Abstract
To gain a wider view of the pathways that regulate mitochondrial function, we combined the effect of heat stress on respiratory capacity with the discovery potential of a genome-wide screen in Saccharomyces cerevisiae. We identified 105 new genes whose deletion impairs respiratory growth at 37°C by interfering with processes such as transcriptional regulation, ubiquitination and cytosolic tRNA wobble uridine modification via 5-methoxycarbonylmethyl-2-thiouridine formation. The latter process, specifically required for efficient decoding of AA-ending codons under stress conditions, was covered by multiple genes belonging to the Elongator (e.g. ELP3) and urmylation (e.g., NCS6) pathways. ELP3 or NCS6 deletants had impaired mitochondrial protein synthesis. Their respiratory deficiency was selectively rescued by overexpression of tRNALysUUU as well by overexpression of genes (BCK1 and HFM1) with a strong bias for the AAA codon read by this tRNA. These data extend the mitochondrial regulome, demonstrate that heat stress can impair respiration by disturbing cytoplasmic translation of proteins critically involved in mitochondrial function and document, for the first time, the involvement in such process of the Elongator and urmylation pathways. Given the conservation of these pathways, the present findings may pave the way to a better understanding of the human mitochondrial regulome in health and disease.
INTRODUCTION
Most energy stored in nutrients is converted into ATP, aerobically, by the mitochondrial oxidative phosphorylation system (OXPHOS). The OXPHOS system consists of four enzymatic complexes that form the mitochondrial respiratory chain and a fifth complex, the FoF1 ATPase that drives the synthesis of ATP. The OXPHOS complexes comprise approximately 100 proteins of dual genetic origin. Some additional hundred nucleus-encoded proteins are required for their biogenesis (1). The complete mitochondrial proteome comprises approximately 800 (yeast) to 1500 (humans) proteins depending on the complexity of the eukaryotic organism (2,3). The vast majority of these proteins are encoded in the nuclear genome, translated by cytoplasmic ribosomes and imported into mitochondria. Only a handful of proteins (8 in yeast and 13 in humans), all essential components of the OXPHOS complexes, are encoded by the mitochondrial genome, translated by mitochondrial ribosomes and assembled with the newly imported proteins to form the OXPHOS system (4,5). Mitochondrial ribosomes and the translation process are closer to those of their bacterial ancestor than to their cytoplasmic counterparts, as demonstrated by the sensitivity to antibiotics and other properties, although two billion years of mitochondrial evolution resulted for example in deviations in the genetic code (6,7) and significant differences in the actual process of translation (8,9). Mitochondria have a simplified decoding system that allows a small set of mitochondrial tRNAs to recognize all the codons (10,11). The 24 mitochondrial tRNAs of S. cerevisiae and the 22 human mitochondrial tRNAs are encoded by the mtDNA genome. However, in some species, tRNAs need to be imported from the cytosol. This requirement is extended to some tRNAs for yeast and human mitochondria under stress conditions such as elevated temperature (12,13).
The study of the functional mitochondrial proteome has traditionally exploited the facultative aerobe/anaerobe growth capacity of the yeast Saccharomyces cerevisiae. This property allowed for the selection of respiratory deficient strains, which remain viable when grown in the presence of fermentable carbon sources such as glucose. Fifty years ago, chemical mutagenesis, followed by selection of respiratory mutants -so called mitochondrial or nuclear petite (pet) mutants- was probably the first successful high-throughput approach towards the characterization of the mitochondrial proteome (reviewed in (14–16)). Because of the conservation of fundamental cellular processes and genes between yeast and humans (17,18), S. cerevisiae is also an invaluable lower eukaryotic model system to identify and characterize human genes involved in mitochondrial diseases (reviewed in (19–21) and references therein). In fact, many human genes responsible for mitochondrial diseases were first identified in yeast (19). Over the last decade, a number of genome-wide approaches, including subcellular localization and in silico prediction studies (22,23), and whole exome sequencing (24) have facilitated the identification of nuclear genes involved in mitochondrial function and disease. However, the availability of deletion mutant collections covering nearly all protein-coding genes in different genetic formats (haploid, homozygous diploid and heterozygous diploid; reviewed in (25)) has offered a powerful means, thus far unique to yeast, to exploit S. cerevisiae as a model organism to pursue the characterization of not only the mitochondrial proteome but also the extended mitochondrial regulome. In particular, systematic screens of the haploid mutant collection searching for mutants able to grow on fermentable carbon sources (glucose) but not on non-fermentable, respiratory carbon sources (ethanol, glycerol or lactate), have led to the identification of novel genes involved in mitochondrion structure and function (26–30). Additionally, recent data indicate that hundreds of non-mitochondrial proteins involved in different (and seemingly disparate) cellular processes, can also play a role in optimizing or regulating OXPHOS function (31), thus contributing to the so called mitochondrial regulome. However, despite research efforts by a growing number of laboratories worldwide, the integration of mitochondrial function with other cellular processes and metabolic networks that can impair respiratory capacity and OXPHOS when mutated is thus still poorly understood and approximately one third of the 800 proteins thought to constitute the yeast mitochondrial proteome and of the 1500 proteins estimated to conform the mammalian mitochondrial proteome remain to be identified (23,32).
In all studies conducted so far, the ability of mutant strains to grow on respiratory carbon sources was monitored at the optimal growth temperature of 30°C. However, deletion of some genes involved in functions as diverse as DNA/RNA interaction (33), OXPHOS complex assembly (34–37) and oxidative stress response (38) leads to a respiratory defect exclusively at high temperature (37°C). Therefore, we anticipated that a deletion mutant collection screening conducted under heat-stress conditions would have the potential to uncover new genes and pathways involved in regulation of mitochondrial biogenesis and function. As a result of this modified screening we discovered 105 previously unidentified genes (67% orthologous to human genes) whose deletion interferes with mitochondrial OXPHOS function. Notably, 10% of these genes are involved in wobble uridine modification of a specific subset of nuclear-encoded tRNAs.
MATERIALS AND METHODS
Yeast strains and media
The MATα deletion library (BY4742 background) was purchased from Open Biosystems (Huntsville, AL, USA) and converted into a 384-well plate format by manual multipinning (39). Strains were grown at 30°C or 37°C in YP (1% yeast extract, 2% peptone) complete standard media or synthetic complete (SC) media (0.19% YNB, 0.5% ammonium sulphate, 1 g/l dropout mix without uracil) as specified in the text; media were supplemented with various carbon sources (2%, w/v each) as indicated.
OXPHOS mutant screening
A total of 4688 haploid deletion mutants were utilized for genomic phenotyping, which was performed as described (39). Individual plates from the deletion mutant collection (384-wells master plates with seven empty control-wells plus the BY4742 rho0 strain in one well) were inoculated with 50 μl of G418 (200 μg/ml) containing liquid YPD medium using a 384 pin tool (VP 384F; V&P Scientific Inc., San Diego). After 2 days at 30°C, cells were transferred to YP-agar (without G418), supplemented with either glucose or ethanol as a carbon source and cultured for 2 days at 30 and 37°C, followed by plate examination for respiratory growth based on relative colony size and digital image recording. Four replicate screens, using independent starter cultures, were carried out for each condition, followed by verification of mutant strains that were scored as sensitive in at least two screens by serial dilution spot assays. These were performed by pre-growing selected candidate mutants at 30°C in YPD medium for 24 h, followed by determination of OD600 with a microplate reader, adjustment of cell density to an OD600 value of 1.0 with YPD medium and serial dilution in ten-fold increments. Aliquots (4 μl) of each dilution were then spotted onto YP-agar plates supplemented with glucose, ethanol, glycerol or lactate and growth was examined after incubation at 30°C and 37°C for 2–3 days. Mutant strains exhibiting a growth reduction on ethanol at the first, second and third or fourth dilution were classified as ‘highly’ (HS), ‘medium’ (MS) or ‘lowly’ (LS) sensitive, respectively (Supplementary Table S1).
