Abstract
GW182 family proteins play important roles in microRNA (miRNA)-mediated RNA silencing. They directly interact with Argonaute (Ago) proteins in processing bodies (P bodies), cytoplasmic foci involved in mRNA degradation and storage. Recently, we revealed that a human GW182 family protein, TNRC6A, is a nuclear-cytoplasmic shuttling protein, and its subcellular localization is regulated by its own nuclear localization signal and nuclear export signal. Regarding the further controlling mechanism of TNRC6A subcellular localization, we found that TNRC6A protein is tethered in P bodies by direct interaction with Ago2 under Ago2 overexpression condition in HeLa cells. Furthermore, it was revealed that such Ago proteins might be strongly tethered in the P bodies through Ago-bound small RNAs. Thus, our results indicate that TNRC6A subcellular localization is substantially controlled by the interaction with Ago proteins. Furthermore, it was also revealed that the TNRC6A subcellular localization affects the RNA silencing activity.
INTRODUCTION
In small RNA-mediated gene silencing, Argonaute (Ago) proteins play key roles by directly binding to microRNAs (miRNAs) or small interfering RNAs (siRNAs) (reviewed in 1,2). GW182 family proteins are shown to be necessary for miRNA-mediated RNA silencing, hereafter referred to as miRNA silencing, in animals (reviewed in 3,4). They interact with Ago proteins via their N-terminal regions containing multiple glycine-tryptophan (GW) repeats (5–10). On the other hand, their C-terminal regions, called as silencing domain, recruit cytoplasmic poly(A)-binding protein 1 (PABPC1), the PAN2-PAN3 and CCR4-NOT deadenylase complexes, leading to translational repression and mRNA degradation of miRNA-targeted mRNAs (4,11–17).
In human HEp-2, HeLa, and many other cell lines, a human GW182 family protein, TNRC6A, is known to be localized mainly in the processing (P) bodies (18–20), which are cytoplasmic foci that contain proteins involved in mRNA degradation, storage, and translational repression (reviewed in 21). However, recently we found a nuclear localization signal (NLS) and a nuclear export signal (NES) in the central region of TNRC6A and showed that it is a nuclear–cytoplasmic shuttling protein and its subcellular localization is regulated by its own NLS and NES (10). In good agreement with our previous result, several of recent reports have suggested that human GW182 proteins might function in the nucleus as well as in the cytoplasm. Chromatin silencing and alternative splicing, or transcriptional control targeting promoter RNA of inflammatory pathway genes is shown to be associated with RNA silencing factors in the nucleus (10,22–24). Furthermore, immunohistochemical analyses of several human cancers, such as gastric, colorectal, prostate and esophageal cancers, show the strong nuclear localization of TNRC6A (25,26). Thus, the actual subcellular localization is different due to the cell types, and the molecular machinery involved in specification of the subcellular localization of TNRC6A remained unknown.
In our previous report, we showed that TNRC6A protein with NES mutations (TNRC6A-NES-mut) is predominantly localized in the nucleus, due to the defect of the cytoplasmic export machinery via its NES (10). Regarding the further controlling mechanism of TNRC6A subcellular localization, in this paper, we show that, when wild-type Ago proteins are overexpressed, TNRC6A-NES-mut proteins are tethered in P bodies by direct binding to abundant amount of Ago proteins, but they are not tethered by Ago2 mutant protein deficient for binding to small RNA and also TNRC6A protein. These results suggest that TNRC6A protein is tethered by the excessive amount of Ago proteins in the P bodies through Ago-bound small RNAs, which is considered to form base-pairing with complementary RNAs in the P bodies. Our results strongly suggest that the nuclear-cytoplasmic shuttling of TNRC6A is mechanistically regulated by its own NLS and NES, but its subcellular localization is substantially determined by the comparative expression level of cellular Ago proteins. Furthermore, it was revealed that the cytoplasmic TNRC6A protein enhances miRNA silencing activity when excessive amount of Ago proteins were expressed, although TNRC6A overexpression repressed both RNA interference (RNAi) and miRNA silencing activity with low level of Ago2 protein in the cytoplasm.
MATERIALS AND METHODS
Plasmid construction
The expression plasmids of pmyc-GFP, a full-length TNRC6A, pmyc-GFP-TNRC6A, and its derivatives were constructed as described previously (10). pIRESneo-FLAG/HA-Ago1, -Ago2, -Ago3, and -Ago4 (27) were kindly provided by Dr Thomas Tuschl through Addgene, and designated as pFLAG/HA-Ago1, -Ago2, and -Ago3, respectively, in this study. For construction of pFLAG/HA-FL, FL cDNA was amplified from firefly luciferase gene in pGL3-Control (Promega) by PCR using primers (5′- AAAAGGAAAAGCGGCCGCATGGAAGACGCCAAAAACATAAAG-3′ and 5′-AAAGGGGAATTCTTACACGGCGATCTTTCCGCCCT-3′). The amplified product was digested with NotI and EcoRI, and ligated with NotI/EcoRI-digested fragment of pFLAG/HA-Ago2. For construction of an empty vector, pFLAG/HA, an untagged Ago2 expression construct, pAgo2, and pFLAG/HA-Ago2-Y529E encoding Ago2 with a substitution from tyrosine to glutamic acid at amino acid residue 529, the linear fragments with such deletions or mutations were amplified from pFLAG/HA-Ago2 by PCR using primers (5′-TGAGAATTCAGTGGATCCACTAGTAACGG-3′ and 5′-GCGGCCGCTAGCGTAATCGGGCACG-3′ for pFLAG/HA, 5′-CATGGCGGCGGCGATATCGATCCG-3′ and 5′- TACTCGGGAGCCGGCCCCGCACTTG-3′ for pAgo2, and 5′-GCCGAGGTCAAGCGCGTGGGAGAC-3′ and 5′- TTCCACGGGCGTCTTGCCGGGCAGGATG-3′ for pFLAG/HA-Ago2-Y529E), and self-ligated.
FLAG/HA-Dcp1 expression plasmid was constructed as follows: At first, both strands of chemically synthesized oligonucleotides for FLAG and HA tags (5′- CTAGCCCACCATGGACTACAAGGACGACGATGACAAGTACCCTTATGACGTGCCCGATTACGCTA-3′ and 5′- AGCTTAGCGTAATCGGGCACGTCATAAGGGTACTTGTCATCGTCGTCCTTGTAGTCCATGGTGGG-3′) were annealed and inserted into NheI and HindIII sites in pcDNA3.1-myc-Dicer (28) to generate pcDNA3.1-FLAG/HA-Dicer. A fragment of Dcp1 CDS region was then amplified using cDNA from HEK293 cells by primers (5′-AAAGGGCTCGAGATGGAGGCGCTGAGTCGAGCTG-3′ and 5′-AAAAGGAAAAGCGGCCGCTCATAGGTTGTGGTTGTCTTTGTTC-3′), and a FLAG/HA-containing vector fragment was amplified from pcDNA3.1-FLAG/HA-Dicer using primers (5′-AAAAGGAAAAGCGGCCGCTCTAGAGGGCCCGTTTAAACCCG-3′ and 5′- AAAGGGCTCGAGAGCGTAATCGGGCACGTCATAAGG-3′) for deleting Dicer region. Both fragments were digested with XhoI and NotI, and ligated. For construction of FLAG/HA-RCK/p54 expression plasmid, pFLAG/HA-RCK/p54, a cDNA fragment of RCK/p54 CDS region was amplified using cDNA purified from HEK293 cells by primers (5′- AAAGGGAAGCTTATGAGCACGGCCAGAACAGAGAAC-3′ and 5′-AAAGGGTCTAGATTAAGGTTTCTCATCTTCTACAGGC-3′). Amplified RCK/p54 fragment was digested with HindIII and XbaI, and ligated with HindIII/XbaI-digested fragment of pcDNA3.1-FLAG/HA-Dicer.
The expression constructs of chemokine (C-X-C motif) receptor 4 (CXCR4) shRNA and control DsRed shRNA, named pSUPER-CXCR4 and pSUPER-DsRed, respectively, were constructed as described previously (10). The plasmid expressing Renilla luciferase harboring a perfectly complementary target site or four target sites with a central bulge for CXCR4 siRNA in its 3′ UTR, psiCHECK-CXCR4-perfect-match (PM) or psiCHECK-CXCR4-mismatch (MM) 4×, was constructed by inserting a pair of annealed oligonucleotides (5′-TCGAGAAGTTTTCACTCCAGCTAACAG-3′ and 5′-AATTCTGTTAGCTGGAGTGAAAACTTC-3′) or two pairs of annealed oligonucleotides to form a tandem insertion with four target sites (5′-TCGAGAAGTTTTCACAAAGCTAACAAGTCAAGTTTTCACAAAGCTAACA-3′ and 5′-TTGACTTGTTAGCTTTGTGAAAACTTGACTTGTTAGCTTTGTGAAAACTTC-3′, and 5′-AGTCAAGTTTTCACAAAGCTAACAAGTCAAGTTTTCACAAAGCTAACAG-3′ and 5′-AATTCTGTTAGCTTTGTGAAAACTTGACTTGTTAGCTTTGTGAAAAC-3′), respectively, into the XhoI/EcoRI sites of psiCHECK-1 (Promega).
Cell culture
HeLa cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS; Sigma) at 37°C with 5% CO2. Transfection was conducted using Lipofectamine 2000 (Invitrogen) or polyethylenimine MAX (Polysciences) (29). For Leptomycin B (LMB; Calbiochem) treatment, 2 days after transfection, HeLa cells were washed twice with DMEM with 10% FBS and incubated in DMEM with 10% FBS containing 10 ng/ml LMB or the same volume of solvent for 4 or 8 h.
