Abstract
The 3′X domain of hepatitis C virus is a strongly conserved structure located at the 3′ terminus of the viral genomic RNA. This domain modulates the replication and translation processes of the virus in conjunction with an upstream 5BSL3.2 stem–loop, and contains a palindromic sequence that facilitates RNA dimerization. Based on nuclear magnetic resonance spectroscopy and gel electrophoresis, we report here that domain 3′X adopts a structure composed of two stem–loops, and not three hairpins or a mixture of folds, as previously proposed. This structure exposes unpaired terminal nucleotides after a double-helical stem and palindromic bases in an apical loop, favoring genomic RNA replication and self-association. At higher ionic strength the domain forms homodimers comprising an intermolecular duplex of 110 nucleotides. The 3′X sequences can alternatively form heterodimers with 5BSL3.2. This contact, reported to favor translation, likely involves local melting of one of the 3′X stem–loops.
INTRODUCTION
Hepatitis C virus (HCV) affects ∼170 million people and is one of the major causes of cirrhosis and liver cancers worldwide. The HCV genome comprises a positive sense, single-stranded RNA molecule coding for a single polyprotein of about 3000 amino acids. The open reading frame (ORF) is flanked by 5′ and 3′ untranslated regions (UTR) that are relatively well conserved and contain structured regions essential for replication, translation and infectivity. At the 5′ UTR, the internal ribosome entry site (IRES) is formed by ∼340 nucleotides (nt) arranged in two major secondary structure domains (Figure 1A). The IRES is involved in the initiation of the synthesis of the viral polyprotein through a mechanism that does not depend on the initiation factors normally used by most cellular mRNAs, and its three-dimensional structure has been extensively studied by nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography and cryo-electron microscopy methods (reviewed in (1,2)).
Figure 1.
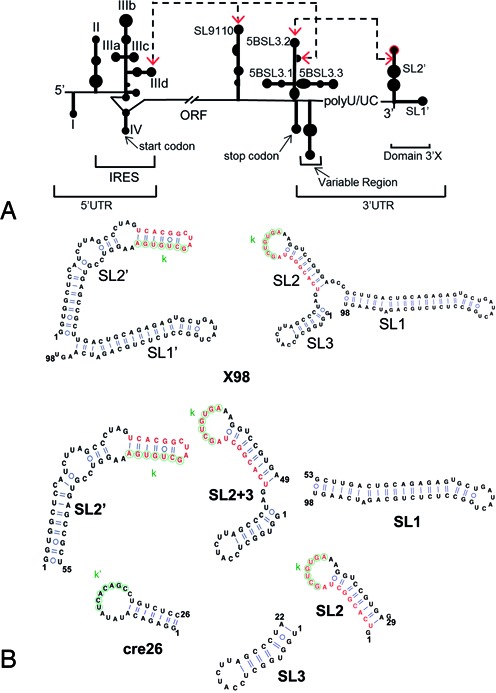
HCV RNA and 3′X terminal domain. (A) Schematic representation of functional cis-acting structures identified in the HCV genome. Distal RNA-RNA interactions proposed to have functional relevance are identified. (B) RNA sequences analysed in this study, together with their predicted secondary structures: X98 (full-length 98-nt 3′X domain), SL1 (extended SL1 subdomain), SL2’ (SL2’ subdomain; also identified as X55 in the literature), SL2+3 (SL2 and SL3 stem–loops), SL2 (SL2 hairpin), SL3 (SL3 hairpin), and cre26 (apical 5BSL3.2 hairpin). Two alternative conformations of the 3′X domain are indicated. These structures expose differently the overlapping functional sequences k (indicated with green circles) and DLS (red nt). Sequence k has been proposed to establish a distal kissing loop interaction with a complementary k’ sequence located in the apical loop of the 5BSL3.2 structure, and DLS is a 16-nt palindromic sequence involved in HCV RNA dimerization.
On the other side of the ORF, the 3′ UTR contains ∼200 nt and comprises a variable segment, a pyrimidine-rich tract and a strongly conserved 98-nt terminal domain called 3′X (3,4) (Figure 1A). Different reports have shown that the 3′X domain is essential for viral replication (5–9). Indeed, this region is recognized by the viral replication complex through an unknown mechanism to start synthesis of the negative polarity strand (10,11). Moreover, a distal interaction between the 3′X domain and a conserved stem–loop (termed 5BSL3.2, or alternatively SL9266) located in the nearby NS5B-coding region of the ORF influences the replication and translation processes (12–16). In turn, two alternative distal RNA-RNA interactions between an internal loop of 5BSL3.2 and upstream sequences in the ORF (SL9110) and the IRES (subdomain IIId) have also been proposed to modulate replication and translation (16–20) (Figure 1A). In addition, the 3′X region contains a 16-nt palindromic sequence (called DLS) that can mediate dimerization of two viral genomes in vitro (21–23) (Figure 1B and Supplementary Figure S1).
So far, enzymatic and chemical footprinting experiments have established that the last 43–46 nt of the 3′X domain form a stable stem–loop termed SL1 (also called SL9601). However, the secondary structure of the remaining 49–55 nt forming the 5′-half of the domain is less well defined. Most reports have concluded that they form two additional stem–loops (SL2 and SL3), but alternative folds comprising one (identified here as SL2’) or three hairpins, or mixtures of these structures, have also been proposed (18,22,24–26) (Figure 1B). This 49–55 nt segment contains the overlapping sequences k and DLS involved in the distal contact with the 5BSL3.2 apical loop and in genomic RNA dimerization, respectively, and is absolutely invariable among the different HCV genotypes (27). Since the different 3′X secondary structures expose to different degrees the k and DLS nt, several authors have proposed that the 3′X domain would act as a dynamic switch signaling the transition between the replication, translation and possibly genome packaging processes of the virus (16,18,20,23,26,28) (Figure 1B).
In contrast to the IRES and despite the functional relevance and high sequence conservation of the 3'X region, there are no structural analyses of the full-length 3'X domain carried out with high-resolution methods. This has been likely due to the conformational flexibility and dimerization propensity of this RNA sequence, which can complicate significantly this type of analyses. Here, we report a study of the secondary structure of the 3′X terminal RNA domain, as well as an examination of its interaction with the 5BSL3.2 stem–loop, based on NMR spectroscopy and gel electrophoresis. The results of these experiments indicate that, in the monomeric state, the preferred structure of the domain contains two stems (SL1’ and SL2’). According to the available functional evidence, this structure would promote viral replication and dimerization. Indeed, at higher ionic strength the domain forms symmetric dimers comprising an extended SL2’ duplex of 110 nt, flanked on both sides by intramolecular SL1’ hairpins. The full-length domain and other 3′X sequence constructs containing the k sequence were also able to form heterodimers with a 5BSL3.2 hairpin. However, we did not detect formation of individual SL2 and SL3 stem–loops by the domain in any of the conditions tested, and the SL2 hairpin was not clearly identified in constructs designed to favor these individual subdomains. The intermolecular interaction with 5BSL3.2 requires some disruption of the two-stem domain structure to unfold the k nt. Our results indicate that this alteration only occurs in the presence of this or other cofactors.