To determine mitochondrial DNA mutation rate, ‘petite’ (pet) frequency, defined as the percentage of colonies displaying a petite phenotype after a 5-day incubation at 37°C, was evaluated as described previously (40); at least 4000 colonies were analyzed for each strain.
Gene ontology analysis was done using Funspec program (GO Biological Process; http://funspec.med.utoronto.ca/).
Cytochrome spectra and mitochondrial respiration analysis
Reduced vs. oxidized spectra were recorded (Varian Cary300 UV-VIS Spectrophotometer) at room temperature on suspensions of cells cultured for 24 h at 37°C in SC medium containing a non-repressing glucose concentration (0.6%, w/v). Oxygen consumption rates were measured at 37°C on the same suspensions using a Clark-type oxygen electrode (Oxygraph System Hansatech Instruments England) containing 1 ml of air-saturated respiration buffer (0.1 M phthalate–KOH, pH 5.0, 10 mM glucose). The reaction was started by adding 20 mg equivalents of wet-weight cells as described previously (41).
In vivo mitochondrial protein synthesis
Mitochondrial gene products were labeled in whole cells at 30 and 37°C for 10 min with 35S-methionine (7 mCi/mmol; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) in the presence of 0.2 mg/ml cycloheximide to inhibit cytoplasmic protein synthesis (42). Equivalent amounts of pulsed total cellular proteins were precipitated with trichloroacetic acid (25% w/v), fractionated by SDS-PAGE (17.5% polyacrylamide gel), transferred to a nitrocellulose membrane and exposed to a Kodak X-OMAT X-ray film. Autoradiographic images were digitalized and densitometry performed with the Quantity One software (Bio-Rad).
Gene overexpression studies
The Escherichia coli XL1-Blue strain was used for plasmid propagation and DNA cloning experiments. The coding sequences of the tRNA genes of interest [tR(UCU)M1, tK(UUU)L, tK(UUU)D, tK(CUU)G1, tT(UGU)G1, tG(UCC)O, tQ(UUG)C, tQ(CUG)M, retrieved from SGD; see the text for further details] and the ELP3 gene were amplified by PCR using genomic DNA from the BY4742 strain as template and the oligonucleotides described in Supplementary Table S2 as primers. Individual amplicons were cloned into the YEp352 vector (URA3 auxotrophic marker) using SalI-KpnI [for tG(UCC)O, tK(CUU)G1, tQ(UUG)C, tR(UCU)M1, tT(UGU)G1 genes], BamHI-SacI [for tK(UUU)L, tK(UUU)D genes] and KpnI–BamHI [for tQ(CUG)M and ELP3 genes] restriction/insertion sites. tE(UUC) as well as independent replicates of the tQ(UUG) and tK(UUU) tRNA gene constructs and a triple combination of these three tRNA genes (tKQE) all cloned in the pRS425 plasmid (LEU2 auxotrophic marker) (43) were kindly provided by Dr. Patrick Pedrioli.
Yeast genes with a high AAA codon usage (SPT7, VPS3, HFM1, CCM1, BCK1, PET309, NAM2, DSS1, FZO1, BEM2; see Supplementary Table S3) and the ELP3 gene cloned in the pGB1805 expression vector under control of a GAL1 promoter were purchased from Thermo Scientific (Yeast ORF Collection). After sequence verification, individual plasmids were used for yeast transformation (39). Galactose, in combination with ethanol, was used as an inducer and a respirable carbon source (44) in these experiments.
Statistical analysis
Data are expressed as mean ± standard deviation (SD) from at least three independent experiments. Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Dunnett's post hoc test (GraphPad Prism 5 Software; **, P-value ≤ 0.01; ***, P-value ≤ 0.001).
RESULTS
Genomic phenotyping analysis of mitochondrial function under thermal stress conditions
We screened the yeast deletion mutant collection (4688 non-essential gene deletion strains in a haploid background) by growing the strains on rich solid medium containing either glucose (YPD), a fermentable carbon source, or ethanol (YPE), a non-fermentable (respiratory) carbon source, at 30°C and at 37°C, followed by fitness scoring and subsequent validation and characterization of the mutants exhibiting a growth defect in YPE.
The primary screening was performed in four replicates and pet mutants in at least two screens were individually verified by serial dilution assays conducted on YPD and YPE as explained in the ‘Methods’ section. We identified a total of 489 mutant strains displaying various degrees of growth impairment when cultured on YPE at 37°C (and/or at 30°C; see Table 1 for a summary of the results). The mutants were classified as high (HS), medium (MS) and low (LS) sensitive based on their serial dilution assay respiratory growth phenotypes (Figure 1A, but see also Table 1 and Supplementary Table S1). One hundred and thirty four mutant strains displayed an attenuated growth on YPE only at 37°C, while 355 mutants exhibited a respiratory defect also at 30°C. Two-hundred sixty-eight of the latter mutants failed to grow on YPE at both 30°C and 37°C, while a phenotype worsening effect at 37°C was observed for the remaining 87 mutants (Table 1).
Table 1. Classification of the 489 mutant strains displaying different degrees of respiratory growth impairment on YPE at 37°C and/or at 30°C.
| Total number and type of mutants | HSa | MSa | LSa | New mutants | Human orthologsb |
|---|---|---|---|---|---|
| 134 mutants respiratory growth-defective only at 37°C | 58 | 52 | 24 | 65 | 52 (80%) |
| 355 mutants respiratory growth-defective also at 30°C: | |||||
| –268 mutants respiratory growth-defective at both 30°C and 37°C | 221 | 26 | 21 | 22 | 16 (73%) |
| –87 mutants displaying a phenotype worsening effect at 37°Cc | 56c | 31c | — | 18 | 8 (44%) |
aMutants were classified as high (HS), medium (MS) and low (LS) sensitive based on their serial dilution assay OXPHOS phenotypes (see also Figure 1A and Supplementary Table S1).
bThe numbers (and percentages) of yeast genes with an orthologous counterpart in the human genome, only refers to the newly identified 105 genes that cause a respiratory growth defect when mutated.
cNumber of mutants displaying a HS or MS OXPHOS phenotype at 37°C compared to a less severe phenotype at 30°C.
Figure 1.

Heat stress-dependent OXPHOS mutants. (A) Representative examples of pet mutant identification by genomic phenotyping (left) and corresponding serial dilution assays (right) used for mutant classification based on OXPHOS phenotype severity (HS, MS and LS: high-, medium- and low -sensitive mutants). A mitochondrial DNA-depleted rho° strain (ρ°) and the wild type (WT) strain were used as positive and negative controls, respectively; the names of the specific gene deletions utilized as representative examples are indicated on the right. Cytochrome spectra (B) and respiratory rates (C) used for further mutant classification (A, B, C, D in a decreasing order of severity).
Comparison of our results with the Saccharomyces Genome Database (SGD, (45)) and with the results of previous mitochondrion-targeted genomic phenotyping studies conducted at 30°C (26–30) showed that 77% (376 out of 489) of the genes retrieved by our screen encode previously characterized mitochondrial proteins (Supplementary Table S1). The remaining 113 genes, which have never been associated to mitochondrial function before, encode mostly non-mitochondrial proteins that could be potentially involved in regulation of mitochondrial processes (referred to as ‘new’ in Table 1 and in Supplementary Table S1).