Fluorescence microscopy
HeLa cells were inoculated on a glass coverslip in a well of a 12-well culture plate at 1 × 105 cells/ml/well at 1 day before transfection. Total amount of transfected plasmids per well was 0.5 or 1.0 μg/well in any combinations of used plasmids. At 2 days after transfection, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde at room temperature for 15 min. The antibody staining was essentially performed as previously reported (30). Briefly, the fixed cells permeabilized with PBS containing 0.2% Triton X-100 were incubated with the first antibody (1:400–800 dilution) in PBS, and this was followed by treatment with the second antibody (1:400 dilution) in PBS. The cells were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen) and observed under a Zeiss Axiovert 200 fluorescence microscope, using a ×63 objective lens. Quantification of fluorescence microscopy images was performed using ImageJ software (NIH). The first antibodies used for staining the cells were as follows: anti-human AGO2 monoclonal antibody, 4G8 (Wako), Living Colors Full-Length Aequorea victoria polyclonal GFP antibody (Clontech), mouse monoclonal antibody to Dcp1a (Abcam; ab57654), anti-Rck/p54 polyclonal antibody (MBL), anti-HA-tag antibody, 561 (MBL) and anti-HA-tag antibody, TANA2 (MBL). The second antibodies were as follows: FITC-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories), FITC-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories), Cy5-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories), and Mouse IgG, Cy5-linked (from goat) (GE Healthcare).
Western blot
Cells were lysed with ice-cold lysis buffer (10 mM Hepes–NaOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 140 mM NaCl, 0.5 mM DTT, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 0.5% NP40, and 1× complete protease inhibitor cocktail [Roche]) on ice for 30 min and centrifuged. Protein concentration of the supernatant was measured using a Bio-Rad protein assay kit with bovine serum albumin as the standard. Samples were separated by SDS-PAGE and the gels were transferred to PVDF membranes using a Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked for 1 h in TBST (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween-20) supplemented with 5% skim milk (Difco) and washed once with TBST. Following incubation with the first antibody (1:500–2000 dilution) in TBST at 4°C overnight, the membranes were washed three times with TBST and treated with the second antibody (1:10 000 dilution) in TBST. After being washed three times with TBST, the membranes were incubated with ECL Prime Western Blotting Detection Reagent (GE Healthcare) and signals were detected by an ImageQuant LAS 4000 (GE Healthcare). The first antibodies used for western blot were as follows: anti-human AGO2 monoclonal antibody, 4G8, Living Colors Full-Length Aequorea victoria polyclonal GFP antibody, monoclonal ANTI-FLAG M2 antibody (Sigma), DYKDDDDK Tag Antibody (Cell Signaling Technology), mouse anti-lamin A/C monoclonal antibody (BD Transduction Laboratories), rabbit anti-GW182 (TNRC6A) antibody (Bethyl; A302-329A), and monoclonal anti-alpha tubulin antibody (ICN/CAPPEL Biomedicals). The second antibodies were as follows: ECL anti-mouse IgG, horseradish peroxidase linked whole antibody (GE Healthcare) and ECL anti-rabbit IgG, horseradish peroxidase linked whole antibody (GE Healthcare).
Cell fractionation
Nuclear and cytoplasmic fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo scientific) according to the manufacturer's recommendation.
Immunoprecipitation
Cells were plated in 9-cm dish at 2 × 106 cells/10 ml one day before transfection. For the experiment of Supplementary Figure S6, 5 μg of pFLAG/HA-FL or pFLAG/HA-Ago2 and 5 μg of pmyc-GFP-TNRC6A-NES-mut were transfected into the cells. Two days after transfection, cells were washed once with PBS and lysed with ice-cold lysis buffer. Lysates were centrifuged at 20 000 g for 10 min at 4°C, and the supernatants were centrifuged at 48 000 g for 30 min at 4°C. Thirty μl of protein A agarose (MILLIPORE) was equilibrated with 500 μl of lysis buffer and incubated with 2.5 μg of an anti-GFP antibody in lysis buffer for 1 h at 4°C. Then the cell lysates were added to the antibody-bound protein A agarose and incubated with constant rotation for 2 h at 4°C. The beads were washed twice with the lysis buffer containing 300 mM NaCl, once with lysis buffer, and resuspended in 40 μl of 2 × SDS sample buffer. Bound proteins were eluted by boiling the beads for 5 min and separated by SDS-PAGE.
For analyses of Ago-bound miRNAs, 10 μg of pFLAG/HA, pFLAG/HA-Ago2-WT or -Y529E were transfected into the cells. Two days after transfection, cells lysates were prepared as described above and incubated with 60 μl ANTI-FLAG M2 affinity gel (Sigma) with constant rotation for 2 h at 4°C. The beads were washed twice with the lysis buffer containing 300 mM NaCl and once with the lysis buffer. Sixty percent of beads were used for RNA extraction, and the remaining beads were used for protein analysis. Bound proteins were eluted by boiling the beads for 5 min and separated by SDS-PAGE. Quantification of Ago-bound miRNAs was performed as previously reported (10).
For the experiment of Figure 6G, 2 μg of pFLAG/HA, pFLAG/HA-Ago2-WT or -Y529E along with 8 μg of the expression construct of myc-GFP tagged proteins were transfected into the cells. Two days after transfection, cells lysates were prepared as described above and incubated with 60 μl ANTI-FLAG M2 affinity gel with constant rotation for 2 h at 4°C. The beads were washed three times with the lysis buffer, and resuspended in 40 μl of 2 × SDS sample buffer.
Figure 6.
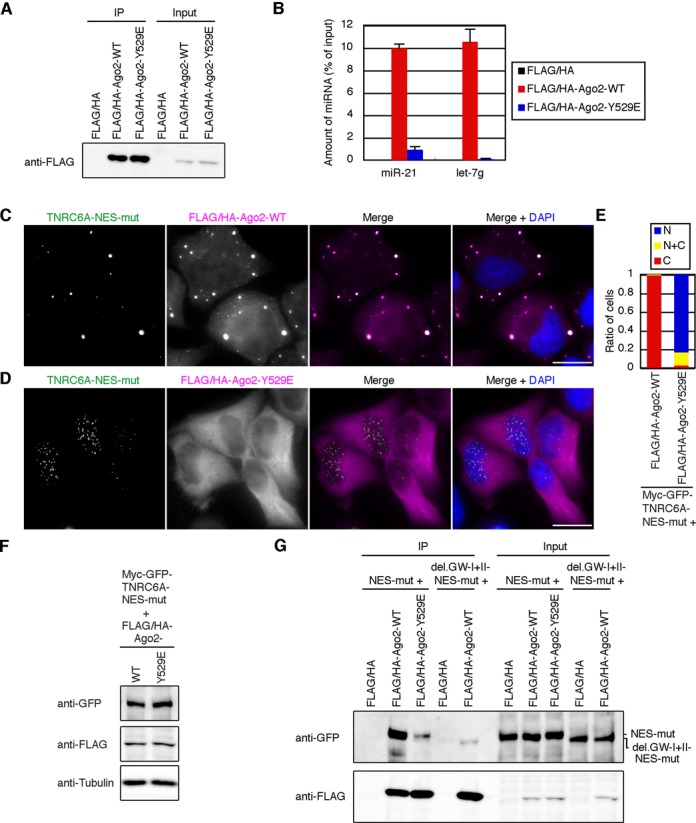
Overexpression of small RNA binding-deficient Ago2 protein does not Inhibit the nuclear transport of TNRC6A.(A) Cell lysates from HeLa cells transfected with empty vector (pFLAG/HA), pFLAG/HA-Ago2-WT, or pFLAG/HA-Ago2-Y529E were immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates (IP) and cell lysates (input) were analyzed by western blot. (B) MiRNAs contained in the immunoprecipitates of (A) was quantified by real-time PCR. Data are the mean and standard deviation of the triplicate measurements. (C and D) Fluorescent microscopy images of HeLa cells expressing myc-GFP-TNRC6A-NES-mut along with FLAG/HA-Ago2-WT (C) or -Y529E (D). Cells were stained with an anti-HA antibody. Bars, 20 μm. (E) The ratio of cells expressing GFP-signal of myc-GFP-TNRC6A-NES-mut exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). (F) Western blot of myc-GFP-TNRC6A-NES-mut and FLAG/HA-Ago2-WT or -Y529E. (G) Cell lysates from HeLa cells expressing myc-GFP-TNRC6A-NES-mut or myc-GFP-TNRC6A-NES-mut-del.GW-I+II along with FLAG/HA, FLAG/HA-Ago2-WT or -Y529E were immunoprecipitated with an anti-FLAG antibody. IPs and inputs were analyzed by western blot.
Transfection of small RNA duplexes
Fifty picomol/well of small RNA duplexes were transfected into HeLa cells simultaneously or one day after transfection with 0.5 μg/well of pmyc-GFP-TNRC6A-NES-mut. RNA oligonucleotides purchased from Sigma were as follows: siControl (5′-CGUACGCGGAAUACUUCGAUU-3′and 5′-UCGAAGUAUUCCGCGUACGUG-3′); miR-200b (5′-UAAUACUGCCUGGUAAUGAUGA-3′and 5′-CAUCUUACUGGGCAGCAUUGGA-3′); and let-7b (5′-UGAGGUAGUAGGUUGUGUGGUU-3′and 5′-CUAUACAACCUACUGCCUUCCC-3′).
Gene silencing activity assay
HeLa cells were plated at 1.0 × 105 cells/well in a well of a 24-well plate at 1 day before transfection. Cells were simultaneously transfected with psiCHECK-CXCR4-PM or -MM 4× (10 ng/well), pGL3-Control (100 ng/well; Promega) encoding firefly luciferase gene, 100 ng/well of shRNA expression construct (pSUPER-CXCR4 or pSUPER-DsRed) or 50 pmol/well of siRNA (siCXCR4 or control siGY441 or siDsRed), the indicated amount of pFLAG/HA or pFLAG/HA-Ago2, pmyc-GFP and/or pmyc-GFP-TNRC6A or its mutant expression constructs. The cells were harvested 48 h after the transfection, and the relative Renilla/firefly luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega). RNA sequences of siRNAs were as follows: siCXCR4 (5′-UGUUAGCUGGAGUGAAAACUU-3′ and 5′-GUUUUCACUCCAGCUAACACA-3′); siGY441 (5′-AUGAUAUAGACGUUGUGGCUG-3′ and 5′-GCCACAACGUCUAUAUCAUGG-3′); siDsRed (5′-UUCUUCUGCAUUACGGGGCGG -3′ and 5′-GCCCCGUAAUGCAGAAGAAGA-3′).
