Abstract
Telomerase, a unique ribonucleoprotein complex that contains the telomerase reverse transcriptase (TERT), the telomerase RNA component (TERC) and the TERC-binding protein dyskerin, is required for continued cell proliferation in stem cells and cancer cells. Here we identify SRSF11 as a novel TERC-binding protein that localizes to nuclear speckles, subnuclear structures that are enriched in pre-messenger RNA splicing factors. SRSF11 associates with active telomerase enzyme through an interaction with TERC and directs it to nuclear speckles specifically during S phase of the cell cycle. On the other hand, a subset of telomeres is shown to be constitutively present at nuclear speckles irrespective of cell cycle phase, suggesting that nuclear speckles could be the nuclear sites for telomerase recruitment to telomeres. SRSF11 also associates with telomeres through an interaction with TRF2, which facilitates translocation of telomerase to telomeres. Depletion of SRSF11 prevents telomerase from associating with nuclear speckles and disrupts telomerase recruitment to telomeres, thereby abrogating telomere elongation by telomerase. These findings suggest that SRSF11 acts as a nuclear speckle-targeting factor that is essential for telomerase association with telomeres through the interactions with TERC and TRF2, and provides a potential target for modulating telomerase activity in cancer.
INTRODUCTION
Telomeres, the specialized nucleoprotein complexes at the ends of linear eukaryotic chromosomes, are essential for maintaining genome integrity and have been implicated in aging and cancer (1,2). Mammalian telomeres consist of long tracts of duplex TTAGGG repeats with 3′ single-stranded G overhangs and are tightly associated with the six-subunit protein complex shelterin that provides telomere protection by preventing chromosome ends from being recognized as DNA damage (3–6). Although homologous recombination-mediated DNA synthesis has been demonstrated for replenishing telomeric DNA (7,8), the maintenance of telomere repeats in most eukaryotic organisms requires the enzyme telomerase which adds telomeric repeats onto the 3′ ends of linear chromosomes by reverse transcription (9,10). Telomerase is upregulated in human cancer cells but repressed in normal somatic cells, suggesting that the activation of telomerase supports tumor proliferation and survival by maintaining functional telomeres (11–13).
Telomerase undergoes a highly elaborate, stepwise process for the assembly and trafficking of the telomerase holoenzyme (14–17). After the preassembly in nucleoli, the telomerase ribonucleoprotein (RNP) is transported to Cajal bodies by the direct interaction of the telomerase RNA component (TERC) CAB box sequence with TCAB1 (18–21). Depletion of TCAB1 does not affect telomerase RNP assembly and telomerase enzymatic activity but reduces telomerase localization to Cajal bodies, resulting in a failure to maintain functional telomeres (22). To elongate telomere repeats, Cajal bodies containing the telomerase RNP transiently associate with telomeric chromatin. It has been recently reported that the OB-fold domain of TPP1 is required for telomerase recruitment to telomeres through the interaction with telomerase reverse transcriptase (TERT), and this interaction is an essential step in telomere length maintenance (23–25). Nonetheless, many open questions remain about the precise molecular mechanisms of telomerase recruitment and how telomerase efficiently finds the site of action in the context of chromatin architecture.
In a search for proteins capable of interacting with TERC using a RNA affinity chromatography, we identify SRSF11 (also named SRp54) as a TERC-interacting factor that localizes to nuclear speckles, subnuclear structures that are enriched in pre-messenger RNA splicing factors (26,27). SRSF11 has been shown to function as a splicing factor that is a member of the highly conserved family of serine/arginine (SR) proteins (28). Previously, SRSF11 was shown to stimulate the exclusion of tau exon 10, competing with Tra2β that enhances exon 10 inclusion for binding to its target site (29). Recently, it was reported that overexpression of SRSF11 increased β-deletion splice variant mRNA levels of human TERT, indicating that the TERT transcript is a target of SRSF11 (30). In this work, we show that SRSF11 associates with active telomerase through the interaction with TERC and directs it to nuclear speckles specifically during S phase. We also show that SRSF11 can associate with telomeres through the interaction with TRF2, which are constitutively present at nuclear speckles. Our data suggest that nuclear speckle is the S phase-specific nuclear site where telomerase is loaded on telomeres, and that SRSF11 functions as a nuclear speckle-targeting factor that is essential for telomerase recruitment to telomeres.
MATERIALS AND METHODS
Cell culture and plasmids
The human cervical carcinoma cell line HeLa S3 and the human breast cancer cell line MCF7 were cultured in Dulbecco's modified Eagle's medium, the human osteosarcoma cell lines U2OS and Saos-2 were cultured in McCoy's modified medium and the human non-small cell lung carcinoma cell line H1299 was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin in 5% CO2 at 37°C. The expression vectors for Flag-SRSF11 were constructed by inserting the full-length and truncated fragments from the SRSF11 cDNA into p3xFlag-CMV 7.1 plasmid (Sigma-Aldrich). The expression vectors for GST fusion proteins were constructed by cloning the cDNAs into pGEX5X-1 plasmid (Amersham Biosciences).
RNA affinity chromatography
The TERC DNA was cloned in the XhoI and EcoRI sites of pBluescript SK (+) (Stratagene), and TERC template was prepared by linearizing with EcoRI. TERC transcripts were prepared by in vitro transcription using a MAXIscript T7/T3 Kit according to the manufacturer's recommendation (Ambion). Transcribed RNA was cleared by MEGAclear Kit (Ambion) and labeled with biotin by RNA 3′-end biotinylation kit according to the manufacturer's recommendation (Thermo Scientific). For purification of TERC binding proteins, HeLa S3 cells were lysed in lysis buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.5% NP-40, 5 mM 2-mercaptoethanol and protease inhibitor cocktail) supplemented with 50 unit/ml RNase inhibitor and incubated for 30 min on ice. Lysates were incubated with biotinylated TERC bound to streptavidin agarose beads for 2 h with rotation at 4°C. The bound proteins were washed three times with lysis buffer and eluted with SDS sample buffer. TERC-interacting proteins were fractionated by SDS-PAGE, excised from the gel and analyzed by LC-MS/MS.
Peptide identification using nano LC-MS/MS
Nano LC-MS/MS analysis was performed with a nano HPLC system (Agilent), and the nano chip column (150 mm × 0.075 mm, Agilent) was used for peptide separation. The mobile phase A for LC separation was 0.1% formic acid in deionized water and the mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was designed for a linear increase from 5% B to 30% B in 25 min, 40% B to 60% B in 5 min, 90% B in 10 min and 5% B in 15 min. The flow rate was maintained at 300 nL/min. Product ion spectra were collected in the information-dependent acquisition (IDA) mode and were analyzed by Agilent 6530 Accurate-Mass Q-TOF (Agilent) using continuous cycles of one full scan TOF MS from 200 to 1500 m/z (1.0 s) plus three product ion scans from 50 to 1800 m/z (1.5 s each). Precursor m/z values were selected starting with the most intense ion, using a selection quadrupole resolution of 3 Da. The rolling collision energy feature was used, which determines collision energy based on the precursor value and charge state. The dynamic exclusion time for precursor ion m/z values was 60 s.
Double thymidine block
Double thymidine block was performed as described previously (31). Briefly, HeLa S3 cells were cultured to logarithmic phase and incubated in medium containing 2 mM thymidine (Sigma-Aldrich). After 19 h, cells were washed twice with phosphate-buffered saline (PBS) and incubated with regular medium for 9 h before a second incubation in 2 mM thymidine for 17 hr. Cells were released and collected at 2 h intervals after release from the second thymidine block. Cell cycle progression was monitored by propidium iodide staining and flow cytometry analysis using a FACScan flow cytometer (BD Biosciences).