To distinguish between a genuine OXPHOS impairment and other possible defects related to ethanol utilization or tolerance, these 113 mutant strains were individually verified by serial dilution assays carried out on rich medium supplemented with lactate, acetate or glycerol as carbon source. One hundred and five strains were found to display a defective growth on at least two different respiratory carbon sources, thus strongly suggesting an OXPHOS impairment. The remaining eight strains that exhibited a defective growth exclusively on ethanol-supplemented media (see Supplementary Table S4) had mutations in genes related to phospholipid biosynthesis, cell wall integrity and stress/heat resistance that probably affect ethanol tolerance, and were thus excluded from further analysis. Out of the 105 new mutants with a specific OXPHOS impairment, 65 had a respiratory growth-defective phenotype only at 37°C, 18 exhibited a phenotype worsening at this temperature, while the remaining 22 mutants were growth-defective even at 30°C (Table 1 and Supplementary Table S5). Most newly identified genes (95%) code for proteins of known function but not previously associated to mitochondrial biogenesis. Two gene products (Ldb16 and Ynl320w) were previously localized to the mitochondrion (46) but not characterized from a functional point of view, while four mutants are deleted in genes coding for as yet uncharacterized open reading frames. Seventy of the 105 newly identified genes that cause OXPHOS impairment when deleted have orthologous counterparts in the human genome (Table 1 and Supplementary Table S1).
Cytochrome spectra and respiration capacity of the newly identified mitochondrial mutants
To gain insight into the specific functional defects of the 105 new mutants identified by our screen, we determined the cytochrome absorption spectra and oxygen consumption rates for each mutant, using the isogenic wild type strain as a reference (see Supplementary Table S5 for the entire list of mutants and associated phenotypes). We also included a subset of 19 mutants previously identified as LS at 30°C, but not fully characterized in other screens (26) (Supplementary Table S6). These analyses highlighted a good correlation between cytochrome spectra and respiratory rates and allowed us to classify the mutant strains into four different classes (A–D) of decreasing phenotype severity (see Figure 1B and C). Class A mutants are characterized by strongly altered cytochrome spectra and highly attenuated oxygen consumption rates; intermediate, but progressively less altered phenotypes are displayed by class B and C mutants, whereas class D mutants differ only slightly from the wild type strain. Mutant classes A-C are the most populated (85% of the entire mutant set; Figure 1C) thus indicating an important direct or indirect role of the corresponding gene products in the function of the OXPHOS system.
The set of newly identified genes is enriched in ‘tRNA wobble uridine modification’ components
As revealed by Gene Ontology (GO) ‘biological process’ analysis (Table 2), the most significant and enriched GO categories matched by the newly identified heat-sensitive pet mutant strains were ‘tRNA wobble uridine modification’ (P-value = 2.1E−10) and ‘protein urmylation’ (P-value = 3.8E−06). The latter is a post-translational modification relying on the ubiquitin-like polypeptide Urm1 that acts on protein substrates (47) but also plays a crucial role in wobble uridine modification of some tRNAs via the thiolating enzyme complex Ncs6–Ncs2 (48–50). Multiple genes coding for proteins covering all steps of tRNA wobble uridine (U34) modification, specifically 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) formation that in yeast requires Elongator (ELP) and urmylation (Urm) pathway components plus the tRNA methyltransferase Trm9, are present in our mutant set (see Figure 2A for a schematic representation of the wobble uridine modification pathway). Additional components of this pathway that were also retrieved among the genes causing respiratory deficiency when deleted, include: Sit4 and Kti12, two proteins that positively influence ELP complex activity (51,52); Urm1, previously identified as mitochondrial function-related (28) and Uba4, both needed for uridine thiolation (48,49). Therefore, the main missing components are Elp5 and Trm112, plus Kti11 and Kti14, which are not present in the mutant collection, and Kti13 which was not found likely due to its relatively modest effect on mcm5s2 modification (53).
Table 2. GO Biological process analysis of new genes involved in mitochondrial biogenesis under stress conditions.
| GO term | P-value |
|---|---|
| tRNA wobble uridine modification [GO:0002098] | 2.1E−10 |
| Regulation of transcription from RNA polymerase II promoter [GO:0006357] | 1.2E−07 |
| Protein urmylation [GO:0032447] | 3.8E−06 |
| Ubiquitin-dependent protein catabolic process via the MVB sorting pathway [GO:0043162] | 5.3E−06 |
| Late endosome to vacuole transport [GO:0045324] | 2.6E−05 |
| Protein processing [GO:0016485] | 5.1E−05 |
| Protein targeting to vacuole [GO:0006623] | 9.7E−05 |
Figure 2.

Outline of respiration-deficient, wobble uridine modification pathway-related mutants and mitochondrial protein synthesis analysis of the elp3Δ and ncs6Δ mutant strains. (A) Schematic representation of the mcm5s2 uridine modification pathway and associated genes involved in this modification that cause respiratory deficiency when deleted. Uridine modifications by Elongator, Urmylation and Trm9–Trm112 pathway components are boxed in blue, yellow and red, respectively. Newly identified genes are shown on a green background encircled by a continuous line; a previously known gene involved in this pathway (URM1) that was also identified in our screen is shown on a green background encircled by a dotted line; two components encoded by genes not present in the deletion mutant collection are shown in gray. (B) Mitochondrial protein synthesis analysis carried out at 30 and 37°C in the wild type strain and in the elp3Δ and ncs6Δ mutant strains. A bar-plot representation of the data obtained at 37°C from three independent experiments for the indicated deletion mutant strains and mitochondrial polypeptides (polypeptide levels in the mutant compared to the wild type strain ± SD) is shown below the gel autoradiography; significant differences (***P-value ≤ 0.001) between mutant and wild type polypeptide levels are indicated.
Other biological processes significantly represented among the newly identified respiration-defective mutants are ‘regulation of transcription from RNA polymerase II promoter’ (P-value = 1.2E−07), ‘ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway’ (P-value = 5.3E−06), ‘protein processing’ (P-value = 5.1E−05) and ‘protein targeting to vacuole’ (P-value = 9.7E−05) (Table 2; see ‘Discussion’).
Defective tRNALysUUU wobble uridine modification indirectly affects mitochondrion integrity under heat stress conditions
Among the multiple relevant pathways identified in our screen, we chose to focus on tRNA wobble uridine modification, the most GO-supported and functionally homogenous class of mutants. We initially examined mitochondrial protein synthesis (MPS) in three mutants, elp3Δ, trm9Δ and ncs6Δ, representative of the three main steps of tRNA U34 modification (54–56) (Figure 2B). Elp3Δ and ncs6Δ were scored as MS mutants based on their growth-phenotype on non-fermentable carbon sources at 37°C, yet displayed fairly strong respiratory chain and oxygen consumption defects and were both classified as class B mutants. Under the same conditions, trm9Δ behaved as an LS mutant with only a slightly defective cytochrome spectrum and respiratory rate (class D mutant; Supplementary Table S5).