MATERIALS AND METHODS
Sequences
The sequences analysed in this study correspond to HCV genotype 1b and were obtained from vector pFK-I389FLUCNS3-3′ET, kindly provided by Dr R. Bartenschlager (University of Heidelberg, Germany).
Secondary structure predictions
The secondary structure predictions were carried out with the ViennaRNA (http://rna.tbi.univie.ac.at/) (29) and/or Mfold (http://mfold.rna.albany.edu) (30) web servers. The structures were drawn using VARNA (http://varna.lri.fr/) (31).
Preparation of RNA samples for NMR spectroscopy and gel electrophoresis experiments
The SL1 (46 nt), SL2 (29 nt), SL3 (22 nt), SL2+3 (49 nt), SL2’ (55 nt) and cre26 (26 nt) subdomain RNA sequences used for NMR and gel electrophoresis were prepared by T7-polymerase in vitro transcription using synthetic oligonucleotide DNA templates. Construct X98 (full-length 98-nt 3′X domain) was transcribed from a pUC19 plasmid containing a ScaI restriction site for linearization and run-off transcription. For the SL1, SL2+3, SL2’ and X98 sequences, we also generated 15N- and/or 13C/15N-labeled transcripts using NTPs obtained from CortecNet. All constructs were purified on denaturing gels containing 20% acrylamide and 8 M urea. After electroelution from the gel, the RNAs were ethanol-precipitated two times and desalted with Sephadex G-25 cartridges. The full-length sequence X98 was also purified by FPLC using a 26/20 anion-exchange column and a NaCl gradient (32), followed by concentration in a diafiltration device. Prior to NMR or gel electrophoresis experiments, all samples were microdialyzed in aqueous solutions containing 10 mM sodium phosphate (pH 6.0) and 0.1 mM EDTA with no added salts, or additionally containing either 100 mM NaCl or 1–5 mM MgCl2. Before each NMR and gel electrophoresis experiment, the RNA samples were heated at 95°C for 5 min and immediately placed on ice for 30 min (for low salt conditions) or cooled down slowly (higher salt conditions), unless otherwise specified.
NMR spectroscopy
The RNA concentration of the NMR samples ranged between 34 and 240 μM. NMR spectra were acquired on 600 MHz (cryoprobe-equipped) and 500 MHz Bruker Avance III spectrometers, and analysed using Topspin 1.3 (Bruker Biospin) and Sparky 3.110 (T.D. Goddard, D.G. Kneller, UCSF USA, 2004). The unlabeled systems were studied using 2D watergate-NOESY (with 150 ms mixing time) and watergate-TOCSY experiments (60 ms mixing time) recorded in 90% H2O/10% D2O at two temperatures (typically 16 and 27°C). TOCSY and NOESY (250 ms) experiments were also acquired in D2O at 27°C. The recycle delays were 1.6 and 2 s for all homonuclear TOCSY and NOESY experiments, respectively.
The 15N-labeled and/or 13C/15N-labeled samples were analysed with two-dimensional 1H-15N HSQC experiments recorded in H2O at 16 and 27°C, as well as HNN COSY experiments allowing detection of hydrogen bonds between bases via two bond N–N couplings (33). For HSQC experiments we acquired 1024 and 256 complex points in the direct and indirect dimensions, respectively, and 64 scans for each indirect experiment. For HNN-COSY experiments, the delay for evolution of the 2JNN coupling was set to 15 ms, and we collected 2048 and 128 complex points in the t2 and t1 dimensions, respectively, with 320 scans for each t1 increment. The recycle delays ranged between 1.0 and 1.3 s for experiments with labeled samples.
Assignments and secondary structure determination
The assignment of subdomain constructs SL1 and SL2’ involved imino and amino signals as well as non-exchangeable aromatic and H1’ protons, and was based on standard analyses (34) of NOESY and TOCSY spectra acquired in H2O and D2O at 16 and 27°C. These assignments allowed the identification of the base-paired stems contained in each structure. Construct SL2+3 exhibited broader NOESY and TOCSY signals and we could only generate tentative assignments of its exchangeable resonances, aided by comparisons with the exchangeable spectra of two smaller SL2 and SL3 constructs (Figure 1B). Determination of the secondary structure of the full-length domain was based on comparisons of the HSQC and NOESY data of X98 with those of subdomains SL1, SL2+3 and SL2’ acquired under identical conditions.
Determination of monomer and dimer formation as a function of ionic strength
This analysis was performed by comparing HNN-COSY spectra obtained with 100% 15N-labeled samples and with mixtures of 50% 15N-labeled and 50% unlabeled sequences (35,36). In conditions that favor the formation of monomeric hairpins, the relative intensity of diagonal peaks and crosspeaks is the same in 100%- and 50%-labeled samples. In contrast, the relative intensities of the crosspeaks generated by dimeric species involving intermolecular base pairs diminish, since most of the HNN signals arise from dimers composed of labeled and unlabeled strands (35,36). In this series of experiments we kept the total RNA concentration constant and used the same acquisition parameters. At higher ionic strength the number of scans per t1 increment was increased from 320 to 368 or 592 to improve the signal-to-noise ratio, since these conditions favored dimeric species with significantly higher molecular weight.
Gel electrophoresis experiments
Native gels were run at 4°C for 24 h under constant voltage (80 V). We used 20% 19:1 acrylamide:bisacrylamide gels, and either 89 mM tris-borate (TB), or 89 mM tris-borate and 1–5 mM MgCl2 (TBM) as running buffers. These experiments involved 10–20 μM RNA samples, prepared as specified above. All gels were stained with methylene blue and destained with water.
