxyl to C7’ position of HygA (Supplementary Figure S1) and their counterparts in the A-site tRNA (indicated by highlights in the chemical structures shown in panels C and F) have a root mean square deviation (RMSD) of 1.57 Å. When these 15 atoms are directly aligned on themselves they have a RMSD of 0.86 Å. Both the aminoacyl moiety of the A-site tRNA (C and D) and HygA (E and F) appear to be coordinated by a similar hydrogen bond network that includes the 23S rRNA residues A2451 (via 2′-OH of the ribose), U2506 (2′-OH of the ribose in the acylated tRNA and O2 of the base in the 50S–HygA structure) and A2602 (via the N6 amino group) while U2585 in the 50S–HygA complex is kept at a position similar to that seen in PDB accession code: 4QCP (25) by forming a symmetric 4-carbonyl-N3 U-U pair with residue U2506. The carbonyl carbon present in both ligands is labeled as C’. Note that in a native aminoacyl-tRNA an ester oxygen rather than an amide nitrogen links the aminoacyl moiety to the tRNA (this position is indicated by an asterisk in panels B, C and F) that can serve as a proton acceptor only. However, an analogous hydrogen bond between the ester oxygen and U2506 can be formed if the uridine moiety is in the enol form having a hydroxyl group at position 2 acting as proton donor toward the electron pair of the aminoacyl ester oxygen.
Despite the chemical similarity of the interactions observed, they differ in their precise spatial arrangement because HygA alters the conformation of 23S nucleotides (U2585, U2506) and itself does not enter as deeply into the A-site, particularly the aminocyclitol moiety, as a fully accommodated A-site substrate (25). Namely it appears that the U2585–U2506 base pair and the conformation of the aminocyclitol moiety prevents HygA from being accommodated exactly like an aminoacyl tRNA (Figure 3A and B). Given the chemical similarity between HygA and the aminoacyl-tRNA, the PTC state observed in the presence of HygA could be characteristic of a canonical state acquired as the aminoacyl-tRNA enters or leaves the A-site that is trapped and exploited by HygA to inhibit protein synthesis or it could be an off-pathway state induced only by the drug.
HygA binding induces structural changes not only in proximal nucleotides but also in distal helices that connect the PTC to the GTPase associated region
Hydroxyl radical rRNA cleavage of E. coli 70S ribosomes bound to HygA supports the description of the HygA binding site presented above. While alteration in the reactivity of A2453, A2572, G2574, G2576 and G2581 (Figure 4A and B) can be explained by their proximity to the HygA binding site (Figure 4C and D), unexpectedly, additional nucleotides in more distant regions were also affected, indicating that HygA binding caused long-range conformational changes in the 23S rRNA. The affected nucleotides cluster in specific regions of H89, H90 and H92/92a (Figure 4D). These helices stem from the central loop region of domain V of the 23S rRNA, where the PTC and thus HygA and the aminoacyl moiety of the A-site tRNA are located, and reach the GTPase associated region (GAR; H43/H44, H95 and the ribosomal proteins L11, L10 and L7/L12) which influences the GTPase activity of translational factors such as the elongation factors EF-Tu and EF-G (Figure 4 D).
Figure 4.
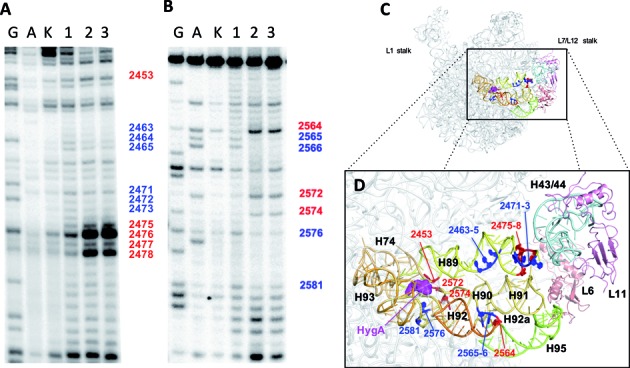
The effect of HygA on the accessibility of the 23S rRNA to hydroxyl radical cleavage. Primer extension analyses was used to define the in situ cleavage pattern of the (A) 2450 and (B) 2550 regions in the absence (lane 1) or presence of 10 μM (lane 2) and 100 μM (lane 3) HygA. The lanes marked G and A are sequencing lanes and lane K contains the control 23S rRNA sample not subjected to cleavage. (C and D) The nucleotides whose accessibility was decreased (blue) or increased (red) by HygA are shown in relation to the entire 50S ribosomal subunit (C) and more closely (D) where specific residues and rRNA helices are labeled. Further details are given in ‘Materials and Methods’ section.
