Abstract
Although post-transcriptional gene silencing (PTGS) has been studied for more than a decade, there is still a gap in our understanding of how de novo silencing is initiated against genetic elements that are not supposed to produce double-stranded (ds)RNA. Given the pervasive transcription occurring throughout eukaryote genomes, we tested the hypothesis that unintended transcription could produce antisense (as)RNA molecules that participate to the initiation of PTGS triggered by sense transgenes (S-PTGS). Our results reveal a higher level of asRNA in Arabidopsis thaliana lines that spontaneously trigger S-PTGS than in lines that do not. However, PTGS triggered by antisense transgenes (AS-PTGS) differs from S-PTGS. In particular, a hypomorphic ago1 mutation that suppresses S-PTGS prevents the degradation of asRNA but not sense RNA during AS-PTGS, suggesting a different treatment of coding and non-coding RNA by AGO1, likely because of AGO1 association to polysomes. Moreover, the intended asRNA produced during AS-PTGS is capped whereas the asRNA produced during S-PTGS derives from 3′ maturation of a read-through transcript and is uncapped. Thus, we propose that uncapped asRNA corresponds to the aberrant RNA molecule that is converted to dsRNA by RNA-DEPENDENT RNA POLYMERASE 6 in siRNA-bodies to initiate S-PTGS, whereas capped asRNA must anneal with sense RNA to produce dsRNA that initiate AS-PTGS.
INTRODUCTION
Genome-wide surveys have revealed the abundance of natural-antisense transcripts (NATs) existing across eukaryotes (1–3). These molecules originate either from the transcription of the DNA strand opposite to a reference gene (cis) or from a distant locus (trans). The simultaneous production of both sense and antisense RNA has been reported to lead to a variety of outcomes but no common pattern seems to emerge from the different examples (3). On the other hand, it has long been known that convergent transcription of pericentromeric repeats in Schizosaccaromices pombe leads to the formation of heterochromatin and silencing (4,5). Converging transcript can indeed be used as a tool for transcriptional silencing not only in fission yeast but also zebrafish as well as mammalian cells (6–8). Also, compelling evidence in mouse oocytes has proven that antisense (as)RNA molecules can regulate genes and transposons at the post-transcriptional level both in cis and in trans (9,10). There is therefore an enormous regulation potential in asRNA that begs further investigation.
As in other eukaryotes, many complementary RNA pairs have been identified in plants (11–13). While some transcriptomic studies have detected negatively correlated expression patterns for RNA pairs in Arabidopsis thaliana (13,14) others have failed to detect any significant trend (15). The question therefore remains as to how these RNA molecules can regulate genes. In plants, the intentional expression of asRNA to interfere with a precise target sequence was commonly used even before the discovery of RNA interference (RNAi) (16,17). This technique was applied to different models with varying efficiencies but the underlying mechanism was never investigated at the genetic level. In contrast, many genetic screens have been set up to decipher the mechanisms of inverted repeat-triggered and sense-triggered post-transcriptional gene silencing (IR-PTGS and S-PTGS). Both involve an initiating double-stranded (ds)RNA being processed by the DICER-LIKE proteins (DCL) into small interfering (si)RNA molecules (18–20). These small molecules are bound by ARGONAUTE (AGO) proteins to form the core of the RNA-induced silencing complex (RISC) that interferes with complementary RNAs. In the case of IR transgenes, dsRNA is automatically formed from a single transcript folding back on itself making silencing highly efficient because the dsRNA molecule is directly processed by DCL2 and/or DCL4 into siRNA molecules (21–23). In the case of sense transgenes, it is assumed that aberrant RNAs are produced, and subsequently converted into dsRNA by RNA-DEPENDENT RNA POLYMERASE (RDR)6 (24,25). This process is thought to occur in cytoplasmic congregations of proteins called siRNA-bodies, which are distinct from P-bodies, and where RDR6 localizes (26).
So far, the reason why a sense transgenes inserted at a given locus triggers S-PTGS while the same transgene at another location does not has remained mysterious. Long ago, it was hypothesized that only some types of transgene arrangements at the insertion site allow enough converging transcription to trigger RNAi (27), but this hypothesis was not formally explored. It gained our interest when examples of endogenous cis-NATs were shown to trigger silencing. Indeed, some stresses induce convergent transcription at precise loci and cause PTGS of complementary transcripts that is necessary for proper response (11,13,28,29). Such a regulatory mechanism has also been reported to be associated with specific developmental phases in Arabidopsis (30) and barley (31). It therefore appears that asRNAs have an important functional role in plants. Importantly, these different studies have shown genetic requirements for this type of regulation that only partially overlap with those required for S-PTGS and IR-PTGS.
To investigate whether asRNAs play a role in the triggering of S-PTGS, well-established Arabidopsis lines carrying sense transgenes were analyzed. We found that asRNA molecules were indeed present at higher level in lines that spontaneously undergo S-PTGS than in lines that stably express the sense RNA. Then, we developed a two-components silencing system involving the independent expression of sense and antisense RNA to decipher the genetic requirements of antisense-triggered PTGS (AS-PTGS). We identified DCL2 and DCL4 as essential factors, while AGO1, RDR6 and SGS3 appear only required for the production of secondary siRNA. Our analyses also revealed a different contribution of AGO1 to the degradation of sense and antisense RNAs, highlighting the differences between S-PTGS and AS-PTGS. We eventually discovered that the unintended asRNA produced during S-PTGS is uncapped, which probably allows its conversion into dsRNA by RDR6, whereas the intended asRNA produced during AS-PTGS is capped and must anneal with the sense RNA to form dsRNA.
MATERIALS AND METHODS
Plant material
Transgenic lines 6b4, L1, L2, JAP3 and SUC-SUL, and ago1-1, ago1-27, dcl2Kas-1, dcl4-5, rdr6sgs2–1, sgs3-1, xrn3-3 and xrn4-5 mutants have been described previously (22,23,25,32–35). Seeds were sterilized with bleach and ethanol and grown on S-Medium from Duchefa (http://www.duchefa-biochemie.com) under 18 h of light and 6 h dark cycles with an average temperature of 22°C.
Cloning and transformation
To generate the p35S:SUG-tCaMV construct, the GUS coding sequence was extracted as an PstI–PstI fragment from plasmid pRAJ260 and cloned between the 35S promoter and terminator of plasmid pLBR19. Then, the p35S:SUG-tCaMV construct was extracted as a SstI-XbaI fragment and cloned into the binary vector pBiB-Hyg before transfer to Agrobacterium. Arabidopsis plants were transformed by floral dipping and transformants selected on medium supplemented with 30 μg/ml hygromycin.
RNA extraction, fractionation, RNA gel blot hybridization and sequencing
Shoots or whole plants were pooled 5, 11 or 17 days after germination on sterile S-medium and grinded into liquid nitrogen. Buffer containing 0.1 M NaCl, 2% SDS, 50 mM Tris–HCl (pH 9), 10 mM EDTA (pH 8) and 20 mM ß-mercaptoethanol was added to the frozen powder and RNA was extracted twice with phenol and recovered by ethanol precipitation. For small RNA analysis, total RNA was separated on a 15% denaturing polyacrylamide gel and stained with ethidium bromide. For sequencing, small RNA bands were cut out of the gel and sent to GATC Biotech (http://www.gatc-biotech.com) for library preparation and sequencing using HiSeq2000 sequencer. For blots, RNA was transferred to nylon membrane, hybridized in Sigma PerfectHyb buffer (https://www.sigmaaldrich.com/) and stripped with boiling 0.1% SDS solution.
To obtain high molecular weight RNA, total RNA was precipitated overnight in 2 M LiCl at 4°C and recovered by centrifugation. The samples were then separated on 1.5% agarose gel running in 20 mM HEPES, 1 mM EDTA and 0.7% formaldehyde. RNA was transferred to nylon membrane by capillarity and hybridized in Sigma PerfectHyb buffer.