RESULTS
Modular strategy for NMR analyses
Initial gel electrophoresis analyses confirmed that the full-length 3′X domain sequence (hereafter identified as X98) was structurally complex and could dimerize in higher ionic strength conditions (Figure 2 and Supplementary Figure S2). To facilitate the analysis by NMR spectroscopy of this 98-nt sequence we followed a modular approach (37), and studied several smaller RNA constructs first. These included a 46-nt sequence (SL1) containing the extended SL1 subdomain present in the three-stem domain conformation (Figure 1B, right), and two sequences comprising the 5′-half of the domain: 49-nt construct SL2+3, which was predicted to form the two hairpin (SL2 and SL3) structure included in the three-stem 3′X fold, and 55-nt construct SL2’, predicted to adopt the extended stem–loop structure (SL2’) contained in the two-stem domain fold (Figure 1B). Like X98, SL2+3 and SL2’ comprised the overlapping k and DLS sequences involved in the distal interaction with the 5BSL3.2 loop and viral RNA dimerization, respectively, but the exposure of these sequences is very different in the structures presumably formed by each construct. In the extended SL2’ hairpin, k is almost completely buried in the upper double-helical stem, whereas DLS nt occupy the apical loop and would be available to initiate dimerization through the formation of a kissing loop with another SL2’ hairpin. In contrast, the two-stem structure predicted for SL2+3 would expose the k sequence in the apical loop of the SL2 hairpin, and initiation of dimer formation would imply the disruption of the SL2 stem (Figure 1B).
Figure 2.
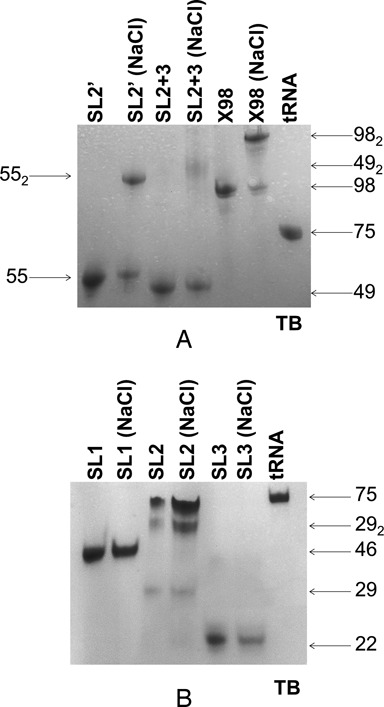
Gel electrophoresis analysis of 3′X domain sequences. (A) Constructs SL2’, SL2+3 and X98 (full-length 3′X domain); (B) constructs SL1, SL2 and SL3. In (A) and (B), the experiments compare the mobility in native conditions of 3′X domain sequences previously folded in the absence or presence of 100 mM NaCl, relative to a tRNA control. Conditions: 20 μM RNA, TB running buffer.
Structure of subdomain SL1
NMR analyses of construct SL1 (Figure 3A), together with gel electrophoresis assays in different ionic conditions (Figure 2B and Supplementary Figure S2B), confirmed that this sequence adopted a single, monomeric stem–loop structure in solution. The NMR 1H–15N HSQC experiments (Figure 3A) were particularly useful to determine the secondary structure of SL1 and other 3′X sequences, since each HN crosspeak indicated the presence of a G or U nt involved in a base pair, and the crosspeak frequencies depended on the nature of the base pairs and their neighbors. The nt giving rise to each crosspeak were assigned with the support NOESY and TOCSY spectra (Supplementary Figure S3) as explained in Materials and Methods. This methodology allowed us to deduce that hairpin SL1 contained three double-helical segments separated by two single-nt bulges (U69 and A92), and was closed by a U72GAUACUG79 apical octaloop comprising a wobble U72:G79 pair at its base (Figure 3B). The hairpin included a terminal G53CU55:A96GU98 double-helical segment involving the last A96, G97 and U98 nt of the HCV genome. These three base pairs are formed in the extended SL1 hairpin contained in the three-stem 3′X domain structure, but absent in the shortened SL1’ subdomain present in the two-stem domain fold (Figure 1B).
Figure 3.
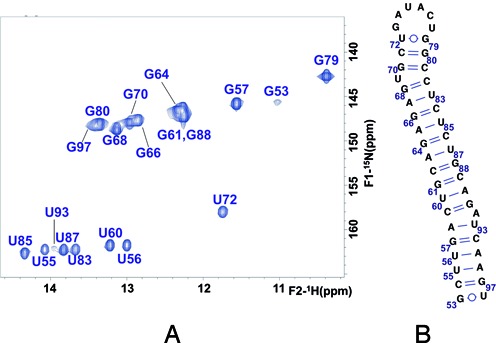
NMR spectroscopy study of subdomain SL1. (A) Assignment of the 1H-15N HSQC spectrum of construct SL1 based on NOESY and TOCSY analyses in H2O and D2O. Conditions: 72 μM SL1, 0 mM NaCl/MgCl2, 27°C. (B) Secondary structure of the extended SL1 subdomain supported by the NMR data.
Structure of the 5′ half of the domain
Sequences SL2+3 and SL2’ contained the 16-nt palindromic DLS tract (Figure 1B) and thus were anticipated to form dimeric species. This was confirmed by gel electrophoresis experiments: in the presence of 100 mM NaCl or 1 mM MgCl2, both constructs formed dimers in addition to monomers, and the proportion of dimers was greater for SL2’ (Figure 2A and Supplementary Figure S2A). In the absence of added salts, however, SL2+3 and SL2’ were monomeric (Figure 2A).
HSQC, HNN-COSY, NOESY and TOCSY analyses of SL2’ in low ionic strength conditions (Figure 4 and Supplementary Figure S4) indicated the formation of a monomeric SL2’ hairpin structure involving all 55 nt and closed by an apical tetraloop containing the palindromic DLS tract C29UAG32 (Figure 4C). This was confirmed by an analysis of HNN-COSY spectra obtained with a 100% 15N-labeled sample and with a mixture of 50% unlabeled and 50% 15N-labeled sequences (35,36), which demonstrated that in these NMR conditions, SL2’ was monomeric (Figure 4B). The NMR data clearly supported the presence of the lower G2UGGCUC8:G48AGCCGC54 and upper C24ACGG28:C33UGUG37 double-helical segments of the structure, each exhibiting diagnostic NOE connectivity patterns involving an A:U and a G:U pair in both cases (Supplementary Figure S4). The base pairs adjacent to the two predicted 6:4 and 4:2 internal loops were not detected or gave rise to significantly weaker signals, suggesting that this region of the SL2’ hairpin was likely dynamic.
Figure 4.
