Abstract
Small RNAs (sRNAs) are important regulators of gene expression during bacterial stress and pathogenesis. sRNAs act by forming duplexes with mRNAs to alter their translation and degradation. In some bacteria, duplex formation is mediated by the Hfq protein, which can bind the sRNA and mRNA in each pair in a random order. Here we investigate the consequences of this random-order binding and experimentally demonstrate that it can counterintuitively cause high Hfq concentrations to suppress rather than promote sRNA activity in Escherichia coli. As a result, maximum sRNA activity occurs when the Hfq concentration is neither too low nor too high relative to the sRNA and mRNA concentrations (‘Hfq set-point’). We further show with models and experiments that random-order binding combined with the formation of a dead-end mRNA–Hfq complex causes high concentrations of an mRNA to inhibit its own duplex formation by sequestering Hfq. In such cases, maximum sRNA activity requires an optimal mRNA concentration (‘mRNA set-point’) as well as an optimal Hfq concentration. The Hfq and mRNA set-points generate novel regulatory properties that can be harnessed by native and synthetic gene circuits to provide greater control over sRNA activity, generate non-monotonic responses and enhance the robustness of expression.
INTRODUCTION
Bacterial small RNAs (sRNAs) play an important role in rapid adaptation of gene expression to environmental stress (1) and pathogenesis (2). Most sRNAs inhibit target gene expression (termed ‘silencing sRNAs’ even if the amount of decrease in expression is modest) by forming sRNA–mRNA duplexes that decrease mRNA translation and/or increase mRNA degradation (3). Less commonly, sRNAs can activate target gene expression (termed ‘activating sRNAs’) by forming sRNA–mRNA duplexes that increase translation and/or decrease mRNA degradation (4,5). Some sRNAs can act both as inhibitors and activators of gene expression depending on the specific target mRNA and conditions (6–11). Rarely, sRNAs not only regulate the translation and degradation of a target mRNA but also code for functionally important proteins that are expressed under some conditions (12).
In many bacteria, including Escherichia coli and Salmonella typhimurium, duplex formation usually requires the hexameric Hfq protein, which acts as an RNA chaperone that unfolds sRNA and mRNA sequences and mediates their annealing (13). In addition, Hfq can help silence gene expression by blocking ribosome binding and recruiting RNase E (14). While Hfq is necessary for duplex formation in some bacteria, the reason for this is enigmatic (15). sRNAs in many gram positive bacteria including Staphylococcus aureus and Listeria monocytogenes do not require Hfq for their activity (16,17). Moreover, even in bacteria where Hfq is typically required for duplex formation, there are sRNAs that can form duplexes independently of Hfq (8,18–21). Given the above, the possibility must be considered that Hfq involvement in duplex formation may not have evolved to overcome barriers to efficient duplex formation, but instead barriers evolved to prevent duplex formation occurring without Hfq (such as secondary structures that prevent Hfq independent duplex formation and short lifetimes for sRNAs when they are not bound to Hfq). That is, there may be functional advantages in some bacteria to having duplex formation dependent on Hfq availability. Requiring Hfq to mediate duplex formation also places additional constraints on the evolution of sRNA and target mRNA sequences (22).
One possible advantage of Hfq-mediated duplex formation may be related to a puzzling and unusual feature of its kinetics: Hfq can bind the sRNAs and mRNAs in any order for at least some cognate pairs (23–25) (Figure 1A). This random-order binding, as opposed to compulsory-order binding, is theoretically inefficient (26). Furthermore, the random-order (bi-uni) reaction scheme for Hfq-dependent duplex formation is not known to occur with other bacterial and eukaryotic proteins involved in RNA guided silencing mechanisms including Argonaute, PIWI, Aubergine and Cas (27–31) (note: these proteins bind the non-coding RNA first).
Figure 1.