Strand-specific qRT-PCR
Sterile plants were pooled 11 days after germination and grinded into liquid nitrogen. RNA was isolated using RNeasy Plant Mini Kit from QIAGEN (https://www.qiagen.com). cDNA were generated form 1 μg of total RNA using the first strand cDNA kit from Thermo Fisher Scientific (http://www.lifetechnologies.com) using the specific primer RT_ASGUS_Linker: CGACTGGAGCACGAGGACACTGAGACTGGCATGAACTTCGGTGAA (bold letters represent linker sequence). PCR amplification was done using a primer containing the Linker sequence, LK: CGACTGGAGCACGAGGACACTGA, and a primer specific to the RbcS region, RbcS1Rev: TCACAGTTCGATAGCGAAAACCGA. SsoAdvanced Universal SYBR Green Supermix from Biorad (http://www.bio-rad.com/) was used in the Mastercycler® ep realplex from Eppendorf (www.eppendorf.com). eIF1a expression was used has amplification control since unspecific annealing of RT primer produces cDNA from highly abundant RNA (36).
Data processing
Datasets were cleaned using bioinformatic tools developed previously (37). Reads ranging from 20 to 25 nucleotides in length were selected using PRINSEQ (38) and aligned using standard Bowtie2 algorithm (39). Aligned reads were sorted with SAMtools (40) and distribution plots were generated using a script from the S-MART tools (37).
RESULTS
Production of siRNAs starts at the 3′ end of GUS in the S-PTGS line L1
A series of plants carrying a p35S-GUS-tRbcS transgene were previously characterized, including line 6b4 that stably expresses GUS and lines L1 and L2, which spontaneously undergo S-PTGS (32). Probing GUS siRNA accumulation in L1 using three large fragments covering the GUS coding sequence (Figure 1A) revealed that siRNAs come from 5′, central and 3′ parts of the gene (41). However, no siRNA were detected using a probe covering the first 300bp at the 5′ end of GUS (42). To determine if the absence of siRNAs at the 5′ end reflects an incomplete spreading of siRNAs from 3′ to 5′, the kinetics of siRNA accumulation was determined using the original 5′, central and 3′ probes (41). The kinetics was followed from day 5 to day 17 during which GUS activity decreases (32), indicating the progressive establishment of L1 S-PTGS. Whereas siRNAs of both polarities accumulated at high level at the 3′ end as early as 5 days after germination (DAG), they accumulated at lower level in the central region and were barely detectable at the 5′ end (Figure 1B). From day 5 to day 17, the accumulation of siRNAs increased at the 5′ end, decreased at the 3′ end while it remained constant in the central part. Together, these results suggest that siRNAs spread from 3′ to 5′ during the establishment of L1 S-PTGS.
Figure 1.
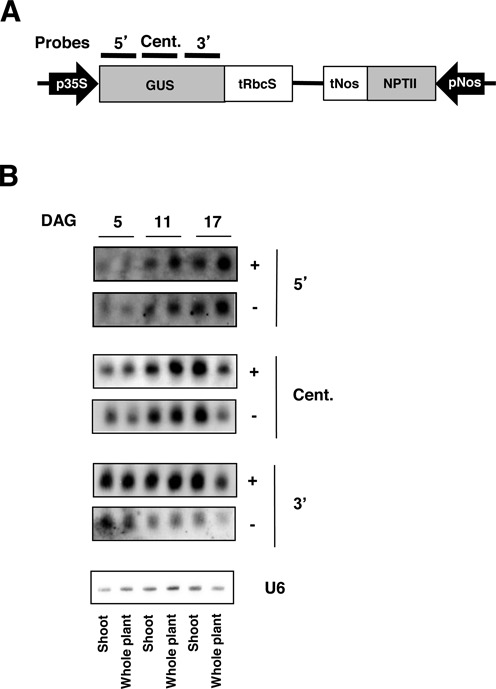
Silencing originates from the 3′ in L1. (A) Scheme of the T-DNA cassette in 6b4, L1 and L2 lines. Regions covered by different probes are represented above the GUS coding sequence. (B) small RNA blots of either shoots or whole L1 plants isolated at different days after germination (DAG) indicated above. The membranes were blotted with different RNA probes with different orientations indicated on the right. A U6 probe was used as loading control for all membranes, one representative blot is shown.
Spontaneous triggering of S-PTGS correlates with the production of asRNA
So far, the reason why lines L1 and L2, but not 6b4, undergo PTGS has remained unexplained. Given the organization of the p35S-GUS-tRbcS and pNos-NPTII-tNos transgenes on the T-DNA (Figure 1A) and the weak efficiency of the Nos terminator (43), transcription read-through that bypasses the Nos terminator of the NPTII transgene could produce an RNA antisense to the 3′ end of GUS (27). Subsequent annealing of the asRNA to GUS RNA could form dsRNA that initiate PTGS, consistent with the first appearance of siRNAs at the 3′ end of GUS. Alternatively, the asRNA could be recognized as an aberrant RNA and transformed into dsRNA by RDR6 in siRNA-bodies (26). Therefore, the presence of such asRNA (hereafter referred to as SUG) was tested by qRT-PCR in 6b4, L1 and L2. To insure strand-specificity, a linker-oligonucleotide was used to prime the cDNA synthesis reaction (36). The level of SUG RNA detected in lines L1 and L2 was two-fold higher than in 6b4 (Figure 2A). However, because lines L1 and L2 undergo PTGS (32), SUG RNA is likely degraded together with GUS RNA, and therefore underestimated in this experiment. To circumvent this issue, SUG RNA levels were tested in the rdr6 mutant background in which S-PTGS is blocked (25). The level of SUG RNA detected in lines L1/rdr6 and L2/rdr6 was 4- to 5-fold higher than in line 6b4/rdr6 (Figure 2B), highlighting a positive correlation between the presence of SUG RNA and the triggering of S-PTGS.
Figure 2.
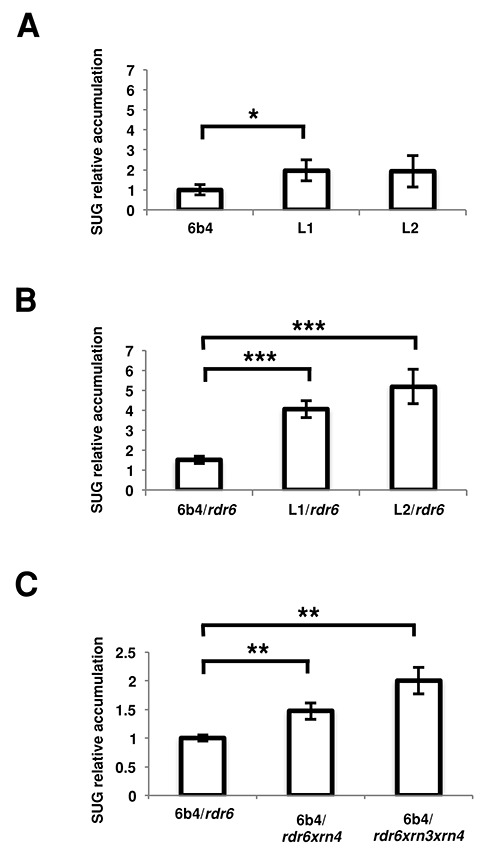
Antisense RNA is more abundant in lines that spontaneously trigger S-PTGS and is degraded by 5′->3′ RNA exonucleases. (A) Relative presence of SUG RNA in different lines in WT background. SUG ΔCt were calculated using eIF1a Ct and expressed as a fold change compared to 6b4 (6b4 = 1). Error bars represent the standard deviation of the biological triplicate. (B) Relative presence of SUG RNA in different lines in rdr6 background. SUG ΔCt were calculated using eIF1a expression as internal control and represented as a fold change compared to 6b4 (6b4 = 1). Error bars represent the standard deviation of the biological triplicate. (C) Relative presence of SUG RNA in different rdr6 xrn backgrounds. SUG ΔCt were calculated using eIF1a Ct and expressed as a fold change compared to 6b4/rdr6 (6b4 = 1). Error bars represent the standard deviation of the biological triplicate. The values obtained were submitted to the Student's t-test to calculate the null hypothesis probability (P). The asterisks represent the different levels of confidence (*P< 0.05, **P< 0.01, ***P< 0.005).