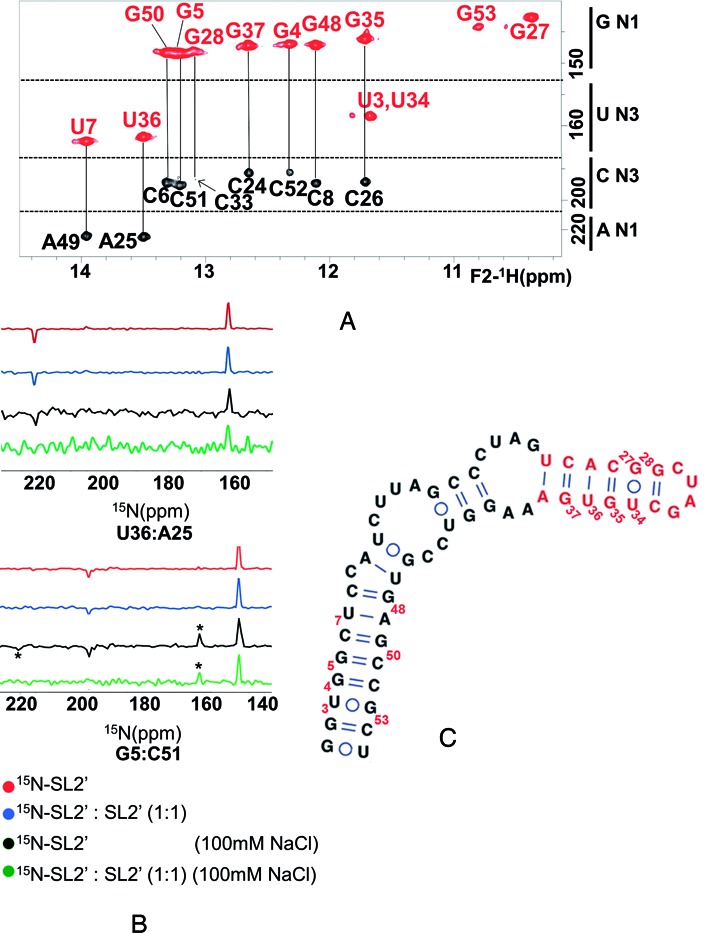
NMR analysis of subdomain SL2’ and impact of ionic strength on monomer and dimer formation. (A) Assignment of an HNN-COSY spectrum of construct SL2’ acquired at low ionic strength, based on NOESY and TOCSY analyses in H2O and D2O. Conditions: 100 μM SL2’, 0 mM NaCl/MgCl2, 27°C. (B) One-dimensional slices along the 15N axis of HNN-COSY spectra of SL2’, comparing the diagonal and crosspeak intensities in fully labeled, 0 mM NaCl (red); 50% labeled and 50% unlabeled, 0 mM NaCl (blue); fully labeled, 100 mM NaCl (black), and 50% labeled and 50% unlabeled, 100 mM NaCl (green) conditions. The data correspond to base pairs U36:A25 and G5:C51, located in the upper and lower stems of SL2’, respectively. In all cases, the experiments were acquired at 27°C with a total SL2’ concentration of 100 μM, and the spectra were normalized with respect to the diagonal peak height. The crosspeaks marked with asterisks and detected in the presence of NaCl correspond to the diagnostic U30:A31 pair of the SL2’ dimer. (C) Secondary structure of the SL2’ monomer supported by the NMR results.
In the presence of 100 mM NaCl, the pattern of crosspeaks in the 1H-15N HSQC spectrum of SL2’ did not undergo significant changes relative to that detected in the absence of added salts, although two new HN crosspeaks with chemical shifts and NOE connectivities typical of Watson–Crick G:C and A:U iminos appeared (Figures 4B and 5A). Examination of HNN-COSY spectra acquired in these conditions with a 100% 15N-labeled sample and with a mixture of 50% unlabeled and 50% 15N-labeled sequences indicated the formation of a dimer species (Figure 4B). The NOE connectivity patterns remained mostly unchanged, confirming the conservation of the base-paired stems described above. Altogether, these observations implied that in the presence of 100 mM NaCl, SL2’ formed the extended 110-nt symmetric dimer shown in Figure 5B. The secondary structure of the SL2’ dimer is similar to that of the monomer, with the exception of the continuous 16-base pair duplex present in the middle of the dimer and formed by the self-complementary DLS nt. This explains the similarity of the HSQC and NOESY spectra at lower and higher ionic strength, and the appearance of the two extra HN crosspeaks in the presence of NaCl, indicating the formation of the new U30:A31 and G32:C29 pairs of the DLS duplex. These bases were located in the hairpin tetraloop and were thus undetectable in the monomeric form (Figures 4 and 5).
Figure 5.
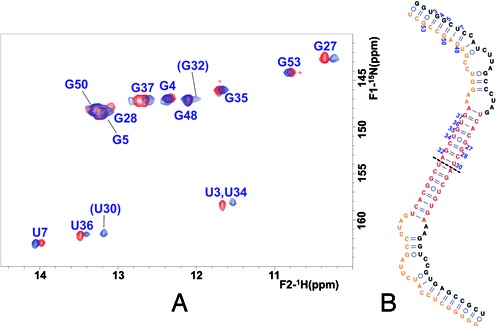
Secondary structure of the SL2’ dimer detected at higher ionic strength. (A) Superposition of 1H–15N HSQC spectra of SL2’ acquired in the absence (red) and presence (blue) of 100 mM NaCl. The two HN imino crosspeaks detected at 100 mM NaCl but absent at lower ionic strength that were assigned to U30 and G32 are indicated in parentheses. Conditions: 61 μM SL2’, 27°C. (B) NMR-supported structure of the SL2’ dimer. The NMR signals of each of the two symmetrical halves of the dimer (separated by a dashed line) are equivalent.
Unlike SL2’, construct SL2+3 was predicted to favor the two-hairpin structure present in the three-stem domain fold (Figure 1B, right). In the absence of added salts, the HSQC spectrum of SL2+3 was different relative to that of SL2’ in the same conditions (Supplementary Figure S5A), confirming that SL2+3 adopted a dissimilar conformation. This was also supported by the slower electrophoretic mobility of SL2+3 relative to the extended stem–loop formed by SL2’ (Figure 2A). Broad signals in the NOESY and TOCSY spectra of SL2+3 meant that we had to rely on two smaller SL2 and SL3 hairpin constructs (Figure 1B) to generate tentative assignments. Comparison of the SL2+3, SL2 and SL3 spectra (Supplementary Figure S5B) indicated that while SL2+3 and SL3 likely formed the SL3 hairpin, the SL2 nt were dynamic in the monomeric SL2 and SL2+3 contexts. Upon the addition of NaCl, significant changes were observed in the spectra of SL2 and SL2+3, and the SL2+3 HSQC became partly similar to that of the SL2’ dimer (Supplementary Figures S5C and S5D). This suggested that SL2+3 underwent a significant base-pair reorganization to form the dimeric species, supporting the conclusion that the monomeric form of this sequence likely adopts an SL2- and SL3-containing conformation where the DLS nt are not self-paired (Supplementary Figure S5E). This was likewise consistent with the reduced dimerization propensity of SL2+3 relative to SL2’ observed in electrophoretic experiments (Figure 2A and Supplementary Figure S2A).