Mitochondrial protein synthesis was analyzed by pulse-labelling experiments carried out at both 30 and 37°C in the presence of 35S-methionine and cycloheximide, an inhibitor of cytosolic translation. Following 35S-methionine incorporation into newly synthesized mitochondrial proteins, labeled cell extracts were gel-fractionated and radioactivity levels associated to individual polypeptide bands in the mutants and in the wild type strain were determined. No significant differences between mutants and wild type were observed at 30°C (Figure 2B). In contrast, a fairly strong reduction in 35S-methionine incorporation was apparent in the ncs6Δ and elp3Δ strains at 37°C. Under the same conditions, 35S-methionine incorporation in the trm9Δ mutant was not significantly different from that of the wild type strain (data not shown).
The mitochondrial translation defect observed in the ncs6Δ and elp3Δ strains at 37°C is not accounted for by mtDNA instability, since the frequency of conversion to rho0/rho− cells in the mutant strains did not differ from wild type (data not shown).
U34 modification by Elp3, Trm9 and Ncs6 is found exclusively on lysine (UUU), glutamine (UUG) and glutamic acid (UUC) tRNAs (43,53), although other ELP pathway-dependent modifications (namely, mcm5U34, ncm5U34 and ncm5U34m) are present on eight additional tRNAs bearing an UXX anticodon (57) (Figure 2A). Mcm5s2 uridine modification has been shown to be dispensable for general translation, but required to positively modulate translation of AAA, CAA, and GAA codon-rich mRNAs under stress conditions through an increased occupancy of the ribosome A-site by the corresponding aminoacyl tRNAs (43,58). Importantly, the mcm5s2-modified glutamine (UUG) tRNA is one of three tRNAs [the others are lysine (CUU) and glutamine (CUG)] that are imported into mitochondria in yeast (12,13). Therefore, a defective ELP-Urm pathway might affect mitochondrial gene expression, specifically mitochondrial translation, by interfering with mcm5s2-modification of the mitochondrially imported glutamine (UUG) tRNA. Alternatively, ELP-Urm mutations might impair the cytosolic synthesis of proteins that are required for mitochondrial gene expression and function under heat stress conditions. These proteins should be encoded by mRNAs with a high content of codons (AAA, CAA, GAA) read by one or more of the U34-modified tRNAs; this contrasts, however, with the generally low content of such codons in the mRNAs for the mitochondrial proteins (shown in Figure 2B) whose synthesis was found to be impaired in the elp3Δ and ncs6Δ mutants under heat-stress conditions (data not shown). We thus reasoned that if the respiratory defect displayed by these mutants is due to a codon-specific cytoplasmic translation impairment, it could be alleviated by overexpression of the corresponding specific tRNAs, as previously observed in other cases of crippled tRNA modification in yeast (58–60). Accordingly, we overexpressed a series of candidate and control tRNA genes into the elp3Δ mutant strain and tested the ability of the resulting transformants to grow on ethanol at 37°C. In a first set of experiments (Figure 3A–C), we tested the rescue capacity of two tRNAs modified by the ELP-Urm pathway (tRNALysUUU and tRNAGlnUUG), the latter of which has also been shown to be imported into mitochondria (12); three tRNAs that are only targeted by ELP-dependent modification (tRNAThrUGU, tRNAGlyUCC and tRNAArgUCU); and the other two mitochondrially imported tRNAs that do not to undergo U34 modification (tRNAGlnCUG, tRNALysCUU). tRNALysCUU, in particular, served as a control for a tRNA that, at variance with the non-mitochondrially imported tRNALysUUU, is internalized into mitochondria in an unmodified form at 37°C (13). As shown in Figure 3A-C, only tRNALysUUU rescued the defective respiratory phenotype of the elp3Δ mutant at 37°C (at levels only slightly lower than those of a ELP3 control transformant) and also restored cytochrome spectra and oxygen consumption rates, while no effect was observed with the mitochondrially imported tRNAGlnUUG. In a second set of experiments (separately carried out with a different plasmid under different selection conditions), we tested the respiratory growth capacity of the elp3Δ mutant strain transformed with tRNAGluUUC, two of the previously tested tRNAs (tRNAGlnUUG and the rescue-proficient tRNALysUUU), plus a combination of the three tRNAs all carried by the same vector. As further shown in Figure 3 (panels D–F), despite a slightly lower quantitative effect compared to the results in Figure 3A-C, also in this case tRNALysUUU proved to be the only one, among the three tested tRNAs, capable of rescuing the respiratory growth defect, cytochrome spectrum and oxygen consumption rate of the elp3Δ mutant strain at 37°C. Curiously, while neither tRNAGlnUUG nor tRNAGluUUC displayed any rescue capacity individually, a higher recovery of mitochondrial function (especially oxygen consumption rate) was observed in the case of the triple transformant. It thus appears that similar to what was previously reported for other Elongator related phenotypes (58,61), tRNALysUUU is the only tRNA capable of rescuing the mitochondrial abnormalities caused by ELP3 disruption. Given the exclusively cytoplasmic localization of tRNALysUUU (13) and the lack of effect of the mitochondrion-imported tRNALysCUU, the unique ability of this tRNA to alleviate the respiratory growth defect of the elp3Δ mutant suggests that wobble uridine modification of tRNALysUUU is required to attain adequate levels of translation of cytosolic proteins that are necessary for mitochondrial gene expression and function.
Figure 3.

tRNALysUUU overexpression rescues the respiratory growth defect of the elp3Δ mutant. (A) OXPHOS phenotypes of the indicated elp3Δ-tRNA gene (YEp352 vector with URA3 as auxotrophic marker) transformants grown at 37°C on glucose (SCD) or ethanol (SCE) synthetic medium (the latter supplemented with a non-repressive 0.005% glucose concentration). Elp3Δ cells transformed with the empty YEp352 vector and the same vector carrying the wild type ELP3 gene were used as negative and positive controls, respectively; two different tRNALysUUU genes [tK(UUU)L, #1; tK(UUU)D, #2] were assayed. Cytochrome spectra and oxygen consumption rates of the indicated elp3Δ-tRNA gene transformants are shown in panels (B) and (C), respectively. (D) OXPHOS phenotype assays as in (A) testing one further tRNA (tRNAGluUUC) plus a triple combination of tRNALysUUU, tRNAGlnUUG and tRNAGluUUC (tKQE), in addition to an independent replicate of the single tRNALysUUU and tRNAGlnUUG, all carried by the pRS425 vector (LEU2 auxotrophic marker). Cytochrome spectra and oxygen consumption rates of the same set of transformants are shown in panels (E) and (F). Error bars represent the mean ± SD; significant differences (**P-value ≤ 0.01) are indicated.
Increased dosage of two AAA codon-rich mRNAs encoded by genes that cause respiratory deficiency when deleted rescues the respiration-defective phenotype of the elp3Δ mutant at 37°C
tRNALysmcm5s2UUU has been shown to preferentially, if not exclusively, read the AAA codon (43,60,62). Accordingly, we hypothesized that translation of AAA codon-rich mRNAs encoded by genes that cause respiratory deficiency when deleted might be particularly sensitive to the defective wobble uridine modification taking place in elp3Δ and thus causally related to the OXPHOS phenotype of this mutant. Accordingly, the associated respiratory deficiency should be suppressed, at least in part, by overexpression of such genes -a predicted outcome that has been experimentally verified in other cases of altered tRNA modification (e.g. (63)). To test this hypothesis, we ranked the 489 genes whose deletion causes respiratory deficiency in our screen according to their AAA-codon usage bias (paying special attention to tandemly repeated AAA codons) and the 10 top-scoring genes (see Supplementary Table S3) were transformed into the elp3Δ mutant and assayed for their ability to rescue the respiratory phenotype of this strain. As shown in Figure 4, overexpression of two genes, BCK1 and HFM1, apparently unrelated to mitochondrial function restored respiratory capacity (panel A) as well as cytochrome spectrum and oxygen consumption rate (panels B and C) of the elp3Δ mutant under heat stress conditions. Additional data reported in Figure 5 show that this rescue effect is comparable to that produced by the wild type version of ELP3 (panel A); that a similar, albeit reduced effect (in the case of BCK1) is elicited in the functionally related ncs6Δ mutant (panel B); while no amelioration of respiratory growth is observed under identical conditions in two mutants (spt7Δ and med2Δ, both displaying an MS respiratory phenotype as elp3Δ and ncs6Δ) functionally unrelated to wobble uridine tRNA modification (panel C). BCK1 codes for a mitogen-activated protein kinase kinase (MAPKK) kinase acting in the protein kinase C (PKC) signalling pathway (64), while HFM1 codes for a DNA helicase involved in modulation of genome integrity (65) (see ‘Discussion’).