GST pulldown, immunoprecipitation and immunoblotting
GST pulldown, immunoprecipitation and immunoblot analysis were performed as previously described (32). The expression vectors were transiently transfected into HeLa S3 cells using Lipofectamine-PLUS reagent according to the manufacturer's protocol (Invitrogen). HeLa S3 cells were lysed in lysis buffer and incubated for 30 min on ice. Lysates were clarified by centrifugation (16 000x g) for 15 min at 4°C to remove insoluble material. For the GST pulldown assay, the supernatants were precleared with glutathione-Sepharose 4B (Amersham Biosciences) and incubated with glutathione-Sepharose beads containing GST fusion proteins or control GST for 2 h at 4°C. GST fusion proteins and His-tagged proteins were expressed in Escherichia coli BL21 cells and purified on glutathione-Sepharose 4B and Ni-NTA Sepharose (GE Healthcare), respectively. For immunoprecipitation, the supernatants were precleared with either mouse or rabbit IgG (Santa Cruz Biotechnology) and protein A-Sepharose beads (Amersham Biosciences) for 30 min at 4°C and then incubated with primary antibodies at 4°C overnight, followed by incubation with protein A-Sepharose beads for 1 h at 4°C. After binding, the beads were washed extensively with lysis buffer and subjected to immunoblot analysis. Immunoprecipitation and immunoblotting were performed using anti-Flag (M2, Sigma-Aldrich), anti-SRSF11 (ab74363, Abcam), anti-TERT (Rockland), anti-TCAB1 (ab99376, Abcam), anti-dyskerin (H-300, Santa Cruz Biotechnology), anti-reptin (H-162, Santa Cruz Biotechnology), anti-pontin (H-100, Santa Cruz Biotechnology), anti-TPP1 (ab57595, Abcam), anti-PCNA (Santa Cruz Biotechnology), anti-His (Santa Cruz Biotechnology), anti-Son (Abnova) and anti-tubulin (TU-02, Santa Cruz Biotechnology) antibodies as indicated.
Telomerase assay
The telomeric repeat amplification protocol (TRAP) was used as previously described (33). Briefly, immunoprecipitated proteins (200 ng) were added to telomerase extension reactions and incubated at 37°C for 20 min. Polymerase chain reaction (PCR) was performed using the HTS primer and HACX primer for 30 cycles (denaturation at 94°C for 30 s, annealing at 62°C for 30 s and extension at 72°C for 30 s). As an internal telomerase assay standard, NT and TSNT primers were added to the PCR mixture as described previously (34). Telomerase products were resolved by electrophoresis on a 12% nondenaturing polyacrylamide gel. Bands were then visualized by staining with SYBR Green (Molecular Probes), and the signal intensity was quantified with a LAS-4000 Plus Image analyzer (Fuji Photo Film).
Establishment of stable cell lines
To produce retroviral supernatants, the retrovirus vectors expressing two different shRNAs targeting SRSF11 were co-transfected with pGP (for gag-pol expression) and pE-ampho (for env expression) into HEK293T packaging cells according to the manufacturer's instructions (Takara). The retrovirus vectors were constructed by cloning the shRNA-expressing oligonucleotides targeting SRSF11 (5′-GATCCCCCGTTGACAGAGCTTTGATATTTTCAAGAGAAATATCAA AGCTCTGTCAACGTTTTTA-3′ for shSRSF11–1; 5′-GATCCCCGGATACCTCTAGTAAAGA ATTTTCAAGAGAAATTCTTTACTAGAGGTATCCTTTTTA-3′ for shSRSF11–2) into pSUPER.retro.puro vector (OligoEngine). After 48 h, the culture supernatants were harvested and filtered through a 0.45 μm filter. HeLa S3 cells were transduced with the viral supernatants containing 4 μg/ml polybrene (Sigma-Aldrich). After selection with 1 μg/ml puromycin (Gibco) for 48 h, multiple independent single clones were isolated and checked for protein expression by immunoblotting with anti-SFRS11 antibody.
RNA interference
The siRNA duplexes were transfected into HeLa S3 cells using RNAiMax transfection reagent (Invitrogen). The siRNA target sequences specific for Son were 5′-GCUGAGCGCUCUAUGA UGU-3′ for siSon-1 and 5′-CAAUGUCAGUGGAGUAUCA-3′ for siSon-2. The control siRNA sequence (5′-AATTCTCCGAACGTGT CACGT-3′) was used as a control and did not correspond to any known gene in the databases.
Immunoprecipitation RT-PCR
Following immunoprecipitation, RNA was isolated using TRIzol (Invitrogen) and subjected to RT-PCR. The reverse transcription reaction was performed for 1 h at 42°C using the M-MLV reverse transcriptase (Promega). The following primers were used: TERC (forward primer, 5′-TCTAACCCTAACTGAGAAGGGCGTAG-3′ and reverse primer, 5′-GTTTGCTCTAGAATGA ACGGTGGAAG-3′) and the human GAPDH gene (forward primer, 5′-TCTGCCCCCTCTGCT GATGC-3′ and reverse primer, 5′-CCACCACCCTGTTGCTGTAG-3′).
Immunofluorescence and telomere fluorescence in situ hybridization (FISH)
Immunofluorescence and FISH were performed by modifying existing protocols (35). Briefly, cells grown on glass coverslips were fixed with 4% paraformadehyde in PBS for 10 min, permeabilized in PBS containing 0.5% Triton X-100 for 10 min and then blocked with PBS containing 0.5% bovine serum albumin and 0.2% cold fish gelatin for 10 min. Cells were incubated with rabbit anti-SRSF11 (800 ng/ml, Abcam), rabbit anti-TERT (500 ng/ml, Rockland), rabbit anti-TCAB1 (200 ng/ml, Abcam), rabbit anti-dyskerin (800 ng/ml, Santa Cruz Biotechnology), rabbit anti-Son (400 ng/ml, Abnova), mouse anti-SC35 (1 μg/ml, Abcam), mouse anti-coilin (2 μg/ml, Abcam), rabbit anti-coilin (200 ng/ml Santa Cruz Biotechnology), mouse anti-nucleolin (200 ng/ml, Santa Cruz Biotechnology), goat anti-TRF1 (2 μg/ml, Santa Cruz Biotechnology), rabbit anti-TRF2 (500 ng/ml, Cell signaling), rabbit anti-RAP1 (500 ng/ml, Bethyl laboratory), rabbit anti-POT1 (2 μg/ml, Abcam), mouse anti-TPP1 (1 μg/ml, Abcam) and mouse anti-TIN2 (1 μg/ml, Abcam) for 16 h at 4°C. After thorough washing with PBS, cells were incubated with Alexa Fluor 488-conjugated anti-rabbit or anti-goat immunoglobulin (green), Alexa Fluor 568-conjugated anti-mouse or anti-rabbit immunoglobulin (red) and Alexa Fluor 350-conjugated anti-mouse immunoglobulin (blue) (Molecular Probes) for 1 h in the dark. The coverslips were mounted on microscope slides using Vectashield mounting medium with DAPI (Vector Laboratories). Immunofluorescence images were captured using a confocal laser-scanning microscope LSM 510 (Carl Zeiss).
For telomere FISH experiments, immunofluorescence was performed as described above using the appropriate primary and secondary antibodies. After wash with PBS, cells were refixed with 4% paraformaldehyde in PBS for 20 min at room temperature (36). The coverslips were dehydrated with 70%, 95% and 100% ethanol for 5 min each and allowed to dry completely. Hybridization solution containing 10 mM Tris-HCl, pH 7.2, 70% formamide, 1 mg/ml blocking reagent (Roche) and Cy3-(CCCTAA)3 peptide nucleic acid probe (Panagene) was added to coverslip, and the coverslips were heated for 10 min at 80°C. After incubation for 12 h in the dark, the coverslips were washed twice in washing solution (10 mM Tris-HCl, pH 7.2, 70% formamide) and twice in PBS.
TERC fluorescence in situ hybridization
Immunofluorescence was carried out first, and TERC FISH was performed essentially as previously described (24), with the following modifications. Cells were refixed with 4% formaldehyde in PBS for 10 min, permeabilized in 70% ethanol for 1 h and rehydrated in 50% formamide in 2X SSC (300 mM NaCl, 30 mM sodium citrate at pH 7.2) for 5 min at room temperature. Prehybridization solution containing 100 mg/ml dextran sulphate, 0.125 mg/ml E. coli tRNA, 1 mg/ml nuclease-free BSA, 0.5 mg/ml salmon sperm DNA, 1 mM vanadyl ribonucleoside complexes, and 50% formamide in 2X SSC and were added to coverslip for 1 h at 37°C in a humidified chamber. Cells were then hybridized in the prehybridization solution containing a mixture of six Quasar 570 labeled oligonucleotide probes (Biosearch) overnight at 37°C. After hybridization, cells were washed twice with 50% formamide in 2X SSC and twice in PBS. Sequences of six oligonucleotide probes for TERC FISH were as follows: probe 1 5′-AAGTCAGCGAGAAAAACAGC-3′; probe 2, 5′-TCTAGAATGAACGGTGGAAG-3′; probe 3, 5′-CCAGCAGCTGACATTTTTTG-3′; probe 4, 5′-GCTGACAGAGCCCAACTCTT-3′, probe 5, 5′-GTCCCACAGCTCAGGGAATC-3′; probe 6, 5′-CATGTGTGAGCCGAGTCCTG-3′.