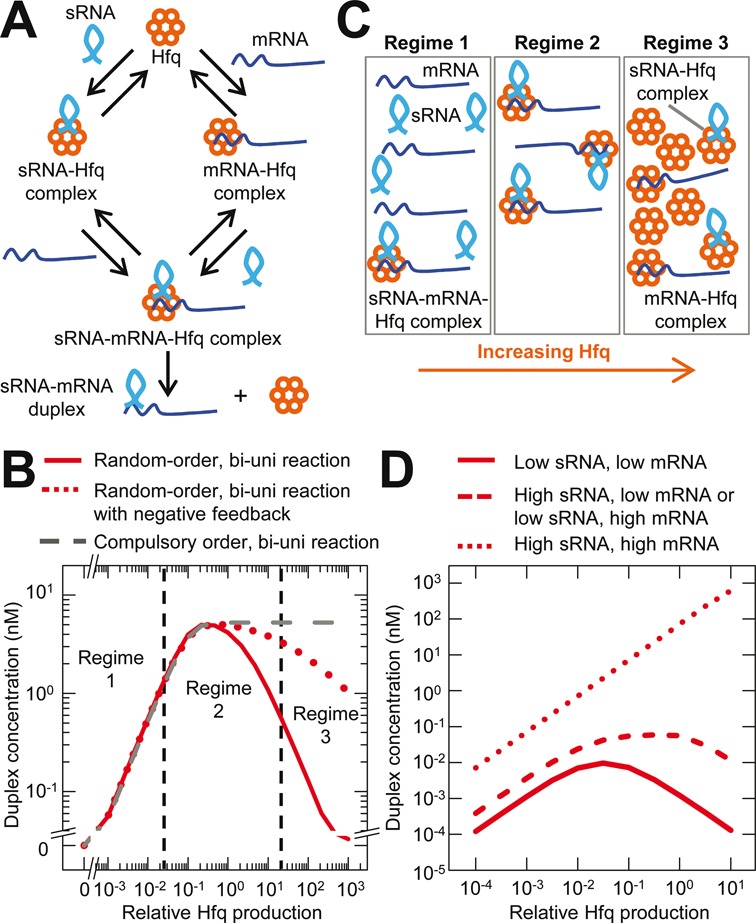
Random-order binding results in an optimal Hfq concentration for maximum sRNA activity (‘Hfq set-point’). (A). Random-order, bi-uni reaction scheme (i.e. two substrates and one product with Hfq being the catalyst) for sRNA–mRNA duplex formation. (B) Simulated duplex formation in bi-uni reaction schemes with: (i) random-order binding; (ii) random-order binding with negative feedback; and (iii) compulsory-order binding. Regimes 1, 2 and 3 are described in panel C and are separated by vertical dash lines. (C) Illustration of the three regimes for duplex formation. Regime 1 has insufficient Hfq for the amounts of sRNA and mRNA. Regime 2 has an optimal amount of Hfq so that it is not a limiting factor. Regime 3 has too much Hfq for the amount of sRNA and mRNA resulting in their sequestration, reduced sRNA–mRNA–Hfq ternary complex formation and consequently reduced duplex formation. (D) Duplex concentration as a function of relative Hfq production at low or high production rates of sRNA and mRNA. Note: the curves with high sRNA and low mRNA production rates and low sRNA and high mRNA production rates overlie one another.
Here we investigate random-order binding of sRNAs and mRNAs to Hfq to determine its functional consequences and thus how Hfq contributes to the properties of sRNA regulation. This study has three parts. First, we experimentally demonstrate that suboptimal sRNA activity not only occurs with too little Hfq but also with too much Hfq because random-order binding causes sequestration of sRNAs and mRNAs in singly bound Hfq complexes. This was shown for two silencing sRNA–mRNA pairs (RyhB–sodB and MicC–ompC) and one activating sRNA–mRNA pair (GlmZ–glmS). The Hfq concentration that results in maximum sRNA activity for a specified level of sRNA and mRNA production is termed the ‘Hfq set-point’. Second, we experimentally demonstrate that physiological Hfq, sRNA and mRNA concentrations can create the conditions for an Hfq set-point and suppression of sRNA activity. Third, we show theoretically that if there is random-order binding of sRNAs and mRNAs to Hfq but the mRNA–Hfq complexes do not lead to sRNA–mRNA–Hfq ternary complexes, then high mRNA concentrations can sequester Hfq in dead-end mRNA–Hfq complexes. This sequestration of Hfq decreases the concentration of sRNA–Hfq complexes, which are the only effective path to duplex formation, resulting in decreased sRNA activity. We obtain experimental evidence that the MicC–ompC pairs form dead-end mRNA–Hfq complexes and as a consequence maximum MicC activity requires an optimal mRNA concentration as well as an optimal Hfq concentration (termed the ‘mRNA set-point’). Together our findings demonstrate that random-order binding to Hfq alters the general requirements and properties of duplex formation, which has important implications for analyzing and harnessing sRNA regulation in gene networks.