Figure 4.
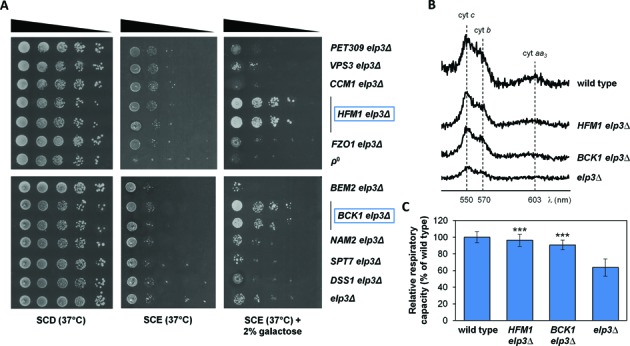
Increased dosage of the AAA codon-rich genes BCK1 and HFM1 rescues respiratory deficiency in the elp3Δ mutant under heat stress conditions. (A) Growth performance of the elp3Δ strain transformed with a multicopy expression vector (pGB1805; URA3 auxotrophic marker, GAL1 promoter) harbouring the indicated AAA codon-rich genes. Transformants were cultured on fermentative (SCD, left panel) or oxidative (SCE supplemented with 0.005% glucose) medium, in the absence (middle panel) or the presence (right panel) of 2% galactose at 37°C. The ρ° and elp3Δ strains served as negative controls; the HFM1 and the BCK1 transformants were assayed in duplicate. Cytochrome spectra and oxygen consumption rates of the elp3Δ-BCK1 and elp3Δ-HFM1 transformants are shown in panels (B) and (C). Significant differences (***P-value ≤ 0.001) between the respiratory capacity of elp3Δ and the BCK1/HFM1 transformants are indicated.
Figure 5.
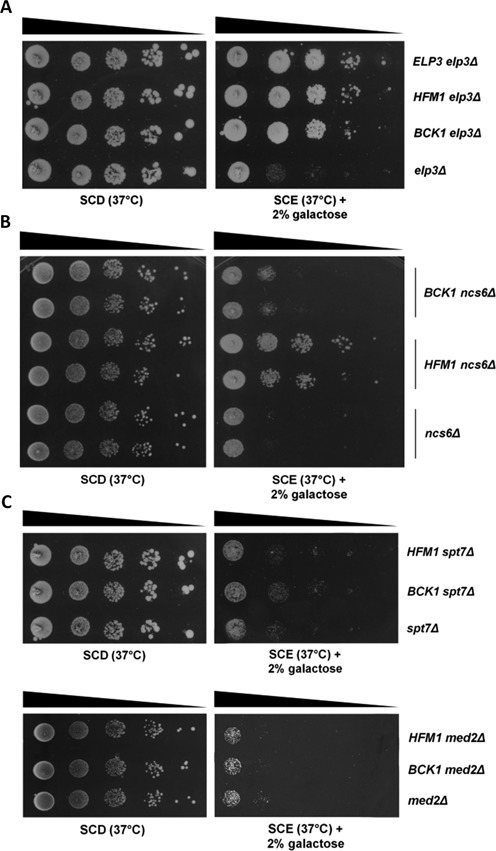
Mutant strain specificity of BCK1- and HFM1-mediated suppression. (A) Respiratory growth performance (37°C) of the elp3Δ strain overexpressing BCK1- and HFM1 compared with that of the same strain transformed with a ELP3 overexpressing plasmid (positive control); elp3Δ served as a negative control for these experiments. Cells cultured at 37°C on glucose (SCD) or ethanol (SCE) plus galactose medium are shown in the left and the right panels, respectively. (B) Respiratory growth serial dilution assays as in (A) conducted on the ncs6Δ mutant. (C) Respiratory growth assays conducted with BCK1- and HFM1 in two mutant strains (spt7Δ and med2Δ) functionally unrelated to wobble uridine modification.
DISCUSSION
Defining the mitochondrial proteome and its regulome under standard and stress conditions is a prerequisite for fully understanding mitochondrial physiology in health and disease. Given the strong evolutionary conservation of essential biochemical pathways from yeast to mammals, the characterization of the mitochondrial regulatory network in humans can benefit from comprehensive high-throughput studies in S. cerevisiae. Here, we combined the negative effect of heat stress on respiratory capacity with the discovery potential of a genome-wide phenotypic screening in yeast to expand the repertoire of genes affecting mitochondrial function. This allowed the identification of 105 new genes and novel pathways, most notably wobble uridine tRNA modification, which cause respiratory deficiency when disrupted. A total of 124 mutants (including all the newly identified genes and a subset of poorly characterized genes with a purported role in mitochondrial biogenesis) were functionally tested for mitochondrial respiratory chain (MRC) integrity and oxygen consumption capacity. Given the good correlation between respiratory growth, oxygen consumption rate and cytochrome spectra, the OXPHOS-negative phenotype of at least 86% of the mutants appears to be causally related to MRC impairment. We cannot exclude, however, the possibility that a mild respiratory impairment might underlie the defective phenotypes of at least a subset of the class D mutants (Supplementary Table S5). A role distinct from cellular respiration must be postulated, instead, for the six class D mutants that displayed a HS OXPHOS phenotype (vps8Δ, rpl23BΔ, erg4Δ, pho86Δ, rav1Δ, srn2Δ; Supplementary Tables S5 and S6).
Most genes whose deletion causes respiratory deficiency at 37°C code for non-mitochondrial proteins that may indirectly impinge on mitochondrion structure or function. These include the CAP-binding complex proteins Cbc2 and Sto1, various transcriptional regulators (e.g. Ume6, Mss11, Gcr2, Smi1, Rim101, Swi6) as well as chromatin remodeling components (e.g. Spt8, Eaf7, Rtt109, Rsc2), for some of which indirect evidence pointing to a role in mitochondrial biogenesis has been reported before (26,66) (Supplementary Table S5). Additional mutants include deletants in four genes (rim20Δ, rim21Δ, rim8Δ and dfg16Δ) involved in the proteolytic activation of the transcriptional repressor Rim101, which regulates cell wall assembly as well as metal ion and alkaline pH tolerance ((39,67,68) and references therein). Potentially related hits comprise 11 components of the Endosomal Sorting Complex Required for Transport (ESCRT) I, II and III complexes (Supplementary Table S1), some of which (e.g. Spt22, Snf7 and Vps20) interact with Rim20 and are required for proteolytic activation of Rim101 (69). Of note, a causal relationship between vacuolar pH, endosomal trafficking and mtDNA stability has been recently reported (70).