RESULTS
Overexpression of Ago proteins inhibits the nuclear transport of TNRC6A
Previously we reported that human TNRC6A is a nuclear–cytoplasmic shuttling protein, and its subcellular localization is intrinsically regulated by its own NLS and NES (10). In accordance with this finding, TNRC6A protein with NES mutation (TNRC6A-NES-mut) was predominantly localized in the nucleus in human HeLa cells (10). However, when Ago proteins were overexpressed with TNRC6A-NES-mut, we noticed that majority of TNRC6A-NES-mut proteins were colocalized with Ago proteins in the cytoplasm, and rarely localized in the nucleus (Figure 1). The result suggests that Ago proteins substantially govern the subcellular localization of TNRC6A.
Figure 1.
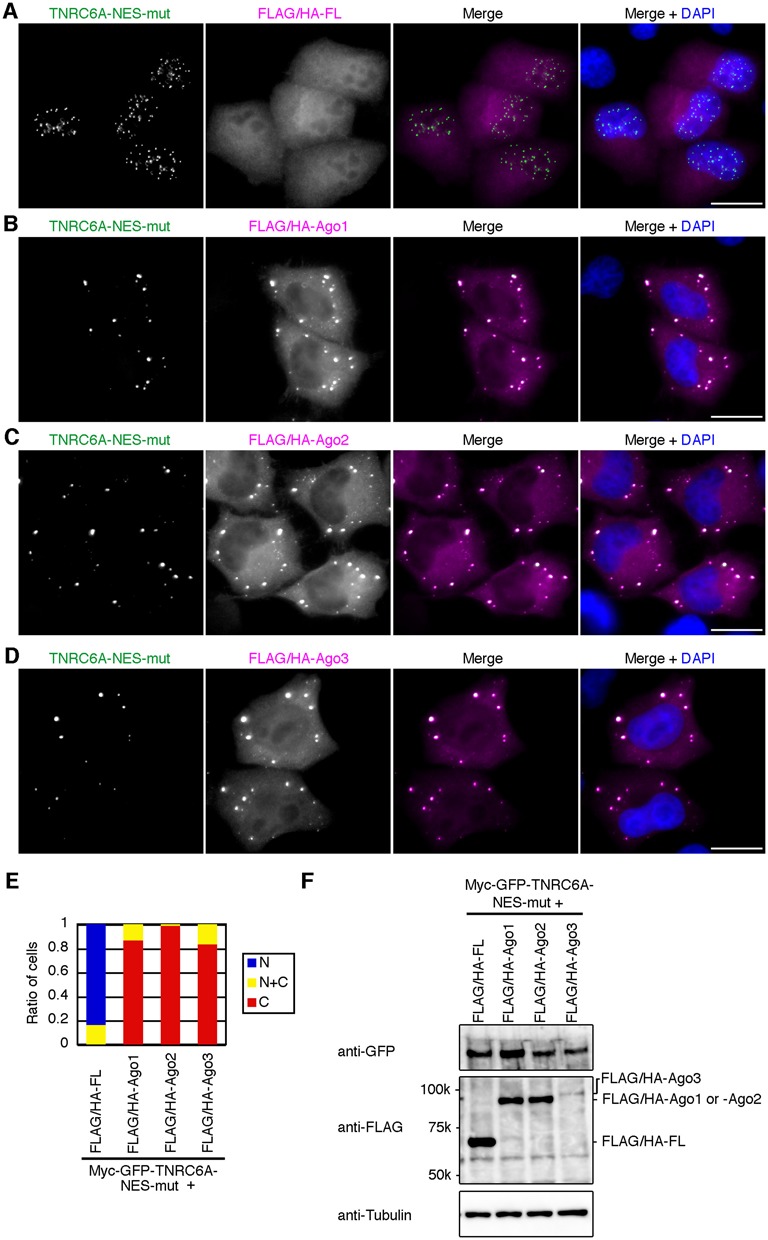
Overexpression of Ago proteins inhibits the nuclear transport of the dotted foci of GFP-signals of myc-GFP-TNRC6A-NES-mut proteins. (A–D) HeLa cells expressing myc-GFP-TNRC6A-NES-mut with either of the indicated proteins, FLAG/HA-FL (A), -Ago1 (B), -Ago2 (C), or -Ago3 (D), were stained with an anti-HA antibody, followed by Cy5-conjugated anti-mouse IgG. Fluorescent images are shown from the left: GFP signals of myc-GFP-TNRC6A-NES-mut; Cy5 signals of the second antibody against anti-HA antibody; the merged images of GFP and Cy5, in which GFP is shown in green, the Cy5 in magenta; the merged images with DAPI (blue). Bars, 20 μm. (E) The ratio of cells expressing the dotted GFP-signals of myc-GFP-TNRC6A-NES-mut exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). (F) Western blot of myc-GFP-TNRC6A-NES-mut and control FLAG/HA-FL, or FLAG/HA-tagged Ago1, Ago2, and Ago3 proteins. An anti-tubulin antibody was used as a loading control.
To investigate the effect of Ago overexpression on the subcellular localization of TNRC6A-NES-mut protein, 0.5 μg/well of the expression construct of myc-GFP-tagged TNRC6A-NES-mut (pmyc-GFP-TNRC6A-NES-mut) was transfected into HeLa cells with 0.5 μg/well of the expression construct of FLAG- and HA-tagged four types of Ago proteins (pFLAG/HA-Ago1–Ago4) or control firefly luciferase (pFLAG/HA-FL) (Figure 1). GFP signals of myc-GFP-TNRC6A-NES-mut in the cells were observed under a fluorescent microscope (Figure 1A–D), and cells were classified into three groups according to the localizations of the ‘dotted foci’ with GFP signals at first, that is, those with the dotted GFP signals (i) exclusively in the nucleus, (ii) exclusively in the cytoplasm, and (iii) in both the nucleus and the cytoplasm (Figure 1E). When control FLAG/HA-FL was coexpressed with myc-GFP-TNRC6A-NES-mut, the dotted GFP-signal of TNRC6A-NES-mut protein was observed exclusively in the nucleus in 80–90% of the cells (Figure 1A and E), as observed in the cells exogenously expressing TNRC6A-NES-mut protein alone (10). In contrast, such a nuclear-specific localization of TNRC6A-NES-mut was not observed under overexpression condition of FLAG/HA-Ago1, -Ago2, or -Ago3 (Figure 1B–E). In >80% of the cells expressing FLAG/HA-Ago1, -Ago2, or -Ago3, GFP-signal of TNRC6A-NES-mut was observed exclusively in the cytoplasmic foci with Ago proteins (Figure 1B–E). Expression level of FLAG/HA-Ago3 was apparently lower than that of FLAG/HA-Ago1 or -Ago2 (Figure 1F), but it might be enough for inhibiting the nuclear transport of TNRC6A-NES-mut protein (Figure 1E). The expression level of FLAG/HA-Ago4 was too low to evaluate the effect of Ago4 overexpression on the subcellular localization of TNRC6A (data not shown). Furthermore, GFP signal of TNRC6A-NES-mut protein was observed exclusively in the cytoplasm in most of cells under the overexpression condition of Ago2 proteins without FLAG/HA-tag (Supplementary Figure S1), suggesting that Ago2 protein itself has certain ability to tether TNRC6A-NES-mut in the cytoplasm even when deleting FLAG/HA-tag. In addition, we also reported that the translocation of TNRC6A protein from nucleus to cytoplasm is inhibited by the treatment with LMB, an inhibitor of Exportin 1-dependent nuclear export of TNRC6A (10). In accordance with such results, when HeLa cells were treated with LMB, clear nuclear accumulation of the dotted GFP-signal of myc-GFP-TNRC6A-WT protein was observed (Supplementary Figure S2B and F). However, in consistent with the result using myc-GFP-tagged TNRC6A-NES-mut (Figure 1C and E), the nuclear transport of TNRC6A-WT was inhibited by Ago2 overexpression (Supplementary Figure S2C and F). These results clearly indicated that Ago2 overexpression inhibits the nuclear transport of myc-GFP-TNRC6A-WT or myc-GFP-TNRC6A-NES-mut.
We further examined the detailed subcellular distribution of the ‘diffused’ TNRC6A proteins. For this purpose, we used anti-GFP antibody, which can detect the weak diffused GFP signals of myc-GFP-TNRC6A-NES-mut with the higher sensitivity (Figure 2A and B; 10), and classified the cells into three groups according to the subcellular localization of the diffused signals (Figure 2C). In almost all cells expressing control FLAG/HA-FL, most of the diffused signals of myc-GFP-TNRC6A-NES-mut proteins were observed in the nucleus in addition to the dotted foci (Figure 2A and C). In Ago2 overexpression condition, most of the dotted foci of myc-GFP-TNRC6A-NES-mut proteins were also detected in the cytoplasm, and those in the nucleus were rarely observed (Figure 2B and C). In about 85% of the cells, the diffused signals of TNRC6A-NES-mut proteins in the cytoplasm were stronger than or equal to those in the nucleus (Figure 2B and C). Consistent with these results, the nuclear transport of the diffused myc-GFP-TNRC6A proteins induced by the LMB treatment was also evidently suppressed in Ago2 overexpression condition (Supplementary Figure S3). Thus, these results indicated that the nuclear transport of the diffused myc-GFP-TNRC6A proteins is inhibited by the overexpression of Ago2 protein as well as that of the dotted myc-GFP-TNRC6A proteins.
Figure 2.