Helices, H89/H90-H92, also form the aa-tRNA accommodation corridor, which the aminoacyl tRNA transits as it is accommodated into the PTC (46). This corridor presents several steric barriers, which recent simulations suggest the tRNA navigates by following multiple PTC entry pathways and the selection of these pathways could be influenced by the conformation of the PTC (46).
HygA allows aminoacyl-tRNA binding to the ribosome but perturbs its subsequent adjustment that allows it to act as a PT substrate
The crystallographic data obtained in this study indicate that HygA binds within the PTC and thereby occludes the aminoacyl-A76 ribose moiety from the catalytic site (Figure 3). Accordingly, experiments were carried out to determine to which extent this competition affects the binding of the entire aminoacyl-tRNA to the ribosome. As seen from the results presented in Figure 5A HygA does not inhibit EF-Tu-dependent binding of Phe-tRNA to poly(U) programmed 70S ribosomes even in the presence of high HygA concentrations when measured by nitrocellulose filtration which assesses the association of the tRNA with ribosome. In comparison, similar concentrations of the known 30S A-site inhibitor, tetracycline, caused almost complete inhibition of the binding (Figure 5A). This finding is in agreement with the results of a previous study (10). However, when the A-site binding was followed by stopped-flow kinetics using fluorescently labeled Phe-tRNAphe (Prf16/17) (Figure 5B), which reflects not only tRNA binding but also its chemical environment, a clear effect of HygA was detected. In fact, unlike in the control samples in which the fluorescence intensity of the labeled tRNA increases and then decreases reflecting an initial binding of the ligand followed by its adjustment on the ribosome, the latter adjustment step was not observed in the samples containing HygA, as the fluorescence intensity remained high (Figure 5B). Together these findings indicated that whereas HygA does not completely prevent tRNA binding to the A site it prevents the tRNA from reaching a fully accommodated state. In agreement HygA inhibits the PT reaction when assayed using both natural amino acid substrates (fMet and Phe) as well as the aminoacyl mimic, puromycin (Figure 5C and D) (10). The HygA dose-response curves show that inhibition of mRNA translation and dipeptide formation are superimposable (Figure 5C) indicating that inactivation of the PT activity can fully account for the block of mRNA translation caused by this antibiotic. Namely HygA appears to be a powerful inhibitor of PT activity, binding tightly to the ribosome with respect to canonical A-site substrates and effectively blocking both the specific PT assay and the more general mRNA translation assay possibly from the first peptide bond that yields the ‘initiation dipeptide’. This could explain why the dose response curves for the inhibition of mRNA translation and dipeptide formation are essentially superimposable. This premise is supported by the observation of Guerrero and colleagues that HygA once bound to the ribosome cannot easily be washed away by sucrose gradients or gel filtration chromatography (10). The binding of HygA into the PTC is further indicated by the observation that HygA competes with the known A-site binding antibiotic chloramphenicol (Figure 6A; (10)) but it responds differently to a 23S rRNA mutation (G2061A) (47) that confers resistance to chloramphenicol. Namely an in vitro translation assay that utilizes ribosomes harboring the G2061A mutation (47) is sensitive to CAM but not HygA (Figure 6B). These results support the crystallographic studies that indicate HygA primarily interferes with the accommodation of the aminoacyl-tRNA's CCA end (acceptor domain) into the PTC thus blocking the PT reaction.
Figure 5.
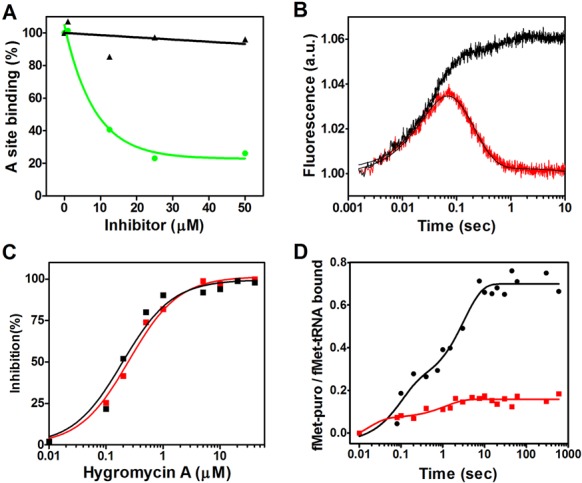
The effect of HygA on aminoacyl-tRNA binding to the A-site, transpeptidation and mRNA translation. (A) The EF-Tu and poly(U)-dependent binding of Phe-tRNA to the A site of Escherichia coli 70S ribosomes is shown as a function of increasing concentrations of HygA (black) or tetracycline (green). (B) The kinetics of EF-Tu-dependent A-site binding of proflavine-labeled Phe-tRNA to MFmRNA-programmed 70S ribosomes carrying P-site bound fMet-tRNA in the absence (red tracing) and presence (black tracing) of 20 μM of HygA. The binding kinetics were followed in a fluorescence stopped-flow apparatus. Further details are given in ‘Materials and Methods’ section. (C) Dose-response of hygromycin A inhibition of in vitro mRNA translation (red) and fMet-Phe dipeptide formation (black). (D) Time course of fMet-puromycin formation in the absence (black) or presence (red) of 20 μM of hygromycin A. Further details are given in ‘Materials and Methods’ section.