Telomere length analysis
To measure the telomere length, genomic DNA was isolated from HeLa S3 cells stably expressing SRSF11 shRNAs or control shRNA using the GenElute kit (Sigma-Aldrich), digested with Hinf1 and Rsa1 and separated on 1% agarose gel as described previously (37). DNA samples were transferred to the positively charged Hybond N+ (Amersham Biosciences) membrane and hybridized with a 32P-labeled (TTAGGG)4 probe.
RESULTS
Identification of SRSF11 as a TERC-interacting protein
To identify proteins that interact with human TERC, we used a RNA affinity assay with biotinylated TERC and HeLa S3 cell extracts. TERC-specific binding proteins were identified by nano-liquid chromatography-tandem mass spectrometry (nano LC-MS/MS) (Supplementary Figure S1). Among the known telomerase components, dyskerin, TCAB1 and nucleolin were enriched in the TERC complexes (Figure 1A) (20,38,39). We also identified SRSF11, an essential splicing factor of the SR protein family (28). In addition, this approach identified Thoc1, an essential component required for RNP biogenesis and RNA processing (40). However, we were unable to identify TERT in this assay possibly due to its low cellular abundance. Because SRSF11 has been shown to localize to nuclear speckles (26,27), we wanted to investigate the role of SRSF11 in the assembly and trafficking of telomerase. We first determined whether TERC and SRSF11 associate in vivo. Endogenous SRSF11 was specifically bound to Flag-TERT that was immunoprecipitated from HeLa S3 cells, as were endogenous TCAB1, dyskerin, reptin, pontin and TERC (Figure 1B). Flag-TERT associations with SRSF11, TCAB1, dyskerin and TERC were disrupted by RNase A treatment of the extract, which degraded TERC. In contrast, Flag-TERT interactions with reptin and pontin were not RNase A-sensitive (41). Reciprocal immunoprecipitation showed that Flag-SRSF11 associates with TERT, TCAB1, dyskerin and TERC (Figure 1C). Flag-SRSF11 associations with endogenous TERT and TERC were RNase A-sensitive. However, Flag-SRSF11 associations with TCAB1 and dyskerin were not disrupted by RNase A treatment, suggesting the protein-protein interactions. Endogenous SRSF11 and TERC were immunoprecipitated by endogenous TERT, and endogenous TERT and TERC were recovered by endogenous SRSF11 (Figure 1D). These associations were disrupted by RNase A treatment but not by DNase I treatment (Supplementary Figure S2A). The fact that SRSF11 associates with TERT through TERC was further verified by the GST pulldown assay (Figure 1E) and immunoprecipitation experiments with telomerase-negative cells (42) (Supplementary Figure S2B). We next examined whether SRSF11 associates with catalytically active telomerase. Lysates from HeLa S3 cells were immunoprecipitated with a SRSF11-specific antibody and analyzed for telomerase activity by the TRAP assay. Immunoprecipitates of endogenous SRSF11 contained telomerase activity that was abolished by RNase A treatment (Figure 1F). Taken together, these data suggest that SRSF11 is a novel TERC-binding protein.
Figure 1.
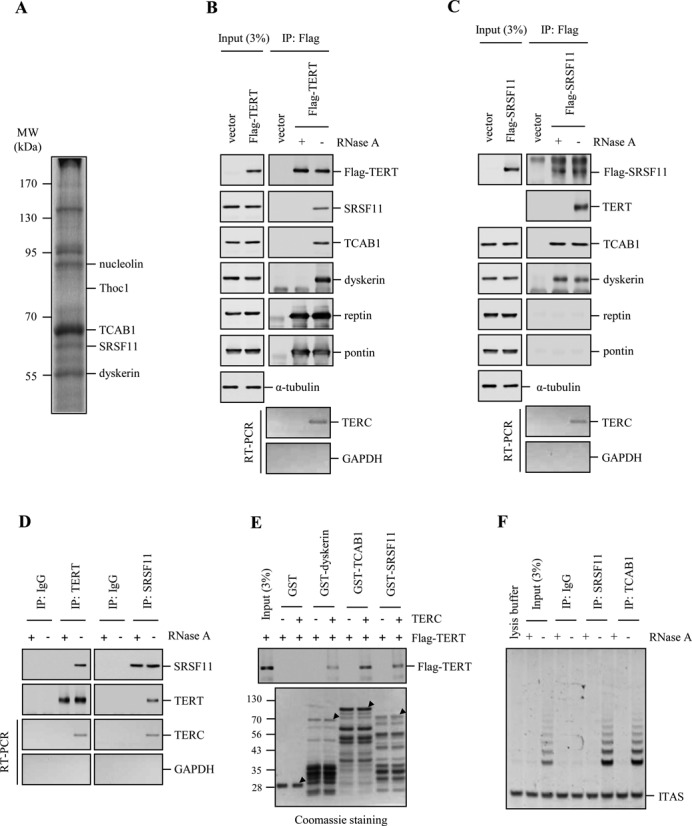
Identification of SRSF11 as a TERC-interacting protein. (A) Lysates from HeLa S3 cells were incubated with biotinylated TERC bound to streptavidin agarose beads and assayed for protein binding by Coomassie staining of SDS-PAGE gel. Binding proteins were identified by nano LC-MS/MS. Molecular size markers are shown in kilodaltons. (B and C) HeLa S3 cells expressing Flag-TERT (B) or Flag-SRSF11 (C) were subjected to immunoprecipitation (IP) with anti-Flag antibody, followed by immunoblotting to detect various telomerase components and quantitative reverse transcriptase-PCR (RT-PCR) to detect TERC. The extracts were treated with 0.1 mg/ml RNase A during immunoprecipitation. (D) Lysates from HeLa S3 cells were immunoprecipitated with anti-TERT and anti-SRSF11 antibodies, followed by immunoblotting with anti-SRSF11 and anti-TERT antibodies and quantitative RT-PCR to detect TERC. IgG was used as a negative control. The extracts were treated with 0.1 mg/ml RNase A during immunoprecipitation. (E) The various GST fusion proteins were affinity-purified and incubated with the lysates from cells expressing Flag-TERT, followed by immunoblotting with anti-Flag antibody. In vitro transcribed TERC was added to the lysates before GST pulldown. The purified GST fusion proteins were visualized by Coomassie staining and indicated with arrowheads. Molecular mass markers are shown in kilodaltons. (F) Immunoprecipitates of endogenous SRSF11 and TCAB1 from HeLa S3 cells were analyzed for telomerase activity by the TRAP assay. IgG was used as a negative control. ITAS, internal telomerase assay standard.
Identification of the domains in SRSF11 and TERT for their interaction
SRSF11 is a member of the highly conserved family of SR proteins which are structurally composed of one or more RNA recognition motifs (RRMs) and an arginine/serine-rich (RS) domain (28). To determine the specificity of the interaction between SRSF11 and TERT, HeLa S3 cells were transfected with Flag-TERT and subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblotting to detect a variety of SR proteins (Figure 2A). Flag-TERT interacts with endogenous SRSF11 but not with other SR proteins such as SRSF1 (SF2/ASF), SRSF2 (SC35), U2AF35 and U2AF65 (Figure 2B). These results suggest that the association between SRSF11 and TERT could be specific.
Figure 2.
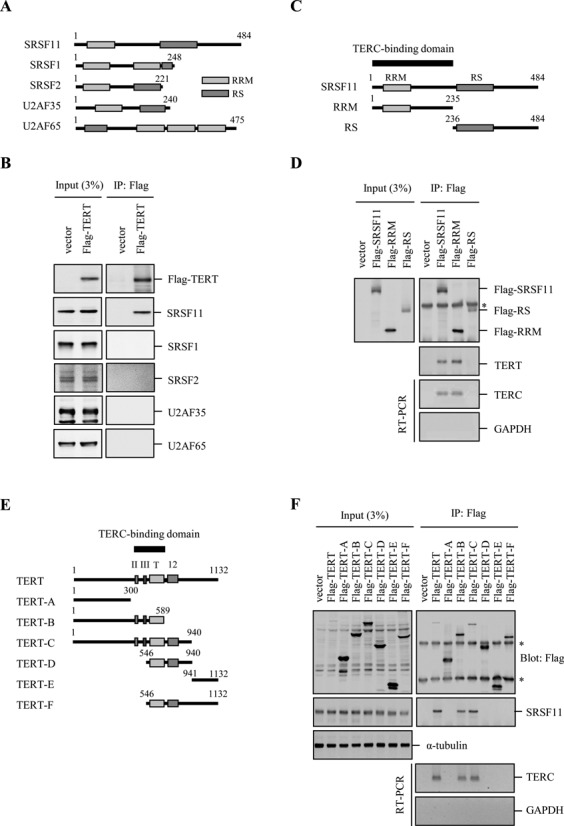
Identification of the domains in SRSF11 and TERT that are required for their interaction. (A) Schematic representation of the human SR protein family. RRM, RNA recognition motif; RS, arginine/serine-rich domain. (B) Lysates from HeLa S3 cells expressing Flag-TERT were immunoprecipitated with anti-Flag antibody, followed by immunoblotting to detect various SR proteins as indicated. (C) Schematic representation of the region of SRSF11 involved in TERC binding. (D) Lysates from HeLa S3 cells expressing Flag-SRSF11, Flag-RRM and Flag-RS were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-TERT antibody and quantitative RT-PCR to detect TERC. The asterisk marks the position of nonspecific immunoglobulin chains. (E) Schematic representation of the region of TERT involved in SRSF11 binding. (F) Lysates from HeLa S3 cells expressing the various Flag-TERT domains were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-SRSF11 antibody and quantitative RT-PCR to detect TERC. The asterisks mark the positions of nonspecific immunoglobulin chains.