Remarkably, lines L1, L1/rdr6, L2 and L2/rdr6 tolerate kanamycin (the selection marker for the NPTII resistance gene) to a level as high as 150μg/ml, whereas lines 6b4 and 6b4/rdr6 exhibit growth defects on 30μg/ml kanamycin and die on 50μg/ml kanamycin. At first, this result indicates that, unlike the p35S-GUS-tRbcS transgene, the pNos-NPTII-tNos transgene does not undergo silencing in lines L1 and L2. Secondly, this result indicates that the NPTII gene is more efficiently transcribed in lines L1 and L2 than in line 6b4. This likely explains why SUG RNA is more abundant in the silenced lines, assuming that the SUG RNA results from a read-through that bypasses the Nos terminator of the pNos-NPTII-tNos transgene. Together, these results suggest that the higher transcription occurs at the genomic location where the T-DNA integrates, the higher is the chance that sufficient amounts of GUS and SUG RNAs are produced to trigger S-PTGS.
Development of an AS-PTGS system
To determine if the production of an asRNA consistently triggers PTGS of a stable GUS reporter, a p35S-SUG-tNos transgene was generated and introduced into wild-type (WT) Arabidopsis plants. The resulting SUG lines were crossed with 6b4 to identify plants in which GUS is efficiently silenced. In three crosses, hybrid plants carrying the 6b4 and SUG transgenes showed significantly reduced GUS activity (Supplemental Figure S1). Hereafter, all experiments were performed with the SUG6 line that showed a 3:1 Mendelian segregation indicating insertion at a single locus. At first, the accumulation of SUG long RNA and siRNA was analyzed in homozygous SUG6 plants. Results indicate that the SUG RNA stably accumulates in line SUG6, and that no siRNA is spontaneously produced in this line (Figure 3). Then, plants homozygous for both 6b4 and SUG6 were produced and analyzed in a similar manner. Results indicate that the 6b4/SUG6 line lacks both GUS and SUG long RNAs, while both sense and antisense siRNA accumulated (Figure 3). This result indicates that silencing in the 6b4/SUG6 line results from post-transcriptional degradation of GUS and SUG long RNAs and not from transcriptional interference between the p35S-GUS-tRbcS and p35S-SUG-tNos transgenes. Hereafter we refer to this system as antisense-triggered PTGS (AS-PTGS).
Figure 3.
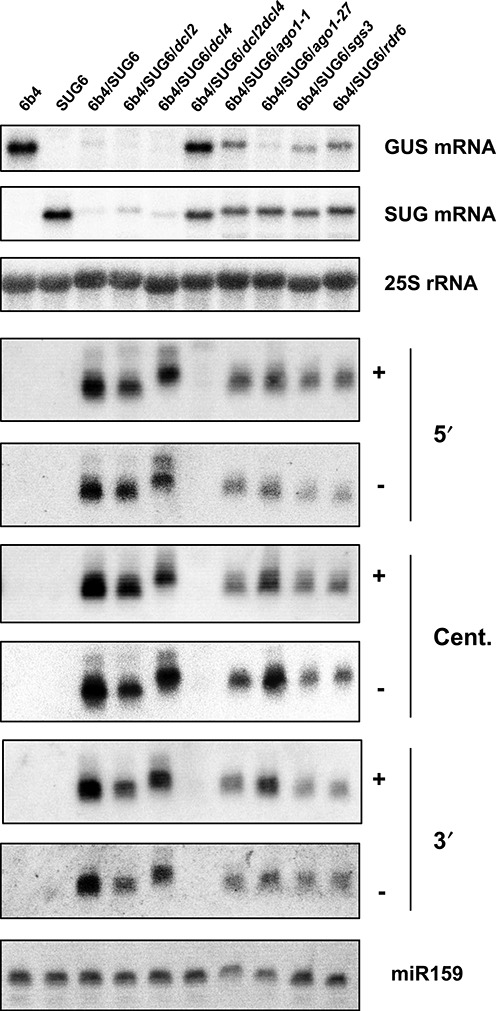
Genetic requirement of AS-PTGS. HMW and LMW RNA blots of whole 11 DAG plants in various backgrounds indicated on top. For GUS and SUG mRNA, the 3′ RNA probe was used in different orientation. Loading was controlled using a 25S RNA probe. For small RNA, 5′, Central and 3′ RNA probes were used in different orientations as indicated on the right. The different membranes were probed with a miR159 oligonucleotide for loading and a representative blot is shown.
Genetic determinants of AS-PTGS
To decipher the genetic requirement for AS-PTGS, line 6b4/SUG6 was crossed to various RNA silencing mutants. Since the most straightforward explanation for AS-PTGS is the spontaneous pairing of GUS and SUG RNA to form dsRNA that is subsequently processed by DCL proteins, we first tested the effect of dcl2 and dcl4 mutations. Indeed, DCL2 and DCL4 have previously been identified as essential factors in both S-PTGS and IR-PTGS as well as in antiviral PTGS (21,23,44,45). We observed that dcl2 and dcl4 single mutations seemingly have no effect on GUS/SUG silencing, and that only the dcl2dcl4 double mutant released AS-PTGS. Concordantly, GUS and SUG RNA accumulated in 6b4/SUG6/dcl2dcl4 plants at the level of 6b4 and SUG6 individual plants, respectively, and lacked siRNAs (Figure 3). These results are similar to those obtained using the 6b4/306 (IR-PTGS) and L1 (S-PTGS) systems (23,42), indicating that S-PTGS, IR-PTGS and AS-PTGS have the same requirements for DCL proteins.
The involvement of AGO1, a central protein in S-PTGS and IR-PTGS (46,47), was tested using the ago1-1 null allele. Analysis of 6b4/SUG6/ago1-1 plants revealed partial release of AS-PTGS. Indeed, GUS and SUG RNAs accumulate, but at levels lower than those observed in 6b4/SUG6/dcl2dcl4 plants (Figure 3). Moreover, sense and antisense siRNAs are still detected, but again at levels lower than those observed in 6b4/SUG6 plants (Figure 3). These results suggest that, in 6b4/SUG6/ago1–1 plants, AS-PTGS is limited to the processing of annealed GUS/SUG dsRNA by DCL2 and DCL4, and that the siRNAs that are detected correspond to primary siRNAs.
RDR6 and SGS3 are required for the production of secondary siRNAs, which are essential components of S-PTGS, but dispensable for IR-PTGS (24,25,34,47,48). Therefore, we tested the effect of rdr6 and sgs3 null alleles on AS-PTGS. Like ago1-1, rdr6 and sgs3 null mutations partially impaired 6b4/SUG6 silencing (Figure 3). Indeed, GUS mRNA accumulates at similar level in 6b4/SUG6/rdr6, 6b4/SUG6/sgs3 and 6b4/SUG6/ago1-1, i.e. below the level observed in 6b4/SUG6/dcl2dcl4 plants. Moreover, both sense and antisense siRNA molecules are still detected in both cases, although at a level lower than in 6b4/SUG6, indicating that partial PTGS is still operating (Figure 3). Analyses of the effect of rdr6 and sgs3 mutations on two other SUG lines (SUG7 and SUG9) yielded similar results. Indeed, GUS activity in 6b4/SUG6/rdr6, 6b4/SUG6/sgs3, 6b4/SUG7/rdr6, 6b4/SUG7/sgs3, 6b4/SUG9/rdr6 and 6b4/SUG9/sgs3 plants was higher than in 6b4/SUG6, 6b4/SUG7 and 6b4/SUG9 plants but lower than in 6b4/rdr6 and 6b4/sgs3 controls (Supplemental Figure S1). This confirms that the partial impairment of AS-PTGS by rdr6 and sgs3 mutations is not specific to the SUG6 locus. These results therefore suggest that the production of secondary siRNAs through RDR6 and SGS3 is generally required to complete RNA degradation during AS-PTGS.