Figure 6.
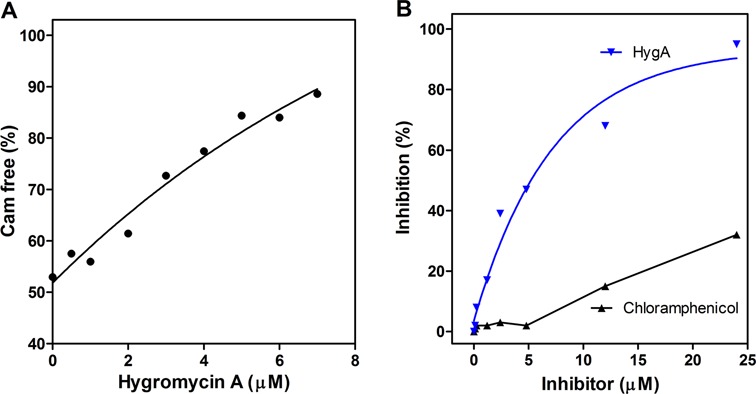
(A) Hygromycin A—chloramphenicol competition for binding to the 50S ribosomal subunit. Release of 50S ribosomes-bound [14C]-chloramphenicol upon addition of increasing concentrations of HygA. Further details are given in ‘Materials and Methods’ section. (B) Comparison of the inhibitory activities of HygA (blue) and chloramphenicol (black) on 027 mRNA translation in a cell free extract of a Thermus thermophilus mutant (23S rRNA G2061A; (47)).
DISCUSSION
The present crystallographic localization of HygA on the 50S ribosomal subunit together with the biochemical data shed light on the ribosomal binding site and on the molecular mechanism of action of this antibiotic. At the molecular level, the data indicate that the primary mechanism by which HygA inhibits protein synthesis resides in its ability to bind the PTC, in a pocket that overlaps with the binding site of the aminoacyl-ribosyl moiety at the 3′end of the A-site tRNA (Figure 3).
As seen in Figure 5A and B we show that although HygA does not prevent the EF-Tu-dependent delivery of the aminoacyl-tRNA to the ribosome, it interferes with the correct positioning of the tRNA on the ribosome. Together we interpret the structural and biochemical data to indicate that HygA allows initial binding of the aminoacyl-tRNA to the 30S A-site but prevents its subsequent accommodation into the 50S A-site by sterically occluding the aminoacyl-ribose moiety from binding within the PTC thus inhibiting peptide bond formation. The interpretation that HygA specifically impedes aminoacyl-tRNA binding into the PTC is in agreement with experiments showing that (i) HygA blocks PT in assays using either the large 50S subunit or the complete 70S ribosome and (ii) HygA inhibits binding to the A site of acceptor mimics to the large subunit, despite the lack of inhibition of larger substrates to the 70S ribosome (Figure 5A) (10,12).
As seen in Figure 3 the region of HygA that is structurally related to the aminoacyl-tRNA, does not bind as deeply into the A-site as an aminoacyl-tRNA, but make several interactions with PTC nucleotides that are comparable to the A-site tRNA (Figure 3C–F) and positions critical PTC nucleotides (i.e. U2506, U2585 and A2602) in conformations distinct from the activated PTC conformation induced by canonical A-site substrates (25,26,45). This HygA induced state of the PTC could be an off-pathway conformation unique to the drug or represent an intermediate state normally acquired when canonical A-site substrates bind or exit the A-site that is stabilized by the drug.