MATERIALS AND METHODS
Strains and plasmids
Details of strains, plasmids and oligonucleotides are in Supplementary Tables S1 and S2 and Figure S1. Strains and plasmids will be available through Addgene. Sources of the plasmids and their components and inserts are reported elsewhere (32,33).
Measurement of GFP fluorescence
Single colonies were inoculated into Luria-Bertani (LB) media with a final concentration of 100 μg/ml of ampicillin and grown overnight at 37°C and shaking at 200 rpm. Cells from the overgrown culture were diluted 1/1000 to 1/10 000 in 3 ml of fresh LB with 100 μg/ml of ampicillin and grown under the same growth conditions. After 3 h, 3–120 μl of culture was inoculated into 3 ml of fresh LB with 100 μg/ml of ampicillin and 0–1000 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown for 2.5 h resulting in a final OD600nm ∼0.01–0.1. Cells were placed on ice and measured by flow cytometry using the Beckman–Coulter EPICS XLMCL (488 nm laser) or FC500 (488 nm laser). Data were analyzed using Flow Explorer 4.1 (R. Hoebe, University of Amsterdam, Amsterdam, The Netherlands) and Matlab software (MathWorks). We assume GFP fluorescence is directly proportional to the GFP concentration.
Measurement of RNA concentrations by quantitative RT-PCR
Cells from overnight cultures were inoculated into 10 ml of LB media with a final concentration of 100 μg/ml of ampicillin and 1 mM of IPTG (except for cultures of HL716 which had no ampicillin or IPTG) and grown at 37°C with shaking at 200 rpm for ∼3–5 h. Two milliliters of culture were collected at OD600nm 0.2–0.4, centrifuged, the supernatant was removed, and the cell pellet frozen on dry ice. Total RNA was extracted from the cell pellet using the RNeasy kit (Qiagen) with DNase treatment (Qiagen). cDNA was synthesized from the total RNA using the iScript Select cDNA Synthesis kit (Bio-Rad) with random primer mix. Identical cDNA reactions with RNA but without reverse transcriptase (−RT) were also made. The cDNA and −RT samples were diluted 1 in 3 with nuclease free water and polymerase chain reaction (PCR) amplified in parallel using the iQ5 Real-Time PCR Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). The cycle threshold (CT) values were automatically specified by the iQ5 software. The relative amount of RNA was calculated using the formula: (2−CTcDNAx−2−CTRTx)/(2−CTcDNA16S−2−CTRT16S), where CTcDNAx is the CT value for the cDNA using oligonucleotides that are specific to the sRNA or mRNA being measured, CTRTx is the CT value measured for the −RT sample using the same oligonucleotides used to measured CTcDNAx, CTcDNA16S is the CT value for the cDNA using oligonucleotides that amplify the 16S rRNA and CTRT16S is the CT value for the −RT sample with the same 16S rRNA oligonucleotides as for CTcDNA16S.
Model and simulations
The simulations used a previously reported model (26,34). In brief, the model uses ordinary differential equations to describe the kinetics of duplex formation and protein production, and we numerically solved the system for steady state values. The equations include the production, degradation, association and/or dissociation reactions for the sRNA, target mRNA, Hfq, sRNA–mRNA duplex, target protein, sRNA–Hfq, mRNA–Hfq and sRNA–mRNA–Hfq. The model assumes that a single Hfq hexamer is sufficient for duplex formation and that Hfq is recycled following duplex formation and release. RNA cycling on and off of Hfq (25,35) is included in the model through the dissociation of sRNAs and mRNAs from singly bound Hfq complexes and sRNA–mRNA–Hfq ternary complexes. The effect of RNA cycling on the Hfq set-point has been previously reported (26).