Further genes whose deletion causes respiratory deficiency even at 30°C code for specific components of the transcription/chromatin remodeling (e.g. Med2, Spt8, Ctf4) and cytosolic translation (e.g., Rpl39, Rps27b) machineries. These include PEX5, a gene coding for a membrane receptor required for peroxisomal matrix protein import whose deletion causes a severe respiratory decline as well as specific defects in respiratory complex assembly (OXPHOS, class A mutant; Supplementary Table S5). Interestingly, mitochondrial respiratory alterations have been reported in a PEX5 knockout mouse model of Zellweger Syndrome with defective peroxisomal biogenesis (71).
The most peculiar and comprehensive class of genes for which we discovered a previously unknown role in mitochondrial biogenesis at 37°C codes for proteins involved in 5-methoxycarbonylmethyl-2-thiouridine formation at the U34 position of some cytoplasmic tRNAs, a modification required for efficient mRNA decoding under stress conditions (43,58). These include ELP and Urm pathway components and the tRNA methyltransferase Trm9. Interestingly, a similarly comprehensive repertoire of wobble uridine tRNA modification components has been retrieved from a screening for genes whose mutation abolishes sensitivity to zymocin, a toxin that cleaves tRNAs only when they bear the U34-mcm5s2modification (53). The first step of this modification, catalysed by the ELP complex, is U34-cm5 formation on 11 UXX tRNAs, that is followed by a second, Trm9-mediated methylation (U34-mcm5) of five tRNAs, three of which (tRNAGlnUUG, tRNALysUUU and tRNAGluUUC) are thiolated by Ncs6-Ncs2 to produce mcm5s2U34. Deletion of genes (ELP3 and NCS6) critically involved in the first and third step of wobble uridine modification strongly impairs MPS, whereas no appreciable MPS defect was observed in the trm9Δ mutant (Figure 2B). This is in agreement with previous results indicating a relatively minor and growth condition-dependent effect of Trm9-catalyzed methylation on the decoding properties of at least some of its tRNA targets (57). Yeast mitochondria encode an apparently complete set of tRNAs, yet three nuclear-encoded tRNAs (tRNALysCUU, tRNAGlnCUG and tRNAGlnUUG) are known to be imported into mitochondria (12,13,72). In particular, it has been shown that tRNALysCUU import is dispensable for oxidative growth at 30°C, but becomes essential at 37°C (13). This is likely explained by the fact that wobble uridine thiolation of the mitochondrially encoded tRNALysUUU is impaired at 37°C, thus preventing efficient decoding of the lysine AAG codon under temperature stress conditions (13). It is less clear, instead, the need for mitochondrial import of the nuclear-encoded tRNAGlnUUG, for which a tRNA bearing the same anticodon is encoded by the mitochondrial genome. The fact that tRNAGlnUUG, one of the tRNAs modified by the ELP-Urm pathway (57), is imported into mitochondria led us to hypothesize that the depressed MPS observed in the elp3Δ and ncs6Δ mutants might result from a hypomodification of this particular tRNA at 37°C. Such an effect, however, seems incompatible with the lack of any significant cognate codon (CAA) enrichment in the mRNAs coding for the mitochondrial polypeptides monitored by the MPS assay (data not shown and Figure 2B). Moreover, the fact that overexpression of tRNAGlnUUG alone failed to rescue respiratory deficiency at 37°C further argues against the hypothesis that a defective ELP-Urm pathway might negatively affect MPS by interfering with wobble uridine modification of this particular tRNA. Instead, our data indicate an indirect effect of tRNA hypomodification on mitochondrial biogenesis caused by impaired translation of a specific subset of cytoplasmic proteins required for optimal mitochondrial function. This is strongly supported by the respiratory growth rescue ability exhibited by plasmid vectors overexpressing only tRNALysUUU, a tRNA with an exclusively cytoplasmic localization (13). It is interesting to note, in this regard, that the peroxide hypersensitivity phenotype displayed by fission yeast elp3 deletants is similarly rescued by tRNALysUUU overexpression (58). So, despite the as yet unexplained increase in the suppression of defective respiration (especially oxygen consumption rate) observed with the triple construct bearing two other tRNAs (tRNAGlnUUG and tRNAGluUUC) in addition to tRNALysUUU, it appears that the latter tRNA is responsible for respiratory growth rescue and that efficient translation of mRNAs with a high content of AAA codons is critically required to optimize mitochondrial function. Further to this point, the yeast genome displays a strong imbalance in gene copy number for the two cytoplasmic tRNALys, with 14 copies of tRNALysCUU but only seven copies of the tRNALysUUU gene (73).
In keeping with the above hypothesis, multicopy suppression-based testing of 10 nuclear genes with a high AAA codon content (see Supplementary Table S3) and causing a respiratory defective phenotype when deleted, identified two genes (BCK1 and HFM1) that specifically rescue the respiratory growth defect of the elp3Δ and ncs6Δ mutants, but not of functionally unrelated mutants, at 37°C. Bck1 is the MAPKK kinase component of a PKC1 signalling pathway involved in the control of cell integrity. Deletion of this gene caused a HS phenotype in our screen but has never been directly related to a respiratory growth defect. It is interesting to note, however, that Slt2, the MAPK component of the PKC1 signalling pathway in which Bck1 is involved and that was also identified as a respiratory deficient mutant (Supplementary Table S1), has recently been shown to impair yeast growth on glycerol when mutated (74). Although the essential nature of PKC1 prevented its examination in the present screen, the above data suggest a generalized involvement of the PKC1-Bck1-Slt2 pathway in the maintenance of mitochondrial function. Other potential links between Bck1 and mitochondrial function have been suggested by two previous genome-wide studies, which revealed a protective role of this kinase against reactive oxygen species in yeast cells treated with farnesol (64) as well as its involvement in mitophagy (75), the process responsible for the removal of unfitted mitochondria. Also notable are the negative genetic synthetic interactions of BCK1 deletion with ELP3, ELP4, ELP6 and NCS6 deletions (76). Our observation may thus be explained by an as yet uncharacterized, but apparently critical involvement of Bck1 in mitochondrial biogenesis/turnover and function.
Hfm1 (also known as Mer3) is a meiosis-specific DNA helicase (65) for which no specific link to mitochondria has been reported so far. The hfm1 disruptant was classified as MS at 37°C, but also exhibited a clear, although less intense, respiratory growth phenotype at 30°C, consistently with another OXPHOS screen conducted at this temperature (29). Evidence for a non-exclusive, meiosis-related role also emerged from two other genome-wide studies, in which HFM1 was retrieved as a gene involved in the utilization of dipeptides, a major nitrogen source for microbial as well as mammalian cells (77), with also a role in the control of chronological life span (78). Interestingly, dipeptide production/utilization is regulated by the ubiquitin proteasome system (79), another well covered biological process that we found to be required for oxidative growth under heat stress (Table 2). What is perhaps even more relevant is that various nuclear yeast and mammalian DNA helicases (e.g., Pif1) are involved in the maintenance of mtDNA integrity (80).
