Abstract
AlgR is a key transcriptional regulator required for the expression of multiple virulence factors, including type IV pili and alginate in Pseudomonas aeruginosa. However, the regulon and molecular regulatory mechanism of AlgR have yet to be fully elucidated. Here, among 157 loci that were identified by a ChIP-seq assay, we characterized a gene, mucR, which encodes an enzyme that synthesizes the intracellular second messenger cyclic diguanylate (c-di-GMP). A ΔalgR strain produced lesser biofilm than did the wild-type strain, which is consistent with a phenotype controlled by c-di-GMP. AlgR positively regulates mucR via direct binding to its promoter. A ΔalgRΔmucR double mutant produced lesser biofilm than did the single ΔalgR mutant, demonstrating that c-di-GMP is a positive regulator of biofilm formation. AlgR controls the levels of c-di-GMP synthesis via direct regulation of mucR. In addition, the cognate sensor of AlgR, FimS/AlgZ, also plays an important role in P. aeruginosa virulence. Taken together, this study provides new insights into the AlgR regulon and reveals the involvement of c-di-GMP in the mechanism underlying AlgR regulation.
INTRODUCTION
The opportunistic pathogen Pseudomonas aeruginosa is associated with severe nosocomial infections and is the leading cause of mortality in patients with cystic fibrosis (1). P. aeruginosa can form a biofilm, which is a sessile community of bacteria that can switch to a motile lifestyle (2). Bacteria in a biofilm are surrounded by an extracellular matrix consisting of extracellular DNA, exopolysaccharides and proteins (3,4), which confers resistance to antibiotic treatment and immune cells (5,6). Biofilms can form on a variety of surfaces in natural, industrial and hospital niches. Biofilms are also responsible for persistent and chronic infections because of their high resistance to antimicrobial agents and the selection of phenotypic variants (7).
P. aeruginosa strains isolated from the lungs of patients with cystic fibrosis synthesize and secrete excessive amounts of alginate (8,9), which enable P. aeruginosa to evade phagocytosis by neutrophils and macrophages (10). In addition, alginate insulates bacteria from reactive oxygen intermediates (11) and hypochlorite generated by the phagocytic cells of the host (12). The overexpression of alginate also contributes to P. aeruginosa biofilm development and function. For example, an algU/T-overproducing strain exhibits enhanced microcolony formation and displays highly structured mature biofilm (13).
In P. aeruginosa, the alginate biosynthesis enzymes are encoded by the alginate biosynthetic operon, algD-alg8-alg44-algKEGXL (14). The operon is controlled by a group of regulators including AmrZ, AlgB, Vfr and CysB (15–18). AmrZ regulates P. aeruginosa biofilm development and virulence (17), which is dependent on the concentrations of intracellular c-di-GMP (19). The production of alginate is tightly controlled by a complex regulatory network (20), that includes MucA, MucB, MucC, MucD, MucP, AlgP (hp1), AlgQ, AlgW, AlgT (AlgU) and ClpXP (21–27).
AlgR/AlgZ is a two-component system that controls alginate production (28). The LytTR-family regulator AlgR activates transcription of algD by binding to three distinct sites in its promoter region, including two located unusually far upstream of the algD transcription start site (29). AlgR also regulates alginate production through algC by binding to its promoter (30). In addition, AlgR globally influences the expression of a broad range of virulence factors including the rhl quorum-sensing system, hydrogen cyanide production, type III secretion system (T3SS) and rhamnolipid production (31–33). In a pulmonary infection model, mice infected with an algR mutant showed a greater survival rate than those infected by the wild-type strain (34). In P. aeruginosa, AlgR, but not FimS/AlgZ, is a key regulator of alginate production (35). Moreover, although no evidence has yet confirmed the direct interaction of, or phosphotransfer between, AlgR and FimS/AlgZ, the phosphorylation of AlgR enhances its affinity to some promoter regions (31).
Although AlgR and FimS/AlgZ have been widely studied for several decades, the direct in vivo targets of AlgR, and the distinct regulatory mechanisms of AlgR and FimS/AlgZ, have not been fully elucidated. Here, we detected 157 loci in the P. aeruginosa genome that were bound by AlgR by using a ChIP-seq (chromatin immunoprecipitation (ChIP) followed by high throughput DNA sequencing) approach. The AlgR consensus binding sequence was identified by using the multiple EM for motif elicitation (MEME) suite and verified by footprint assays in vitro, which uncovered several unprecedented regulatory pathways including AlgR-CzcR and AlgR-MucR. In addition, many AlgR-bound genes were involved in pathogenesis, indicating the importance of AlgR in P. aeruginosa virulence. In sum, the ChIP-seq data provided several new insights into the molecular regulatory mechanism of AlgR.
The presence of an AlgR-MucR pathway is consistent with previous reports showing that ΔalgR mutant produces lesser biofilm than does the wild-type PAO1 strain (32,36). We demonstrated that AlgR positively regulates mucR, which encodes a diguanylate cyclase (DGC). MucR has a DGC (GGDEF) domain that is responsible for c-di-GMP biosynthesis. Overexpression of mucR led to increased concentrations of intracellular c-di-GMP. This observation suggests that compromised c-di-GMP level in the ΔalgR mutant accounted for the reduction in biofilm production, which is consistent with previous reports showing that c-di-GMP positively regulates biofilm production (37,38). Overall, this work provides new insights into the molecular mechanisms used by AlgR to control P. aeruginosa pathogenesis.
MATERIALS AND METHODS
Bacterial stains and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The P. aeruginosa PAO1 strain and derivatives were grown at 37°C on LB agar plates or in LB broth with shaking at 200 rpm. Antibiotics were used at the following concentrations: for Escherichia coli, gentamicin (39) at 15 μg/ml, ampicillin at 100 μg/ml and tetracycline 10 μg/ml; for P. aeruginosa, gentamicin (39) at 50 μg/ml in LB or 150 μg/ml in PIA (Pseudomonas Isolate Agar); tetracycline at 150 μg/ml in LB or 300 μg/ml in PIA; carbenicillin at 500 μg/ml in LB.
ChIP-seq analysis
ChIP was performed as previously described with minor changes (40,41). The wild-type P. aeruginosa containing either empty pAK1900 or pAK1900-AlgR-VSV was cultured in LB medium supplemented with ampicillin until the mid-log phase (OD = 0.6, Optical Density), before it was treated with 1% formaldehyde for 10 min at 37°C. Crosslinking was stopped by addition of 125 mM glycine. Bacterial pellets were washed twice with a Tris buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl), and then resuspended in 500 μl IP buffer (50 mM HEPES-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, mini-protease inhibitor cocktail (Roche)) and the DNA was sonicated (Fisher 505 Sonic Dismembrator) to sizes of 100–300 bp (20% total output, 20-second on, 30-second off, for 2 min on ice). Insoluble cellular debris was removed by centrifugation and the supernatant used as input sample in IP experiments. Both control and IP samples were added into protein A beads (General Electric), and then incubated with 50 μl agarose-conjugated anti-VSV antibodies (Sigma) in IP buffer. Washing, crosslink reversal and purification of the ChIP DNA were conducted by following previously published protocols (40). DNA fragments (150–250 bp) were selected for library construction and sequencing libraries prepared using the NEXTflex™ ChIP-Seq Kit (Bioo Scientific). The libraries were sequenced using the HiSeq 2000 system (Illumina). ChIP-seq reads were mapped to the P. aeruginosa genomes, using TopHat (Version 2.0.0) with two mismatches allowed (42). Only the uniquely mapped reads were kept for the subsequent analyses. The enriched peaks were identified using MACS software (version 2.0.0) (43), which was followed by MEME analyses to generate the AlgR-binding motif (44). The ChIP-seq data files have been deposited in National Center of Biotechnology Information's Gene Expression Omnibus (GEO) and can be accessed through GEO Series accession number GSE65356.