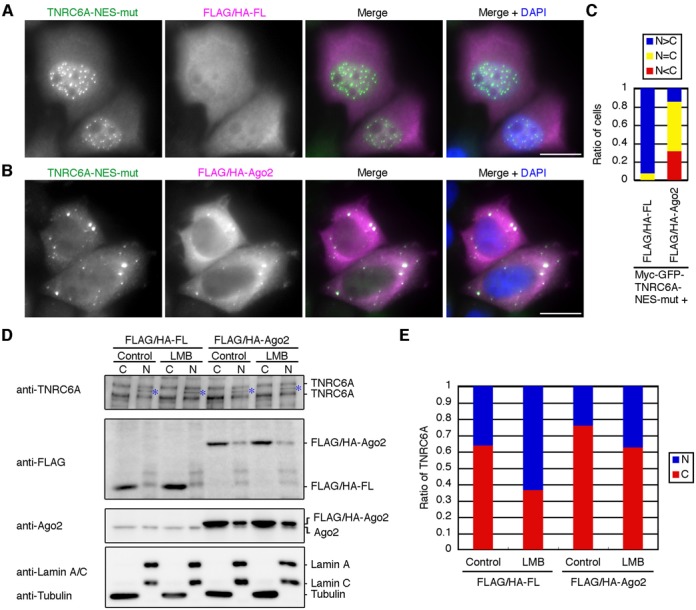
Overexpression of Ago proteins inhibits the nuclear transport of the diffused signals of myc-GFP-TNRC6A-NES-mut and endogenous TNRC6A proteins. (A and B) HeLa cells expressing myc-GFP-TNRC6A-NES-mut with FLAG/HA-FL (A) or -Ago2 (B) were stained with an anti-GFP antibody and an anti-HA antibody, followed by FITC-conjugated anti-rabbit IgG and Cy5-conjugated anti-mouse IgG. Fluorescent images are shown from the left: FITC signals of the second antibody against anti-GFP antibody; Cy5 signals of the second antibody against anti-HA antibody; the merged images of FITC and Cy5, in which FITC is shown in green, the Cy5 in magenta; the merged images with DAPI (blue). Bars, 20 μm. (C) The ratio of cells in which the diffused GFP signals of myc-GFP-TNRC6A-NES-mut in the nucleus were stronger than (N>C, blue), equal to (N = C, yellow), or weaker than (N<C, red) those in the cytoplasm. (D) HeLa cells were inoculated in a well of a 6-well culture plate at 4 × 105 cells/2ml/well at 1 day before transfection. The cells were transfected with FLAG/HA-FL or FLAG/HA-Ago2 plasmid (2.0 μg/well), and re-inoculated at 8 × 105 cells/2ml/well at 1 day after transfection. At 1 day later, the cells were treated with LMB for 8 hr, and fractionated. Fifteen μg of the cytoplasmic fraction (C) or 2.5 μg of the nuclear fraction (N) were analyzed by western blot using an anti-TNRC6A, anti-FLAG, and anti-Ago2 antibodies. Note that HeLa cells endogenously express two isoforms of TNRC6A, ∼210 kDa TNRC6A (NP_055309) and ∼182 kDa TNRC6A (AAK62026) proteins, and that the cytoplasmic fraction was loaded about 1.6-fold compared with the nuclear fraction in each sample. Anti-tubulin and anti-Lamin A/C antibodies were used as a cytoplasmic and a nuclear marker, respectively. Asterisks indicate nonspecific bands. (E) The ratio of signal intensities of TNRC6A proteins between the nucleus (N, blue) and the cytoplasm (C, red). Signal intensities of the bands corresponding to ∼210 kDa and ∼182 kDa TNRC6A in D were measured and added up in each fraction and converted into total signal intensities based on the volume of the loaded fraction, and the nuclear-cytoplasmic ratio of TNRC6A was calculated. The similar results were obtained by two independent experiments.
Next we examined whether overexpression of Ago2 protein inhibits the nuclear transport of endogenous TNRC6A. However, since the amount of endogenous TNRC6A proteins might be too small to detect by immunohistochemical procedures using anti-TNRC6A antibody, we carried out western blot using subcellular fractions of HeLa cells transfected with control pFLAG/HA-FL and pFLAG/HA-Ago2 constructs with or without LMB treatment, and the nuclear-cytoplasmic ratio of the signal intensities of sum of ∼210 kDa and ∼182 kDa TNRC6A in each sample was estimated (Figure 2D and E). In the cells expressing control FLAG/HA-FL, about 35% of total TNRC6A proteins were detected in the nuclear fraction by western blot. However, an apparent increase up to >60% of endogenous TNRC6A protein was observed in the nuclear fraction with the LMB treatment. On the other hand, only ∼25% of endogenous TNRC6A protein was detected in the nuclear fraction from the cells expressing FLAG/HA-Ago2 without LMB treatment, suggesting that overexpression of Ago2 inhibits the nuclear-cytoplasmic shuttling of endogenous TNRC6A protein. Although the nuclear ratio of TNRC6A protein in the cells expressing FLAG/HA-Ago2 increased after the treatment with LMB up to ∼35%, the ratio was obviously low in comparison with the ratio of the cells expressing FLAG/HA-FL with LMB treatment (∼60%). In the cells transfected with pFLAG/HA-Ago2, overexpressed Ago2 proteins detected by anti-FLAG and anti-Ago2 antibodies were predominantly increased in the cytoplasmic fractions compared to the nucleus fractions (Figure 2D). These results indicated that the nuclear transport of endogenous TNRC6A was also inhibited by Ago2 overexpression.
These results also indicate that overexpression of Ago proteins inhibits the nuclear transport of TNRC6A.
Inhibition of TNRC6A nuclear transport is dependent on Ago2 expression level
Nuclear transport of TNRC6A protein was inhibited by the overexpression of Ago proteins. Thus, it was considered that the expression levels of Ago proteins may control TNRC6A subcellular localization. To test the effects of the amount of Ago protein for TNRC6A subcellular localization, the different amount of pFLAG/HA-Ago2 was transfected into HeLa cells with a definite amount of pmyc-GFP-TNRC6A-NES-mut (0.5 μg), and the subcellular localization of myc-GFP-TNRC6A-NES-mut was examined (Figure 3A and Supplementary Figure S4). Although we could not distinguish endogenous Ago2 protein from FLAG/HA-Ago2 by western blot due to the indistinguishable overlapped band, Ago2 level detected by an anti-Ago2 antibody was upregulated according to the increased amount of the transfected plasmid, pFLAG/HA-Ago2 (Figure 3A; the first panel). The increase in Ago2 protein is assumed to be originated from the transfected plasmid, pFLAG/HA-Ago2, since Ago2 protein detected by anti-FLAG antibody was also upregulated according to the increased amount of the transfected pFLAG/HA-Ago2 (Figure 3A; the second panel). In well-agreement with the upregulation of Ago2 protein, the ratio of cells, in which GFP-signal of TNRC6A-NES-mut was localized predominantly in the nucleus, decreased and that, in which that of TNRC6A-NES-mut was observed in the cytoplasm, increased (Figure 3A; the lowest panel and Supplementary Figure S4). This result suggests that inhibition of TNRC6A nuclear transport is subjected by Ago2 expression level.
Figure 3.
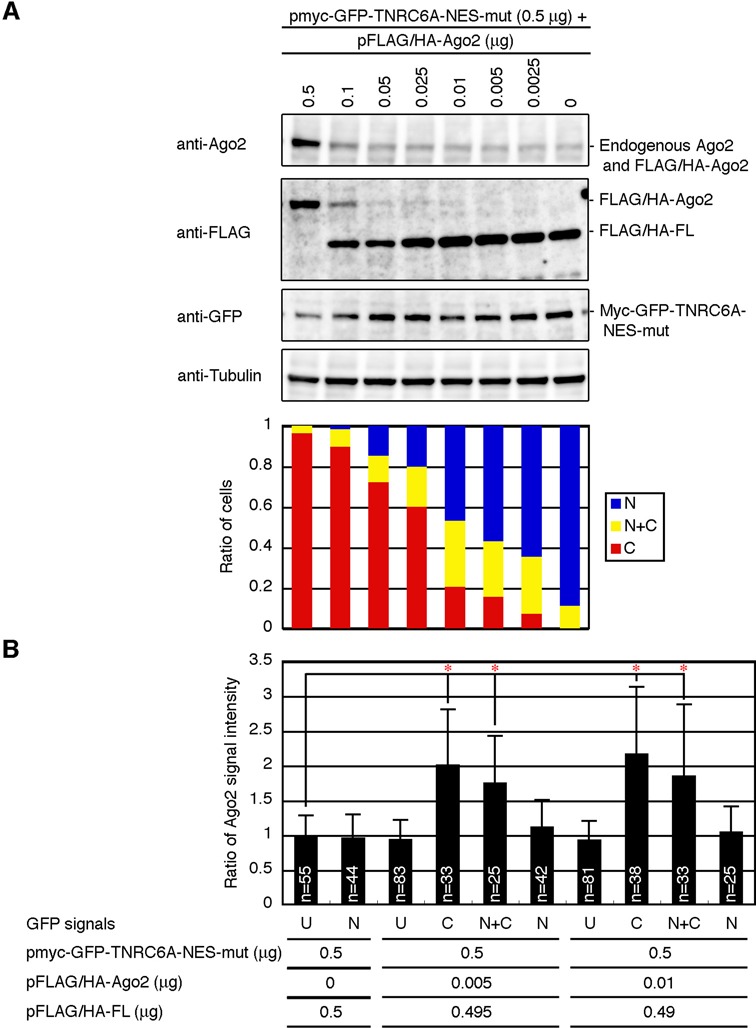
Effects of different expression levels of Ago2 on the subcellular localization of TNRC6A-NES-mut protein. (A) Upper four panels, HeLa cells were transfected with pmyc-GFP-TNRC6A-NES-mut (0.5 μg) and the mixture of pFLAG/HA-Ago2 and pFLAG/HA-FL (total amount = 0.5 μg). Cell lysates were analyzed by western blot. Note that endogenous Ago2 (∼97 kDa) and FLAG/HA-Ago2 (∼100 kDa) proteins were detected as one overlapped band in the first panel, since they could not be separated in a 4–20% gel. Lowest panel, the ratio of cells expressing GFP-signal of myc-GFP-TNRC6A-NES-mut exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). Typical fluorescent microscopy images are shown in Supplementary Figure S4. (B) Estimation of Ago2 protein level by fluorescent microscopy images. HeLa cells were transfected with pmyc-GFP-TNRC6A-NES-mut (0.5 μg) and the mixture of pFLAG/HA-Ago2 and pFLAG/HA-FL (total amount = 0.5 μg). Cells were stained with an anti-Ago2 antibody. Cells, in which no GFP signals were observed, were categorized as undetected (U), and others were classified by the subcellular localization of GFP-signal of myc-GFP-TNRC6A-NES-mut; exclusively in the nucleus (N), cytoplasm (C), or both (N+C). Data represents the mean ratio of Ago2 signal intensities ± standard deviation. The average signal intensity of Ago2 in the cells in which no GFP signals were not detected in the sample without transfection of pFLAG/HA-Ago2 (pFLAG/HA-Ago2, 0 mg, U) was set to 1. *P < 0.0001, t-test. n, the number of quantified cells. Typical fluorescent microscopy images are shown in Supplementary Figure S5.