To map the region in SRSF11 that is responsible for TERC binding, we generated deletion constructs lacking an amino-terminal RRM or a carboxy-terminal RS domain (Figure 2C). Endogenous TERT and TERC were immunoprecipitated by Flag-SRSF11 and Flag-RRM but not by Flag-RS (Figure 2D), indicating the binding ability of RRM to TERC. To determine the domain in TERT that is required for the interaction with SRSF11, we assessed binding of endogenous SRSF11 by immunoprecipitating a series of deletion fragments of TERT (Figure 2E). SRSF11 was immunoprecipitated only by the TERT fragments containing amino acid residues 301–589 (Figure 2F). TERC was also found to be immunoprecipitated by the same TERT fragments. Since this region is essential for TERC binding, these results suggest that the interaction of TERT with SRSF11 could be dependent on TERC. We also showed that removing the TERC-binding domain on TERT abolished TERC binding and SRSF11 association (Supplementary Figure S3).
SRSF11 associates with catalytically active telomerase in a cell cycle-dependent manner
Because telomerase is restricted to act on telomeres during S phase (31,43), it is likely that the binding of SRSF11 to telomerase is regulated in a cell cycle-dependent manner. To test this possibility, HeLa S3 cells were synchronized at G1/S transition using double thymidine blockade, and cell cycle progression was monitored by FACS analysis at 2 h intervals following release of the thymidine blockade (31). We investigated the interaction of SRSF11 with telomerase components during the cell cycle. Combined immunoprecipitation-immunoblot analysis revealed that the amount of TERC associated with immunoprecipitated SRSF11 peaked in mid/late S phase, as observed for TERT, TCAB1 and dyskerin (Figure 3A). In the TRAP assay with SRSF11 immunoprecipitates, telomerase activity also peaked in mid/late S phase (Figure 3B). These results demonstrate that the association of SRSF11 with active telomerase is S-phase specific.
Figure 3.
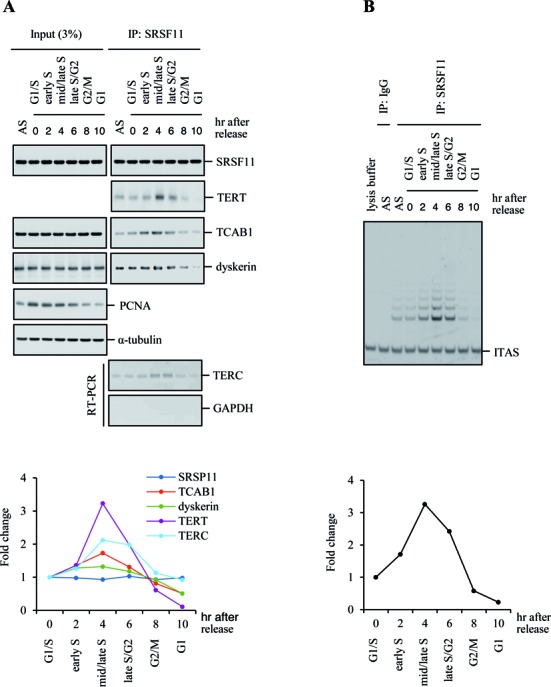
SRSF11 associates with catalytically active telomerase in a cell cycle-dependent manner. (A) SRSF11 associates with active telomerase in a cell cycle-dependent manner. HeLa S3 cells were synchronized by double thymidine block and harvested at 2 h intervals over a 10 h time course after release. The 0 h time point corresponds to blocked cells before release. Immunoprecipitates of endogenous SRSF11 from synchronized cells were subjected to immunoblotting and quantitative RT-PCR as indicated. Immunoblot for proliferation cell nuclear antigen (PCNA) was used for an independent marker of S phase. Band intensities were quantified and displayed as fold change in the accompanying graph. AS, asynchronous cells. (B) Immunoprecipitates of endogenous SRSF11 from synchronized cells were analyzed for telomerase activity by the TRAP assay. IgG was used as a negative control. Telomerase products were quantified and displayed as fold change in the accompanying graph. ITAS, internal telomerase assay standard.
To determine the nuclear localization of endogenous SRSF11 during the cell cycle, we performed indirect immunofluorescence with synchronized HeLa S3 cells and found that SRSF11 localizes to nuclear speckles throughout most of the cell cycle as indicated by dual staining with a SC35-specific antibody (Figure 4A). SRSF11 overlapped with the Cajal body marker coilin during mid/late S phase but rarely co-localized in other cell phases (Figure 4B), suggesting that telomerase-containing Cajal bodies are targeted to nuclear speckles specifically during S phase. However, SRSF11 was excluded from the nucleoli as indicated by staining with a nucleolin-specific antibody (Figure 4A). Interestingly, approximately 70% of telomeres co-localized with both SRSF11 and SC35 at the perimeter of nuclear speckles irrespective of cell cycle phase (Figure 4C and D), suggesting that a subset of telomeres is constitutively present at nuclear speckles. This was further supported by the findings that the shelterin components associate with nuclear speckles (Supplementary Figure S4). We also found that the telomerase holoenzyme components including TERT, dyskerin, TCAB1 and TERC co-localize to nuclear speckles during mid/late S phase (Figure 4E and F). S phase-specific co-localization of TERC with nuclear speckles was also observed in other telomerase-positive cells (Supplementary Figure S5). These observations suggest that nuclear speckles may serve as the S phase-specific nuclear sites for telomerase loading on telomeres.
Figure 4.
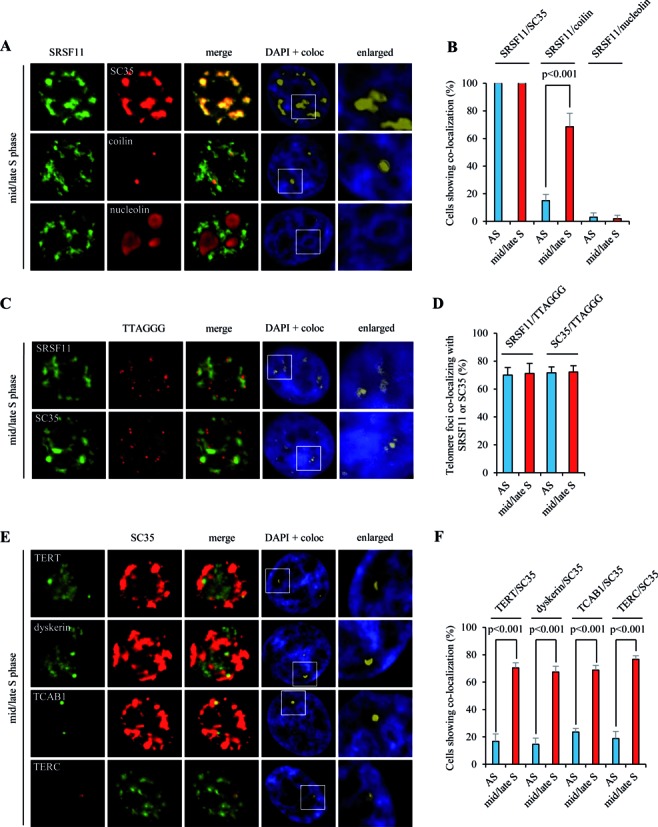
Telomerase localizes to nuclear speckles specifically during S phase. (A) HeLa S3 cells were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of SRSF11 with SC35 (nuclear speckle marker), coilin (Cajal body marker) or nucleolin (nucleolus marker). Co-localization channels were calculated using Meta Imaging SeriesR MetaMorph software and appear in white in all figures. DNA was stained with DAPI. Coloc, co-localization. (B) The average percentage of cells showing co-localization of SRSF11 with SC35, coilin or nucleolin and standard deviation were determined by analyzing >100 nuclei for each cell cycle phase. Statistical analyses were performed using a two-tailed Student's t test. AS, asynchronous cells. (C) HeLa S3 cells were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of SRSF11 or SC35 with telomeric sites marked by TTAGGG-specific FISH probe. (D) The average percentage of telomere foci co-localizing with SRSF11 or SC35 per cell and standard deviation were determined by analyzing >100 nuclei for each cell cycle phase. (E) HeLa S3 cells were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of TERT, dyskerin, TCAB1 or TERC with SC35. (F) The average percentage of cells showing co-localization of TERT, dyskerin, TCAB1 or TERC with SC35 and standard deviation were determined by analyzing >100 nuclei for each cell cycle phase.