Expression and purification of AlgR protein
AlgR protein was expressed and purified as previously described (36,45). Briefly, 10 ml of overnight pre-cultures grown from a single colony was inoculated into 1 l of autoclaved LB medium containing 100 μg/ml ampicillin. The cells were grown at 37ºC, 250 rpm to OD600 ∼0.6 and then the temperature was reduced to 16ºC. Protein expression was induced with 1 mM IPTG (Isopropyl β-D-1-Thiogalactopyranoside). The overnight culture was harvested at 4ºC by centrifugation at 6300 g for 8 min. All subsequent steps were performed at 4ºC. The pellet was suspended in 20 ml TMBG buffer (50 mM Tris-HCl [pH 7.6], 5 mM MgCI2, 5 mM 2-mercaptoethanol, 10% glycerol) containing 1 mM PMSF (Phenyl-methanesulfonyl fluoride). The cells were lysed by sonication and centrifuged at 12 000 rpm for 25 min. The supernatant was filtered through a 0.45 μm filter and applied to a heparin-agarose column equilibrated with TMBG buffer. After washing with TMBG buffer, the column was eluted with a 300-ml linear gradient of 0–1.2 M NaCl prepared in TMBG buffer. Peak fractions were pooled and verified by sodium dodecyl sulphate polyacrylamide gel electrophoresis (Supplementary Figure S1A).
Electrophoretic mobility shift assays (EMSA)
Various amounts of AlgR proteins were incubated with different DNA probes (Supplementary Table S2) in 20 μl of the gel shift-loading buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5 mM dithiothreitol, 10% glycerol and 3 μg/ml sheared salmon sperm DNA). After incubation at room temperature for 20 min, the samples were analyzed by 6% polyacrylamide gel electrophoresis in 0.5 X TBE (Tris/Boric Acid/EDTA) buffer at 90 V for 90 min. The gels were stained by SYBR GOLD dye (Life Technologies) and subjected to a gel-doc system (Tanon 5500).
Dye primer-based DNase I footprint assay
The DNA footprint assay was followed as previously described (46). Briefly, a 301-bp promoter fragment of the mucR promoter region that encompasses bases from –246 to +55 was generated by polymerase chain reaction (PCR) with primers mucRgf (carrying 6-FAM at the 5′) and mucRgr. Forty nanomolar of 6-FAM-labeled mucR promoter probe was incubated with varying amounts of AlgR protein ranging from 0 to 2 μM in gel-shift loading buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 10% Glycerol and 3 μg/ml sheared salmon sperm DNA). After several optimization experiments, the nuclease digestion was found to work best with 0.05 units of DNase I (New England Biolabs, NEB) per 20 μl reaction for 5 min at 25°C. The reaction was stopped with 0.25 M EDTA and extracted with phenol-chloroform-isoamylalcohol (25:24:1). Control digestions with the mucR promoter probe were done with 10 μM of bovine serum albumin instead of AlgR. The DNA fragments were purified with the QIAquick PCR Purification kit (Qiagen) and eluted in 15 μl distilled water. About 5 μl of digested DNA was added to 4.9 μl HiDi formamide (Applied Biosystems) and 0.1 μl GeneScan-500 LIZ size standards (Applied Biosystems). The samples were analyzed with the 3730 DNA Analyzer. Results were analyzed with Peak Scanner (Applied Biosystems). The identical footprint assay protocol was applied to the czcR promoter.
Construction of complemented vectors
Plasmids p-algR, p-algR-VSV, p-fimS, p-mucR, P-mucRDGC, p-czcR were constructed respectively by amplifying corresponding fragments with the primer pairs (Supplementary Table S2) pAK-algR-S/pAK-algR-A, pAK-fimS-S/pAK-fimS-A, pAK-mucR-S/pAK-mucR-A, pAK-czcR-S/pAK-czcR-A, pAK-mucRDGC-S/pAK-mucRDGC-A, pAK-sadC-S/pAK-sadC-A by PCR. The PCR products were digested with the indicated enzymes and cloned into PAK1900 (47). For construction of Mini-CTX-algR, the Mini-CTX-algR-S/Mini-CTX-algR-A primer was used to amplify the algR gene. The digested PCR products were cloned into the corresponding enzymes sites (BamHI/HindШ) in Mini-CTX-lacZ to generate Mini-CTX-algR. All constructs were verified by sequencing.
Construction of P. aeruginosa ΔalgR, ΔfimS, ΔmucR, ΔalgRΔmucR, ΔfimSΔmucR mutants
For gene replacement, a SacB-based strategy (48) was employed as described in our previous studies (41). To construct the algR null mutant (ΔalgR), PCRs were performed to amplify an upstream fragment (1978 bp) and a downstream fragment (1966 bp) of algR by using primers pEX-algR-up-S/pEX-algR-up-A (containing BamHI/XbaI) and pEX-algR-down-S/pEX-algR-down-A (containing XbaI/HindIII), respectively (Supplementary Table S2). The two PCR products were digested and then cloned into BamHI/HindIII-digested gene replacement vector pEX18Ap, yielding pEX18Ap-algR. A 0.9 kb gentamicin resistance cassette cut from pPS858 with XbaI was cloned into pEX18Ap::algRSA, yielding pEX-algR-Gm. The resultant plasmid, pEX-algR, was electroporated into the wild-type PAO1 strain with selection for gentamicin resistance. Colonies were screened for gentamicin resistance, carbenicillin sensitivity and loss of sucrose sensitivity (5%), which typically indicates a double crossover event and thus of gene replacement occurring. The ΔalgR mutant was further confirmed by PCR.
For deletion of fimS gene in PAO1, the upstream fragment (1935 bp) of the intended deletion was amplified with primer pair pEX-fimS-up-S/pEX-fimS-up-A (BamHI/XbaI) (Supplementary Table S2) while the downstream fragment (1960 bp) was amplified with primer pair pEX-fimS-down-S/pEX-fimS-down-A (XbaI /HindIII). The 0.9 kb gentamicin resistance cassette was cloned into pEX18Ap-fimS, yielding pEX-fimS-Gm. For deletion of algR/fimS gene in PAO1, the upstream fragment (1935 bp) of the fimS gene was amplified with primer pair pEX-fimS-up-S/pEX-fimS-up-A (BamHI/XbaI) and the downstream fragment (1966 bp) of algR gene was amplified with primer pair pEX-algR-down-S/pEX-algR-down-A (XbaI /HindIII). The 0.9 kb gentamicin resistance cassette was cloned into pEX18Ap-fimS/algR, yielding pEX-fimS/algR-Gm. For deletion of mucR gene in PAO1, the upstream fragment (1611 bp) of the mucR gene was amplified with primer pair pEX-mucR-up-S/pEX-fimS-up-A (BamHI/XbaI) while the downstream fragment (1962 bp) was amplified with primer pair pEX-mucR-down-S/pEX-mucR-down-A (XbaI/HindIII). The 1.5 kb tetracycline resistance cassette was cloned into pEX18Ap-mucR, yielding pEX-mucR-Tc.
In order to make an algR/mucR double deletion strain (ΔalgRΔmucR), the resultant plasmid, pEX-mucR-Tc, was electroporated into ΔalgR strain with selection for tetracycline resistance. Colonies were screened for tetracycline resistance and loss of sucrose sensitivity (5%). The ΔalgRΔmucR mutant was further confirmed by PCR. The mucR gene in ΔfimS mutant was deletion by a similar strategy with plasmid pEX-mucR-Tc.
Construction of the promoter-reporter plasmids
The plasmid pMS402 carrying a promoterless luxCDABE reporter gene cluster was used to construct promoter-lux fusions as reported previously (49). The promoter region was amplified by PCR using the corresponding primers (Supplementary Table S2): pKD-mucR-S/pKD-mucR-A, pKD-czcR-S/pKD-czcR-A, pKD-cdrA-S/pKD-cdrA-A, pKD-4396-S/pKD-4396-A, pKD-purL-S/pKD-purL-A, pKD-2189-S/pKD-2189-A, pKD-2867-S/pKD-2867-A, pKD-1736-S/pKD-1736-A. These promoter regions were cloned into the BamHI-XhoI sites upstream of the lux genes in pMS402. The construct was transformed into the PAO1 strains by electroporation. Cloned promoter sequences were confirmed by DNA sequencing.
Luminescence screening assays
Expression of lux-based reporters from bacteria grown in liquid culture was measured as counts per second of light production in a Synergy 2 Plate Reader (Biotek) (50). Overnight cultures of the reporter strains were diluted to an A600 of 0.2 and cultivated for an additional 2 h before use. The cultures were inoculated into parallel wells of a black 96-well plate with a transparent bottom. A 5-μl volume of the fresh cultures was inoculated into the wells containing a total volume of 95 μl medium plus other components, and the A600 value in the wells was adjusted to around 0.07. A 60-μl volume of filter-sterilized mineral oil was added to prevent evaporation during the assay. Promoter activities were measured every 30 min for 24 h. Bacterial growth was monitored at the same time by measuring the OD at 595 nm in a Synergy 2 Plate Reader (BioTek).