The ELP complex was originally identified as an RNA polymerase II-associated transcription elongation factor, and subsequently shown to be involved also in wobble uridine tRNA modification ((58,59) and references therein). The latter function appears to be central to ELP action and may indirectly explain other cellular defects associated to ELP mutations in various organisms through an attenuated and aberrant translation of proteins playing critical roles in specific cellular processes or stress responses. These include temperature, chemical and starvation stress sensitivity, transcriptional/chromatin remodeling, telomeric silencing, DNA damage response and cell division defects in yeast ((60,81) and references therein), neurological alterations in C. elegans and in humans (e.g. familial dysautonomia, amyotrophic lateral sclerosis and Rolandic epilepsy (82–85)) as well as altered neuron migration caused by defective α-tubulin acetylation (86) and impaired zygotic paternal genome demethylation (87). Our results extend the range of ELP roles to mitochondrial function and demonstrate an equally important involvement in this process of the Urm complex responsible for wobble uridine tRNA thiolation (49). All the components of the yeast wobble uridine tRNA modification machinery as well as the AAA codon-rich suppressor genes BCK1 and HFM1 are orthologous to human genes. Beyond these pathways, approximately 70% of the new genes revealed by our screen are orthologous to human genes. Also worth of note is the known causal association between mutations interfering with wobble base modification of some mitochondrial tRNA genes and human neurodegenerative mitochondriopathies such as MERF (Myoclonic Epilepsy with Ragged Red Fibers) and MELAS (Myopathy, Encephalopathy, Lactic Acidosis, and Stroke-like episodes) (88). This opens the possibility that wobble base modification impairing mutations targeting tRNALysUUU, including ELP/Urm complex mutations, may be similarly involved in as yet poorly understood human mitochondrial pathologies. Therefore, future studies mining the findings presented in this work are expected to facilitate a better understanding of the human mitochondrial regulome in health and mitochondrial disease as well as in diseases of broad social impact such as age-related neurodegenerative disorders.
Supplementary Material
Acknowledgments
We thank Iliana Ferrero (Dept. of Life Sciences, University of Parma) for insightful advice and discussions, and Alizia Ongarini (a former graduate student) for her competent help with the respiratory growth screening in an early phase of this work. We are also very grateful to Patrick Pedrioli and Kshitiz Tyagi (The Scottish Institute for Cell Signalling, University of Dundee) for the kind gift of the tK(UUU), tQ(UUG), tE(UUC) and the triple tKQE tRNA constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Education University and Research [PRIN 2009, 200975W76L to C.Do.]; Regione Emilia-Romagna [DGR no. 1631/2009 to S.O]; National Institutes of Health [RO1 grants GM105781 and GM071775 to A.B.]; American Heart Association [a Development Grant to F.F.] and Stanley Glaser Award [to F.F.]. Funding for open access charge: PRIN 2009 (Italian Ministry of Education University and Research [200975W76L]; Regione Emilia-Romagna [DGR no. 1631/2009]; NIH Research Project grants [GM105781 and GM071775]; American Heart Association Grant and the Stanley Glaser Award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Koopman W.J., Distelmaier F., Smeitink J.A., Willems P.H. OXPHOS mutations and neurodegeneration. EMBO J. 2012;32:9–29. doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reinders J., Zahedi R.P., Pfanner N., Meisinger C., Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 3.Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzagoloff A., Myers A.M. Genetics of mitochondrial biogenesis. Annu. Rev. Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 5.Stuart R.A. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 2008;40:411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- 6.Osawa S., Ohama T., Jukes T.H., Watanabe K., Yokoyama S. Evolution of the mitochondrial genetic code. II. Reassignment of codon AUA from isoleucine to methionine. J. Mol. Evol. 1989;29:373–380. doi: 10.1007/BF02602907. [DOI] [PubMed] [Google Scholar]
- 7.Jukes T.H., Osawa S. The genetic code in mitochondria and chloroplasts. Experientia. 1990;46:1117–1126. doi: 10.1007/BF01936921. [DOI] [PubMed] [Google Scholar]
- 8.Towpik J. Regulation of mitochondrial translation in yeast. Cell. Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- 9.Christian B.E., Spremulli L.L. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonitz S.G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F.G., Nobrega M.P., Thalenfeld B.E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 1980;77:3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipinski K.A., Kaniak-Golik A., Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome–from genetics to evolution and systems biology. Biochim. Biophys. Acta. 2010;1797:1086–1098. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Rinehart J., Krett B., Rubio M.A., Alfonzo J.D., Soll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamenski P., Kolesnikova O., Jubenot V., Entelis N., Krasheninnikov I.A., Martin R.P., Tarassov I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol. Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 14.Tzagoloff A., Dieckmann C.L. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger K.H., Yaffe M.P. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 2000;8:508–513. doi: 10.1016/s0966-842x(00)01862-x. [DOI] [PubMed] [Google Scholar]
- 16.Contamine V., Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassett D.E. Jr, Boguski M.S., Hieter P. Yeast genes and human disease. Nature. 1996;379:589–590. doi: 10.1038/379589a0. [DOI] [PubMed] [Google Scholar]
- 18.Foury F. Human genetic diseases: a cross-talk between man and yeast. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 19.Barrientos A. Yeast models of human mitochondrial diseases. IUBMB life. 2003;55:83–95. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- 20.Smith M.G., Snyder M. Yeast as a model for human disease. Curr. Protoc. Hum. Genet. 2006 doi: 10.1002/0471142905.hg1506s48. Chapter 15, Unit 15.16. [DOI] [PubMed] [Google Scholar]
- 21.Rinaldi T., Dallabona C., Ferrero I., Frontali L., Bolotin-Fukuhara M. Mitochondrial diseases and the role of the yeast models. FEMS Yeast Res. 2010;10:1006–1022. doi: 10.1111/j.1567-1364.2010.00685.x. [DOI] [PubMed] [Google Scholar]
- 22.Perocchi F., Jensen L.J., Gagneur J., Ahting U., von Mering C., Bork P., Prokisch H., Steinmetz L.M. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2006;2:e170. doi: 10.1371/journal.pgen.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elstner M., Andreoli C., Klopstock T., Meitinger T., Prokisch H. The mitochondrial proteome database: MitoP2. Methods Enzymol. 2009;457:3–20. doi: 10.1016/S0076-6879(09)05001-0. [DOI] [PubMed] [Google Scholar]
- 24.Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L., Smertenko T., Alston C.L., Neeve V.C., Best A., et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014;312:68–77. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever G., Nislow C. The Yeast Deletion Collection: A Decade of Functional Genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz L.M., Scharfe C., Deutschbauer A.M., Mokranjac D., Herman Z.S., Jones T., Chu A.M., Giaever G., Prokisch H., Oefner P.J., et al. Systematic screen for human disease genes in yeast. Nat. Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 27.Dimmer K.S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luban C., Beutel M., Stahl U., Schmidt U. Systematic screening of nuclear encoded proteins involved in the splicing metabolism of group II introns in yeast mitochondria. Gene. 2005;354:72–79. doi: 10.1016/j.gene.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Merz S., Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess D.C., Myers C.L., Huttenhower C., Hibbs M.A., Hayes A.P., Paw J., Clore J.J., Mendoza R.M., Luis B.S., Nislow C., et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livnat-Levanon N., Glickman M.H. Ubiquitin-proteasome system and mitochondria – reciprocity. Biochim. Biophys. Acta. 2011;1809:80–87. doi: 10.1016/j.bbagrm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Elstner M., Andreoli C., Ahting U., Tetko I., Klopstock T., Meitinger T., Prokisch H. MitoP2: an integrative tool for the analysis of the mitochondrial proteome. Mol. Biotechnol. 2008;40:306–315. doi: 10.1007/s12033-008-9100-5. [DOI] [PubMed] [Google Scholar]