Solution structure of the full-length 3′X domain
As observed for SL2+3 and SL2’, gel electrophoresis experiments indicated that X98 formed a mixture of monomeric and dimeric structures in the presence of 100 mM NaCl or 1 mM MgCl2 (Figure 2A and Supplementary Figure S2A), but adopted a monomeric conformation at low ionic strength (Figure 2A). Armed with the information obtained from the SL1, SL2+3 and SL2’ subdomain constructs, we undertook the NMR study of the full-length domain. Under low ionic strength conditions and at low RNA concentration (34 μM), the HSQC data indicated that X98 adopted a single, well defined conformation (Figure 6A). The electrophoretic mobility of X98, together with the HNN-COSY results previously obtained with construct SL2’, indicated that in these conditions the observed conformation was most likely monomeric. Moreover, comparison of the HSQC spectrum of X98 with those of SL1, SL2+3 and SL2’ acquired under the same conditions immediately established that the structure adopted by the 3′X sequence corresponded to the two-stem domain conformation (Figure 6). First, there was an excellent match with the crosspeaks of construct SL2’ but no match with the SL2+3 crosspeaks (Figure 6A and Supplementary Figure S5A). Second, the 3′X domain crosspeaks also had an excellent correspondence with those of construct SL1 with the exception of the HN imino crosspeaks corresponding to the terminal G53:U98, C54:G97 and U55:A96 pairs, which were not detected (Figure 6A). These base pairs are present in the extended SL1 stem–loop formed by the three-stem domain structure, but absent in the shortened SL1’ subdomain of the two-stem domain fold, where G53, C54 and U55 are paired at the base of the SL2’ stem, and the terminal A96, G97 and U98 nt form an unpaired tail (Figures 1B and 6). Taking into account these exceptions, the NOE crosspeak pattern of X98 was also similar to those of SL1 and SL2’. Thus, the NMR data clearly indicated that X98 adopted the two-stem domain structure. We did not detect any evidence supporting the presence of the three-stem fold, or of a mixture of secondary structures.
Figure 6.
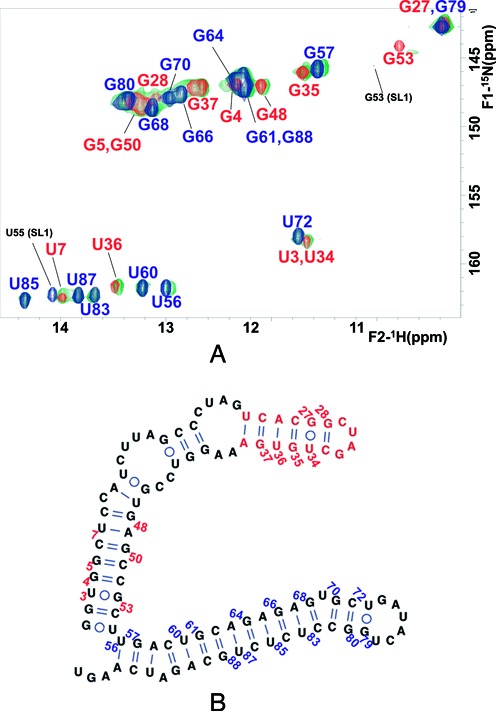
NMR analysis of full-length 3′X domain of HCV. (A) 1H–15N HSQC spectrum of X98 at low ionic strength (green), superposed with those of the SL1 (blue) and SL2’ (red) constructs acquired under the same temperature and ionic conditions. The assignments appear in blue and red for crosspeaks of nt belonging to subdomains SL1’ and SL2’, respectively. The SL1 G53 and U55 HN crosspeaks that do not match 3′X signals are marked with black assignments. Conditions: 34 μM X98, 0 mM NaCl/MgCl2, 27°C. (B) Secondary structure of the 3′X domain monomer supported by the NMR data.
In the presence of 100 mM NaCl, the HSQC crosspeaks of X98 broadened, suggesting the formation of a larger species. However, as observed for SL2’ (Figure 5A) the patterns of HSQC and NOESY crosspeaks did not undergo significant changes with respect to those detected in low-salt conditions (Supplementary Figure S6A). Moreover, there was an excellent chemical shift match relative to the HSQC crosspeaks of SL1 and SL2’ detected in the same ionic conditions (data not shown). In addition, two additional HN imino crosspeaks were detected, matching those assigned to the G32:C29 and U30:A31 pairs in the middle of the SL2’ DLS duplex. Altogether, these observations indicated that in these conditions the 3′X domain formed the symmetric dimer structure shown in Supplementary Figure S6B. The structure of the 3′X dimer is similar to that of the SL2’ dimer, but contains two SL1’ stem–loops on both sides, each containing an unpaired A96GU98 3′ terminal tail.
Effect of magnesium
The Mg2+ ions stabilize RNA tertiary structure to a higher degree than monovalent ions (see e.g. (38,39)). If the three-stem domain fold contained more tertiary interactions than the two-stem domain fold, the presence of Mg2+ would stabilize that structure, which was not observed at low ionic strength or with 100 mM NaCl. We thus evaluated the effect of adding different MgCl2 concentrations on the NMR spectra of constructs SL1, SL2’, SL2+3 and X98, using folding conditions that favored monomeric species (Supplementary Figure S7). Upon addition of Mg2+, weak additional peaks with unusual chemical shifts appeared in the HSQC spectra of all constructs, as well as DLS U30 and G32 crosspeaks indicating some dimer formation by SL2’, SL2+3 and X98 (Supplementary Figure S7A). However, these conditions failed to modify the main HN crosspeak pattern detected in each system, and the Mg2+-induced crosspeaks of the full-length domain matched the Mg2+-induced crosspeaks of SL1 and SL2’, but did not match those of construct SL2+3 (Supplementary Figure S7B). This confirmed that Mg2+ did not alter the secondary structure adopted by the full-length domain or the subdomain constructs. Rather, the spectral changes observed in each system probably corresponded to the stabilization of local tertiary structure (likely involving bases within or near loops), or to changes in the global three-dimensional shape of each secondary structure species.