Next, we estimated the expression level of Ago2 protein necessary for anchoring TNRC6A in the cytoplasm in individual cells, because the amount of Ago2 protein is presumed to differ in each cell according to the subcellular localization of TNRC6A-NES-mut protein. When cells were transfected with 0.01 or 0.005 μg/well of pFLAG/HA-Ago2, GFP-signal of TNRC6A-NES-mut protein was exclusively observed in the nucleus in about 45% or 55% of the cells, respectively, exclusive cytoplasmic localization of GFP-signal of TNRC6A-NES-mut protein was observed in 20% or 15% of the cells, and in the remaining 35% or 30% of the cells, GFP-signal of TNRC6A-NES-mut was localized in both nucleus and cytoplasm (Figure 3A; the lowest panel). These cells were stained with an anti-Ago2 antibody, and Ago2 signal intensities of individual cells were quantified from fluorescent microscopy images: The Ago2 signal intensity was counted by subtracting the average signal intensity of the control cells stained with a secondary antibody alone from the signal intensity of the cells stained with the anti-Ago2 antibody, and the ratio of mean Ago2 signal intensity relative to that in the control cells was calculated. For control cells, we used cells transfected with neither pmyc-GFP-TNRC6A-NES-mut nor pFLAG/HA-Ago2, which may represent the cells with no GFP signals (Figure 3B and Supplementary Figure S5). The ratios of average Ago2 signal intensities among the cells without GFP signals even when pFLAG/HA-Ago2 was transfected at 0.01 or 0.005 μg/well were almost similar level at about ‘1’ relative to the control cells (Figure 3B; pFLAG/HA-Ago2, 0 μg, 0.005 μg and 0.01 μg, U), suggesting that both of pmyc-GFP-TNRC6A-NES-mut and pFLAG/HA-Ago2 plasmids were not successfully introduced into these cells. The ratio of average signal intensity of Ago2 in the cells in which TNRC6A-NES-mut protein is localized in the cytoplasm increased approximately 2-fold compared to those in the control cells (Figure 3B; pFLAG/HA-Ago2, 0.005 μg and 0.01 μg, C). Modest increases were observed in the cells in which TNRC6A-NES-mut protein was localized both in the nucleus and cytoplasm (Figure 3B; pFLAG/HA-Ago2, 0.005 μg and 0.01 μg, N+C). In contrast, such an increase was not observed in the cells in which GFP signal of TNRC6A-NES-mut was exclusively observed in the nucleus (Figure 3B; pFLAG/HA-Ago2, 0 μg, 0.005 μg and 0.01 μg, N). Average signal intensity of Ago2 proteins in all the cells used for transfection with 0.005 μg/well of pFLAG/HA-Ago2 was 1.31-fold compared with that of the untransfected cells, consistent with the estimation by western blot (1.38 ± 0.08-fold, two independent experiments), indicating that the Ago2 signal intensity quantified from fluorescent microscopy images reflected the Ago2 protein expression level. These results suggest that Ago2 protein increased to at least ∼2-fold may exhibit the inhibitory ability of nuclear transport of TNRC6A in HeLa cells.
Ago-binding GW motif in TNRC6A is necessary for cytoplasmic anchoring of TNRC6A protein
It has been reported that N-terminal GW-repeats of GW182 family proteins requires for their interaction with Ago proteins (5–10). Human TNRC6A has three GW-repeated Ago-binding motifs, GW-I, -II, and -III (Figure 4A), and GW-I and -II interact with Ago2 with strong affinities compared to GW-III (9,10). A TNRC6A mutant lacking both GW-I and -II motifs (TNRC6A-del.GW-I+II) dramatically reduces the interaction with Ago2, and its mutant lacking three GW motifs (TNRC6A-del.GW-I+II+III) shows no or little binding to Ago2 (10).
Figure 4.
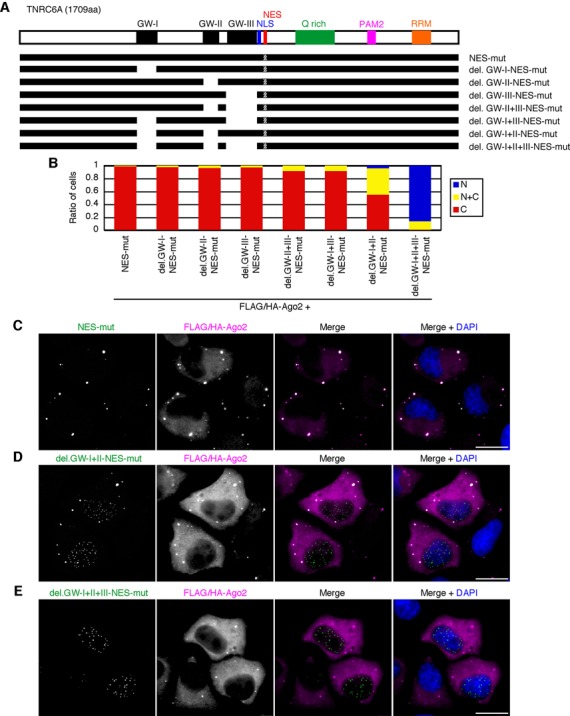
TNRC6A is anchored in the cytoplasmic foci via its Ago-binding GW motifs. (A) Schematic representation of the domain structure of TNRC6A and its mutant proteins. GW-I, -II, and -III (black boxes), GW-repeated Ago-binding motifs. Q-rich (a green box), a glutamine-rich region. PAM2 (a magenta box), a poly(A)-binding protein binding motif 2 (PAM2). RRM (an orange box), RNA recognition motif. (B) The ratio of cells expressing GFP-signal of the indicated myc-GFP-TNRC6A mutant proteins exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). (C–E) Fluorescent microscopy images of HeLa cells expressing the indicated myc-GFP-TNRC6A mutant protein with FLAG/HA-Ago2. Cells were stained with an anti-HA antibody. Bars, 20 μm.
At first, to investigate whether interaction of TNRC6A with Ago2 in the normal condition without Ago2 overexpression is saturated or not, the amounts of Ago2 protein associated with TNRC6A-NES-mut in the normal and Ago2 overexpression conditions in HeLa cells were determined by immunoprecipitation. Under the Ago2 overexpression condition, more abundant amount of Ago2 protein was immunoprecipitated with TNRC6A-NES-mut compared to the normal condition without Ago2 overexpression (Supplementary Figure S6), suggesting that the amount of TNRC6A-interacting Ago2 protein is not saturated and more abundant Ago2 protein can interact with TNRC6A.
Then, to investigate that the interaction between TNRC6A and Ago via GW motifs is required for the anchoring TNRC6A in the cytoplasm by Ago overexpression, we observed the subcellular localization of myc-GFP-TNRC6A-NES-mut lacking each or combinations of GW-I, -II, and -III motifs under overexpression condition of Ago2. In the cells coexpressing myc-GFP-TNRC6A-NES-mut proteins deleting GW-I, -II, -III, -II+III, or -I+III (myc-GFP-TNRC6A-del.GW-I, -II, -III, -II+III, or -I+III-NES-mut) and FLAG/HA-Ago2, GFP signals were mainly observed in the cytoplasm in more than 90% of the cells (Figure 4A-C). However, when pmyc-GFP-TNRC6A-del.GW-I+II-NES-mut, the expression construct of myc-GFP-TNRC6A-NES-mut protein deleting both GW-I and -II, was transfected with pFLAG/HA-Ago2, GFP signals were detected in the cytoplasm in ∼50% of the cells (Figure 4B and D). Furthermore, myc-GFP-TNRC6A-del.GW-I+II+III-NES-mut (Figure 4B and E), in which all of three GW motifs, GW-I, -II, and -III, were deleted, were extremely observed in the nucleus in about 90% of the cells, even when FLAG/HA-Ago2 was coexpressed. Similar results were observed by the transfection of pmyc-GFP-TNRC6A lacking each or combinations of GW-I, -II, and -III motifs with pFLAG/HA-Ago2, followed by LMB treatment (Supplementary Figure S2A and D–F). These results indicate that TNRC6A can be anchored in the cytoplasm via GW motif(s) under the overexpression condition of Ago2. And at least one of three GW motifs is enough for tethering TNRC6A protein in Ago2 overexpression condition, although the ability of GW-III to tether TNRC6A in the cytoplasm is weaker than those of others probably due to its weak binding ability to Ago2 protein.