Quantification of c-di-GMP by LC-MS
Extraction of c-di-GMP from P. aeruginosa cells as previously described (51). Briefly, P. aeruginosa was grown overnight in VBMM (Vogel-Bonner minimal medium: 0.2 g/l MgSO4. 7H2O, 2.0 g/l citric acid, 3.5 g/l NaNH4HPO4. 4H2O, 10 g/l K2HPO4) and subcultured the next day to an OD600 of 0.02 in 250 ml shake flasks containing 30 ml of VBMM. When cells reached an OD600 of ∼0.6, 1.5 ml was removed, centrifuged for 1 min at 14 000 × g and the supernatant was removed. The cell pellet was resuspended in 150 μl 0.6 M perchloric acid. Samples were incubated on ice for 30 min, and cell debris was removed by centrifugation at 4°C for 5 min. Supernatants (150 μl) were removed and samples were neutralized by addition of 30 μl 2.5 M KHCO3. The resulting precipitate was removed by centrifugation at 4°C. These neutralized supernatants were stored at −80°C until analyzed by LC-MS. For protein determinations, three 1.5-ml samples of each culture were removed and centrifuged to pellet cells. These cell pellets were resuspended in 150 μl 1 M NaOH and boiled in a water bath for 10 min. Samples were then stored on ice. The total protein level was measured by the Bio-Rad Protein Assay kit, which is then used for normalization of c-di-GMP value.
Protocols for c-di-GMP separations and analysis by mass spectrometry were based on previous studies (19,51). Compounds were separated on an Acuity UPLC equipped with a C18 Guard Cartridge (Phenomenex) and using a 2.0 × 50 mm Synergi Hydro RP column (Phenomenex). A gradient system was used starting with 98% aqueous (10 mM ammonium formate, pH 4.0) and 2% organic (acetonitrile). The aqueous concentration was adjusted to 70% at 2 min, 20% at 2.5 min, 100% at 3 min and finally held at 98% from 5 to 7.5 min. The m/z 691>152 transition was used for the identification of c-di-GMP. For a standard curve, 50, 100, 250, 500 and 1000 fmol pure c-di-GMP (Invivogen, San Diego) were analyzed by the above method. The c-di-GMP levels are normalized to total protein per ml of culture. Data represent averages of three independent cultures.
Biofilm formation assay
Biofilm formation was measured in a static system as previously described (52) with minor modifications. Visualization of biofilm formation was carried out in 15-ml borosilicate tubes. Briefly, bacteria from overnight cultures were inoculated at 1:100 dilutions into LB medium supplemented with appropriate antibiotics and grown at 30°C for 10 h. Biofilms were stained with 0.1% crystal violet (CV) and tubes were washed with water to remove unbound dye. Quantification of biofilm formation was performed in 24-well polystyrene microtiter plates. LB and appropriate antibiotics was inoculated to a final OD600nm of 0.01. The plates were incubated for 8 h or 20 h at 30°C. CV was added to each tube and stained for 15 min prior to removal by aspiration. Wells were rinsed three times by submerging the tubes in distilled water, and the remaining CV was dissolved in 1 ml of 95% ethanol. A 1-ml portion of this solution was transferred to a new polystyrene tube, and the absorbance was measured at 600 nm.
Twitching motility assay
Bacterial twitching motility was assessed as described previously (53). Overnight cultures were stab inoculated to LB plates containing 1% agar with a sharp toothpick to the bottom of the Petri dish. Twitch plates were incubated at 37°C for 24 h. Photographs were taken with the Tanon 2500 imaging system.
Measurement of pyocyanin production
Pyocyanin was extracted from culture supernatants and measured using previously reported methods (54). Briefly, 3-ml chloroform was added to 5 ml culture supernatant. After extraction, the chloroform layer was transferred to a fresh tube and mixed with 1 ml 0.2 M HCI. After centrifugation, the top layer was removed and its A520 was measured. Pyocyanin concentrations, expressed as μg produced (ml culture supernatant) −1, were determined by multiplying the A520 by 17.072 (54).
RESULTS
ChIP-seq analysis reveals the direct targets of AlgR in the P. aeruginosa genome
Previous studies have demonstrated that AlgR directly binds to the promoters of the algD and fimU genes, and have identified the binding sequences by footprinting assays (29,55). To identify in vivo targets of AlgR, we performed ChIP-seq assays (40). VSV-tagged full length AlgR was overexpressed from plasmid pAK1900 and then transformed into a wild-type strain. To verify the activity of the AlgR-VSV in vivo, we tested the twitching phenotype in the wild-type PAO1 strain, the ΔalgR mutant and the complementary strains (ΔalgR/p-algR and ΔalgR/p-algR-VSV). Consistent with a previous study showing that a ΔalgR strain loses twitching ability (35), the ΔalgR strains carrying p-algR or p-algR-VSV had their twitching abilities restored to the wild-type level (Supplementary Figure S1), indicating that AlgR-VSV is as functional as the wild-type AlgR. Next, chromatin-bound to AlgR was crosslinked, sheared, purified and sequenced. Sequence reads were obtained from two independent ChIP-seq assays by using VSV-specific antibody and mapped to the P. aeruginosa genome. Using the MACS software (43), we identified 157 enriched loci (P-value = e-5) harboring AlgR-binding peaks (Supplementary Table S3), that were enriched >1.5-fold, but were absent in control samples using the wild-type PAO1 strain containing an empty pAK1900 vector without a VSV tag. Consistent with the previous studies, algD and fimU promoters were significantly enriched compared to input DNA (3.22- and 2.97-fold, respectively), whereas the lasR promoter showed no significant enrichment compared to input DNA. Moreover, these 157 loci were located across the genome and were situated both in intergenic regions (45%) and within coding regions (55%), suggesting that AlgR is a global transcriptional regulator in P. aeruginosa (Figure 1A).
Figure 1.
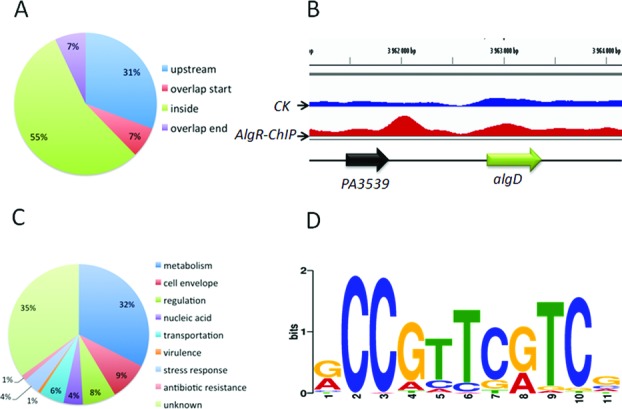
ChIP-seq assay reveals 157 in vivo binding sites of AlgR in the P. aeruginosa genome. (A) The positions of the AlgR-binding peaks are presented in a pie chart. (B) AlgR binds to the promoter region of algD. CK is the control sample. (C) A pie chart presenting the percentage of AlgR targets with functional categories defined in the Pseudomonas database (http://pseudomonas.com). (D) The most significant motif identified by ChIP-seq using the MEME tool is shown. All peaks were used to define the binding motif. The height of each letter presents the relative frequency of each base at different positions in the consensus sequence.
To further determine the biological processes and molecular functions of the AlgR directed target genes, they were categorized based on gene ontology. Protein products encoded by AlgR-bound genes had different functions including those related to metabolism (32%), cell envelope (9%), regulation (8%), nucleic acid (4%), transportation (6%), virulence (1%), stress response (4%), antibiotic resistance (1%) and unknown functions (35%) (Figure 1C). However, we noted that some reported AlgR-bound promoter regions, such as hcnA and rhlA (31), were absent from these 157 loci, perhaps owing to high but nonspecific peaks in the loci from the control sample, or to unsuitable test conditions. Collectively, this newly identified list of target genes strongly suggests that AlgR is at the center of multiple virulence and metabolic pathways.