A hypomorphic ago1 mutation uncouples the degradation of sense and antisense RNA
The ago1-27 hypomorphic allele exhibits minor morphologic defects compared with the ago1-1 null allele, indicating that it retains enough AGO1 activity to allow plants to go through their life cycle (33). Nevertheless, the ago1-27 mutation suppresses S-PTGS as efficiently as ago1 null alleles (33), indicating that S-PTGS is highly sensitive to AGO1 perturbation. In contrast, the ago1-27 mutation does not affect IR-PTGS triggered by 35S-driven transgenes (48), and only partially suppresses IR-PTGS specifically triggered in the companion cells of the phloem, which leads to silencing in a layer of 15–20 cells surrounding the phloem (Supplemental Figure S2). Introduction of the 6b4/SUG6 system into ago1-27 revealed that GUS mRNA is still degraded in 6b4/SUG6/ago1-27 plants (Figure 3), which contrasts with the complete abolition of S-PTGS in L1/ago1-27 (33,41). Nevertheless, ago1-27 did have an effect on AS-PTGS because SUG RNA accumulates at a level similar to that observed in 6b4/SUG6/ago1-1 (Figure 3). Moreover, siRNAs accumulated at lower levels in 6b4/SUG6/ago1-27 than in 6b4/SUG6 controls (Figure 3). Together, these results, suggest a different treatment of coding and non-coding RNA molecules by the mutant ago1-27 protein during AS-PTGS, which may be due to the fact that AGO1 associates with polysomes (49).
Small RNA populations in silenced lines
The different effect of ago1-27 on S-PTGS and AS-PTGS prompted us to take a closer look at the siRNA populations present in WT and mutants carrying the 6b4/SUG6 or L1 transgenes. Small RNA eluted from acrylamide gels were used to generate libraries for short-reads RNA sequencing using Illumina HiSeq 2000. At first, the 20- to 25-nt reads matching the GUS/SUG transgenes were analyzed. The total aligned read count per million (RPM) is given in Table 1, and the ratio to WT reveals the extent of the decreased siRNA levels in the different mutant backgrounds. Results with the L1 locus are in line with published northern blot analyses. Indeed, as recently reported, the dcl2 mutation reduces siRNA production and silencing efficiency, while the dcl4 mutation stimulates siRNA production and silencing because of the role of DCL2 in transitivity, and only the dcl2dcl4 double mutation abolishes siRNA production and PTGS (23). In addition to dcl2dcl4, ago1-1, ago1-27 and rdr6 abolish siRNA production and PTGS as previously reported (41). The amount of GUS/SUG siRNAs in 6b4/SUG6 was lower than that in L1, consistent with the reduced transcription level of 6b4 compared with L1 (32). In 6b4/SUG6/dcl2dcl4, GUS/SUG siRNAs were below detection levels, consistent with GUS and SUG long RNA accumulating at the level of 6b4 and SUG6 plants taken individually (Figure 3). GUS/SUG siRNA levels were low but not absent in 6b4/SUG6/rdr6 and 6b4/SUG6/ago1-1, in line with the partial impairment of silencing observed in these mutants. Consistent with northern blot analysis, GUS/SUG siRNA levels were higher in 6b4/SUG6/ago1–27 than in 6b4/SUG6/ago1-1, but still lower than in 6b4/SUG controls.
Table 1. Number of GUS/SUG siRNA reads found in different backgrounds.
| 6b4/SUG6 | L1 | |||
|---|---|---|---|---|
| RPM | Ratio to WT | RPM | Ratio to WT | |
| WT | 15 605 | 1.00 | 48 865 | 1.00 |
| dcl2 | 8251 | 0.53 | 23 525 | 0.48 |
| dcl4 | 9630 | 0.62 | 57 253 | 1.17 |
| dcl2dcl4 | 528 | 0.03 | 511 | 0.01 |
| ago1–27 | 4523 | 0.29 | 426 | 0.01 |
| ago1-1 | 3808 | 0.24 | 474 | 0.01 |
| rdr6 | 4120 | 0.26 | 514 | 0.01 |
20 to 25 nucleotides long reads aligning to transgene sequences are reported in read per million (RPM). The ratio to the WT background sample of the different series (6b4/SUG6 and L1) is given on the right.
The distribution of aligned reads was then plotted on the transgene transcript sequences. Note that the SUG transgene carries the full GUS ORF but that its 3′UTR differs from that of the GUS transgene. Figure 4 shows the distribution of sense and antisense siRNAs along the GUS sequence and the RbcS 3′UTR in the various backgrounds. As seen in previous studies, discrete populations of small RNA molecules are overrepresented compared to others (50,51), suggesting a preferential production/accumulation of certain siRNAs. However, such hot-spots have been shown to often result from the preferential ligation of certain siRNAs by a given set of adaptors (52,53). Given that this ligation bias is the same in every backgrounds, the relative intensity of each peak can still be compared between in mutants and WT. Beside the lower level of GUS/SUG siRNAs in 6b4/SUG6 compared with L1, the major difference between these two systems consists in the presence of siRNA all over the GUS coding sequence in 6b4/SUG6 whereas they are absent at the 5′ end of the GUS gene in L1 (Figure 4), consistent with a previous study (42). Given that L1 produces more siRNAs than 6b4/SUG6 (Table 1), the absence of siRNAs in the 5′ region cannot be attributed to a lack of coverage. Moreover, siRNAs corresponding to this region are not reduced in 6b4/SUG6/rdr6 and 6b4/SUG6/ago1-1, whereas siRNAs from the rest of the gene are reduced by ago1-1 and rdr6 (Figure 4), suggesting that this region only produces primary siRNAs.
Figure 4.
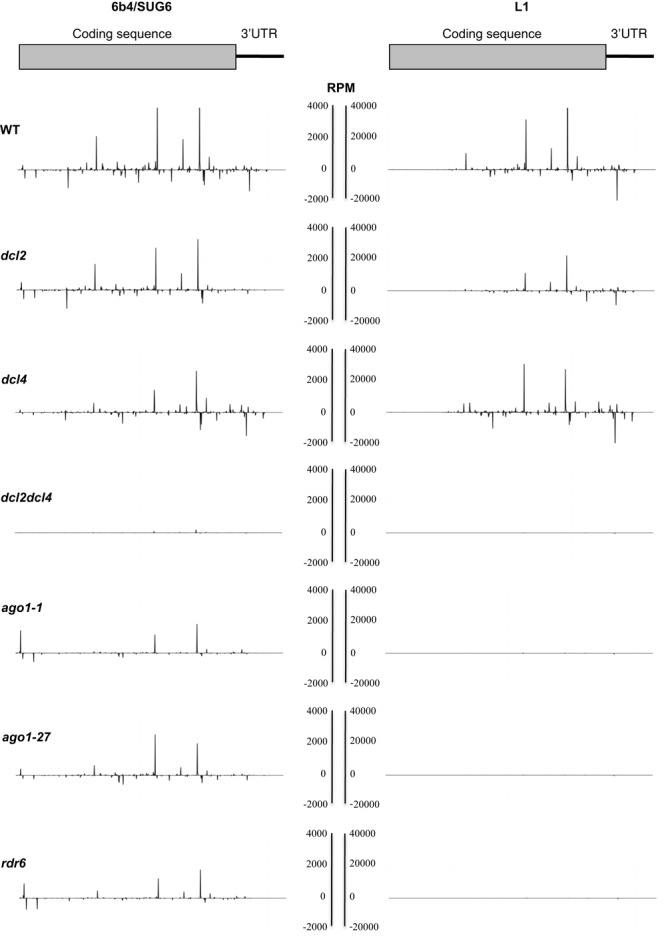
Distribution of GUS siRNA reads in WT and mutant backgrounds. Distribution of the normalized aligned reads along the GUS coding sequence and the RbcS 3′UTR for 6b4 (left column) and L1 (right column). The background is indicated on the left and the ladder for the graphics in represented in the middle as RPM.