SRSF11 is required for telomerase recruitment to telomeres and telomere elongation
To examine an involvement of SRSF11 in telomerase recruitment to telomeres, the expression of endogenous SRSF11 was depleted using short hairpin RNA (shRNA) (Figure 5A). In mid/late S phase cells, depletion of SRSF11 diminished the number of TERC foci at nuclear speckles (Figure 5B and 5C) without affecting overall TERC RNA levels d (Figure 5A), indicating that SRSF11 is essential for the association of telomerase with nuclear speckles. We also found that the levels of telomerase components including TERT, dyskerin and TCAB1 were not affected by SRSF11 depletion (Figure 5A). We next examined the requirement of SRSF11 for associating telomerase with telomeres using triple immunofluorescence staining for co-localization of TERC with coilin and TRF2. We found that TERC co-stained with both coilin and TRF2 during mid/late S phase in control shRNA cells (Figure 5D–F). Depletion of SRSF11 did not affect co-localization of TERC with coilin (Figure 5E) but led to reduced co-localization of TERC with TRF2 (Figure 5F). These results suggest that SRSF11 plays a critical role in telomerase loading on telomeres at nuclear speckles during S phase. Because telomeres still localizes to nuclear speckles after depletion of SRSF11 (Supplementary Figure S6), SRSF11 depletion-dependent reduction in co-localization of TERC with TRF2 could not be due to the absence of telomeres at nuclear speckles.
Figure 5.
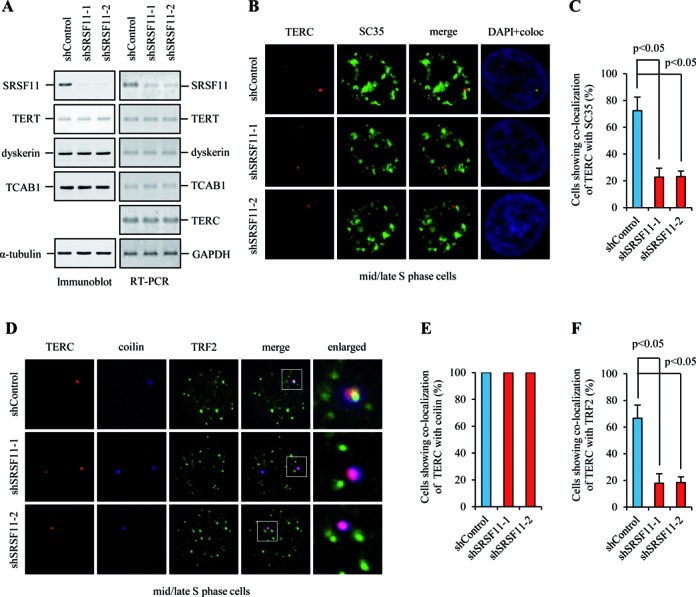
SRSF11 is essential for telomerase loading on telomeres at nuclear speckles during S phase. (A) HeLa S3 cells expressing control shRNA (shControl) or SRSF11 shRNAs (shSRSF11–1 and shSRSF11–2) were subjected to immunoblotting to measure the protein levels of telomerase components and quantitative RT-PCR to detect the mRNA levels of telomerase components and TERC. (B) HeLa S3 cells expressing shControl or shSRSF11 were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of TERC and SC35. Co-localization channels were calculated using Meta Imaging SeriesR MetaMorph software and appear in white in all figures. DNA was stained with DAPI. Coloc, co-localization. (C) The average percentage of cells showing co-localization of TERC with SC35 and standard deviation were determined by analyzing >100 nuclei for each condition. Statistical analyses were performed using a two-tailed Student's t-test. (D) HeLa S3 cells expressing shControl or shSRSF11 were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of TERC with coilin and TRF2. (E and F) The average percentage of cells showing co-localization of TERC with coilin (E) or TRF2 (F) and standard deviation were determined by analyzing >100 nuclei for each condition.
It has been recently documented that TERT is involved in alternative splicing events such as the β-deletion variant (30). If depletion of SRSF11 affects the expression of TERT splice variants, SRSF11-dependent telomerase loading onto telomeres might be an indirect effect of altered TERT levels rather than directly due to impaired recruitment. To test this possibility, HeLa S3 cells were transfected with SRSF11 shRNA or Myc-SRSF11 and subjected to quantitative RT-PCR to measure the levels of TERT variant mRNAs. We found that the amounts of TERT mRNA and β-deletion transcript were not significantly changed upon either depletion or overexpression of SRSF11 (Supplementary Figure S7). Furthermore, the levels of SRSF11 did not affect the amounts of telomerase activity. Because SRSF11 has been previously described as a tau exon 10 splicing repressor (44), we performed the control experiments indicating that depletion of SRSF11 affects tau exon 10 alternative splicing. The results showed that depletion of SRSF11 increases tau exon 10 inclusion whereas overexpression of SRSF11 suppresses tau exon 10 inclusion (Supplementary Figure S8).
To determine whether SRSF11 is required for telomere elongation by telomerase, we assayed telomere lengths in HeLa S3 cells stably expressing SRSF11 shRNA or control shRNA. SRSF11-depleted cells maintained the reduced levels of SRSF11 until population doubling (PD) 99 (Figure 6A). Depletion of SRSF11 did not affect the levels of telomerase components (Figure 6A) and shelterin proteins (Figure 6B). Telomerase activity was not influenced by the level of SRSF11 (Figure 6C), indicating that SRSF11 has no direct regulatory effect on telomerase activity. Whereas cells expressing control shRNA maintained a stable telomere length throughout the duration of the experiments, two different shRNAs targeting SRSF11 led to progressive telomere shortening with cell passage (Figure 6D and E). These results suggest that SRSF11 is essential for telomere length maintenance in telomerase-positive cells.
Figure 6.
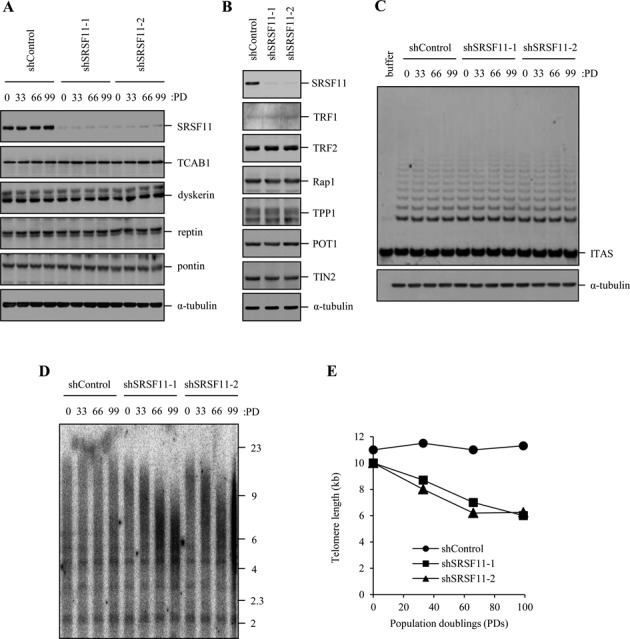
SRSF11 is required for telomere elongation. (A) HeLa S3 cells stably expressing control shRNA (shControl) or SRSF11 shRNAs (shSRSF11–1 and shSRSF11–2) were harvested at the indicated population doublings (PDs) and subjected to immunoblotting to measure the levels of various telomerase components. (B) HeLa S3 cells stably expressing shControl or shSRSF11 were harvested at PD 99 and subjected to immunoblotting to measure the levels of various shelterin proteins. (C) HeLa S3 cells stably expressing shControl or shSRSF11 were harvested at the indicated PDs and analyzed for telomerase activity by the TRAP assay. ITAS, internal telomerase assay standard. (D) HeLa S3 cells stably expressing shControl or shSRSF11 were harvested at the indicated PDs, and genomic DNA was digested with Hinf1 and Rsa1, followed by Southern blot analysis using a telomere repeat probe. (E) Graphical representation of average telomere length versus population doubling number.