The ChIP-seq data allowed us to further define the consensus-binding site of AlgR. By using MEME tool on the identified peaks (44), we determined an 11-bp AlgR consensus sequence (Figure 1D). This sequence was predominated by 5′-CCGTTCGTC-3′ in positions 2–10, with a cytosine at position 2, which matches the motif revealed in previous footprint assays on algD and fimU promoters (29,55). Moreover, we searched for this motif in a previously published ΔalgR microarray dataset (56) and found that the AlgR binding motif is overrepresented in the differentially expressed genes, confirming its accuracy.
AlgR directly regulates genes associated with the cell envelope and virulence
The ChIP-seq results suggested that AlgR directly binds to 14 genes that are associated with the cell envelope (Figure 1C). Eight of these 14 genes carry an AlgR binding site within 800 bp from the translation start of the genes (Table 1, Supplementary Table S3). Two AlgR targets, algD and fimU, have been previously characterized (55). Furthermore, AlgR binds to the upstream of genes that are potentially, or are known to be related to P. aeruginosa virulence, including amrZ, mucR, opgG, czcC, PA1048 and PA0499. amrZ encodes a transcriptional regulator involved in P. aeruginosa biofilm development and virulence (17). A recent study further showed that AmrZ regulates biofilm formation and chronicity by influencing c-di-GMP synthesis (19). MucR is annotated as a c-di-GMP-synthesizing enzyme, whose overexpression in P. aeruginosa leads to increased alginate production, wrinkled colony morphology and highly structured biofilm formation (57). The dysregulation of c-di-GMP signaling may account for the lower levels of biofilm production in the ΔalgR mutant, than in the wild-type PAO1 strain (32).
Table 1. ChIP-seq revealed that AlgR directly bound to 14 genes involved in cell envelope and virulence.
| Gene ID | Gene name | Gene product | ChIP-seq fold enrichment | Position |
|---|---|---|---|---|
| PA0499 | putative pili assenbly chaperone | 2.77 | Upstream | |
| PA1048 | putative membrane protein | 2.92 | Upstream | |
| PA1727 | mucR | Cyclase | 3.0 | Upstream |
| PA2522 | czcC | Membrane protein precursor CzcC | 3.16 | Upstream |
| PA2824 | sagS | Surface attachment and growth sensor hybrid | 2.93 | Overlap start |
| PA2960 | pilZ | Type 4 fimbrial biogenesis protein PilZ | 2.81 | Inside |
| PA3064 | pelA | PelA | 3.06 | Inside |
| PA3385 | amrZ | Alginate regulator Z | 2.48 | Upstream |
| PA3540 | algD | GDP-mannose 6-dehydrogenase | 3.22 | Upstream |
| PA4541 | lepA | Hypothetical protein | 2.73 | Inside |
| PA4550 | fimU | type 4 fimbrial biogenesis protein | 2.97 | Upstream |
| PA4625 | cdrA | cyclic diguanylate-regulated TPS partner A | 2.24 | Inside |
| PA5040 | pilQ | Type 4 fimbrial biogenesis outer membrane protein PilQ precursor | 2.71 | Inside |
| PA5078 | opgG | glucans biosynthises | 2.06 | Upstream |
Validation of AlgR ChIP-seq results in vitro and in vivo
To verify the enriched loci harboring AlgR-binding peaks in the P. aeruginosa genome (Supplementary Table S3), we performed EMSA assays on selected targets. To this end, we expressed and purified AlgR protein as previously described (36,45) (Supplementary Figure S2A). Given that a typical transcriptional factor binds to gene promoter regions, six promoter regions of AlgR target genes (PA4396, PA2189, PA1736, purL, PA2867 and amrZ) were selected and tested using EMSA in vitro, and the lasR promoter was used as a negative control. The results showed that AlgR efficiently binds to all but the amrZ probe in a concentration-dependent manner, while the negative control lasR promoter remained unbound at the highest concentration (0.4 μM) (Figure 2A–G).
Figure 2.
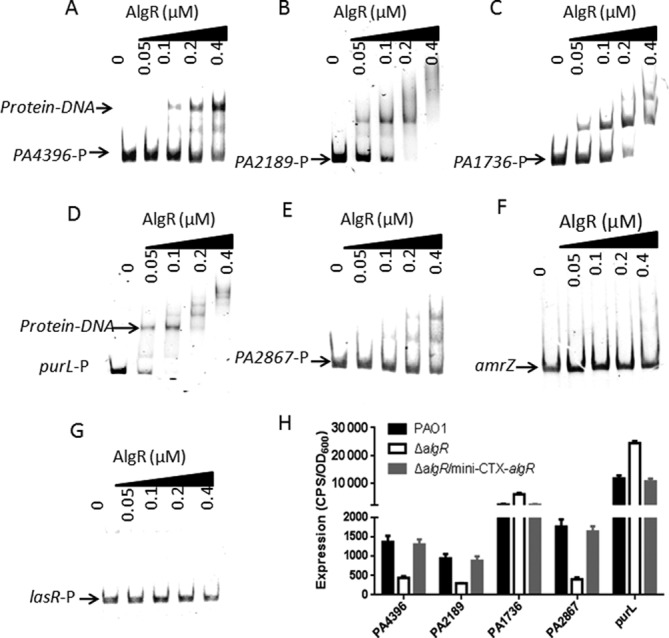
(A) AlgR binds to the selected target regions in vitro. The chosen promoter regions (for PA4395/PA4396, PA2189, PA1736, purL and PA2867) and EMSA analyses, were described in Materials and Methods. PCR products containing the indicated fragments were added to the reaction mixtures at ∼50 nM each. AlgR protein was added to the reaction buffer in lanes 2–5 with 0.05, 0.1, 0.2, 0.4 μM, respectively. No protein was added in lane 1. The negative control (lasR promoter region) showed no binding to AlgR. (B) The expression of selected genes (PA4396, PA2189, PA1736, PA2867 and purL) was evaluated in the wild-type PAO1, the ΔalgR mutant and the complemented (ΔalgR/Mini-CTX-algR) strains. Data are shown as mean ± SEM from three experiments.
To further confirm the AlgR-binding peaks in vivo, we next investigated the gene expression levels of several newly identified AlgR targets in vivo. We generated an algR deletion strain in the wild-type PAO1 background (ΔalgR) and the ΔalgR complemented (ΔalgR/mini-CTX-algR) strain as described in Materials and Methods. Subsequently, the expression of these selected genes (PA4396, PA2189, PA1736, purL and PA2867) was evaluated in the wild-type PAO1 strain, the ΔalgR strain and the complemented (ΔalgR/mini-CTX-algR) strain. As shown in Figure 2H, the expression of PA4396, PA2189, PA1736, purL and PA2867 was differentially affected by the deletion of algR. However, the expression level of amrZ in the ΔalgR strain was the same as the wild-type PAO1 strain (Supplementary Figure S2B), indicating that AlgR is not required for amrZ expression.
AlgR regulates pyocyanin production by regulating CzcR expression
AlgR specifically binds to the intergenic region between czcR and czcC genes (Figure 3A). The membrane-bound CzcCBA protein complex and its cognate two-component system CzcR-CzcS are involved in heavy metal and carbapenem resistance in P. aeruginosa (58). CzcR directly binds to the promoter region of phzA1 and thus negatively regulates pyocyanin synthesis (59), which led us to test whether AlgR regulated the production of pyocyanin via CzcR. To test this hypothesis, we performed an EMSA assay and found that AlgR directly binds to the czcR promoter region (Figure 3B). We further mapped an AlgR-binding site in the promoter region of czcR using a dye-primer-based DNase I footprinting assay (Figure 3C). Using Peak Scanner Software (Applied Biosystems), we compared the electropherograms generated with and without AlgR, and uncovered a specific AlgR protected region extending from nucleotides −445 to −437 (CCGTTCATC) relative to the czcR start codon (Figure 3C). This site matches the consensus sequence identified from the ChIP-seq data, which confirms that the motif is crucial to the DNA-binding ability of AlgR.
Figure 3.