AS-PTGS against endogenous mRNA
To test if the genetic requirements described for 6b4/SUG6 silencing are specific to this system or if it generally applies to AS-PTGS, the coding sequence of the PHYTOENE DESATURASE (PDS) gene was placed behind the 35S promoter in inverted orientation to produce an antisense SDP RNA. PDS was selected as a target because the silencing of this gene produces an obvious visual phenotype (21,34). The p35S-SDP-tNos transgene was introduced into WT, dcl2dcl4 and rdr6 backgrounds. Primary transformants were visually inspected at 14 DAG and classified as severely or moderately silenced for the endogenous PDS gene (Table 2). Eighty three percent of the WT transformants showed a pds phenotype, confirming that antisense transgenes can efficiently silence certain endogenous genes. Given the results obtained with the 6b4/SUG6 system, and knowing that, unlike transgenes, endogenous genes are incapable of producing secondary siRNAs (54,55), we expected the rdr6 mutation to very moderately affects PDS AS-PTGS. Indeed, the frequency of transformants exhibiting a pds phenotype was only slightly reduced in rdr6 compared with WT (Table 2). In contrast, the percentage of silenced plants was dramatically reduced in the dcl2dcl4 mutant. In particular, no dcl2dcl4 transformant exhibited the severe pds phenotype that was observed in 42% of the WT transformants. Nevertheless, 22% of the dcl2dcl4/p35S-SDP-tNos transformants still exhibited a moderate pds phenotype, suggesting that DCL1 and/or DCL3 can partially substitute to DCL2 and DCL4 in the dcl2dcl4 mutant, as previously shown for IR-PTGS (47).
Table 2. Number of plants silenced for PDS gene.
| WT-like | moderate | severe | total | |
|---|---|---|---|---|
| WT | 33 (23) | 51 (36) | 59 (41) | 143 (100) |
| dcl2dcl4 | 146 (78) | 42 (22) | 0 (0) | 188 (100) |
| rdr6 | 57 (32) | 56 (32) | 63 (36) | 176 (100) |
Transformed plants from different indicated backgrounds were scored for having severe, moderate or no silencing phenotype. Percentage of total plants for each category is indicated inside parenthesis.
Uncapped asRNA likely is the aberrant RNA molecule that is converted into dsRNA by RDR6 to initiate S-PTGS
The results presented above indicate that antisense transgenes can efficiently silence homologous sense transgenes or endogenous genes through the action of siRNAs processed by DCL2 or DCL4. However, these results do not prove that the asRNA detected in L1 and L2 lines during S-PTGS acts as the asRNA intentionally produced in AS-PTGS. Indeed, although both S-PTGS and AS-PTGS produce siRNAs and are suppressed in the dcl2dcl4 double mutant, there are more differences than similarities between these two pathways. First, dcl2 and dcl4 single mutations reduce and enhance S-PTGS respectively (23), whereas they have no effect on AS-PTGS (Figure 3). This is the consequence of DCL2 producing secondary siRNAs more efficiently than DCL4, but being obscured by DCL4 in WT plants. Secondly, ago1, rdr6 and sgs3 null mutations abolish S-PTGS but only reduce the efficiency of AS-PTGS. Thirdly, the hypomorphic ago1-27 mutation suppresses S-PTGS as efficiently as the null ago1-1 mutation whereas it does not restore GUS mRNA accumulation on AS-PTGS. These differences may be explained by the very little amount of asRNA produced during S-PTGS whereas larger amounts of asRNA are produced during AS-PTGS. Indeed, even in the rdr6 background where PTGS is impaired, asRNA derived from L1 can only be detected by RT-PCR (Figure 2), whereas the amount of asRNA produced in the SUG6 line is detectable by northern blot (Figure 3). Using a DNA probe that detects GUS and SUG with equal efficiency reveals that the SUG6 line produces less SUG than the 6b4 line produces GUS (Supplemental Figure S3). Nevertheless, the amount of SUG intentionally produced by the p35S-SUG-tNos transgene is clearly more abundant than what is accidentally produced due to the organization of the p35S-GUS-tRbcS and pNos-NPTII-tNos transgenes on the T-DNA in L1.
The higher abundance of SUG in the 6b4/SUG6 system compared with L1 likely makes the amplification step more important, if not crucial, for S-PTGS. However, qualitative aspects of the asRNA may also play an important role. We detected SUG RNA in lines L1 and L2 (Figure 2A and B), which likely results from transcription read-through bypassing the weak Nos terminator of the NPTII transgene, resulting in a chimeric NPTII-SUG RNA. If this RNA were marked as aberrant and transformed into dsRNA by RDR6 in siRNA-bodies, this would cause silencing of both GUS and NPTII, resulting in the loss of kanamycin resistance. However, lines L1 and L2 are highly resistant to kanamycin, ruling out this hypothesis. Moreover, our qRT-PCR experiments succeeded at amplifying internal fragments within the SUG sequence but failed at amplifying an NPTII-SUG chimeric RNA. Therefore, we performed 5′ RACE experiments on L1/rdr6 plants to characterize the 5′ end of the SUG RNAs. We identified five different uncapped 5′ ends, which are all located within the RbcS terminator sequence (Supplemental Figure S4). To explain this result, we propose that the 3′ end maturation of the NPTII-SUG chimeric RNA, which involves cleavage at the tNos cleavage/polyadenylation site followed by the addition of a poly(A) tail, results in the production of a capped and polyadenylated NPTII mRNA and of an uncapped SUG RNA. Translation of the capped and polyadenylated NPTII mRNA promotes high levels of kanamycin resistance, while the uncapped SUG RNA cleavage product likely is degraded by 5′-to-3′ exonucleases, resulting in a series of molecules which 5′ ends are located downstream the tNos cleavage/polyadenylation site.
The uncapped SUG RNA liberated by 3′ end maturation of the NPTII transcript makes a very good candidate for the long-searched aberrant RNA that upon transformation into dsRNA by RDR6 in siRNA-bodies initiates S-PTGS specifically against the GUS transgene. To test this hypothesis and further determine if the level of uncapped SUG RNA directly conditions the entry into S-PTGS, we examined the effect of the xrn3 and xrn4 mutations on the level of SUG RNA. Indeed, xrn3 and xrn4 mutants are compromised in 5′-to-3′ degradation of uncapped RNA in the nucleus and cytoplasm, respectively, and we previously reported that both mutations allow line 6b4 to trigger PTGS (35). Because xrn3 only has a weak inducing effect on S-PTGS, we analyzed the effect of either xrn4 alone or in combination with xrn3, which results in 100% triggering of 6b4 S-PTGS. Since both GUS and SUG RNA are degraded when S-PTGS is triggered (see Figure 2A vs B), the level of SUG RNA was examined in 6b4/xrn4rdr6 and 6b4/xrn3xrn4rdr6 plants and compared to that in 6b4/rdr6 plants in which rdr6 prevents S-PTGS. Results presented in Figure 2C indicate that the amount of SUG RNA is the highest when both nuclear and cytoplasmic 5′-to-3′ degradation of uncapped RNA are prevented, thus supporting the hypothesis that the more uncapped SUG RNAs escape degradation, the more they have a chance to enter siRNA-bodies and trigger S-PTGS.