Nuclear speckle is the nuclear site for telomerase recruitment to telomeres
If nuclear speckle is an important nuclear compartment for telomere regulation, we would expect disruption of nuclear speckle structure to impair telomerase recruitment and telomere length control. To examine whether nuclear speckle is required for telomerase recruitment or simply a place that SRSF11-telomerase complex passes through, nuclear speckle structure was disrupted by depleting a large scaffolding protein Son that is essential for proper nuclear organization of pre-mRNA processing factors in nuclear speckles (45). Immunofluorescence staining of HeLa S3 cells with a Son-specific antibody showed co-localization with SC35 in nuclear speckles (Figure 7A). Depletion of Son by two independent siRNAs led to a significant reduction in Son immunofluorescence signal. SC35 localization in nuclear speckles was altered to a round-shape nuclear structure after depletion of Son (Figure 7A), suggesting that Son is required to maintain nuclear speckle organization (45). We next examined the effect of Son depletion on co-localization of TERC and TRF2. Whereas TERC mostly co-stained with TRF2 in control siRNA cells, Son depletion caused a clear reduction in co-localization of TERC and TRF2 (Figure 7B and C). These results suggest that proper nuclear speckle organization is required for telomerase recruitment to telomeres. In addition, we found that Son depletion does not affect the levels of telomerase components (Figure 7D).
Figure 7.
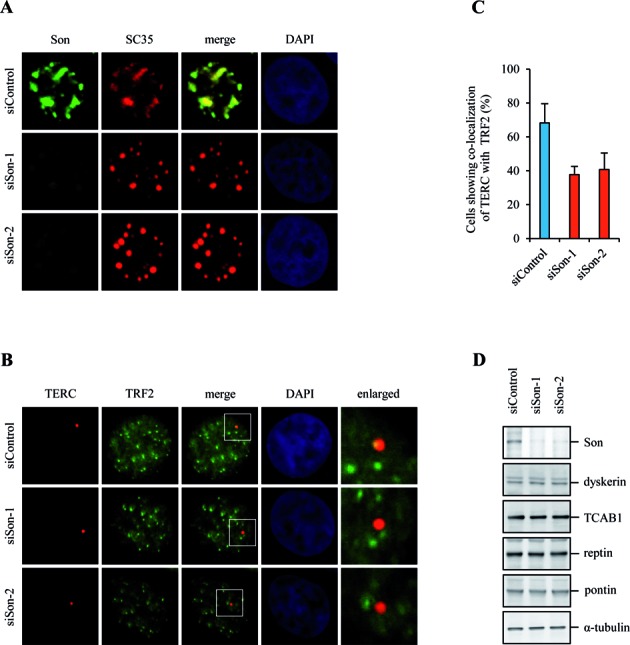
Nuclear speckle organization is required for telomerase recruitment to telomeres. (A) HeLa S3 cells expressing control siRNA (siControl) or Son siRNAs (siSon-1 and siSon-2) were analyzed by indirect immunofluorescence for co-localization of Son and SC35. DNA was stained with DAPI. (B) HeLa S3 cells expressing siControl or siSon were analyzed by indirect immunofluorescence for co-localization of TERC and TRF2. (C) The average percentage of cells showing co-localization of TERC and TRF2 and standard deviation were determined by analyzing >100 nuclei for each condition. (D) HeLa S3 cells expressing siControl or siSon were subjected to immunoblotting to measure the protein levels of telomerase components as indicated.
SRSF11 interacts with TRF2 irrespective of cell cycle phase
Although telomerase is targeted to nuclear speckles through the interaction between SRSF11 and TERC, it is uncertain how telomerase efficiently finds the telomeres within nuclear speckles. To address this issue, we examined whether SRSF11 interacts with the shelterin components and found that endogenous SRSF11 can form a stable complex with endogenous TRF2 but not with other shelterin components in HeLa S3 cells (Figure 8A). Endogenous SRSF11 was also immunoprecipitated by endogenous TRF2 (Figure 8B), indicating that SRSF11 interacts with TRF2 in mammalian cells. The GST pulldown assay further verified that SRSF11 directly interacts with TRF2 (Figure 8C). We next determined the specificity of the interaction between SRSF11 and TRF2. The results showed that Flag-TRF2 interacts with SRSF11 and U2AF35 but not with other SR proteins including SRSF1, SRSF2 and U2AF65 (Figure 8D). It will be interesting to investigate the physiological significance of the interaction of TRF2 with U2AF35.
Figure 8.
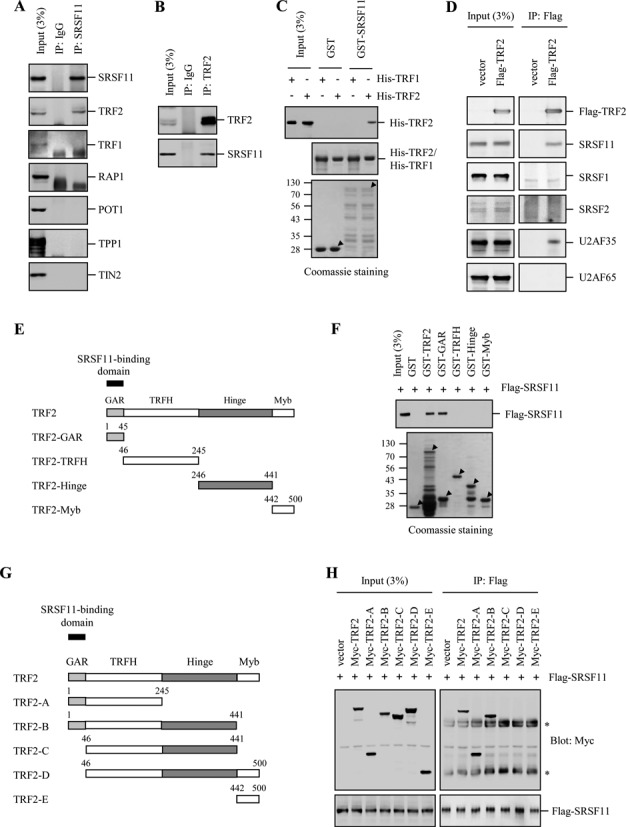
SRSF11 interacts with the GAR domain of TRF2. (A) Lysates from HeLa S3 cells were immunoprecipitated with anti-SRSF11 antibody, followed by immunoblotting to detect various shelterin components. IgG was used as a negative control. (B) Lysates from HeLa S3 cells were immunoprecipitated with anti-TRF2 antibody, followed by immunoblotting with anti-SRSF11. (C) GST and the GST-SRSF11 fusion proteins were affinity-purified and incubated with His-TRF2 (or His-TRF1) that was purified on Ni-NTA beads, followed by immunoblotting with anti-HA antibody. The purified GST fusion proteins were visualized by Coomassie staining and indicated with arrowheads. Molecular mass markers are shown in kilodaltons. (D) Lysates from HeLa S3 cells expressing Flag-TRF2 were immunoprecipitated with anti-Flag antibody, followed by immunoblotting to detect the various SR proteins as indicated. (E) Schematic representation of the TRF2 domains fused to GST. (F) GST and the various truncated GST-TRF2 fusion proteins were affinity-purified and incubated with lysates from HeLa S3 cells expressing Flag-SRSF11, followed by immunoblotting with anti-Flag antibody. The purified GST fusion proteins were visualized by Coomassie staining and indicated with arrowheads. (G) Schematic representation of the TRF2 domains involved in SRSF11 binding. (H) HeLa S3 cells were co-transfected with Flag-SRSF11 and various truncated Myc-TRF2 and subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblotting with anti-Myc antibody. The various truncated Myc-TRF2 proteins were detected by immunoblotting with anti-Myc antibody. The asterisks mark the positions of nonspecific immunoglobulin chains.