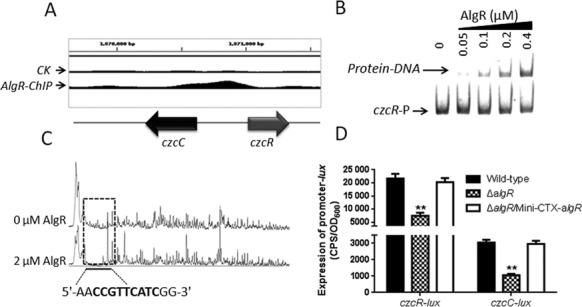
AlgR directly binds to the czcR promoter region and controls its activity. (A) AlgR binds to the czcR-czcC intergenic region according to the ChIP-seq analyses. (B) An EMSA experiment showed that AlgR directly binds to the promoter region of czcR. A PCR product containing the czcR promoter region was added to each reaction mixtures at 30 nM. AlgR protein was added to reaction buffer in lanes 2–5 with 0.4, 0.2, 0.1, 0.05 μM, respectively. No protein was added in lane 1. (C) AlgR binds to the motif (CCGTTCATC) in the czcR promoter region. Electropherograms showed the protection pattern that included the czcR promoter region after digestion with DNase I following incubation in the absence or presence of 2.0 μM AlgR. The identified protected region showed a significant reduction in the peak pattern compared with the control. (D) The activity of czcC and czcR was activated by AlgR in vivo. The expression of czcC and czcR was tested in the wild-type PAO1, the ΔalgR mutant and the complemented (ΔalgR/Mini-CTX-algR) strains. Data are shown as mean ± SEM from three experiments.
Given the direct interaction between AlgR and the czcR-czcC promoter region, we next attempted to determine whether the expression of czcR-czcC is regulated by AlgR in vivo. We constructed czcR and czcC promoter-lux fusions (czcR-lux and czcC-lux, Supplementary Table S1) and then measured their activities in the wild-type PAO1 strain, the ΔalgR strain and its complemented strain (ΔalgR/Mini-CTX-algR). As shown in Figure 3D, the relative activities of czcR-lux or czcC-lux were ∼3-fold lower in the ΔalgR strain than in the wild-type strain. Introduction of the plasmid Mini-CTX-algR into the ΔalgR deletion strain restored their activities to the wild-type levels. This observation led us to investigate if AlgR contributes to pyocyanin production. After 24 h of growth, the ΔalgR mutant produced more green pigment than the wild-type PAO1 or the complemented strain (ΔalgR/Mini-CTX-algR). In addition, the expression of the czcR gene in the ΔalgR strain restored pigment production to the wild-type level (Supplementary Figure S3A). As expected, the expression of phzA1 was higher in the ΔalgR strain than in the wild-type PAO1 (Supplementary Figure S3B). Taken together, these results clearly demonstrated that AlgR negatively regulates pyocyanin production by controlling CzcR expression.
AlgR directly regulates the expression of mucR
The ChIP-seq analysis indicated that AlgR binds to the promoter region of mucR (Figure 4A), which was confirmed by EMSA assay in vitro (Figure 4B). We also located a specific AlgR-binding site in the promoter region of mucR (mucR-p) by using a dye-based DNase I footprinting assay. As shown in Figure 4C, a protected region is identified from nucleotides −99 to −90 (CCGTTGGTC) relative to the mucR start codon, which matched the conserved motif identified in the ChIP-seq analysis. We next sought to determine if the motif sequence was important for the binding of AlgR to the mucR promoter. We performed an EMSA using a 362 bp mucR-D DNA fragment (from nucleotides −461 to −100 of the start codon of mucR, and therefore missing of the conserved motif), and a mucR-M DNA fragment in which the sequence CCGTTCGTC was mutated to TAGTTCGTC (mutated nucleosides underlined). As shown in Supplementary Figure S4A, neither the mucR-D nor mucR-M DNA fragment was bound by AlgR, indicating that the motif is critical for AlgR binding.
Figure 4.
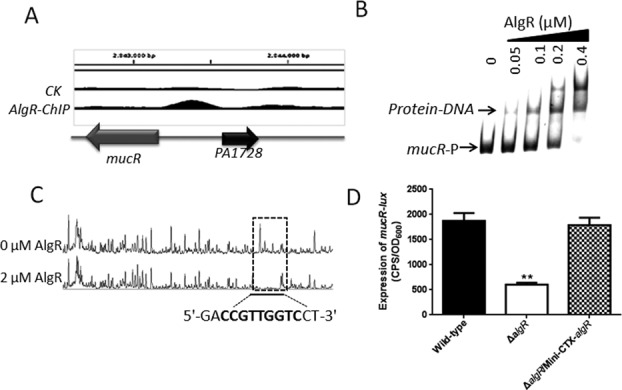
AlgR directly binds to the mucR promoter region and controls its activity. (A) AlgR binds to the promoter region of mucR according to the ChIP-seq analyses. (B) An EMSA experiment showed that AlgR directly binds to the promoter region of mucR. PCR products containing the mucR promoter region were added to the reaction mixtures at 40 nM each. AlgR protein was added to reaction buffer in lanes 2–5 with 0.4, 0.2, 0.1, 0.05 μM, respectively. No protein was added in lane 1. (C) AlgR binds to the motif (CCGTTGGTC) in the mucR promoter region. Electropherograms showed the protection pattern of the mucR promoter region after digestion with DNase I following incubation in the absence or presence of 2.0 μM AlgR. The protected region identified showed significant reduction in the peak pattern compared with the control. (D) The expression of mucR-lux was evaluated in the wild-type PAO1, the ΔalgR mutant and the complemented (ΔalgR/Mini-CTX-algR) strain. Data are shown as mean ± SEM from three experiments.
To further investigate if AlgR regulates mucR, we measured the activity of the mucR promoter in the ΔalgR mutant. As expected, the expression of mucR was ∼3-fold lower in the ΔalgR mutant than in the wild-type strain. Constitutive expression of algR in the ΔalgR mutant restored the activity of the mucR promoter to the wild-type level (Figure 4D). In addition, the mutation in the AlgR motif (mucR-M) abolished the promoter activity, and it was insensitive to the deletion of algR (Supplementary Figure S4B), suggesting that this motif is required for the full activity of the mucR promoter.
AlgR controls intracellular c-di-GMP level
MucR carries a DGC (GGDEF), a phosphodiesterase (PDE) (EAL) and a conserved integral membrane-sensing MHYT domain, that is annotated as a c-di-GMP synthase. Since AlgR regulates MucR, we speculated that AlgR controls the intracellular level of c-di-GMP. To this end, the level of c-di-GMP was measured in the ΔalgR mutant using the PcdrA-lux reporter fusion, a cdrA promoter-lux reporter that is responsive to intracellular level of c-di-GMP in P. aeruginosa (Supplementary Table S1). As shown in Figure 5A, the ΔalgR mutant displayed reduced level of c-di-GMP compared to the wild-type PAO1 strain, and this reduced level was restored by expression of algR in the ΔalgR mutant. Moreover, we measured the concentration of intracellular c-di-GMP via LC-MS/MS, which showed that the ΔalgR mutant produced 3-fold less c-di-GMP than the wild type and the ΔalgR complemented (ΔalgR/p-algR) strains (Figure 5B).
Figure 5.
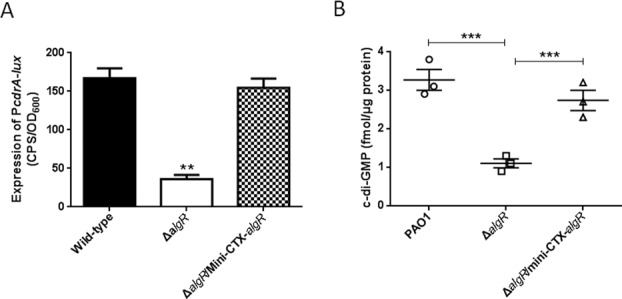
AlgR controls intracellular c-di-GMP level. (A) The expression of cdrA-lux was lower in the ΔalgR mutant than in the parental PAO1 and the complemented (ΔalgR/Mini-CTX-algR) strain. Data are shown as mean ± SEM from three experiments (**P < 0.01). (B) The ΔalgR mutant exhibited a decreased intracellular c-di-GMP concentration. Measurement of c-di-GMP by LC-MS/MS indicated that the ΔalgR mutant produced less intracellular c-di-GMP than the wild-type PAO1 (***P < 0.005).