DISCUSSION
The overwhelming presence of antisense RNA (asRNA) in eukaryote cells raises important questions as to the biological implications of such molecules. In fission yeast, convergent transcripts processed by Dicer leads to the production of siRNA molecules that guide heterochromatin formation at pericentromeric regions (4,5). This mechanism however appears to extend beyond the centromere organization as it is also used to self-regulate the RNAi genes in S. pombe (56). In mouse oocytes, Dicer-dependent degradation of RNA pairs is thought to be essential for the proper regulation of female meiosis (9,10). Hence, this conserved RNAi-dependent mechanism can be used to artificially target silencing of genes in fission yeast and mammalian cells (7) as well as in zebrafish (8). It is therefore likely that the biological significance of asRNA molecules has not yet been fully unraveled.
In plants, transgene silencing phenomenon mediated by sense transgenes (S-PTGS) or inverted repeat transgenes (IR-PTGS) have been the subject of numerous studies that have shed light on antiviral and endogenous RNAi pathways (reviewed in (18–20)). However, little is known about PTGS triggered by transgenes that intentionally produce asRNA (AS-PTGS). To better understand how asRNA can trigger silencing, we set up two systems, one directed against an endogenous gene and one directed against a stably expressed sense transgene. Firstly, expressing an asRNA complementary to the endogenous PDS gene results in efficient PDS silencing (Table 2). AS-PTGS against PDS is strongly reduced by the combination of dcl2 and dcl4 mutations and not by the rdr6 mutation (Table 2), similar to IR-PTGS directed against PDS or SUL (34,47). Secondly, crossing a line stably expressing an exogenous GUS mRNA with a line stably expressing SUG, an asRNA complementary to the GUS mRNA, causes the degradation of GUS and SUG RNAs with concomitant appearance of 21- and 22-nt siRNAs, indicating efficient AS-PTGS. Again, the combination of dcl2 and dcl4 mutations neutralizes AS-PTGS (Figure 3), just like it does for IR-PTGS against the GUS mRNA (23). However, whereas IR-PTGS against the GUS mRNA occurs normally in sgs3 and rdr6 mutants (48), AS-PTGS is partially impaired in sgs3 and rdr6 mutants (Figures 3 and 4). Likely, during AS-PTGS, DCL2/DCL4-mediated processing of annealed GUS/SUG RNAs generates primary siRNA molecules that direct the cleavage of unpaired GUS and SUG RNAs, similar to primary siRNA molecules generated from the hairpin RNA during IR-PTGS. However, the frequency of intermolecular annealing between GUS and SUG RNAs to form dsRNA during AS-PTGS is certainly lower than the frequency of intramolecular annealing of the inverted-repeat RNA produced during IR-PTGS. Therefore, we can speculate that the amount of primary siRNAs produced during AS-PTGS is insufficient to catalyze the cleavage of all unpaired GUS and SUG RNAs. This contrasts the amount of primary siRNAs produced during IR-PTGS, which is likely sufficient to catalyze the cleavage of all target RNAs. This makes IR-PTGS independent of RDR6 and SGS3. However, efficient AS-PTGS can only be completed if some cleaved products of GUS/SUG RNAs are protected by SGS3 and transformed into dsRNA by RDR6, allowing the production of secondary siRNAs by DCL2 and DCL4, which complete the degradation of all unpaired GUS and SUG RNAs.
The independence of IR-PTGS on SGS3 and RDR6, and the partial dependence of AS-PTGS on these two components contrast the complete impairment of S-PTGS in sgs3 and rdr6 mutants. It has been hypothesized that S-PTGS is triggered by an accidentally produced aberrant RNA that is somehow converted to dsRNA. Because the initial aberrant RNA trigger is not abundant, it leads to the production of very few primary siRNAs, thus making S-PTGS totally dependent on SGS3 and RDR6 for the production of secondary siRNAs that execute the cleavage GUS target mRNAs. However, little is known about this aberrant RNA trigger. A positive correlation has been observed between S-PTGS efficiency and transgene transcription rate and/or improperly terminated mRNA molecules (32,43,57) but the nature of the aberrant RNA trigger remains unsolved. The hypothesis that antisense transcription could provide the initial trigger for S-PTGS (26) has been much less explored than the other avenues and we sought to correct this.
The S-PTGS system that our group has been using has an intrinsic predisposition to produce asRNA given the converging orientation of the GUS and NPTII transgenes on the T-DNA (Figure 1A). In line with this, examination of small RNA blots in silenced lines revealed that siRNA molecules are first generated from the 3′ of the GUS mRNA and then spread 5′ (Figure 1B). However, silencing is not bidirectional, and the NPTII transgene does not undergo PTGS. Rather, lines exhibiting GUS S-PTGS are four to five times more resistant to kanamycin than non-silenced lines. Consistently, four to five times more SUG asRNA are found in silenced lines compared with non-silenced lines (Figure 2B). We therefore hypothesized that read-through transcription of the NPTII transgene reaches to the GUS sequence because the Nos terminator is a weak terminator in plants (43). Consequently, T-DNAs integrated at genomic location that do not promote high levels of transcription, would produce insufficient amount of NPTII-SUG to trigger GUS S-PTGS and only confer low levels of resistance to kanamycin. Inversely, T-DNAs integrated at genomic location that promote high levels of transcription would produce high amounts of NPTII-SUG thereby triggering GUS S-PTGS and conferring a high level of resistance to kanamycin. The opposite effects observed on GUS and NPTII rules out the possibility that the chimeric NPTII-SUG RNA is the aberrant RNA that enters into siRNA-bodies for conversion into dsRNA by RDR6 because this would result in both GUS and NPTII silencing. Moreover, our incapacity to detect the chimeric NPTII-SUG RNA suggests that it is very labile. Consistently, RACE experiments revealed SUG 5′ ends located downstream of the NPTII sequence, suggesting that the read-through NPTII-SUG transcript is cleaved at the tNos or tRbcS polyadenylation sites, resulting in the production of a capped and polyadenylated NPTII mRNA that confers kanamycin resistance and an uncapped SUG RNA. We propose that this uncapped SUG RNA is the long-searched aberrant RNA trigger that enters into siRNA-bodies where it is protected by SGS3 and transformed into dsRNA by RDR6, thus specifically initiating S-PTGS against the GUS transgene.
Despite the attractiveness of the above hypothesis, it remains possible that the read-through NPTII-SUG transcript is not detected because, upon annealing with the GUS mRNA, it forms dsRNA that is immediately processed into GUS/SUG siRNAs by DCL2 and DCL4, thus specifically initiating PTGS against the GUS transgene, and letting the NPTII transgene unsilenced. If it were the case, S-PTGS should in fact correspond to a form of AS-PTGS that strongly requires SGS3 and RDR6 because the amount of primary siRNAs is very low. However, S-PTGS is enhanced by xrn3 and xrn4 mutations, which impair 5′-to-3′ degradation of uncapped RNA in the nucleus and cytoplasm, respectively, indicating that the S-PTGS trigger likely is uncapped. Since the read-through NPTII-SUG transcript is capped, it should not be susceptible to degradation by XRN3 and XRN4. Therefore, S-PTGS efficiency should not be affected by xrn3 and xrn4 if the read-through NPTII-SUG transcript is the trigger. In contrast, the SUG RNA resulting from the maturation of the read-through NPTII-SUG transcript is uncapped, and thus susceptible to degradation by XRN3 and XRN4, supporting the hypothesis that it is the actual S-PTGS trigger.