When the accessibility of the 23S rRNA nucleotides to hydroxyl radical is probed in the presence and absence of HygA, it becomes evident that the binding of HygA affects not just the cleavage pattern of nucleotides located in its vicinity, but also alters the chemical reactivity of nucleotides distant from the binding pocket of HygA (Figure 4). Although at this point we do not have any indication if the alterations observed in the chemical probing experiments may have a secondary role in HygA activity as in fact our results indicate that the main mechanism of action of HygA is to sterically clash with the accommodating A-site tRNA, the altered reactivity of 23S rRNA nucleotides distant from the binding pocket of HygA (i.e. in H89/90/92) could be a side effect of it stabilizing an intermediate state normally transitioned by canonical A-site substrates. In this respect, it is noteworthy that the nucleotides displaying significant changes in reactivity cluster in strategic regions of four helices, H89/H90/H92/H92a (Figure 4) which on the basis of biochemical (48), mutagenesis (49) and molecular dynamics studies (50), have been suggested to function as a communication channel between the PTC and GAR allowing for coordination of the elongation factors EF-Tu and EF-G as a function of the substrates present in and conformation of the PTC (Supplementary Figure S4). A structural basis for the alternation in hydroxyl radical accessibility cannot be extrapolated from the presented structure given that differences can be due to slight changes in the conformation or dynamics of the bases that are not easily assessed in X-ray crystallographic maps at the current resolution.
Previous experiments indicated that the binding site of HygA overlaps and/or is incompatible with the binding of other A-site targeting antibiotics like chloramphenicol, lincomycin and macrolides that possess a disaccharide at position 5 of the lactone ring, while the interaction of HygA was found to be compatible with the binding of macrolides that possess a monosaccharide at position 5 of the lactone ring and do not inhibit the PT reaction (10,11). The 50S–HygA structure presented here allows one to rationalize the compatibility between these various drugs.
For example, it has been suggested that HygA shares a binding site with chloramphenicol and lincomycin (10). When the 50S–HygA structure is aligned with that of chloramphenicol (CAM; PDB accession code: 1YJN) (51) and clindamycin (CLY), a semisynthetic derivative of lincomycin (PDB accession code: 3OH5) (52,53), CAM and CLY clash with the furanosyl and cinnamic acid moieties of HygA (Supplementary Figure S3A and B). When interactions of CAM with the ribosome are compared with those of HygA, the latter outnumbers the former both in polar and non-polar contacts (Supplementary Figure S3B–E), keeping in line with the finding that HygA binds more strongly to the ribosome and is a more potent inhibitor than CAM (Figure 6A, (10)). Interestingly, although HygA and CAM completely overlap for their binding pocket (Supplementary Figure S3B), they differ in the specific interactions with the rRNA (Supplementary Figure S3C–E) such that a 23S rRNA mutation, G2061A (47), confers resistance to CAM but not HygA (Figure 6B).
Concerning the macrolide antibiotics it has been observed that they interfere with the binding of HygA to a greater extent if a disaccharide is linked to position 5 of the lactone ring (11). Superpositioning the 50S–HygA structure with that of representative macrolides (Supplementary Figure S5) (54), allows one to rationalize these observations (11). For example, the mycarose moiety in the disaccharide extension of both carbomycin and spiramycin (PDB accession codes: 1K8A and 1KD1, respectively; (54)) encroaches on the HygA binding site (Supplementary Figure S5B and C). In the case of spiramycin the overlap mainly concerns the furanose moiety while in the case of carbomycin the overlap extends to both the furanosyl and cinnamic acid moieties of HygA (Supplementary Figure S5B and C). This is in good agreement with the observed ability of spiramycin to reduce the binding of HygA while carbomycin completely prevents it although both drugs contain a disaccharide in position 5 of the lactone ring (11). On the contrary, erythromycin, which has only a monosaccharide at the position 5 of the lactone ring and therefore does not extend into the PTC, does not overlap with HygA (PDB accession code: 3OFR; Supplementary Figure S5D; (53)) and shows compatible binding with HygA (Supplementary Figure S5D and (11)). This understanding is important for designing new antibiotics based on the HygA and/or macrolide backbone.
The presented HygA structure is largely consistent with previous work detailing the drugs structure-activity relationships. Namely, such studies show that the furanose moiety can be substituted with short lipophilic chains (8,55,56), which reflects the fact that although contributing to the interaction of the drug with the ribosome this moiety does not overlap with the A-site substrate and thus does not directly play a role in drug's inhibiting activity. If the furanose moiety is maintained, the tight packing of the 2″ and 3″ hydroxyl groups with the walls of the pocket is in agreement with previous work showing that the 3″ OH is essential (55) and the 2″ OH is phosphorylated in the HygA producing organism in one of several possible mechanisms of self-resistance (14). Furthermore the acetyl group of the furanose moiety is amendable to modification (as seen with compounds 5″-dihydrohygromycin A) (57), which is in agreement with the fact that this group extends into the lumen of the ribosomal tunnel. In the cinnamic acid moiety the strict steric requirements imposed on active substituents (8,58,59) is supported by the structure which indicates this moiety is tightly surrounded on one face by the 23S rRNA and on the other by the P-site bound tRNA (as inferred by aligning structures harboring P-site substrates to that of the HygA structure). The observation by Palaniappan et al. (60) that desmethylhygromycin and methoxyhygromycin display an in vitro activity comparable to HygA suggests that the ring structure of the methylenedioxy in the aminocyclitol moiety is not strictly required for ribosome interaction. In fact the potential hydrogen bonds observed between O5 with the amine N4 of C2573 and between O4 with the amine N2 of G2583 in the HygA-50S structure would be similarly possible for the desmethylhygromycin and methoxyhygromycin derivatives.