Among the AlgR-targets, three genes associated with c-di-GMP synthesis (PA1433, arr and sagS) (60) carry an AlgR-binding site inside their coding regions. The EMSA results revealed that AlgR directly binds to PA1433, but not to arr and sagS (Supplementary Figure S5A). However, the expression of PA1433 in the ΔalgR strain was similar to that in the wild-type strain (Supplementary Figure S5B), suggesting that AlgR does not regulate PA1433. Given that AlgR does not interact with other c-di-GMP-related genes in the ChIP-seq targets, our results suggest that AlgR modulates intracellular c-di-GMP levels by controlling levels of mucR.
MucR is a DGC
MucR contains a GGDEF and an EAL domain, which encode a DGC and a PDE, respectively. However, MucR only shows DGC activity (60). As shown in Figure 6A, the overexpression of mucR in the wild-type strain resulted in accumulation of 5-fold more c-di-GMP than the parental strain, as determined using a cdrA-lux reporter. A similar result was also observed for the expression of mucRDGC (the DGC domain of MucR only) in the wild-type PAO1 strain. Moreover, we measured the concentration of intracellular c-di-GMP via LC-MS/MS, which showed that the overexpression of mucR or mucRDGC caused the accumulation of more c-di-GMP than the wild-type strain carrying an empty vector (Figure 6B).
Figure 6.
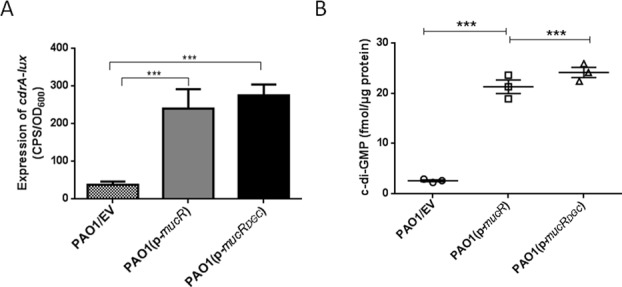
mucR encodes a DGC (A) The expression of cdrA-lux was induced in the wild-type PAO1 strain carrying multiple copies of a plasmid expressing mucR or mucRDGC. (B) The overexpression of mucR or mucRDGC increased the intracellular level of c-di-GMP. LC-MS/MS measurement of c-di-GMP revealed that the PAO1 strain containing p-mucR or p-mucRDGC accumulates significantly more intracellular c-di-GMP than the PAO1 strain containing the empty vector. Data are shown as mean ± SEM from three experiments. Statistical significance was determined using Student's t-test (***P < 0.001). EV represents empty vector.
AlgR regulates biofilm production via mucR and c-di-GMP
Although AlgR regulates biofilm production by repressing the rhl quorum-sensing system (32), the detailed regulatory mechanism has not been described. Since AlgR directly regulates mucR, we hypothesized that AlgR regulates c-di-GMP concentrations in vivo, and thereby controls the level of biofilm formation. To verify this, we grew 24-hour static cell biofilms of the wild-type PAO1 strain, the ΔalgR mutant, the ΔalgR complemented strain (ΔalgR/p-algR), the ΔalgRΔmucR double mutant and the ΔalgR mutant overexpressing mucR (p-mucR). We reasoned that if reduced biofilm production results from reduced expression of mucR and a lower level of c-di-GMP in the ΔalgR mutant, a ΔalgRΔmucR double mutant should produce less biofilms than the parental strain, whereas the overexpression of mucR in the ΔalgR mutant should restore the biofilm production to the level in the wide-type strain. Consistent with this hypothesis, the ΔalgRΔmucR double mutant produced significantly lesser biofilm than did the ΔalgR single mutant, whereas the ΔalgR mutant overexpressing mucR produced more biofilm than did the wild-type PAO1 (Figure 7A). In addition, we observed that the ΔalgRΔmucR double mutant produced significantly lesser c-di-GMP than did the ΔalgR mutant, and the overexpression of mucR in the ΔalgR mutant accumulated more c-di-GMP (Figure 7B). These results suggested that the reduced mucR expression and intracellular c-di-GMP levels contributed to the compromised biofilm production in the ΔalgR mutant.
Figure 7.
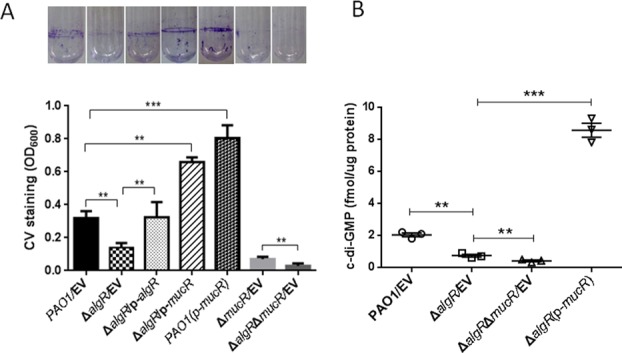
The regulation of biofilm formation by AlgR is dependent on mucR and c-di-GMP. (A) Quantification of CV staining of biofilms grown in microtiter plates for 14 h. Results represent means ± SEM, and data are representative of three independent experiments. Photos of the tubes from the binding assay were taken. (B) The reduced intracellular levels of c-di-GMP in the ΔalgR mutant were restored by overexpressing mucR. Results represent means ± SEM, and data are representative of three independent experiments. Statistical significance was determined using Student's t-test (***P < 0.005, **P < 0.01). EV represents empty vector.
The impacts of AlgR and AlgZ/FimS on P. aeruginosa pathogenicity
As previously reported, the two-component system AlgZ(FimS)/AlgR controls alginate production, twitching and swarming (35). Here we demonstrated that the ΔalgR strain had reduced intracellular levels of c-di-GMP and thus affected biofilm formation. This observation led us to test whether the loss of fimS results in a similar phenotype. To this end, we constructed a single fimS deletion mutant (ΔfimS) as previously described. As shown in Supplementary Figure S6, the ΔfimS strain showed reduced adherence compared to the parental PAO1 strain under static conditions. Moreover, ectopic expression of a p-mucR, a p-mucRDGC or a p-sadC suppressed the biofilm formation phenotype of the fimS mutant (Supplementary Figure S6), indicating that the cognate sensor FimS regulates biofilm formation by controlling levels of c-di-GMP.
We also tested the pathogenicity of the ΔfimS strain in a mouse model. C57BL/6 mice were infected intranasally with ∼1×107 cells of either the wild-type PAO1 or the ΔfimS strain. The Kaplan–Meier survival analysis showed that the loss of fimS significantly improved mouse survival, as compared to mice infected by the wild-type PAO1 strain (Figure 8). Interestingly, mice infected by the ΔalgRΔmucR strain, but not the ΔfimSΔmucR strain, showed greater survival than mice infected with respective ΔalgR or ΔfimS single mutants (Figure 8), indicating that factors other than c-di-GMP are involved in fimS-mediated virulence regulation. Overall, these data demonstrated that both AlgR and its cognate sensor FimS are positive regulators of P. aeruginosa pathogenicity.
Figure 8.
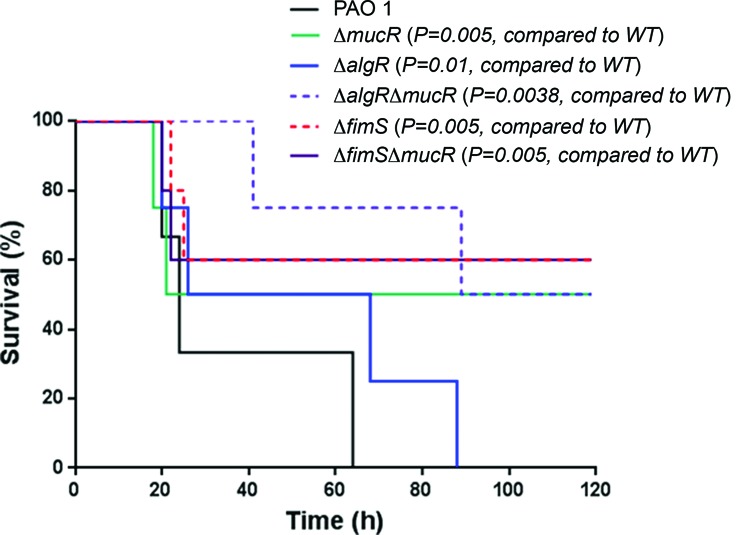
The lack of either algR or fimS leads to reduced virulence of P. aeruginosa. C57BL6 mice were intranasally challenged with ΔmucR, ΔalgR, ΔfimS, ΔalgRΔmucR, ΔfimSΔmucR or wild-type PAO1 strains, at 107 cfu in 50 μl PBS, and moribund mice were killed to obtain survival data (Kaplan–Meier Curve with Log-Rank test, n = 6).