To identify the region in TRF2 that is responsible for SRSF11 binding, a series of TRF2 fragments were fused to GST and used in the in vitro binding assay (Figure 8E). GST-TRF2 and GST-GAR bound efficiently to SRSF11 whereas other GST-fusion proteins had no detectable binding activity (Figure 8F). To confirm the binding ability of the GAR domain to SRSF11, we generated deletion constructs lacking the GAR and Myb domains (Figure 8G). When HeLa S3 cells cells were co-transfected with Flag-SRSF11 and various Myc-TRF2 fragments, only TRF2 fragments containing the GAR domain was immunoprecipitated by Flag-SRSF11 (Figure 8H).
Because SRSF11 associates with active telomerase through TERC specifically during S phase, we investigated whether the interaction between SRSF11 and TRF2 is regulated by the cell cycle. HeLa S3 cells expressing Flag-SRSF11 and TRF2-V5 were synchronized in mid/late S phase and analyzed by immunoprecipitation. The results showed that the amounts of TRF2 associated with immunoprecipitated SRSF11 were not changed during the cell cycle (Supplementary Figure S9A), indicating that SRSF11 interacts with TRF2 in a cell cycle-independent manner. To determine co-localization of SRSF11 with TRF2 during the cell cycle, we explored the nuclear distribution of endogenous SRSF11 and TRF2 of synchronized cells. Many of TRF2 foci co-localized to SRSF11 irrespective of cell cycle phase (Supplementary Figure S9B and SC), further supporting that telomeres are constitutively present at nuclear speckles.
Both SRSF11-TERC interaction and SRSF11-TRF2 interaction are essential for telomerase recruitment to telomeres at nuclear speckles
To map the domain of SRSF11 that mediates the interaction with TRF2, we accessed binding of TRF2 by immunoprecipitating two SRSF11 deletion mutants, RRM and RS, in HeLa S3 cells (Figure 9A). Intriguingly, the results showed that TRF2 interacts with the RS domain, which does not overlap with the TERC-binding RRM domain (Supplementary Figure S10A). Based on these results, we hypothesized that both interactions are required for telomerase recruitment to telomeres at nuclear speckles. To test this possibility, Myc-RRM and Myc-RS were expressed in HeLa S3 cells and assayed for their localization. Whereas Myc-RRM was detected in a diffused pattern throughout the nucleus and cytoplasm, Myc-RS showed strong co-localization with nuclear speckles (Supplementary Figure S10B). We next analyzed the ability of these deletion mutants to target telomerase to nuclear speckles. Overexpression of RS in mid/late S phase cells had little or no effect on TERC association with nuclear speckles compared to vector control (Figure 9B and C). However, overexpression of RRM led to reduced localization of TERC in nuclear speckles, suggesting that RRM acts as a dominant negative form to target telomerase to nuclear speckles. We also examined the effect of deletion mutations on the association of telomerase with telomeres using triple immunofluorescence staining. Overexpression of RRM or RS in mid/late S phase cells did not affect co-localization of TERC with coilin (Figure 9D and E) but significantly inhibited TERC association with TRF2 (Figure 9D and F). These results suggest that both SRSF11 RRM-TERC interaction and SRSF11 RS-TRF2 GAR interaction are required for telomerase recruitment to telomeres at nuclear speckles. Because the interaction between SRSF11 and TRF2 is essential for telomerase association with telomeres, we examined whether TRF2 is also required for telomere localization to nuclear speckles. The results showed that depletion of TRF2 does not disrupt co-localization of telomeres with nuclear speckles (Supplementary Figure S11), suggesting that telomeres are constitutively present at nuclear speckles even in the absence of TRF2.
Figure 9.
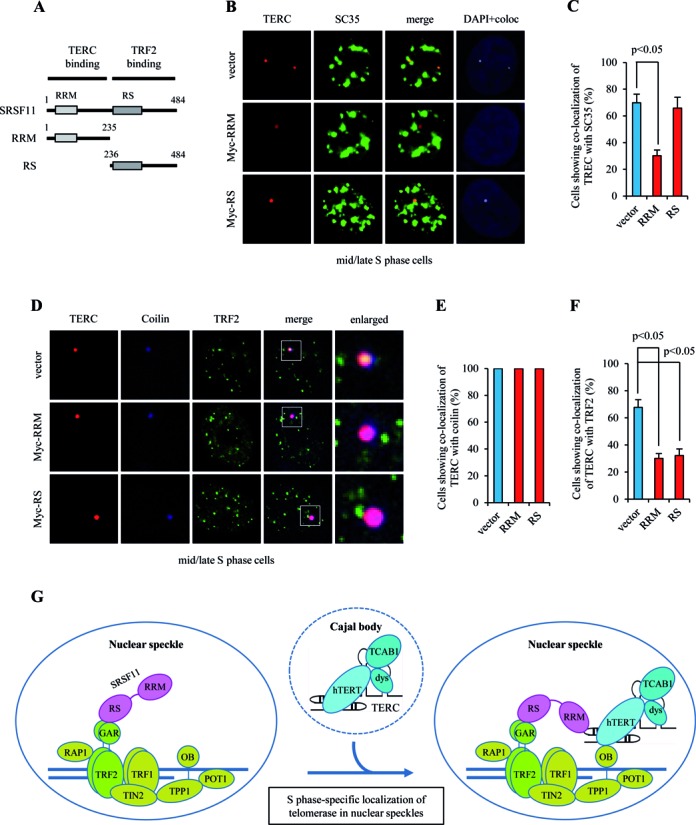
SRSF11 associates with telomeres through the interaction with TRF2. (A) Schematic representation of the RRM and RS domains in SRSF11. RRM, RNA recognition motif; RS, arginine/serine-rich domain. (B) HeLa S3 cells expressing Myc-RRM or Myc-RS were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of TERC with SC35. Co-localization channels were calculated using Meta Imaging SeriesR MetaMorph software and appear in white in all figures. DNA was stained with DAPI. Coloc, co-localization. (C) The average percentage of cells showing co-localization of TERC with SC35 and standard deviation were determined by analyzing >100 nuclei for each condition. Statistical analyses were performed using a two-tailed Student's t-test. (D) HeLa S3 cells expressing Myc-RRM or Myc-RS were synchronized in mid/late S phase and analyzed by indirect immunofluorescence for co-localization of TERC with coilin and TRF2. (E and F) The average percentage of cells showing co-localization of TERC with coilin (E) or TRF2 (F) and standard deviation were determined by analyzing >100 nuclei for each condition. (G) Model for SRSF11 function in the telomere elongation pathway.
DISCUSSION
Telomere elongation by telomerase in human cancer cells involves multiple steps including telomerase RNP biogenesis, holoenzyme assembly, TCAB1-dependent telomerase accumulation in Cajal bodies and the redistribution of telomerase to telomeric chromatin (46–48). Although much has been recently progressed about the roles and mechanisms of telomerase components in these multiple processes, it is still uncertain how telomerase efficiently finds the site of action in the context of chromatin architecture and what mechanisms ensure that telomerase specifically acts on telomeres in S phase. It has been recently reported that the OB-fold domain of TPP1 is required to recruit telomerase to telomeres by binding to TERT (23–25). This process should be regulated to restrict telomere elongation to S phase (31,43). In this study, we have identified SRSF11 as a novel TERC-binding factor that localizes to nuclear speckles. SRSF11 associates with active telomerase enzyme and directs it to nuclear speckles. Importantly, the SRSF11-telomerase complex is regulated by the cell cycle and peaks during each S phase. SRSF11 also associates with telomeres through the interaction with TRF2. Taken together, these data demonstrate that nuclear speckle is the S phase-specific nuclear site where telomerase is loaded on telomeric chromatin.
Depletion of SRSF11 causes a clear reduction in telomere length with cell passage, suggesting that SRSF11 might represent a general mechanism for maintaining telomere length. The critical question that remains to be answered is how SRSF11 regulates telomere length. It is likely that SRSF11 may act as a molecular link for the recruitment of telomerase to telomeres. Based on the results presented in this work, we propose a model to explain how telomerase is recruited to chromosome ends specifically during S phase (Figure 9G). After the preassembly of telomerase RNA and proteins in nucleoli (22), the telomerase RNP is transported to Cajal bodies by the direct interaction of the TERC CAB box with TCAB1 (18–21). Telomerase-containing Cajal body is then targeted to nuclear speckles through SRSF11 binding to TERC. Although SRSF11 had no direct regulatory effect on telomerase activity, depletion of SRSF11 reduced telomerase localization to nuclear speckles, resulting in a failure to maintain telomere length. Because co-localization of telomerase with Cajal bodies was not influenced by SRSF11 depletion, the SRSF11-dependent trafficking of telomerase to nuclear speckles occurs at a step after the telomerase localization to Cajal bodies. Importantly, co-localization of SRSF11 with telomerase occurs specifically during S phase, suggesting that SRSF11 is a S phase-specific telomerase holoenzyme component.