Remarkably, AS-PTGS and IR-PTGS are not abolished in ago1 mutants. Indeed, neither the null ago1-1 nor the hypomorphic ago1-27 mutations suppress AS-PTGS (Figure 3) or IR-PTGS triggered by 35S-driven transgenes (48). Moreover, the hypomorphic ago1-27 mutation only slightly reduces IR-PTGS specifically triggered around the phloem (Supplemental Figure S2), suggesting that another AGO is at play during AS-PTGS and IR-PTGS, at least when AGO1 is impaired. AGO2, which has been implicated in antiviral PTGS together with AGO1 (58), is a good candidate but its role in AS-PTGS and IR-PTGS remains to be tested. The impairment of S-PTGS in both the null ago1-1 and the hypomorphic ago1-27 mutants contrasts the mild or null effect of these mutations on AS-PTGS and IR-PTGS, and indicates an absolute dependence of S-PTGS on AGO1. Given that S-PTGS is also absolutely dependent on the production of secondary siRNAs, it follows that AGO1 is essential for the production or action of secondary siRNAs. To reconcile these results, we propose that not only the ago1-27 mutation reduces AGO1 cleavage activity (59), but it also impairs AGO1 capacity to engage the production of secondary siRNAs after cleavage. The Arabidopsis ago1-27 mutation affects the Alanine at position 994. In Arabidopsis, this Alanine is conserved in AGO5 and AGO10, but not in other AGOs. Given that AGO2, which carries a Glycine at the position equivalent to 994 in AGO1, is incapable to initiate the production of secondary siRNAs (60), it is possible that ago1-27 cannot produce secondary siRNAs because it is mutated at position 994. Therefore, AS-PTGS and IR-PTGS still function, at least partially, in ago1-27 because primary siRNAs are abundant enough to guide the cleavage of target mRNA through both the partially functioning ago1-27 protein and through another AGO, which could be AGO2 (58,60). In contrast, S-PTGS does not function in ago1-27 because primary siRNAs are scarce and secondary siRNAs are absent. Likely, the low abundance of SUG asRNA arising from read-through transcription of the NPTII transgene results in low amounts of primary siRNAs, which are incapable of significantly impacting the accumulation of the GUS target mRNA. Secondary siRNAs are therefore essential, but neither the mutant ago1-27 protein nor the other AGO at play is capable of producing them, resulting in S-PTGS impairment in ago1-27. Testing the effect of ago2 and mutations in other AGO proteins will help clarifying how many AGOs are at play during S-PTGS, AS-PTGS and IR-PTGS.
RNA gel blots and small RNA sequencing revealed that during S-PTGS siRNAs are not produced from the 5′ end of the GUS mRNA ((42) and Figure 4), suggesting that the AGO1/RDR6 module is incapable of completing the spreading of siRNAs from the 3′ to the 5′ end. In contrast, siRNAs produced during AS-PTGS span the entire GUS/SUG sequence (Figure 4). Given that siRNAs at the 5′ end of GUS are not eliminated in ago1–1 and rdr6 plants whereas siRNAs from the rest of the gene are reduced in these mutants (Figure 4, Table 1 and Supplemental Table S1), it is tempting to speculate that siRNAs coming from the 5′ end of GUS during AS-PTGS are primary siRNAs deriving from DCL2/DCL4-mediated processing of annealed GUS/SUG dsRNA, and not secondary siRNAs produced by AGO1/RDR6. Of course, it is impossible to determine if secondary siRNAs come from GUS or SUG. However, knowing that the AGO1/RDR6 module is incapable to complete the spreading of secondary siRNAs from the 3′ to the 5′ end of GUS during S-PTGS, the presence of primary siRNAs at the 5′ end of GUS (3′ end of SUG) at similar level in ago1-1 and rdr6 and in WT plants during AS-PTGS strongly suggests that the AGO1/RDR6 module is incapable of initiating the production of secondary siRNAs from the 3′ of SUG during AS-PTGS. In contrast, the AGO1/RDR6 module is capable of producing secondary siRNAs from the 3′ of GUS during both S-PTGS and AS-PTGS because AGO1/RDR6-dependent secondary siRNAs coming from the 3′UTR of the GUS transgene are found in both L1 and 6b4/SUG6 plants (Figure 4). What could explain the incapacity of the AGO1/RDR6 module to produce secondary siRNAs from the 3′ end of SUG during AS-PTGS? Contrary to GUS mRNA, SUG RNA does not have a proper reading frame and might therefore be treated differently. This may be because AGO1 is interacting with polysomes (49). Therefore, AGO1 bound to primary siRNAs originating from the GUS/SUG dsRNA during AS-PTGS would be recruited primarily to sense/coding GUS mRNAs but only marginally to antisense/non-coding SUG RNAs, leading to RDR6-dependent production of secondary siRNAs from GUS but not SUG. Eventually, AGO1 bound to secondary siRNAs would cleave both GUS and SUG RNAs in WT plants because of the abundance and dual polarity of secondary siRNAs. Supporting this hypothesis, the hypomorphic ago1-27 mutation prevents SUG degradation but has little effect on GUS degradation during AS-PTGS (Figure 3), likely because the mutant ago1–27 protein still exhibit enough activity to cleave GUS but cannot produce secondary siRNAs that are required to complete the degradation of SUG.
Could complementary RNAs deriving from endogenous genes trigger S-PTGS or AS-PTGS? Natural antisense transcripts (NATs) can derive from opposite strands of the same locus (cis-NATs) or from separate loci (trans-NATs), and they can involve protein-coding genes as well as non-protein coding genes. Mining Arabidopsis RNAseq datasets revealed 4080 cis-NATs and 2491 trans-NATs, and of these 6571 loci, 5385 produce siRNAs (11). The number of NATs is probably underestimated because of limited knowledge on the possibility that genes transcribe 3′ extension under certain circumstances. Indeed, genome-wide analysis of Arabidopsis fry1 mutants, which exhibit decreased XRN2, XRN3 and XRN4 activities, revealed read-through transcription at ∼2000 endogenous genes (61). Moreover, the number of NATs that are capable of producing siRNAs is probably underestimated because one of the two genes of the pair is generally expressed only under certain specific conditions, at least in the few cases analyzed (62). Indeed, despite the high number of NATs, very few examples of actual regulation involving nat-siRNAs have been documented (28,29). If NATs are capped and polyadenylated, nat-siRNA mediated regulation should resemble AS-PTGS. However, the processing of nat-siRNAs usually implicates DCL1, DCL2 or DCL3 but not DCL4 (11,13), suggesting a different pathway than AS-PTGS. Moreover, our results suggest that if a NAT pair involves at least one read-through transcript, maturation at the cleavage/polyadenylation site liberates an uncapped 3′ extension RNA that has the capacity to trigger S-PTGS on complementary mRNA. Also, supporting a similarity with S-PTGS, siRNAs originating from outside of the overlapping regions are found in ∼80% of the cis-NAT pairs (13), suggesting that siRNAs originating from the overlapping region guide primary cleavage, and that at least one cleavage product is transformed into dsRNA by a cellular RDR to produce secondary siRNAs from outside of the overlapping region (13). Several models of NAT regulation have been evoked (62), however a case-by-case analysis is likely required to decipher the regulatory mechanism at each NAT. We anticipate that the transgene S-PTGS and AS-PTGS models developed here will help understanding the regulation of gene expression by endogenous asRNA.
Supplementary Material
Acknowledgments
The authors thank Olivia Mendivil Ramos for her help in analyzing RNAseq data. The authors also thank the lab members for fruitful discussions.
Authors Contribution: H.V. and J.S.P. designed the experiments. J.S.P., V.J., N.B. and C.B. performed the experiments. J.S.P., M.H., M.Z. and H.V. analyzed results. J.S.P. and H.V. wrote the paper.