DISCUSSION
The two-component system AlgR/FimS is a global regulator of P. aeruginosa virulence and plays important roles in switching bacteria from acute to chronic infections. Although several studies provide insights into the function of AlgR, few genes regulated by AlgR possess a known AlgR binding site (56), this suggests an indirect mechanism of AlgR regulation or the existence of additional binding sequences of AlgR. In this study, we performed a ChIP-seq assay that identified 157 in vivo binding sites of AlgR in the P. aeruginosa genome. This technique has been widely used as a powerful tool to investigate functions of bacterial transcription factors (19,61). We recently developed and applied this method to demonstrate the regulatory mechanism of the quorum-sensing regulator VqsM (41). The combination of the present ChIP-seq results and previous microarray data of algR should provide a much better understanding of the regulatory mechanism(s) of AlgR.
Among the 157 AlgR-bound regions, most possess an AlgR-binding site that was identified in previous studies (29,55,56). The majority of the AlgR-bound-genes are involved in metabolism and nucleic acid biosynthesis, indicating important roles of AlgR in the regulation of a variety of metabolic pathways. Other major AlgR targets include a group of genes encoding transcriptional factors and virulence factors. However, some previously known direct targets (such as algD, algC, hcnA, rhlA) of AlgR were not identified in the present work. There are three explanations for omission of these genes from our ChIP-seq results. First, we observed high but nonspecific peaks corresponding to these genes in the control sample without a VSV-tag, indicating an intrinsic disadvantage of using VSV antibody in our ChIP-seq procedure. Second, the interaction between AlgR and these missing targets might not be strong enough under the conditions used in the ChIP-seq protocol (i.e. using the pAK1900 vector, a VSV-tagged AlgR and mid-log phase culture). Third, AlgR may need other co-factors to bind to these targets. Although further optimization of the ChIP-seq method is needed for future experiments, the combination of the ChIP-seq analyses and biochemical assays enabled us to pinpoint most AlgR targets in vivo. We have validated that AlgR binds to only one of the three genes that contain binding sites within the open reading frame (ORF) (Supplementary Figure S5A), which suggests that the real ratio of AlgR binding sites in intergenic regions is higher than 45%.
In the present study, AlgR functioned as an activator for five genes (PA4396, PA2189, PA2867, czcR and mucR) and as a repressor for two genes (PA1736 and purL) (Figures 2H–4). Like other transcription factors, the potential two activities of AlgR are best indicated by its binding position in relation to a promoter region. The position of a response-regulator binding site relative to the sigma-factor binding site is crucial to its regulatory activities. In general, response regulators that bind upstream of the sigma-factor binding site positively regulate gene transcription, and function as repressors when they bind downstream of the sigma-factor binding site (62). We also noticed supershifts when AlgR binds to tested targets (Figure 2), which is reminiscent of a previous report showing that increasing concentrations of a cold inhibitor (nonradioactive labeled probe) resulted in different binding patterns of AlgR (32). These results suggest that multiple AlgR-binding sites are present in these promoters.
AlgR directly binds to the intergenic region of czcR-czcC, the encoded proteins of which are involved in heavy metal and antibiotic resistance in P. aeruginosa (58). A recent study showed that the production of quorum-sensing 3-oxo-C12-HSL and C4-HSL autoinducer molecules is impaired in a czcR mutant. CzcR also negatively regulates the expression of phzA1 by directly binding to its promoter (59). We observed that the ΔalgR mutant produced a deeper blue pigment compared to that produced by the parental strain (Supplementary Figure S3A), which suggests that AlgR controls pigment production by directly regulating the expression of czcR. As expected, the activity of czcR was lower in the ΔalgR mutant than in the wild-type PAO1 (Figure 3D). We therefore concluded that AlgR positively regulates czcR, which then negatively controls pyocyanin production. Unlike a typical regulation of pyocyanin production by a quorum-sensing system (63), this result provided new insights into the complicated regulatory network underlying pyocyanin biosynthesis. Our ChIP-seq results should represent a useful database for characterization of AlgR-regulated pathways in the future. The mucR gene, encoding a proposed DGC, emerged as a key AlgR target. The ΔalgR mutant harboring additional copies of mucR displayed higher intracellular concentration of c-di-GMP than the wide-type strain (Figure 7B). To the best of our knowledge, this is the first report showing that AlgR modulates c-di-GMP, which provides a molecular explanation for our present findings and the earlier studies on the role of AlgR in biofilm formation. C-di-GMP is an important bacterial second messenger that regulates biofilm formation, virulence, differentiation and other processes (64). In an acute pulmonary infection model, the ΔalgRΔmucR double mutant allowed for higher levels of mouse survival than the ΔalgR single mutant (Figure 8), indicating a role of c-di-GMP in bacterial pathogenicity.
There are many DGC and PDE enzymes in P. aeruginosa, suggesting a complex signal transduction network influenced by levels of c-di-GMP. The present work focused on the regulation of expression of mucR that plays important roles in c-di-GMP production and multiple cellular responses. Besides the DGC activity described here and in an earlier study, MucR also contains a PDE (EAL) and a conserved integral membrane-sensing MHYT domain, which is proposed to sense oxygen, CO or NO (65). Interestingly, our ChIP-seq result revealed that AlgR also targets a group of other c-di-GMP-related genes, such as amrZ, arr, sagS and PA1433 (Supplementary Table S3). However, the EMSA results showed that AlgR only binds to PA1433, the expression level of which did not differ between the wild-type strain and the ΔalgR mutant (Supplementary Figure S5). Taken together, we conclude that AlgR regulates c-di-GMP synthesis via mucR.
Overall, the present work provides a deeper understanding of the roles of AlgR in virulence regulation and c-di-GMP synthesis (Figure 9). Moreover, we demonstrated that FimS also plays a role in P. aeruginosa pathogenicity. By binding to algD, fimU, czcR, mucR and other loci in the genome, AlgR and its sensor FimS play a central role mediating a large group of cellular and molecular pathways, including c-di-GMP production, biofilm formation and pathogenicity. Our ChIP-seq data provides a useful database that will allow us to investigate more functions and AlgR targets in the future.
Figure 9.
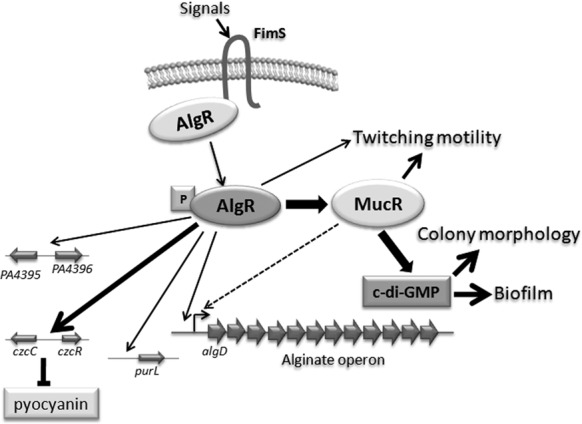
Schematic of the proposed AlgR regulatory mechanism. AlgR controls biofilm formation, twitching motility and alginate synthesis. The present work revealed that c-di-GMP contributes to the regulation of biofilm formation by AlgR. In addition, we have expanded the AlgR regulon to include several new systems, such as pyocyanin production, metabolic processes (PurL) and stress response (PA4395-PA4396).
Supplementary Material
Acknowledgments
We kindly thank Dr Min Wu at University of North Dakota (USA) for performing the animal assays and Dr Daniel J. Wozniak for providing the plasmid pJK223R1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation for Young Scientists of China (31000049); Natural Science Basis Research Plan in ShaanXi Province of China (2010JQ3008); Program for the academic backbone of excellent young of Northwest University (338050069). Project of Tianjin, China (13TXSYJC40100).
Conflict of interest statement. None declared.