- 33.Grandin N., Charbonneau M. Dbf2 is implicated in a Cbt1-dependent pathway following a shift from glucose to galactose or non-fermentable carbon sources in Saccharomyces cerevisiae. Mol. Gen. Genet. 1999;261:402–407. doi: 10.1007/s004380050981. [DOI] [PubMed] [Google Scholar]
- 34.Phillips J.D., Trumpower B.L. QCR9, the nuclear gene encoding a small subunit of the mitochondrial cytochrome bc1 complex, maps to the right arm of chromosome VII in Saccharomyces cerevisiae. Yeast. 1993;9:95–97. doi: 10.1002/yea.320090112. [DOI] [PubMed] [Google Scholar]
- 35.Yang M., Trumpower B.L. Deletion of QCR6, the gene encoding subunit six of the mitochondrial cytochrome bc1 complex, blocks maturation of cytochrome c1, and causes temperature-sensitive petite growth in Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:1270–1275. [PubMed] [Google Scholar]
- 36.Lefebvre-Legendre L., Vaillier J., Benabdelhak H., Velours J., Slonimski P.P., Rago J.P. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J. Biol. Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 37.Spinazzola A., Viscomi C., Fernandez-Vizarra E., Carrara F., D'Adamo P., Calvo S., Marsano R.M., Donnini C., Weiher H., Strisciuglio P., et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 38.Medina-Silva R., Barros M.P., Galhardo R.S., Netto L.E., Colepicolo P., Menck C.F. Heat stress promotes mitochondrial instability and oxidative responses in yeast deficient in thiazole biosynthesis. Res. Microbiol. 2006;157:275–281. doi: 10.1016/j.resmic.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Ruotolo R., Marchini G., Ottonello S. Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 2008;9:R67. doi: 10.1186/gb-2008-9-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baruffini E., Ferrero I., Foury F. In vivo analysis of mtDNA replication defects in yeast. Methods. 2010;51:426–436. doi: 10.1016/j.ymeth.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 41.Goffrini P., Ercolino T., Panizza E., Giache V., Cavone L., Chiarugi A., Dima V., Ferrero I., Mannelli M. Functional study in a yeast model of a novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) diagnosed in a patient affected by a glomus tumor. Hum. Mol. Genet. 2009;18:1860–1868. doi: 10.1093/hmg/ddp102. [DOI] [PubMed] [Google Scholar]
- 42.Barrientos A., Zambrano A., Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagunas R. Energy metabolism of Saccharomyces cerevisiae discrepancy between ATP balance and known metabolic functions. Biochim. Biophys. Acta. 1976;440:661–674. doi: 10.1016/0005-2728(76)90049-9. [DOI] [PubMed] [Google Scholar]
- 45.Issel-Tarver L., Christie K.R., Dolinski K., Andrada R., Balakrishnan R., Ball C.A., Binkley G., Dong S., Dwight S.S., Fisk D.G., et al. Saccharomyces Genome Database. Methods Enzymol. 2002;350:329–346. doi: 10.1016/s0076-6879(02)50972-1. [DOI] [PubMed] [Google Scholar]
- 46.Reinders J., Zahedi R.P., Pfanner N., Meisinger C., Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 47.Goehring A.S., Rivers D.M., Sprague G.F. Jr. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell. 2003;14:4329–4341. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakai Y., Nakai M., Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 49.Leidel S., Pedrioli P.G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 50.Schlieker C.D., Van der Veen A.G., Damon J.R., Spooner E., Ploegh H.L. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jablonowski D., Fichtner L., Stark M.J., Schaffrath R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell. 2004;15:1459–1469. doi: 10.1091/mbc.E03-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frohloff F., Fichtner L., Jablonowski D., Breunig K.D., Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B., Lu J., Byström A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang B., Johansson M.J., Byström A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 56.Noma A., Sakaguchi Y., Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansson M.J., Esberg A., Huang B., Björk G.R., Byström A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Vazquez J., Vargas-Perez I., Sanso M., Buhne K., Carmona M., Paulo E., Hermand D., Rodriguez-Gabriel M., Ayte J., Leidel S., et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013;9:e1003647. doi: 10.1371/journal.pgen.1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esberg A., Huang B., Johansson M.J., Byström A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 60.Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., Hermand D. Translational control of cell division by Elongator. Cell Rep. 2012;1:424–433. doi: 10.1016/j.celrep.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Björk G.R., Huang B., Persson O.P., Byström A.S. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zinshteyn B., Gilbert W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fairn G.D., Macdonald K., McMaster C.R. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J. Biol. Chem. 2007;282:4868–4874. doi: 10.1074/jbc.M610575200. [DOI] [PubMed] [Google Scholar]
- 65.Mazina O.M., Mazin A.V., Nakagawa T., Kolodner R.D., Kowalczykowski S.C. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- 66.Ruotolo R., Tosi F., Vernarecci S., Ballario P., Mai A., Filetici P., Ottonello S. Chemogenomic profiling of the cellular effects associated with histone H3 acetylation impairment by a quinoline-derived compound. Genomics. 2010;96:272–280. doi: 10.1016/j.ygeno.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Castrejon F., Gomez A., Sanz M., Duran A., Roncero C. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell. 2006;5:507–517. doi: 10.1128/EC.5.3.507-517.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamb T.M., Mitchell A.P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu W., Smith F.J. Jr, Subaran R., Mitchell A.P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell. 2004;15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garipler G., Dunn C.D. Defects associated with mitochondrial DNA damage can be mitigated by increased vacuolar pH in Saccharomyces cerevisiae. Genetics. 2013;194:285–290. doi: 10.1534/genetics.113.149708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baumgart E., Vanhorebeek I., Grabenbauer M., Borgers M., Declercq P.E., Fahimi H.D., Baes M. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse) Am. J. Pathol. 2001;159:1477–1494. doi: 10.1016/S0002-9440(10)62534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 73.Percudani R., Pavesi A., Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 74.Bode M., Longen S., Morgan B., Peleh V., Dick T.P., Bihlmaier K., Herrmann J.M. Inaccurately assembled cytochrome c oxidase can lead to oxidative stress-induced growth arrest. Antioxid. Redox Signal. 2013;18:1597–1612. doi: 10.1089/ars.2012.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanki T., Wang K., Baba M., Bartholomew C.R., Lynch-Day M.A., Du Z., Geng J., Mao K., Yang Z., Yen W.L., et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai H., Kauffman S., Naider F., Becker J.M. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics. 2006;172:1459–1476. doi: 10.1534/genetics.105.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burtner C.R., Murakami C.J., Olsen B., Kennedy B.K., Kaeberlein M. A genomic analysis of chronological longevity factors in budding yeast. Cell cycle. 2011;10:1385–1396. doi: 10.4161/cc.10.9.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai H., Hauser M., Naider F., Becker J.M. Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:1805–1813. doi: 10.1128/EC.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding L., Liu Y. Borrowing Nuclear DNA Helicases to Protect Mitochondrial DNA. Int. J. Mol. Sci. 2015;16:10870–10887. doi: 10.3390/ijms160510870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen C., Huang B., Eliasson M., Ryden P., Byström A.S. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7:e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallis D., Russell H.F., Muenke M. Review: Genetics of attention deficit/hyperactivity disorder. J. Pediatr. Psychol. 2008;33:1085–1099. doi: 10.1093/jpepsy/jsn049. [DOI] [PubMed] [Google Scholar]
- 84.Chen C., Tuck S., Byström A.S. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pal D.K., Greenberg D.A. In: Jasper's Basic Mechanisms of the Epilepsies. 4th edn Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. 2010. [Google Scholar]
- 86.Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S., et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 87.Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kirino Y., Suzuki T. Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA biol. 2005;2:41–44. doi: 10.4161/rna.2.2.1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