Footnotes
Present addresses:
Jean-Sébastien Parent, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11743, USA.
Vincent Jauvion, Vilmorin, Route du Manoir, 49250 La Ménitré, France.
Christophe Béclin, Aix-Marseille Université, CNRS, IBDM, UMR7288, Parc Scientifique de Luminy, 13288 Marseille Cedex 9, France.
Matthias Zytnicki, UMIAT, UR-875, INRA, 31326 Castanet-Tolosan, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds de Recherche du Québec – Nature et Technologies (FQRNT) (to J.S.P.); European Molecular Biology Organization (EMBO) (to J.S.P.); French Agence Nationale pour la Recherche [ANR-10-LABX-40 and ANR-11-BSV6-007]; Fondation Louis D. de l'Institut de France. Funding for open access charge: Prix Scientifique Louis D. de l'Institut de France 2009.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Faghihi M.A., Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I.S., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart B.J., Bartel D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 6.Gullerova M., Proudfoot N.J. Transcriptional interference and gene orientation in yeast: noncoding RNA connections. Cold Spring Harb. Symp. Quant. Biol. 2011;75:299–311. doi: 10.1101/sqb.2010.75.048. [DOI] [PubMed] [Google Scholar]
- 7.Gullerova M., Proudfoot N.J. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat. Struct. Mol. Biol. 2012;19:1193–1201. doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews O.E., Cha D.J., Wei C., Patton J.G. RNAi-mediated gene silencing in zebrafish triggered by convergent transcription. Sci. Rep. 2014;4:5222. doi: 10.1038/srep05222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 11.Yuan C., Wang J., Harrison a. P., Meng X., Chen D., Chen M. Genome-wide view of natural antisense transcripts in Arabidopsis thaliana. DNA Res. 2015;22:233–243. doi: 10.1093/dnares/dsv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Liu X.S., Liu Q.R., Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Xia J., Lii Y., Barrera-Figueroa B., Zhou X., Gao S., Lu L., Niu D., Liang W., Chen Z., et al. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H., Vacic V., Girke T., Lonardi S., Zhu J.K. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol. Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubko E., Kunova A., Meyer P. Sense and antisense transcripts of convergent gene pairs in Arabidopsis thaliana can share a common polyadenylation region. PLoS One. 2011;6:e16769. doi: 10.1371/journal.pone.0016769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourque J.E. Antisense strategies for genetic manipulations in plants. Plant Sci. 1995;105:125–149. [Google Scholar]
- 17.Waterhouse P.M., Graham M.W., Wang M.-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodersen P., Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Parent J.-S., Martínez de Alba A.E., Vaucheret H. The origin and effect of small RNA signaling in plants. Front. Plant Sci. 2012;3:179. doi: 10.3389/fpls.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bologna N.G., Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 21.Fusaro A.F., Matthew L., Smith N.A., Curtin S.J., Dedic-Hagan J., Ellacott G.A., Watson J.M., Wang M.B., Brosnan C., Carroll B.J., et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunoyer P., Himber C., Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 23.Parent J.-S., Bouteiller N., Elmayan T., Vaucheret H. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015;81:223–232. doi: 10.1111/tpj.12720. [DOI] [PubMed] [Google Scholar]
- 24.Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 25.Mourrain P., Béclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Jouette D., Lacombe A.M., Nikic S., Picault N., et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 26.Martínez de Alba A.E., Moreno A.B., Gabriel M., Mallory C., Balzergue S., Aubourg S., Maizel A., Gautheret D., Crespi M.D. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015;43:2902–2913. doi: 10.1093/nar/gkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grierson D., Fray R.G., Hamilton A.J., Smith C.J.S., Watson C.F. Does co-suppression of sense genes in transgenic plants involve antisense RNA? Trends Biotechnol. 1991;9:122–123. [Google Scholar]
- 28.Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A., Jr, Zhu J.K., Staskawicz B.J., Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ron M., Alandete Saez M., Eshed Williams L., Fletcher J.C., McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Held M.A., Penning B., Brandt A.S., Kessans S.A., Yong W., Scofield S.R., Carpita N.C. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20534–20539. doi: 10.1073/pnas.0809408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elmayan T., Balzergue S., Beon F., Bourdon V., Daubremet J., Guenet Y., Mourrain P., Palauqui J.C., Vernhettes S., Vialle T., et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morel J.-B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lepère G., Bétermier M., Meyer E., Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zytnicki M., Quesneville H. S-MART, A software toolbox to aid RNA-seq data analysis. PLoS One. 2011;6:6–8. doi: 10.1371/journal.pone.0025988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutet S., Vazquez F., Liu J., Béclin C., Fagard M., Gratias A., Morel J.B., Crété P., Chen X., Vaucheret H. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mlotshwa S., Pruss G.J., Peragine A., Endres M.W., Li J., Chen X., Poethig R.S., Bowman L.H., Vance V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One. 2008;3:e1755. doi: 10.1371/journal.pone.0001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Z., Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouché N., Lauressergues D., Gasciolli V., Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blevins T., Rajeswaran R., Shivaprasad P. V, Beknazariants D., Si-Ammour A., Park H.S., Vazquez F., Robertson D., Meins F., Jr, Hohn T., et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagard M., Boutet S., Morel J.B., Bellini C., Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 48.Béclin C., Boutet S., Waterhouse P., Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 49.Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Paoli E., Dorantes-acosta A., Zhai J., Accerbi M., Jeong D., Park S., Meyers B.C., Jorgensen R.A., Green P.J. Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA. 2009;15:1965–1970. doi: 10.1261/rna.1706109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wroblewski T., Matvienko M., Piskurewicz U., Xu H., Martineau B., Wong J., Govindarajulu M., Kozik A., Michelmore R.W. Distinctive profiles of small RNA couple inverted repeat-induced post-transcriptional gene silencing with endogenous RNA silencing pathways in Arabidopsis. RNA. 2014;20:1987–1999. doi: 10.1261/rna.046532.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szittya G., Moxon S., Pantaleo V., Toth G., Rusholme Pilcher R.L., Moulton V., Burgyan J., Dalmay T. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010;6:e1000838. doi: 10.1371/journal.ppat.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorefan K., Pais H., Hall A.E., Kozomara A., Griffiths-Jones S., Moulton V., Dalmay T. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lunerová-Bedřichová J., Bleys A., Fojtová M., Khaitová L., Depicker A., Kovařík A. Trans-generation inheritance of methylation patterns in a tobacco transgene following a post-transcriptional silencing event. Plant J. 2008;54:1049–1062. doi: 10.1111/j.1365-313X.2008.03475.x. [DOI] [PubMed] [Google Scholar]
- 56.Gullerova M., Moazed D., Proudfoot N.J. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25:556–568. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert D., Lechtenberg B., Forsbach A., Gils M., Bahadur S., Schmidt R. Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell. 2004;16:2561–2572. doi: 10.1105/tpc.104.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X.B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X., Gasciolli V., Vaucheret H., Ding S.W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaucheret H., Vazquez F., Crété P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurihara Y., Schmitz R.J., Nery J.R., Schultz M.D., Okubo-Kurihara E., Morosawa T., Tanaka M., Toyoda T., Seki M., Ecker J.R. Surveillance of 3′ noncoding transcripts requires FIERY1 and XRN3 in Arabidopsis. G3 Genes|Genomes|Genetics. 2012;2:487–498. doi: 10.1534/g3.111.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X., Lii Y., Wu Z., Polishko A., Zhang H., Chinnusamy V., Lonardi S., Zhu J.K., Liu R., Jin H. Mechanisms of small RNA generation from Cis-NATs in response to environmental and developmental cues. Mol. Plant. 2013;6:704–715. doi: 10.1093/mp/sst051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