In addition to its binding to TERC, SRSF11 can interact with TRF2 via the carboxy-terminal RS domain which does not overlap with the TERC-binding RRM domain. These findings suggest that SRSF11 binding to TERC and TRF2 may not be mutually exclusive. Indeed, SRSF11 was shown to associate with a subset of telomeres through the interaction with the GAR domain of TRF2, which are constitutively present at nuclear speckles irrespective of cell cycle phase. In this mode, SRSF11 may act as a scaffold to bring telomerase (via TERC-binding) to telomeres (via TRF2-binding). Once telomerase is loaded on telomeres in a SRSF11-dependent manner, the TERT-TPP1 interaction could be then required to complete telomerase recruitment for high processivity telomere synthesis (23,24). This model predicts that recruitment of telomerase by SRSF11 to telomeres precedes TPP1. To test this possibility, we determined the effect of SRSF11 depletion on the ability of TPP1 to pull down TERT and telomerase activity. The results showed that the interaction between TPP1 and TERT was significantly reduced by SRSF11 depletion (Supplementary Figure S12A). The levels of telomerase activity immunoprecipitated by TPP1 were also reduced when SRSF11 was depleted (Supplementary Figure S12B). These results suggest that SRSF11-dependent telomerase recruitment occurs in a step separate from and prior to the interaction between TPP1 and TERT. We also found that depletion of TRF2 does not disrupt co-localization of telomeres with nuclear speckles, suggesting that TRF2 might be dispensable for telomere localization to nuclear speckles. Nonetheless, TRF2 is necessary to facilitate targeting of SRSF11-telomerase complex to telomeres at nuclear speckles because the interaction between SRSF11 and TRF2 is required for telomerase association with telomeres.
Telomerase has been implicated in human cancer and therefore considered as a potential target for cancer therapy. Functional inhibition of SRSF11 disrupts telomerase recruitment to telomeres and abrogates telomere elongation without interfering with telomerase enzyme activity. The essential role of SRSF11 in the telomerase recruitment pathway suggests the potential for novel strategies to inhibit telomerase activity in human cancer. In humans, loss-of-function mutations in telomerase components including dyskerin, TERC, TERT and TCAB1 have been associated with a number of diseases including dyskeratosis congenita, aplastic anemia, pulmonary fibrosis and multiple types of cancer (49–52). Thus, it is likely that mutations in SRSF11 may also be found in patients with these diseases.
Overall, our results provide an insight into the new cellular function of SRSF11 in addition to its function as a pre-mRNA splicing factor (28). SRSF11 acts as a nuclear speckle-targeting factor that is essential for cell cycle-specific recruitment of telomerase to telomeres and represents a new route for telomere maintenance by modulating telomere length homeostasis.
Supplementary Material
Acknowledgments
We are grateful to Yoon Ra Her, Joonyoung Her, Yu Young Jeong and Jeong Hee Kim for helpful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2013M3A9B6076431 to I.K.C.]; Yonsei University Internal Grant [2014-22-0096 to I.K.C.]. Funding for open access charge: National Research Foundation of Korea [NRF-2013M3A9B6076431 to I.K.C.]; Yonsei University Internal Grant [2014-22-0096 to I.K.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 3.Liu D., O'Connor M.S., Qin J., Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 5.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 6.Sfeir A., de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 8.Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 9.Autexier C., Lue N.F. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi A., Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol. Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 12.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 13.Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., Weinberg R.A. Creation of human tumor cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 14.Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R.H., Meier U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan E.D., Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard P., Kiss A.M., Darzacq X., Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristofari G., Adolf E., Reichenbach P., Sikora K., Terns R.M., Terns M.P., Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Tycowski K.T., Shu M.D., Kukoyi A., Steitz J.A. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venteicher A.S., Abreu E.B., Meng Z., McCann K.E., Terns R.M., Veenstra T.D., Terns M.P., Artandi S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venteicher A.S., Artandi S.E. TCAB1: driving telomerase to Cajal bodies. Cell Cycle. 2009;8:1329–1331. doi: 10.4161/cc.8.9.8288. [DOI] [PubMed] [Google Scholar]
- 22.Lee J.H., Lee Y.S., Jeong S.A., Khadka P., Roth J., Chung I.K. Catalytically active telomerase holoenzyme is assembled in the dense fibrillar component of the nucleolus during S phase. Histochem. Cell Biol. 2014;141:137–152. doi: 10.1007/s00418-013-1166-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhong F.L., Batista L.F., Freund A., Pech M.F., Venteicher A.S., Artandi S.E. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar J., Bell C.F., Weidenfeld I., Zaug A.J., Leinwand L.A., Cech T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M.S., Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 26.Lamond A.I., Spector D.L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 27.Huang S., Spector D.L. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl. Acad. Sci. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J.Y., Kar A., Kuo D., Yu B., Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol. Cell. Biol. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Listerman I., Sun J., Gazzaniga F.S., Lukas J.L., Blackburn E.H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013;73:2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.H., Khadka P., Baek S.H., Chung I.K. CHIP promotes human telomerase reverse transcriptase degradation and negatively regulates telomerase activity. J. Biol. Chem. 2010;285:42033–42045. doi: 10.1074/jbc.M110.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee G.E., Yu E.Y., Cho C.H., Lee J., Muller M.T., Chung I.K. DNA-protein kinase catalytic subunit-interacting protein KIP binds telomerase by interacting with human telomerase reverse transcriptase. J. Biol. Chem. 2004;279:34750–34755. doi: 10.1074/jbc.M401843200. [DOI] [PubMed] [Google Scholar]
- 33.Kim J.H., Kim J.H., Lee G.E., Lee J.E., Chung I.K. Potent inhibition of human telomerase by nitrostyrene derivatives. Mol. Pharmacol. 2003;63:1117–11124. doi: 10.1124/mol.63.5.1117. [DOI] [PubMed] [Google Scholar]
- 34.Kim N.W., Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abreu E., Terns R.M., Terns M.P. Visualization of human telomerase localization by fluorescence microscopy techniques. Methods Mol. Biol. 2011;735:125–137. doi: 10.1007/978-1-61779-092-8_12. [DOI] [PubMed] [Google Scholar]
- 36.Her Y.R., Chung I.K. p300-mediated acetylation of TRF2 is required for maintaining functional telomeres. Nucleic Acids Res. 2013;41:2267–2283. doi: 10.1093/nar/gks1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristofari G., Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 39.Khurts S., Masutomi K., Delgermaa L., Arai K., Oishi N., Mizuno H., Hayashi N., Hahn W.C., Murakami S. Nucleolin interacts with telomerase. J. Biol. Chem. 2004;279:51508–51515. doi: 10.1074/jbc.M407643200. [DOI] [PubMed] [Google Scholar]
- 40.Li Y., Wang X., Zhang X., Goodrich D.W. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol. Cell. Biol. 2005;25:4023–4033. doi: 10.1128/MCB.25.10.4023-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., Artandi S.E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jegou T., Chung I., Heuvelman G., Wachsmuth M., Görisch S.M., Greulich-Bode K.M., Boukamp P., Lichter P., Rippe K. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol. Biol. Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson R.L., Ziegler T.D., Supakorndej T., Terns R.M., Terns M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J.Y., Kar A., Kuo D., Yu B., Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol. Cell. Biol. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma A., Takata H., Shibahara K., Bubulya A., Bubulya P.A. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson R.L., Abreu E.B., Ziegler T., Ly H., Counter C.M., Terns R.M., Terns M.P. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol. Biol. Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y., Tomlinson R.L., Lukowiak A.A., Terns R.M., Terns M.P. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jády B.E., Bertrand E., Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vulliamy T., Marrone A., Goldman F., Dearlove A., Bessler M., Mason P.J., Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 50.Savage S.A., Alter B.P. The role of telomere biology in bone marrow failure and other disorders. Mech. Ageing Dev. 2008;129:35–47. doi: 10.1016/j.mad.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calado R.T., Regal J.A., Kajigaya S., Young N.S. Erosion of telomeric single-stranded overhang in patients with aplastic anaemia carrying telomerase complex mutations. Eur. J. Clin. Invest. 2009;39:1025–1032. doi: 10.1111/j.1365-2362.2009.02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong F., Savage S.A., Shkreli M., Giri N., Jessop L., Myers T., Chen R., Alter B.P., Artandi S.E. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