In conclusion this study indicates that HygA has a distinct ribosomal interaction, induces a unique PTC conformation and thus employs a distinctive molecular strategy to block protein synthesis. This information is critical to developing new antibiotics with improved pharmacokinetics and reduced sensitivities to existing resistance mechanisms by building on existing anti-infective frameworks.
NOTES ADDED IN PROOF
During the revision of the present article, a study describing the structures and mechanism of action of HygA on the ribosome was reported (61). The general conclusion of the two independent articles is essentially the same concerning HygA's mechanism of action. Moreover, a comparison of the HygA-bound structures of the D. radiodurans 50S subunit (this study) and E. coli 70S ribosome in two different states (70S alone, PDB accession code: 4Z3R and 70S plus mRNA, A- and P-site tRNAs, PDB accession code: 4Z3Q; Polikanov et al. (61)) reveals that the binding site of the antibiotic is nearly identical independent of the organism used for the ribosome preparation, the use of 50S subunits or 70S ribosomes or the presence of mRNA and tRNAs (Supplementary Figure S6). One difference, however, concerns the conformation of A2602 and A2062. As these residues are positioned differently in the structures, e.g. A2602 acquires a different conformation in each of the models reported, while HygA retains the same binding position through all three complexes, they likely play a less important role for the binding of the antibiotic and their different positioning rather reflects their high sensitivity to the presence of PTC substrates (25,62).
The only main difference between the two studies concerns the specific stereochemistry of the aminocyclitol moiety of HygA (Supplementary Figure S6). This moiety has been modeled in a neo configuration in our study and in a myo configuration in the work of Polikanov et al. (Supplementary Figure S6; (61)). From the discovery and initial characterization of HygA in the 1950's chemical and spectroscopic analysis have indicated that the aminocyclitol moiety is in the neo-configuration (5–7,38,63–67) as depicted in Supplementary Figure S1 and in the final model presented in this study. Indeed the neo-configuration is also presented in the schematic drawing of HygA in Figure 2E of Polikanov et al. (61) but it is not maintained in the deposited structures (PDB accession code: 4Z3Q and 4Z3R). After communicating and sharing with Polikanov et al., the HygA structure we are reporting in this study, they acknowledged the appropriateness of the neo configuration and will accordingly update the deposited PDB entries as the neo configuration and indeed the HygA structure we are reporting nicely fits and results in good agreement also with their experimental data (Polikanov, Wilson and Blanchard, personal communication). It is important to note that when compared to the myo configuration, the neo-configuration in our structure yields a different geometry and a slightly different pattern for the specific interactions of HygA with the ribosome; most obviously the potential interactions observed in this study between G2583 (N2) and C2573 (N4) with the oxygen atoms in the methylenedioxy ring of the aminocyclitol moiety (Figure 2C) which, are not in hydrogen bonding distance with any ribosomal element in the original model reported by Polikanov et al. (61). In this context, we would like to comment that as indicated by Polikanov et al. (61) initial studies on the function of the methylenedioxy ring in HygA have suggested that this bridge could be important for the antimicrobial activity of HygA due to its ability to bias the aminocyclitol ring toward the twisted boat conformation (58,59,64). This hypothesis had been inferred from (i) spectroscopic and chemical analysis of HygA and its derivatives, which support the twisted boat conformation for HygA in solution (38) and (ii) the observation that methoxyhygromycin, which differs from HygA by the opening of the methylendioxy ring, acquires in solution a chair conformation and has an overall lower antimicrobial activity compared to HygA (68). In our complex, HygA readily acquires a chair like conformation. This result indirectly supports more recent in vivo and in vitro studies on the function of the methylenedioxy ring which confirm its importance for the potency of the drug but points at its function in affecting the antibiotic uptake into the cell rather than the inhibition of the peptidyltransferase center (60).