REFERENCES
- 1.Elkin S., Geddes D. Pseudomonal infection in cystic fibrosis: the battle continues. Expert Rev. Anti Infect. Ther. 2003;1:609–618. doi: 10.1586/14787210.1.4.609. [DOI] [PubMed] [Google Scholar]
- 2.Coggan K.A., Wolfgang M.C. Global regulatory pathways and cross-talk control pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012;14:47–70. [PubMed] [Google Scholar]
- 3.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Mann E.E., Wozniak D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkawash M.A., Soothill J.S., Schiller N.L. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 6.Evans D.J., Brown M.R., Allison D.G., Gilbert P. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J. Antimicrob. Chemother. 1990;25:585–591. doi: 10.1093/jac/25.4.585. [DOI] [PubMed] [Google Scholar]
- 7.Wei Q., Ma L.Z. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013;14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govan J.R., Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen S.S. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 1992;28:1–79. [PubMed] [Google Scholar]
- 10.Speert D.P., Gordon S. Phagocytosis of unopsonized Pseudomonas aeruginosa by murine macrophages is a two-step process requiring glucose. J. Clin. Invest. 1992;90:1085–1092. doi: 10.1172/JCI115924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson J.A., Smith S.E., Dean R.T. Alginate may accumulate in cystic fibrosis lung because the enzymatic and free radical capacities of phagocytic cells are inadequate for its degradation. Biochem. Mol. Biol. Int. 1993;30:1021–1034. [PubMed] [Google Scholar]
- 12.Learn D.B., Brestel E.P., Seetharama S. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 1987;55:1813–1818. doi: 10.1128/iai.55.8.1813-1818.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentzer M., Teitzel G.M., Balzer G.J., Heydorn A., Molin S., Givskov M., Parsek M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic V., Gill J.F., Chakrabarty A.M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delic-Attree I., Toussaint B., Garin J., Vignais P.M. Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol. Microbiol. 1997;24:1275–1284. doi: 10.1046/j.1365-2958.1997.4121799.x. [DOI] [PubMed] [Google Scholar]
- 16.DeVault J.D., Hendrickson W., Kato J., Chakrabarty A.M. Environmentally regulated algD promoter is responsive to the cAMP receptor protein in Escherichia coli. Mol. Microbiol. 1991;5:2503–2509. doi: 10.1111/j.1365-2958.1991.tb02096.x. [DOI] [PubMed] [Google Scholar]
- 17.Waligora E.A., Ramsey D.M., Pryor E.E. Jr, Lu H., Hollis T., Sloan G.P., Deora R., Wozniak D.J. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 2010;192:5390–5401. doi: 10.1128/JB.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozniak D.J., Ohman D.E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones C.J., Newsom D., Kelly B., Irie Y., Jennings L.K., Xu B., Limoli D.H., Harrison J.J., Parsek M.R., White P., et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Salazar J.M., Moreno S., Najera R., Boucher J.C., Espin G., Soberon-Chavez G., Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J. Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershberger C.D., Ye R.W., Parsek M.R., Xie Z.D., Chakrabarty A.M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E) Proc. Natl Acad. Sci. U.S.A. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu D., Eisinger V.M., Rowen D.W., Yu H.D. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. U.S.A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konyecsni W.M., Deretic V. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J. Bacteriol. 1990;172:2511–2520. doi: 10.1128/jb.172.5.2511-2520.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boucher J.C., Martinez-Salazar J., Schurr M.J., Mudd M.H., Yu H., Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu D., Eisinger V.M., Head N.E., Pier G.B., Yu H.D. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deretic V., Konyecsni W.M. Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J. Bacteriol. 1989;171:3680–3688. doi: 10.1128/jb.171.7.3680-3688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deretic V., Dikshit R., Konyecsni W.M., Chakrabarty A.M., Misra T.K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohr C.D., Leveau J.H., Krieg D.P., Hibler N.S., Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielinski N.A., Chakrabarty A.M., Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]
- 31.Okkotsu Y., Tieku P., Fitzsimmons L.F., Churchill M.E., Schurr M.J. Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J. Bacteriol. 2013;195:5499–5515. doi: 10.1128/JB.00726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morici L.A., Carterson A.J., Wagner V.E., Frisk A., Schurr J.R., Honer zu Bentrup K., Hassett D.J., Iglewski B.H., Sauer K., Schurr M.J. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 2007;189:7752–7764. doi: 10.1128/JB.01797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carterson A.J., Morici L.A., Jackson D.W., Frisk A., Lizewski S.E., Jupiter R., Simpson K., Kunz D.A., Davis S.H., Schurr J.R., et al. The transcriptional regulator AlgR controls cyanide production in Pseudomonas aeruginosa. J. Bacteriol. 2004;186:6837–6844. doi: 10.1128/JB.186.20.6837-6844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lizewski S.E., Lundberg D.S., Schurr M.J. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2002;70:6083–6093. doi: 10.1128/IAI.70.11.6083-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitchurch C.B., Alm R.A., Mattick J.S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. U.S.A. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitchurch C.B., Erova T.E., Emery J.A., Sargent J.L., Harris J.M., Semmler A.B., Young M.D., Mattick J.S., Wozniak D.J. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J. Bacteriol. 2002;184:4544–4554. doi: 10.1128/JB.184.16.4544-4554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickman J.W., Tifrea D.F., Harwood C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl Acad. Sci. U.S.A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen L.D., van Gennip M., Rybtke M.T., Wu H., Chiang W.C., Alhede M., Hoiby N., Nielsen T.E., Givskov M., Tolker-Nielsen T. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic Di-GMP level in the bacteria. Infect. Immun. 2013;81:2705–2713. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmann F., Selesi D., Weinmaier T., Tischler P., Rattei T., Meckenstock R.U. Genomic insights into the metabolic potential of the polycyclic aromatic hydrocarbon degrading sulfate-reducing Deltaproteobacterium N47. Environ. Microbiol. 2011;13:1125–1137. doi: 10.1111/j.1462-2920.2010.02391.x. [DOI] [PubMed] [Google Scholar]
- 40.Blasco B., Chen J.M., Hartkoorn R., Sala C., Uplekar S., Rougemont J., Pojer F., Cole S.T. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 2012;8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang H., Deng X., Li X., Ye Y., Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 2014;42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato J., Chakrabarty A.M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. U.S.A. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F.R. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 2006;17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 47.Jansons I., Touchie G., Sharp R., Almquist K., Farinha M.A., Lam J.S., Kropinski A.M. Deletion and transposon mutagenesis and sequence analysis of the pRO1600 OriR region found in the broad-host-range plasmids of the pQF series. Plasmid. 1994;31:265–274. doi: 10.1006/plas.1994.1028. [DOI] [PubMed] [Google Scholar]
- 48.Hoang T.T., Karkhoff-Schweizer R.R., Kutchma A.J., Schweizer H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 49.Duan K., Dammel C., Stein J., Rabin H., Surette M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 50.Liang H., Li L., Dong Z., Surette M.G., Duan K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 2008;190:6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hickman J.W., Harwood C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 53.Rashid M.H., Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. U.S.A. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurachi M. Studies on the biosynthesis of pyocyanine. Isolation and determination of pyocyanine. Bull. Inst. Chem. Res. Kyoto Univ. 1958;36:163–173. [Google Scholar]
- 55.Belete B., Lu H., Wozniak D.J. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 2008;190:2023–2030. doi: 10.1128/JB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lizewski S.E., Schurr J.R., Jackson D.W., Frisk A., Carterson A.J., Schurr M.J. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 2004;186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hay I.D., Remminghorst U., Rehm B.H. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009;75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perron K., Caille O., Rossier C., Van Delden C., Dumas J.L., Kohler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 59.Dieppois G., Ducret V., Caille O., Perron K. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PloS One. 2012;7:e38148. doi: 10.1371/journal.pone.0038148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulasakara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., Lee D.G., Neely A.N., Hyodo M., Hayakawa Y., et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl Acad. Sci. U.S.A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies B.W., Bogard R.W., Mekalanos J.J. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl Acad. Sci. U.S.A. 2011;108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bijlsma J.J., Groisman E.A. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11:359–366. doi: 10.1016/s0966-842x(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 63.Winson M.K., Camara M., Latifi A., Foglino M., Chhabra S.R., Daykin M., Bally M., Chapon V., Salmond G.P., Bycroft B.W., et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. U.S.A. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galperin M.Y., Gaidenko T.A., Mulkidjanian A.Y., Nakano M., Price C.W. MHYT, a new integral membrane sensor domain. FEMS Microbiol. Let. 2001;205:17–23. doi: 10.1111/j.1574-6968.2001.tb10919.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