Interaction with the 5BSL3.2 loop
The seven-nt k sequence of domain 3′X overlaps with the DLS and has been shown to establish a distal interaction with a complementary k’ sequence located in the apical loop of stem–loop 5BSL3.2 (12–16,20,28). It has been proposed that this distal interaction involves the three-stem fold of domain 3′X and particularly hairpin SL2, since in this context the k bases are exposed in the apical loop of SL2 and available to form kissing loop pairs with the k’ nt (12–14,20,28) (Figure 1B).
Hairpin cre26 encompasses the apical loop of 5BSL3.2 containing sequence k’ (Figure 1B). We first used gel electrophoresis experiments to assess the interaction between this system and all 3′X constructs containing the k sequence. In the presence of 1 mM MgCl2 and under folding conditions favoring monomeric species, all SL2+3, SL2’ and X98 constructs formed heterodimers with cre26 (Figure 7A). This was in agreement with a previous report, showing that the interaction between SL2’ and a 5BSL3.2 stem–loop required Mg2+ ions (20).
Figure 7.
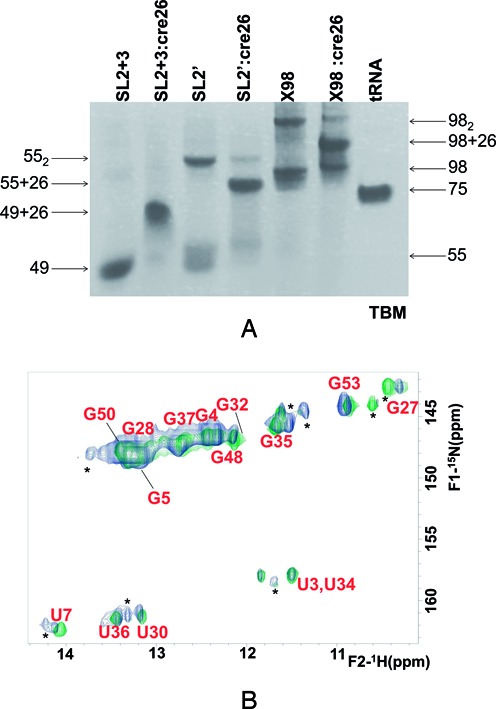
Electrophoretic and spectroscopic analysis of the distal interaction formed between domain 3′X and 5BSL3.2 hairpin cre26. (A) Native TBM gel comparing the electrophoretic mobility of constructs SL2+3, SL2’ and X98 in the absence and presence of one molar equivalent of cre26, relative to a tRNA control. In all cases, the samples were snap-cooled in the absence of NaCl or MgCl2, and then incubated with 1 mM MgCl2 for 150 minutes at 25°C, with or without cre26. Conditions: 10–20 μM RNA, TBM running buffer (1 mM MgCl2). (B) Superposition of 1H-15N HSQC spectra of SL2’, acquired in the absence (green) and presence (blue) of one molar equivalent of an unlabeled cre26 sequence. In both cases, construct SL2’ was snap-cooled in the absence of NaCl or MgCl2 and then incubated with 2 mM MgCl2 for 150 minutes at 25°C, with or without cre26. Since the experiment used unlabeled cre26, all of the HN crosspeaks correspond to SL2’. Conditions: 49 μM RNA, 27°C.
We then utilized NMR spectroscopy to monitor the interaction between an 15N-labeled SL2’ construct and cre26 in conditions similar to those used in the electrophoresis assays (Figure 7B). In the presence of cre26, weak additional crosspeaks appeared in the HSQC spectrum of SL2’, and the U7 and U36 HN signals broadened relative to the Mg2+-containing HSQC spectrum of the isolated construct. These observations confirmed the establishment of the SL2’:cre26 intermolecular interaction. However, the main crosspeaks detected in the SL2’:cre26 mixture matched those detected in the Mg2+-containing isolated SL2’ sample (Figure 7B), and did not correspond with the HSQC signals of construct SL2+3.
DISCUSSION
The strongly conserved (27,40) 3′X tail of HCV RNA has been shown to play key roles in the life cycle of the virus. In this report, we have used NMR and gel electrophoresis experiments to study the solution structure of the 3′X domain and to monitor its interaction with the 5BSL3.2 stem–loop. The 3′X RNA is structurally complex, since it contains a 16-nt palindromic DLS sequence promoting dimerization, and can potentially adopt alternative secondary structures that expose two functional sequences differently. Our analysis was facilitated by the separate study of three smaller SL1, SL2+3 and SL2’ constructs, the first two forming the subdomains present in the three-stem domain fold, and the last one adopting the extended SL2’ stem–loop structure formed by the 5′ half of the domain in the alternative, two-stem domain fold (Figure 1B).
NMR spectroscopy and gel electrophoresis experiments indicated that at low ionic strength and low RNA concentration, the full-length domain sequence X98 formed a single monomeric structure comprising two stem–loops, SL2’ and SL1’. This was revealed by the excellent match with the HSQC spectra of constructs SL1 and SL2’ acquired under identical ionic conditions (Figure 6). The absence of correspondence with SL2+3 signals and with the crosspeaks of the three terminal pairs of SL1, further supported the conclusion that X98 adopted the two-stem fold including an extended SL2’ hairpin as well as a shortened SL1’ subdomain where those base pairs are not formed (Figure 6).
In the presence of 100 mM NaCl, SL2’ and X98 formed dimeric structures (Figure 5 and Supplementary Figure S6). This was demonstrated by gel electrophoresis experiments (Figure 2A), and by HNN-COSY analyses of SL2’ in the absence and presence of salt (Figure 4B). The SL2’ dimer was symmetric and comprised a 16-nt self-complementary DLS duplex (Figure 5). The full-length domain dimer was analogous, but included two SL1’ intramolecular hairpins on each side (Supplementary Figure S6). In terms of secondary structure, the SL2’ and X98 dimers were very similar to the corresponding monomers (Figures 4 and 6). SL2’ and X98 dimer formation was also indicated by the detection at 100 mM NaCl of two diagnostic HN crosspeaks indicating the formation of two symmetric couples of A31:U30 and G32:C29 pairs in the middle of the self-complementary DLS duplex (Figure 5 and Supplementary Figure S6). These pairs are formed by nt that were located in the SL2' hairpin tetraloop and were thus undetectable in the monomeric forms. In this regard, the formation of SL2’ dimers has been shown to involve a kissing intermediate (23), likely formed by two palindromic C29UAG32 SL2’ apical loops.