Supplementary Material
Acknowledgments
We are particularly grateful to Prof. Juan Pedro Ballesta (Madrid) for his kind gift of Hygromycin A that made this work possible and to our colleagues, in particular Dr. Oscar Millet and Dr. Luca Unione at CIC bioGUNE, for thoughtful discussions. These studies could not have been performed without the expert assistance of the staff at the X06SA, X06SA, ID29 and BL13 beam lines (Swiss Light Source [SLS], European Synchrotron Radiation Facility [ESRF] and ALBA, respectively).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bizkaia:Talent and the European Union's Seventh Framework Program (Marie Curie Actions; COFUND; to S.C., A.S., T.K.); Marie Curie Actions Career Integration Grant (PCIG14-GA-2013-632072 to P.F.); Ministerio de Economía Y Competitividad (CTQ2014-55907-R to P.F., S.C.); FIRB Futuro in Ricerca from the Italian Ministero dell'Istruzione, dell'Universitá e della Ricerca (RBFR130VS5_001 to A.F.); Peruvian Programa Nacional de Innovación para la Competitividad y Productividad (382-PNICP-PIBA-2014 (to P.M. and A.F.)). Funding for open access charge: Institutional funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pittenger R.C., Wolfe R.N., Hoehn M.M., Marks P.N., Daily W.A., Mc G.J. Hygromycin. I. Preliminary studies on the production and biologic activity of a new antibiotic. Antibiot. Chemother. 1953;3:1268–1278. [PubMed] [Google Scholar]
- 2.Mann R.L., Gale R.M., Van Abeele F.R. Hygromycin. II. Isolation and properties. Antibiot. Chemother. 1953;3:1279–1282. [PubMed] [Google Scholar]
- 3.Umezawa H. Index of antibiotics from actinomycetes. Tokyo: Tokyo University; 1967. [Google Scholar]
- 4.Habib el S.E., Scarsdale J.N., Reynolds K.A. Biosynthetic origin of hygromycin A. Antimicrob. Agents Chemother. 2003;47:2065–2071. doi: 10.1128/AAC.47.7.2065-2071.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chida N., Ohtsuka M., Nakazawa K., Ogawa S. Total synthesis of antibiotic hygromycin A. J. Org. Chem. 1991;56:2976–2983. [Google Scholar]
- 6.Donohoe T.J., Flores A., Bataille C.J., Churruca F. Synthesis of (-)-hygromycin A: application of Mitsunobu glycosylation and tethered aminohydroxylation. Angew. Chem. 2009;48:6507–6510. doi: 10.1002/anie.200902840. [DOI] [PubMed] [Google Scholar]
- 7.Lo H.J., Chang Y.K., Yan T.H. Chiral pool based efficient synthesis of the aminocyclitol core and furanoside of (-)-hygromycin A: formal total synthesis of (-)-hygromycin A. Org. Lett. 2012;14:5896–5899. doi: 10.1021/ol3028237. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi S.F., Norcia L.J., Seibel S.B., Silvia A.M. Structure-activity relationships of hygromycin A and its analogs: protein synthesis inhibition activity in a cell free system. J. Antibiot. 1997;50:514–521. doi: 10.7164/antibiotics.50.514. [DOI] [PubMed] [Google Scholar]
- 9.Maffioli S.I., Fabbretti A., Brandi L., Savelsbergh A., Monciardini P., Abbondi M., Rossi R., Donadio S., Gualerzi C.O. Orthoformimycin, a selective inhibitor of bacterial translation elongation from Streptomyces containing an unusual orthoformate. ACS Chem. Biol. 2013;8:1939–1946. doi: 10.1021/cb4004095. [DOI] [PubMed] [Google Scholar]
- 10.Guerrero M.D., Modolell J. Hygromycin A, a novel inhibitor of ribosomal peptidyltransferase. Eur. J. Biochem. 1980;107:409–414. doi: 10.1111/j.1432-1033.1980.tb06044.x. [DOI] [PubMed] [Google Scholar]
- 11.Poulsen S.M., Kofoed C., Vester B. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 2000;304:471–481. doi: 10.1006/jmbi.2000.4229. [DOI] [PubMed] [Google Scholar]
- 12.Polacek N., Swaney S., Shinabarger D., Mankin A.S. SPARK–a novel method to monitor ribosomal peptidyl transferase activity. Biochemistry. 2002;41:11602–11610. doi: 10.1021/bi026040s. [DOI] [PubMed] [Google Scholar]
- 13.Kirst H.A., Dorman D.E., Occolowitz J.L., Jones N.D., Paschal J.W., Hamill R.L., Szymanski E.F. The structure of A201A, a novel nucleoside antibiotic. J. Antibiot. 1985;38:575–586. doi: 10.7164/antibiotics.38.575. [DOI] [PubMed] [Google Scholar]
- 14.Dhote V., Gupta S., Reynolds K.A. An O-phosphotransferase catalyzes phosphorylation of hygromycin A in the antibiotic-producing organism Streptomyces hygroscopicus. Antimicrob. Agents Chemother. 2008;52:3580–3588. doi: 10.1128/AAC.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie S., Li W., Yu B. Total synthesis of nucleoside antibiotic A201A. J. Am. Chem. Soc. 2014;136:4157–4160. doi: 10.1021/ja501460j. [DOI] [PubMed] [Google Scholar]