Cytoplasmic TNRC6A and Ago2 protein are co-localized with P body markers
In the cells expressing abundant amount of Ago proteins, TNRC6A-NES-mut was co-localized with Ago proteins in the cytoplasmic foci (Figure 1B–E). The size of such foci increased according to the amount of transfected Ago2 expression constructs (Supplementary Figures S1A–C and S4). Previously we showed that cytoplasmic myc-GFP-TNRC6A is colocalized with P body marker proteins, Dcp1 and RCK/p54 (10). To investigate whether the foci containing both TNRC6A-NES-mut and FLAG/HA-Ago proteins are P bodies, the cells transfected with the expression plasmids of myc-GFP-TNRC6A-NES-mut and FLAG/HA-Ago2 were stained by the antibody of Dcp1 or RCK/p54 (Supplementary Figure S7). All of the foci containing myc-GFP-TNRC6A-NES-mut proteins were revealed to be colocalized with FLAG/HA-Ago2, Dcp1 or RCK/p54, suggesting that the cytoplasmic foci containing both TNRC6A-NES-mut and Ago2 are P bodies.
Next, we tested the effect of Dcp1 or RCK/p54 overexpression on the subcellular localization of TNRC6A-NES-mut. In any cells transfected with Dcp1 or RCK/p54 expression construct, the exclusive cytoplasmic localization of GFP-signal of TNRC6A-NES-mut was not observed (Figure 5). In this condition, endogenous Ago2 was localized in the cytoplasmic foci with Dcp1 and RCK/p54 (date not shown). The result suggests that Dcp1 or RCK/p54 is not able to inhibit the nuclear transport of TNRC6A unlike Ago proteins.
Figure 5.

Overexpression of Dcp1 or RCK/p54 does not inhibit the nuclear transport of TNRC6A. (A and B) Fluorescent microscopy images of HeLa cells expressing myc-GFP-TNRC6A-NES-mut along with FLAG/HA-Dcp1 (A) or FLAG/HA-RCK/p54 (B). Cells were stained with an anti-HA antibody, followed by Cy5-conjugated anti-mouse IgG. Bars, 20 μm. (C) The ratio of cells expressing GFP-signal of myc-GFP-TNRC6A-NES-mut exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). (D) Western blot of myc-GFP-TNRC6A-NES-mut and FLAG/HA-tagged proteins. An anti-tubulin antibody was used as a loading control.
Small RNA binding-deficient Ago2 is not able to anchor TNRC6A protein in the cytoplasm
The amount of Ago2 protein interacted with TNRC6A was revealed to be increased by Ago2 overexpression (Supplementary Figure S6), and TNRC6A is considered to be tethered via interaction with Ago2 in the cytoplasmic P bodies (Figure 4 and Supplementary Figure S2). Thus, it was speculated that Ago2 proteins are anchored to the P bodies by unidentified interacting molecule(s) other than TNRC6A, which could assemble Ago2 proteins as foci. One of the possible mechanisms may be small RNA-mediated interaction with other molecules, such as mRNAs. To investigate such possibility, we used Ago2 mutant (Ago2-Y529E), which is shown to be impaired in small RNA binding and localization in P body by the substitution of tyrosine to glutamic acid at position 529 amino acid, which is located in the binding pocket of Ago2 with 5′-end of small RNA (31,32). We purified RNAs from immunoprecipitates of the cells transfected with wild-type Ago2 or Ago2-Y529E expression constructs (pFLAG/HA-Ago2-WT or pFLAG/HA-Ago2-Y529E), and performed RT-PCR for measuring the amount of typical miRNAs, miR-21 and let-7g. As previously described, the amount of miRNAs associated with Ago2-Y529E was low at less than 10% compared to that with Ago2-WT (Figure 6A and B), and Ago2-Y529E was not localized in the P bodies (Supplementary Figure S8). Under overexpression condition of Ago2-Y529E, GFP-signal of TNRC6A-NES-mut was observed exclusively in the nucleus in >80% cells, although TNRC6A-NES-mut was predominantly localized in the cytoplasm when the expression level of Ago2-WT was similar to that of Ago2-Y529E (Figure 6C–F). Although immunoprecipitation experiments revealed that the interaction of TNRC6A-NES-mut with Ago2-Y529E was evidently reduced compared to that with Ago2-WT, this interaction was slightly stronger than the interaction between TNRC6A-del.GW-I+II-NES-mut and Ago2-WT (Figure 6G). Furthermore, in the absence of TNRC6A overexpression, the overexpression of Ago2-WT formed large P-bodies but Ago2-Y529E could not form such P-bodies (Supplementary Figure S8). Thus, it is considered that predominant nuclear localization of TNRC6A-NES-mut under overexpression condition of Ago2-Y529E should be mainly due to impaired binding of Ago2-Y529E to small RNAs, although a negligible effect of weak interaction between TNRC6A-NES-mut and Ago2-Y529E may be included. Thus, our results strongly suggest that the cytoplasmic anchoring of TNRC6A by Ago2 overexpression may require small RNAs loaded on Ago2 proteins.
Transfection of small RNAs does not affect the subcellular localization of TNRC6A-NES-mut
Above result indicates that interactions between Ago2 and small RNAs are important for the anchoring of TNRC6A in the cytoplasmic P bodies. Therefore, we investigated whether transfection of small RNAs affect the subcellular localization of TNRC6A-NES-mut. One day after transfection with pmyc-GFP-TNRC6A-NES-mut, small RNA duplexes were transfected into the cells (Supplementary Figure S9). Transfection of siRNA or miRNA duplexes did not affect the subcellular localization of GFP-signal of TNRC6A-NES-mut. Same results were obtained by the simultaneous transfection with pmyc-GFP-TNRC6A-NES-mut and small RNA duplexes (data not shown). These results suggest that, since the intracellular machinery that processes small RNAs is considered to be saturated (33,34), the transfection of small RNAs does not increase the amount of small RNAs loaded on Ago proteins in the cells.
The counterbalance of the expression levels of Ago2 and TNRC6A proteins may modulate miRNA silencing activity
Next we examined the effect of Ago2, TNRC6A or TNRC6A-NES-mut overexpression on the activities of siRNA-based RNAi for mRNAs with perfectly complementary sequences and miRNA silencing for mRNAs with partial complementarities. RNAi and miRNA silencing are considered to be performed through different silencing mechanisms: the former requires only Ago2 protein and small RNA in vitro (35) but the latter requires GW182 family proteins in addition to Ago/small RNA (3,4).
To analysis the respective activities, we used short hairpin RNA (shRNA) against CXCR4 with Renilla luciferase reporter constructs containing a perfectly complementary site or four bulged target sites, respectively, for CXCR4 shRNA in its 3′UTR (Figure 7A and B; 36). Consistent with the previous reports (37,38), Ago2 overexpression enhanced RNAi activity in a dose dependent manner (Figure 7C), and such RNAi-enhancing effect by Ago2 was similarly or slightly observed when TNRC6A-WT or any types of its mutant proteins were co-expressed (Figure 7D). Contrary, overexpression of TNRC6A-WT weakened RNAi activity dose-dependently (Figure 7C). Such RNAi-reducing effect was evidently reduced by the overexpression of TNRC6A-NES-mut (Figure 7C), which is localized predominantly in the nucleus. Overexpression of TNRC6A-WT or -NES-mut did not affect the expression level of endogenous Ago2 protein (Supplementary Figure S10). These results suggest that the reduction of RNAi activity may be dependent on the cytoplasmic machinery. The similar results were obtained by siRNA against CXCR4 (Supplementary Figure S11A). When the empty vector of Ago2 protein, pFLAG/HA, was transfected with TNRC6A-WT or TNRC6A-NES-mut expression constructs, unambiguous reduction of RNAi activity was also observed, although the reduction level is rather low for TNRC6A-NES-mut compared to that for TNRC6A-WT (Figure 7D). However, such RNAi-reducing effects were marginal when the TNRC6A proteins deleting Ago-binding motifs were overexpressed (Figure 7D; TNRC6A-del.GW-I+II+III and TNRC6A-del.GW-I+II+III-NES-mut). These results may indicate that the excessive amount of cytoplasmic TNRC6A proteins repress the RNAi activity by the endogenous Ago2 proteins via direct or indirect interaction with Ago proteins mainly in the cytoplasm.
Figure 7.
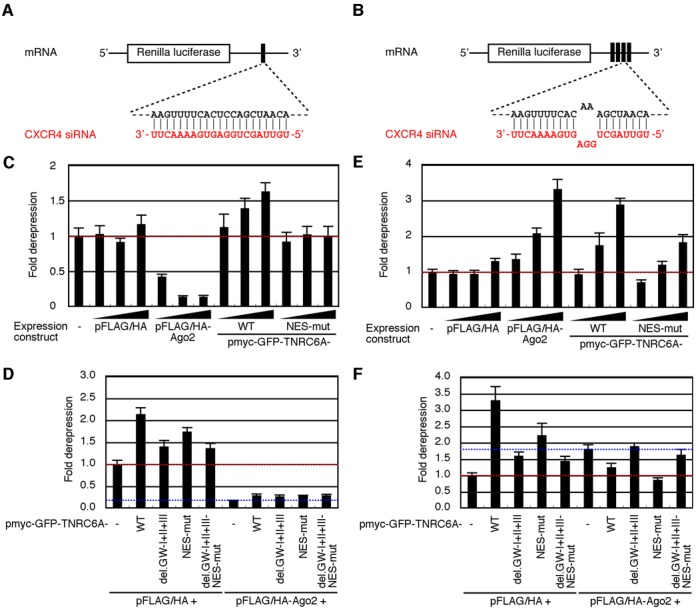
TNRC6A overexpression modulates RNAi and miRNA silencing activities induced by shRNA. (A and B) The schematic base-pairing patterns between CXCR4 siRNA and Renilla luciferase mRNA harboring a perfectly complementary target site for CXCR4 siRNA (A) or four target sites with a central bulge (B) in its 3′UTR, which was encoded by psiCHECK-CXCR4-PM or -MM 4x, respectively. (C and E) HeLa cells were transfected with a mixture of the following four plasmids: psiCHECK-CXCR4-PM (C) or -MM 4× (E), pGL3-Control, pSUPER-CXCR4 or control pSUPER-DsRed, without or with 2.5, or 25, 250 ng/well of pFLAG/HA, pFLAG/HA-Ago2, pmyc-GFP-TNRC6A-WT, or pmyc-GFP-TNRC6A-NES-mut. Two day after transfection, luciferase activities were measured. The relative luciferase activity of pSUPER-CXCR4-transfected cells against that of control pSUPER-DsRed-transfected cells was set to ‘1’. The data were shown as the mean and standard deviation of the triplicate measurements. The actual values of relative luciferase activities of the control cells transfected with shRNA expression constructs alone were 3.4 ± 0.36% (C), and 2.1 ± 0.14% (E), respectively. (D and F) HeLa cells were transfected with a mixture of the following five plasmids: psiCHECK-CXCR4-PM (D) or -MM 4× (F), pGL3-Control, pSUPER-CXCR4 or -DsRed, 250 ng/well of pFLAG/HA or pFLAG/HA-Ago2, without or with 250 ng/well of the indicated TNRC6A expression constructs. Two day after transfection, luciferase activities were measured. The relative luciferase activity of pSUPER-CXCR4-transfected cells against that of pSUPER-DsRed-transfected cells with pFLAG/HA transfection was set to ‘1’. The actual values of relative luciferase activities of the cells transfected with pFLAG/HA alone were 3.4 ± 0.29% (D), and 3.1 ± 0.23% (F), respectively.