Many reports have proposed that the 3′X domain of HCV adopts a conformation containing three separate stems (SL1, SL2 and SL3), where the k sequence is exposed in the apical loop of SL2 and can establish a distal interaction with 5BSL3.2 (12,14,16,18,22,24–26). However, we have not obtained NMR evidence supporting the adoption of the three-stem structure by the isolated 3′X domain. Construct SL2+3 was predicted to favor the two-hairpin (SL2 and SL3) fold present in the three-stem domain (Figure 1B, right), and exhibited broader NMR signals and significantly different HSQC and NOESY crosspeak patterns relative to SL2’. Addition of NaCl induced significant changes in the HSQC data of this construct, which became partly similar to that of the SL2’ dimer. Together with spectral comparisons with SL2 and SL3 hairpin constructs, these observations supported the conclusion that the monomeric form of SL2+3 adopted an SL3- and SL2-containing conformation (Supplementary Figure S5). The NMR spectra of the full-length domain X98 and construct SL2’, however, did not match those of construct SL2+3 at low ionic strength or in the presence of 100 mM NaCl (Supplementary Figure S5). Likewise, addition of Mg2+ ions failed to promote secondary structure changes in X98 and SL2’ (Supplementary Figure S7).
A weaker band was observed immediately below the main monomer bands of constructs SL2’ and X98 in gel electrophoresis experiments (Figure 7A). The weaker band below the main band of construct SL2’ has been proposed to correspond to a structure containing isolated SL2 and SL3 subdomains (20). However, the electrophoretic bands generated by secondary structures containing additional stems would be expected to appear above the main monomer bands (41), and not below them as experimentally observed. A more plausible explanation would be that the minor monomeric bands are not related to the presence of additional stems, but instead result from a different tertiary conformation of the same secondary structure. This possibility is supported by the spectral changes detected in all constructs upon addition of Mg2+ (Supplementary Figure S7).
The 7-nt sequence k of domain 3′X has been proposed to pair with a complementary k’ sequence located in the apical loop of 5BSL3.2 (12,14–16,20,28). In the secondary structures detected by NMR for SL2’ and X98, the k nt are hidden in the upper base-paired stem of SL2’ (Figures 1B, 4 and 6). In spite of this fact, gel electrophoresis experiments revealed that all SL2+3, SL2’ and X98 constructs containing the k sequence were able to interact with 5BSL3.2 hairpin cre26 in the presence of Mg2+ (Figure 7A). We examined by NMR spectroscopy a mixture of SL2’ and cre26 in the conditions used for electrophoresis. New weak crosspeaks appeared, and other crosspeaks broadened relative to the Mg2+-containing HSQC spectrum of the isolated SL2’ construct, confirming the existence of the intermolecular interaction (Figure 7B). Several authors have proposed that the k and k’ nt establish a kissing-loop interaction necessarily involving hairpin SL2 (12–14,20,28). However, we did not obtain evidence supporting the formation of subdomains SL2 and SL3 in these conditions. In fact, formation of SL2 and SL3 by construct SL2’ is unlikely, as it would imply disruption of five base pairs in the lower SL2’ stem to leave a 6-nt unpaired tail (see Figure 1B). Moreover, two different groups detected increased exposure of nt presumably forming the double-helical stem of SL2 upon 5BSL3.2 complex formation (16,18), disfavoring the hypothesis that SL2 is needed for that interaction. These changes, however, would be consistent with the disruption of the upper stem of SL2’, allowing the k bases to pair with k’ nt. We also noted that the SL2 nt were flexible in the NMR spectra of monomeric SL2+3 and SL2 species designed to favor the formation of this hairpin (Supplementary Figure S5), and that a wild-type SL2 construct had to be stabilized by mutagenesis to simplify the SPR sensorgrams detected upon interaction with the 5BSL3.2 loop (28). A previous NMR study failed to detect the contact between a similar SL2 construct and 5BSL3.2 (12), but this observation could have been due to the homodimerization avidity of this sequence and/or to the absence of Mg2+ in those experiments.
In conclusion, the preferred secondary structure of the 3′X domain of HCV comprises two stem–loops (SL2’ and SL1’), and hairpins SL3 and SL2 are not formed in the absence of cofactors. The preferred 3′X structure contains three unpaired nucleotides at the 3′ end of the SL1’ stem, being ideally suited for de novo initiation of RNA synthesis by the viral NS5B polymerase (11) (Figure 8A). This structure is thus likely involved in replication, and probably promotes, with sufficient viral RNA concentration, dimerization and packaging of viral genomes (20,23) through the formation of SL2’ duplexes (Figure 8B). The 5BSL3.2 stem–loop has been proposed to engage the 3′X k sequence through a distal contact, forming a ‘closed’ viral RNA configuration that facilitates translation (15), and we have detected this interaction using NMR spectroscopy (Figure 8C). This contact necessarily entails changes in the 3′X SL2’ stem–loop, allowing sequence k to pair with k’. Our results indicate that this interaction does not occur by selecting one of two stable 3′X structures present in solution, i.e. the 3′X tail does not function as a riboswitch in this manner. Rather, the 3′X structure permitting the contact with 5BSL3.2 only forms in the presence of this last cofactor. Complex formation likely involves intermediate steps that may include local melting of the upper stem of SL2’, (probably aided by the observed flexibility of the SL2’ internal loops), and might conclude with an extended SL1 stem where the tailing nucleotides at the 3′-terminus are base-paired and thus disfavor replication. Further work is in progress to study the structural details of this interaction as well as the three-dimensional shape of the 3′X domain.
Figure 8.