- 16.Barrasa M.I., Tercero J.A., Jimenez A. The aminonucleoside antibiotic A201A is inactivated by a phosphotransferase activity from Streptomyces capreolus NRRL 3817, the producing organism. Isolation and molecular characterization of the relevant encoding gene and its DNA flanking regions. Eur. J. Biochem. 1997;245:54–63. doi: 10.1111/j.1432-1033.1997.00054.x. [DOI] [PubMed] [Google Scholar]
- 17.Suhadolnik R.J. Nucleosides as biological probes. NY; Chichester; Bisbane; Toronto: John Wiley & Sons; 1979. pp. 205–206. [Google Scholar]
- 18.Barrasa M.I., Tercero J.A., Lacalle R.A., Jimenez A. The ard1 gene from Streptomyces capreolus encodes a polypeptide of the ABC-transporters superfamily which confers resistance to the aminonucleoside antibiotic A201A. Eur. J. Biochem. 1995;228:562–569. doi: 10.1111/j.1432-1033.1995.tb20295.x. [DOI] [PubMed] [Google Scholar]
- 19.Epp J.K., Allen N.E. 16th Interscience Conference on Antimicrobial Agentsand Chemotherapy. Chicago: American Society for Microbiology; 1976. A201A, a new antibioticproduced by Streptomyces capreolus, IV. Mode of action studies. [Google Scholar]
- 20.Wilson D.N. On the specificity of antibiotics targeting the large ribosomal subunit. Ann. N.Y. Acad. Sci. 2011;1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 21.Harris R.J., Symons R.H. On the molecular mechanism of action of certain substrates and inhibitors of ribosomal peptidyl transferase. Bioorg. Chem. 1973;2:266–285. [Google Scholar]
- 22.Beringer M., Rodnina M.V. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Schmeing T.M., Seila A.C., Hansen J.L., Freeborn B., Soukup J.K., Scaringe S.A., Strobel S.A., Moore P.B., Steitz T.A. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 24.Schmeing T.M., Huang K.S., Kitchen D.E., Strobel S.A., Steitz T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Polikanov Y.S., Steitz T.A., Innis C.A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voorhees R.M., Weixlbaumer A., Loakes D., Kelley A.C., Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandi L., Fabbretti A., Milon P., Carotti M., Pon C.L., Gualerzi C.O. Methods for identifying compounds that specifically target translation. Methods Enzymol. 2007;431:229–267. doi: 10.1016/S0076-6879(07)31012-4. [DOI] [PubMed] [Google Scholar]
- 28.Brandi L., Dresios J., Gualerzi C.O. Assays for the identification of inhibitors targeting specific translational steps. Methods Mol. Med. 2008;142:87–105. doi: 10.1007/978-1-59745-246-5_8. [DOI] [PubMed] [Google Scholar]
- 29.Fabbretti A., Pon C.L., Hennelly S.P., Hill W.E., Lodmell J.S., Gualerzi C.O. The real-time path of translation factor IF3 onto and off the ribosome. Mol. Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Pape T., Wintermeyer W., Rodnina M.V. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 31.Schlünzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 32.Wilson D.N., Schluenzen F., Harms J.M., Starosta A.L., Connell S.R., Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms J.M., Wilson D.N., Schluenzen F., Connell S.R., Stachelhaus T., Zaborowska Z., Spahn C.M., Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A. 1986;42:140–149. [Google Scholar]
- 38.Kakinuma K., Sakagami Y. Nuclear magnetic resonance and structure of hygromycin (Homomycin)1) Agric. Biol. Chem. 1978;42:279–286. [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou F.C., Sripakdeevong P., Dibrov S.M., Hermann T., Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods. 2013;10:74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keating K.S., Pyle A.M. RCrane: semi-automated RNA model building. Acta Crystallogr. D Biol. Crystallogr. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart O.S., Womack T.O., Sharff A., Flensburg C., Keller P., Paciorek W., Vonrhein C., Bricogne G. http://www.globalphasing.com. 2011 doi: 10.1107/S0907444911007773. GRADE, version 1.2.7. Cambridge, United Kingdom, Global PhasingLtd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurel G., Blaha G., Moore P.B., Steitz T.A. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 2009;389:146–156. doi: 10.1016/j.jmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 46.Whitford P.C., Geggier P., Altman R.B., Blanchard S.C., Onuchic J.N., Sanbonmatsu K.Y. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory S.T., Carr J.F., Rodriguez-Correa D., Dahlberg A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 2005;187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lancaster L., Lambert N.J., Maklan E.J., Horan L.H., Noller H.F. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA. 2008;14:1999–2012. doi: 10.1261/rna.1202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burakovsky D.E., Sergiev P.V., Steblyanko M.A., Konevega A.L., Bogdanov A.A., Dontsova O.A. The structure of helix 89 of 23S rRNA is important for peptidyl transferase function of Escherichia coli ribosome. FEBS Lett. 2011;585:3073–3078. doi: 10.1016/j.febslet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 50.Besseova I., Reblova K., Leontis N.B., Sponer J. Molecular dynamics simulations suggest that RNA three-way junctions can act as flexible RNA structural elements in the ribosome. Nucleic Acids Res. 2010;38:6247–6264. doi: 10.1093/nar/gkq414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu D., Blaha G., Moore P.B., Steitz T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Bulkley D., Innis C.A., Blaha G., Steitz T.A. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunkle J.A., Xiong L., Mankin A.S., Cate J.H. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen J.L., Ippolito J.A., Ban N., Nissen P., Moore P.B., Steitz T.A. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 55.Jaynes B.H., Cooper C.B., Hecker S.J., Blair K.T., Elliott N.C., Lilley S.C., Minich M.L., Schicho D.L., Werner K.M. Synthesis and vitro antibacterial activity of hygromycin a analogs modified at the C4′ aryl position. Bioorg. Med. Chem. Lett. 1993;3:1531–1536. [Google Scholar]
- 56.Jaynes B.H., Elliott N.C., Schicho D.L. Semisynthetic hygromycin A analogs: synthesis and anti-bacterial activity of derivatives lacking the furanose moiety. J. Antibiot. 1992;45:1705–1707. doi: 10.7164/antibiotics.45.1705. [DOI] [PubMed] [Google Scholar]
- 57.Dhote V., Starosta A.L., Wilson D.N., Reynolds K.A. The final step of hygromycin A biosynthesis, oxidation of C-5’’-dihydrohygromycin A, is linked to a putative proton gradient-dependent efflux. Antimicrob. Agents Chemother. 2009;53:5163–5172. doi: 10.1128/AAC.01069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hecker S.J., Cooper C.B., Blair K.T., Lilley S.C., Minich M.L., Werner K.M. Semisynthetic modification of hygromycin A. 2. synthesis and antibacterial activity of aryl analogs. Bioorg. Med. Chem. Lett. 1993;3:289–294. [Google Scholar]
- 59.Hecker S.J., Minich M.L., Werner K.M. Semisynthetic modification of hygromycin A. 1. Synthesis and antibacterial activity of vinyl methyl and amide analogs. Bioorg. Med. Chem. Lett. 1992;2:533–536. [Google Scholar]
- 60.Palaniappan N., Dhote V., Ayers S., Starosta A.L., Wilson D.N., Reynolds K.A. Biosynthesis of the aminocyclitol subunit of hygromycin A in Streptomyces hygroscopicus NRRL 2388. Chem. Biol. 2009;16:1180–1189. doi: 10.1016/j.chembiol.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 61.Polikanov Y.S., Starosta A.L., Juette M.F., Altman R.B., Terry D.S., Lu W., Burnett B.J., Dinos G., Reynolds K.A., Blanchard S.C., et al. Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A. Mol. Cell. 2015;58:832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen J.L., Schmeing T.M., Moore P.B., Steitz T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allen G.R. The synthesis of certain degradation products of the antibiotic 1703–18B. The synthesis of neo-Inosamine-2. J. Am. Chem. Soc. 1956;78:5691–5692. [Google Scholar]
- 64.Chida N., Nakazawa K., Ohtsuka M., Suzuki M., Ogawa S. Total synthesis of Methoxyhygromycin and Its 5-Epimer. Chem. Lett. 1990;19:423–426. [Google Scholar]
- 65.Patrick J.B., Williams R.P., Waller C.W., Hutchings B.L. A new inosamine from an antibiotic. J. Am. Chem. Soc. 1956;78:2652–2652. [Google Scholar]
- 66.Chida N., Ohtsuka M., Nakazawa K., Ogawa S. Total synthesis of hygromycin A. J. Chem. Soc. Chem. Commun. 1989:436–438. [Google Scholar]
- 67.Gurale B.P., Shashidhar M.S., Gonnade R.G. Synthesis of the Aminocyclitol Units of (−)-Hygromycin A and Methoxyhygromycin from myo-Inositol. J. Org. Chem. 2012;77:5801–5807. doi: 10.1021/jo300444b. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida M., Takahashi E., Uozumi T., Beppu T. Hygromycin A and Methoxyhygromycin, Novel Inhibitors of K88 Antigen Synthesis of Enterotoxic Escherichia coli Strain. Agric. Biol. Chem. 1986;50:143–149. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