Unlike the effect on RNAi activity, Ago2 overexpression attenuated miRNA silencing activity in a dose dependent manner (Figure 7E). The similar results were also obtained by siRNA against CXCR4 (Supplementary Figure S11B). In addition, Ago2 overexpression did not affect the expression level of endogenous TNRC6A protein (Supplementary Figure S10). This contradictory result for RNAi and miRNA silencing by the overexpression of Ago2 proteins might be interpreted by the increased level of small RNA-Ago2 complexes. When Ago2 protein is overexpressed, free small RNAs may be loaded on the newly synthesized Ago2 proteins. Such small RNA–Ago2 complexes in the cytoplasm is known to act for enhancement of RNAi activity (35). However, since GW182 family proteins are necessary for miRNA silencing in addition to Ago proteins, actual miRNA silencing activities decreased by Ago2 overexpression. Consistent with this interpretation, when TNRC6A-WT or TNRC6A-NES-mut protein was co-expressed with Ago2 protein, the activities of miRNA silencing were enhanced compared to the overexpression condition of Ago2 alone (Figure 7F). In addition, the necessity of interaction between TNRC6A and Ago2 proteins for enhancement of miRNA silencing activity was also confirmed by the result that the transfection of expression constructs for TNRC6A-del.GW-I+II+III or TNRC6A-del.GW-I+II+III-NES-mut could not increase the miRNA silencing activities (Figure 7F).
The overexpression of either of TNRC6A-WT or TNRC6A-NES-mut, also attenuated miRNA silencing activity, although the attenuating effect was weaker for TNRC6A-NES-mut than that for TNRC6A-WT (Figure 7E and F, and Supplementary Figure S11B), suggesting that the event for reducing miRNA silencing activity by TNRC6A overexpression may occur mainly in the cytoplasm. TNRC6A is known to associate with various cytoplasmic components essential for miRNA silencing, such as PABPC1, PAN2-PAN3 and CCR4-NOT deadenylase complexes. Most of the excessively synthesized TNRC6A proteins is also assumed to interact with these cytoplasmic components, but their interaction with Ago proteins are over-saturated and all of the TNRC6A proteins interacting with these components could not necessarily interact with Ago proteins in HeLa cells. Such imbalance of protein levels among Ago, TNRC6A, and other components might result in the reduction of miRNA silencing activity. This was also speculated by the results of the overexpression of TNRC6A proteins without Ago-binding motifs. The overexpression of TNRC6A-del.GW-I+II+III or TNRC6A-del.GW-I+II+III-NES-mut proteins also reduced the miRNA silencing activities (Figure 7F), probably because they also captured the TNRC6A-associated components.
DISCUSSION
In this paper, we found that TNRC6A protein is tethered in the cytoplasm under overexpression condition of Ago proteins (Figures 1–3 and Supplementary Figures S1–S5). Furthermore, myc-GFP-tagged TNRC6A-WT or TNRC6A-NES-mut lacking all of three Ago binding GW-motifs was not able to be tethered in the cytoplasm and predominantly localized in the nucleus even under overexpression condition of Ago2 (Figure 4 and Supplementary Figure S2), strongly suggested that the direct interaction between TNRC6A and Ago2 is essential for cytoplasmic anchoring of TNRC6A by Ago2 overexpression. TNRC6A has three independent Ago binding motifs (9,10), meaning that one molecule of TNRC6A protein is potentially able to bind three molecules of Ago proteins. However, multiple Ago-binding to TNRC6A was not necessary for the inhibition of nuclear transport of TNRC6A under overexpression condition of Ago, because TNRC6A mutants harboring only one GW-motif, either of GW-I or GW-II, were able to be tethered in the cytoplasm (Figure 4 and Supplementary Figure S2). The result indicated that the binding of one molecule of Ago protein with TNRC6A is necessary and enough for cytoplasmic localization of TNRC6A-NES-mut protein. In our previous study (10), TNRC6A-NES-mut proteins were transported into the nucleus along with Ago2 proteins and miRNAs under normal condition in HeLa cells when Ago2 protein was not overexpressed (10). This might be contradictory paradox for this study. However, although we could not determine whether all TNRC6A-NES-mut proteins observed in the nucleus evidently associated with Ago2 proteins or not in our previous report (10), it was revealed that Ago2 proteins interacting with TNRC6A-NES-mut proteins increased by Ago2 overexpression in this study (Supplementary Figure S6). This means that free TNRC6A-NES-mut proteins without Ago2 interaction abundantly exist in the cells without Ago2 overexpression. Furthermore, Ago2-binding of TNRC6A was not essential for translocation of TNRC6A into the nucleus, because TNRC6A-del.GW-I+II+III-NES-mut can translocate into the nucleus without interaction with Ago2 (10). Thus, although a part of TNRC6A-NES-mut protein may be certainly translocated into the nucleus with Ago2 and miRNAs, the remaining may be moved to the nucleus without interaction with Ago2 proteins in the normal condition. While this manuscript was being revised, Schraivogel et al. reported that Ago proteins as well as TNRC6 proteins are able to shuttle between the cytoplasm and the nucleus and Ago expression level affects nuclear import of TNRC6A (39). They demonstrated that TNRC6 proteins are imported into the nucleus by the Importin-β pathway but Ago2 is not. Thus, their results advocate our previous results that TNRC6A proteins are not necessarily translocated into the nucleus with direct interaction with Ago2 proteins.
In the Ago2 overexpression conditions, the cytoplasmic anchoring of TNRC6A proteins was revealed to be strongly associated with Ago2 anchoring in the P bodies. TNRC6A-NES-mut was colocalized with Ago proteins in the cytoplasmic foci, which size increased according to the Ago overexpression (Supplementary Figures S1 and S4). Such cytoplasmic foci were marked by P body markers, such as Dcp1 and RCK/p54, even under Ago2 overexpression condition (Supplementary Figure S7), indicating that such foci may be P bodies. However, overexpression of P body marker proteins, Dcp1 and RCK/p54, could not anchor TNRC6A proteins in the cytoplasm and did not form P bodies similar to those formed by Ago2 overexpression (Figure 5). Furthermore, the overexpression of TNRC6A did not form such large foci (10), demonstrating that the increasing of cytoplasmic P body size may mainly due to the accumulation of increased amount of Ago proteins by their overexpression. It is known that untranslating mRNAs accumulate in the P bodies (40), suggesting that the associations between Ago2-bound small RNAs and the untranslating mRNAs in the P bodies might cause the Ago2 aggregation in P body. This was clearly indicated by the results of Ago2-Y529E protein (Figure 6). The overexpression of wild type Ago2 inhibited the nuclear transport of TNRC6A-NES-mut protein, but that of Ago2-Y529E did not. The reason was considered to be the marginal amount of small RNAs can loaded on Ago2-Y529E proteins, so that Ago2-Y529E could not sufficiently associate with untranslating mRNAs in the P bodies (Figure 6).
Taken together, although Ago2 overexpression might not be the actual and ideal situation, the following model about the anchoring of TNRC6A in the cytoplasmic P bodies by Ago overexpression can be considered (Figure 8): excessive amount of Ago proteins increases the size of P bodies by their accumulation in the P bodies and is strongly tethered through interaction between Ago-bound small RNAs and their target RNAs in the P bodies, and then TNRC6A is anchored in the P bodies via its direct interaction with Ago. Schraivogel et al. showed that Ago overexpression inhibits the binding of Importin-β to TNRC6A (39). This result does not conflict with our model, because the previous report demonstrated that Importin-β is not colocalized with P bodies (41), suggesting that it might not be able to access the P bodies where TNRC6A is anchored by Ago proteins.
Figure 8.
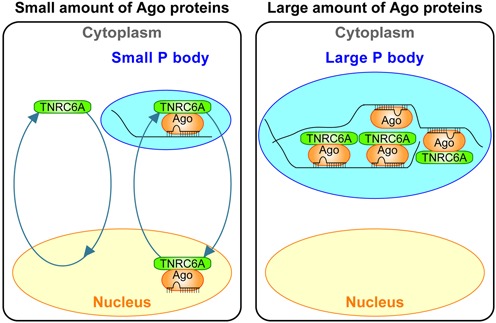
Hypothetic model of contol of the subcellular localization of TNRC6A proteins by Ago proteins. In human HeLa cells expressing a small amount of Ago proteins (left), most of free TNRC6A proteins are distributed in the cytoplasm and shuttles between the nucleus and the cytoplasm using its own NLS and NES, and a part of them may brings Ago-small RNA complexes into the nucleus from the cytoplasmic small P bodies. In the cells expressing a large amount of Ago proteins (right), they are strongly tethered in the P bodies probably through interaction between Ago-bound small RNAs and untranslating mRNAs, and enlarges the size of P bodies to form large foci. Most of TNRC6A proteins are then anchored in the P bodies via its direct interaction with abundant amount of Ago proteins.