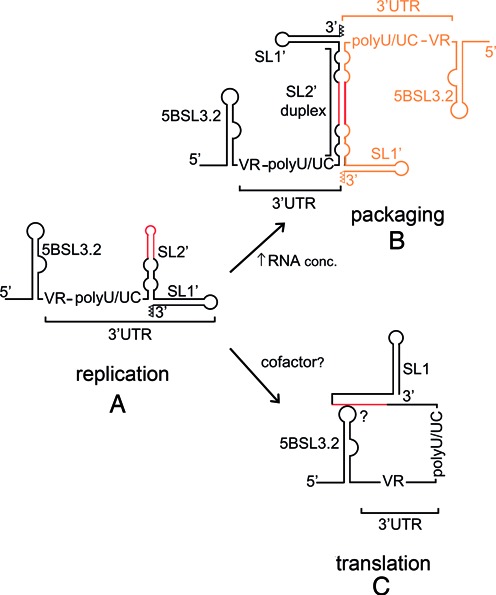
Scheme summarizing the structural features of domain 3′X supported by this study, and their possible impact on the HCV life cycle. (A) Preferred structure of the domain monomers, comprising stem–loops SL2’ and SL1’. This structure contains unpaired nt (represented as open triangles) at the 3′ terminus of stem SL1’ and promotes viral RNA replication. (B) With sufficient RNA concentration domain 3′X forms homodimers comprising an extended 110-nt SL2’ duplex flanked by intramolecular SL1’ hairpins. These structures probably mediate self-association and packaging of viral genomes. (C) The 3′X RNA can interact with an upstream 5BSL3.2 stem–loop. This contact, reported to favor translation, involves local melting of the upper stem of SL2’ and may be aided by protein cofactors. In (A), (B) and (C), the DLS tract is colored red; in (B), the second HCV genome is represented in orange.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr C. Richter for help with the acquisition of SL1 NMR data at BMRZ (Frankfurt), and Dr S. Prado and Prof. R. Flores for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitividad of Spain [BFU-2012-30770 to J.G.]; Universidad Católica de Valencia of Spain [2011-011-006 and 2012-011–-03 to J.G.]; [predoctoral contract 2012–030-001 assigned to A.C-C]; European Union [FP7 Bio-NMR 261863]. Funding for open access charge: Ministerio de Economía y Competitividad of Spain [BFU-2012-30770].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lukavsky P.J. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piñeiro D., Martinez-Salas E. RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis. Viruses. 2012;4:2233–2250. doi: 10.3390/v4102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolykhalov A.A., Feinstone S.M., Rice C.M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka T., Kato N., Cho M.J., Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 5.Kolykhalov A.A., Mihalik K., Feinstone S.M., Rice C.M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friebe P., Bartenschlager R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 2002;76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi M., Lemon S.M. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 2003;77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi M., Lemon S.M. Structure-function analysis of the 3′ stem–loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9:331–345. doi: 10.1261/rna.2144203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masante C., Jaubert C., Palau W., Plissonneau J., Besnard L., Ventura M., Di Primo C. Mutations of the SL2 dimerization sequence of the hepatitis C genome abrogate viral replication. Cell Mol. Life Sci. 2015 doi: 10.1007/s00018-015-1893-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi S.T., Lai M.M.C. In: Hepatitis C Viruses: Genomes and Molecular Biology. Tan SL, editor. Norfolk: Horizon Bioscience; 2006. [PubMed] [Google Scholar]
- 11.Lohmann V. Hepatitis C virus RNA replication. Curr. Top. Microbiol. Immunol. 2013;369:167–198. doi: 10.1007/978-3-642-27340-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friebe P., Boudet J., Simorre J.P., Bartenschlager R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murayama A., Weng L., Date T., Akazawa D., Tian X., Suzuki T., Kato T., Tanaka Y., Mizokami M., Wakita T., et al. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog. 2010;6:e1000885. doi: 10.1371/journal.ppat.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You S., Rice C.M. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J. Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuplin A., Struthers M., Cook J., Bentley K., Evans D.J. Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3′ X-tail. Nucleic Acids Res. 2015;43:2914–2926. doi: 10.1093/nar/gkv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuplin A., Struthers M., Simmonds P., Evans D.J. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012;40:6908–6921. doi: 10.1093/nar/gks370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero-López C., Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero-López C., Barroso-Deljesus A., García-Sacristán A., Briones C., Berzal-Herranz A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X-tail region. Nucleic Acids Res. 2014;42:567–582. doi: 10.1093/nar/gkt841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diviney S., Tuplin A., Struthers M., Armstrong V., Elliott R.M., Simmonds P., Evans D.J. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008;82:9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shetty S., Stefanovic S., Mihailescu M.R. Hepatitis C virus RNA: molecular switches mediated by long-range RNA-RNA interactions. Nucleic Acids Res. 2013;41:2526–2540. doi: 10.1093/nar/gks1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanyi-Nagy R., Kanevsky I., Gabus C., Lavergne J.P., Ficheux D., Penin F., Fossé P., Darlix J.L. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shetty S., Kim S., Shimakami T., Lemon S.M., Mihailescu M.R. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–925. doi: 10.1261/rna.1960410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blight K.J., Rice C.M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T., Lai M.M. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutkiewicz M., Ciesiolka J. Structural characterization of the highly conserved 98-base sequence at the 3′ end of HCV RNA genome and the complementary sequence located at the 5′ end of the replicative viral strand. Nucleic Acids Res. 2005;33:693–703. doi: 10.1093/nar/gki218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada N., Tanihara K., Takada A., Yorihuzi T., Tsutsumi M., Shimomura H., Tsuji T., Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 28.Palau W., Masante C., Ventura M., Di Primo C. Direct evidence for RNA-RNA interactions at the 3′ end of the Hepatitis C virus genome using surface plasmon resonance. RNA. 2013;19:982–991. doi: 10.1261/rna.037606.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darty K., Denise A., Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koubek J., Lin K.F., Chen Y.R., Cheng R.P., Huang J.J. Strong anion-exchange fast performance liquid chromatography as a versatile tool for preparation and purification of RNA produced by in vitro transcription. RNA. 2013;19:1449–1459. doi: 10.1261/rna.038117.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dingley A.J., Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 34.Varani G., Tinoco I. RNA structure and NMR spectroscopy. Q Rev Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 35.Duszczyk M.M., Zanier K., Sattler M. A NMR strategy to unambiguously distinguish nucleic acid hairpin and duplex conformations applied to a Xist RNA A-repeat. Nucleic Acids Res. 2008;36:7068–7077. doi: 10.1093/nar/gkn776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotoya H., Matsugami A., Ikeda T., Ouhashi K., Uesugi S., Katahira M. Method for direct discrimination of intra- and intermolecular hydrogen bonds, and characterization of the G(:A):G(:A):G(:A):G heptad, with scalar couplings across hydrogen bonds. Nucleic Acids Res. 2004;32:5113–5118. doi: 10.1093/nar/gkh849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzakos A.G., Grace C.R., Lukavsky P.J., Riek R. NMR techniques for very large proteins and rnas in solution. Annu. Rev. Biophys. Biomol. Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 38.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puglisi E.V., Puglisi J.D. Probing the conformation of human tRNA(3)(Lys) in solution by NMR. FEBS Lett. 2007;581:5307–5314. doi: 10.1016/j.febslet.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Drexler J.F., Kupfer B., Petersen N., Grotto R.M., Rodrigues S.M., Grywna K., Panning M., Annan A., Silva G.F., Douglas J., et al. A novel diagnostic target in the hepatitis C virus genome. PLoS Med. 2009;6:e31. doi: 10.1371/journal.pmed.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lilley D.M. Analysis of branched nucleic acid structure using comparative gel electrophoresis. Q. Rev. Biophys. 2008;41:1–39. doi: 10.1017/S0033583508004678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