We showed that RNAi activity for cytoplasmic mRNAs by Ago2 overexpression enhanced RNAi activity in a dose dependent manner (Figure 7C) in accordance with the previous reports (37,38). For RNAi activity, since only Ago2 protein and small RNA are necessary for induction of RNAi (35), the absolute amount of Ago2 protein may regulate RNAi activity in the cells. However, RNAi activity is inhibited by the overexpression of TNRC6A-WT more severely than TNRC6A-NES-mut (Figure 7C and D). Since TNRC6A-WT is predominantly localized in the cytoplasm but TNRC6A-NES-mut is in the nucleus (10), TNRC6A-WT may have a profound effect on the RNAi activity in the cytoplasmic mRNA, in this case Renilla luciferase mRNAs. One possible mechanism for inhibition of RNAi in the cytoplasm is inhibition of processing and/or Ago loading of small RNA. It is reported that PIWI domains of Ago proteins interacts with either GW182 family proteins (5,8) or Dicer (42,43), which required for inducing RNAi by processing of shRNA into small RNA (44) and promoting small RNA loading onto Ago2 (45). The excessive amount of cytoplasmic TNRC6A may competitively disturb the interaction of Dicer with Ago2 and may result in the inhibition of RNAi activity. However, siRNA but not shRNA against CXCR4, which is not necessary for Dicer cleavage, also showed the similar effects (Supplementary Figure S11), suggesting that the inhibition of RNAi effect is not due to the inhibition of direct interaction between Ago and Dicer by the overexpression of TNRC6A protein. Therefore, other possible mechanism(s) of inhibition of RNAi and miRNA silencing induced by overexpression of TNRC6A protein is that the interaction between TNRC6A protein with Ago2 protein inhibits RNAi and miRNA silencing by inhibiting siRNA loading into Ago2 or interaction with target mRNA.
In addition, we also showed that Ago2 overexpression attenuated miRNA silencing (Figure 7E and F). This result is consistent with the recent report which showed that Ago2 overexpression reduces off-targeting effects of siRNA (38), because the mechanism of siRNA-based off-target effect is considered to be similar to that of miRNA silencing (46–48). Not only Ago2 but also TNRC6A or TNRC6A-NES-mut overexpression attenuated miRNA silencing under normal condition (Figure 7E and F). However, previous reports showed that TNRC6s overexpression enhanced miRNA silencing (14,49). Such contradictory result may be explained by our experiments that TNRC6A or TNRC6A-NES-mut overexpression enhanced miRNA silencing under overexpression condition of Ago2 whereas their overexpression reduced miRNA silencing activity under normal condition (Figure 7E and F). Therefore, the relative expression levels between Ago and TNRC6 proteins might determine the effect of TNRC6 proteins on the miRNA silencing. These suggest that the counterbalance in the expression levels of Ago and GW182 family proteins might regulate miRNA silencing activity: the excessive amount of either Ago or TNRC6 proteins may competitively inhibit function of Ago proteins and reduce the miRNA silencing activity. In our experimental conditions, it is unambiguously revealed that the proportions between Ago proteins and a human GW182 protein, TNRC6A, in association with other components may be important for regulating miRNA silencing activity. The subcellular localization of one of the essential components for miRNA silencing, TNRC6A, may play a key part in modulation of silencing efficiently. However, as all our results about RNAi and miRNA silencing were dependent on the experiments of the overexpression of tagged proteins, we cannot exclude the possibility that these might not necessarily be results from actual and ideal conditions.
Our results showed that at least ∼2-fold increase of Ago2 protein inhibited the nuclear transport of TNRC6A in HeLa cells (Figure 3). It was reported that the amount of Ago2 protein is about 60% of total cellular Ago family proteins (Ago1∼4) in HeLa cells (50). Thus, ∼2-fold increase of Ago2 protein corresponds approximately ∼1.6-fold increase of total cellular Ago proteins, meaning that such small amount of increase might inhibit the nuclear transport of TNRC6A if all Ago proteins had same activities for cytoplasmic anchoring of TNRC6A. The expression levels of Agos differ in various types of tissues or cell lines, and are affected by tumorigenesis and cell signaling (27,51–57). Therefore, it is considered that cytoplasmic tethering of TNRC6A by excessive amount of Ago proteins might occur in some types of tissues or cell lines.
One possible biological significance of cytoplasmic tethering TNRC6A by Ago overexpression might promote the cytoplasmic function of Ago and/or TNRC6A or inhibit nuclear function(s) of TNRC6A. In accordance with our results, recent reports have suggested that human GW182 proteins might function in the nucleus as well as in the cytoplasm (10,22–24). However, precise function(s) of nuclear GW182 proteins are not well understood. Future analyses will uncover the significance of the regulation of TNRC6A subcellular localization.
Supplementary Material
Acknowledgments
We thank Drs Thomas Tuschl and Patrick Provost for kindly providing plasmids and Ai Nishi for technical assistance of plasmid construction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); Cell Innovation Project (MEXT to K.U.-T.). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT to K.U.-T.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ender C., Meister G. Argonaute proteins at a glance. J. Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 2.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 3.Eulalio A., Tritschler F., Izaurralde E. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun J.E., Huntzinger E., Izaurralde E. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb. Perspect. Biol. 2012;4:a012328. doi: 10.1101/cshperspect.a012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 6.Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazzaretti D., Tournier I., Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lian S.L., Li S., Abadal G.X., Pauley B.A., Fritzler M.J., Chan E.K. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takimoto K., Wakiyama M., Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishi K., Nishi A., Nagasawa T., Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17–35. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 13.Zekri L., Huntzinger E., Heimstädt S., Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntzinger E., Braun J.E., Heimstädt S., Zekri L., Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M., Fabian M.R., Coyle S.M., Sonenberg N., Doudna J.A. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun J.E., Huntzinger E., Fauser M., Izaurralde E. GW182 proteins recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eystathioy T., Jakymiw A., Chan E.K., Séraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Rivas F.V., Wohlschlegel J., Yates JR, III., Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franks T.M., Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.C., et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 23.Matsui M., Chu Y., Zhang H., Gagnon K.T., Shaikh S., Kuchimanchi S., Manoharan M., Corey D.R., Janowski B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M.S., Oh J.E., Kim Y.R., Park S.W., Kang M.R., Kim S.S., Ahn C.H., Yoo N.J., Lee S.H. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 2010;221:139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 26.Yoo N.J., Hur S.Y., Kim M.S., Lee J.Y., Lee S.H. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS. 2010;118:271–276. doi: 10.1111/j.1600-0463.2010.02588.x. [DOI] [PubMed] [Google Scholar]
- 27.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas M., Lu J.J., Ge Q., Zhang C., Chen J., Klibanov A.M. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishi K., Saigo K. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J. Biol. Chem. 2007;282:27503–27517. doi: 10.1074/jbc.M703810200. [DOI] [PubMed] [Google Scholar]
- 31.Rüdel S., Wang Y., Lenobel R., Körner R., Hsiao H.H., Urlaub H., Patel D., Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pare J.M., López-Orozco J., Hobman T.C. MicroRNA-binding is required for recruitment of human Argonaute 2 to stress granules and P-bodies. Biochem. Biophys. Res. Commun. 2011;414:259–264. doi: 10.1016/j.bbrc.2011.09.068. [DOI] [PubMed] [Google Scholar]
- 33.Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata Y., Shimizu E., Hibio N., Ui-Tei K. Fluctuation of global gene expression by endogenous miRNA response to the introduction of an exogenous miRNA. Int. J. Mol. Sci. 2013;14:11171–11189. doi: 10.3390/ijms140611171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 36.Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diederichs S., Jung S., Rothenberg S.M., Smolen G.A., Mlody B.G., Haber D.A. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Börner K., Niopek D., Cotugno G., Kaldenbach M., Pankert T., Willemsen J., Zhang X., Schürmann N., Mockenhaupt S., Serva A., et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 2013;41:e199. doi: 10.1093/nar/gkt836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schraivogel D., Schindler S.G., Danner J., Kremmer E., Pfaff J., Hannus S., Depping R., Meister G. Importin-β facilitates nuclear import of human GW proteins and balances cytoplasmic gene silencing protein levels. Nucleic Acids Res. 2015;43:7447–7461. doi: 10.1093/nar/gkv705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Chang W.L., Tarn W.Y. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doi N., Zenno S., Ueda R., Ohki-Hamazaki H., Ui-Tei K., Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 43.Tahbaz N., Kolb F.A., Zhang H., Jaronczyk K., Filipowicz W., Hobman T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ota H., Sakurai M., Gupta R., Valente L., Wulff B.E., Ariyoshi K., Iizasa H., Davuluri R.V., Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin X., Ruan X., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ui-Tei K., Naito Y., Nishi K., Juni A., Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S., Wang L., Fu B., Berman M.A., Diallo A., Dorf M.E. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6970–6975. doi: 10.1073/pnas.1322545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petri S., Dueck A., Lehmann G., Putz N., Rüdel S., Kremmer E., Meister G. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA. 2011;17:737–749. doi: 10.1261/rna.2348111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki T., Shiohama A., Minoshima S., Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 52.Adams B.D., Claffey K.P., White B.A. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang S.S., Smith I., Glazer C., Hennessey P., Califano J.A. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J. Otorhinolaryngol. Relat. Spec. 2010;72:337–343. doi: 10.1159/000320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Yu C., Gao H., Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10:38. doi: 10.1186/1471-2407-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sand M., Skrygan M., Georgas D., Arenz C., Gambichler T., Sand D., Altmeyer P., Bechara F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012;51:916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 56.Li W., Liu M., Feng Y., Xu Y.F., Che J.P., Wang G.C., Zheng J.H., Gao H.J. Evaluation of Argonaute protein as a predictive marker for human clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2013;6:1086–1094. [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F.Q., Huang J.H., Liu M., Yang F.P., Li W., Wang G.C., Che J.P., Zheng J.H. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int. J. Clin. Exp. Pathol. 2014;7:340–347. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