The equations and parameter values for the simulation have been reported (26,34) but are also included in the Supplementary Methods. The specific values of the rate constants for the association of sRNAs and mRNAs with Hfq and the dissociation of the Hfq complexes (26) affect the properties of the Hfq set-point (see ‘Discussion’ section) (26) but they do not affect whether the Hfq set-point is present or not (so long as there is random-order binding). Therefore we chose arbitrary values for the rate constants and kept the values the same across simulations (unless otherwise stated) so that the results within and between simulations could be directly compared. The values for the rate constants were the same for sRNAs and mRNAs, therefore there is no positive or negative cooperativity for RNA binding in these simulations.
The degradation rate constants for the sRNA and mRNA were also equal, and furthermore they were the same in the free and Hfq bound states. While this does not often occur physiologically, we set the values to be the same so that it was easier to show that changes in target protein concentrations were due to changes in the proportion of duplex formed and not due to changes in the total sRNA or mRNA concentration in the system. Furthermore, we previously showed that the properties of the Hfq set-point are essentially the same whether the degradation rate constants for free sRNAs and Hfq bound sRNAs are equal or the degradation rate constant for free sRNAs is 10-fold greater than for Hfq bound sRNAs (26) (also see ‘Discussion’ section).
RESULTS
Silencing sRNAs (RyhB and MicC) have an Hfq set-point
We sought to test our theoretical prediction (26) that random-order binding of sRNAs and mRNAs to Hfq results in the need for an optimal Hfq concentration to achieve maximum sRNA activity at fixed sRNA and mRNA production rates (Figure 1B). This optimal Hfq concentration (or range of Hfq concentrations) required for maximum sRNA activity is referred to as the ‘Hfq set-point’. The Hfq set-point occurs because there is minimal duplex formation at both relatively low Hfq concentrations due to insufficient Hfq and at relatively high Hfq concentrations due to the low free sRNA and free mRNA concentrations because of their sequestration within sRNA–Hfq and mRNA–Hfq complexes (Figure 1C). Our simulations show that the Hfq set-point does not occur with compulsory-order duplex formation (Figure 1B), and is not abolished by negative feedback (Figure 1B) (note: see ‘Materials and Methods’ section and Supplementary Material for further details of the model and simulations).
We designed the following experimental system to specifically investigate the Hfq set-point. In this system, we used the RyhB and MicC sRNAs, which decrease translation and increase degradation of the sodB and ompC target mRNAs respectively (10,32,36) (Figure 2A). Because the Hfq set-point will be most apparent when the sRNA and mRNA concentrations are low compared to the Hfq concentration (Figure 1D) we transcribed the sRNAs and mRNAs from weak promoters. RyhB and MicC were transcribed from PConM8 (a synthetic promoter derived from the synthetic PCon promoter with mutations at the −35 and −10 sites that decrease transcription by ∼10-fold; Supplementary Figure S2A) in strains with chromosomal hfq and ryhB or micC deleted. The parts of the target mRNA sequences that are necessary for sRNA activity were fused to the gfp gene (Figure 2A) as described in other studies (32,37,38). These fusions allow target protein concentrations (also referred to as gene ‘expression’) to be measured in vivo by GFP fluorescence and they minimize the physiological effects of expressing full-length target proteins. The target mRNAs were transcribed from PLtetO-1M9 (32), which is a mutated version of the synthetic PLtetO-1 promoter with ∼10-fold less transcription (Supplementary Figure S2B). Hfq production was controlled by transcribing the hfq gene from the inducible PLlacO-1 promoter with the very strong T710 ribosome binding sequence (RBS) (Supplementary Figure S2C). Hfq production was varied by adding different IPTG concentrations to the media.
Figure 2.
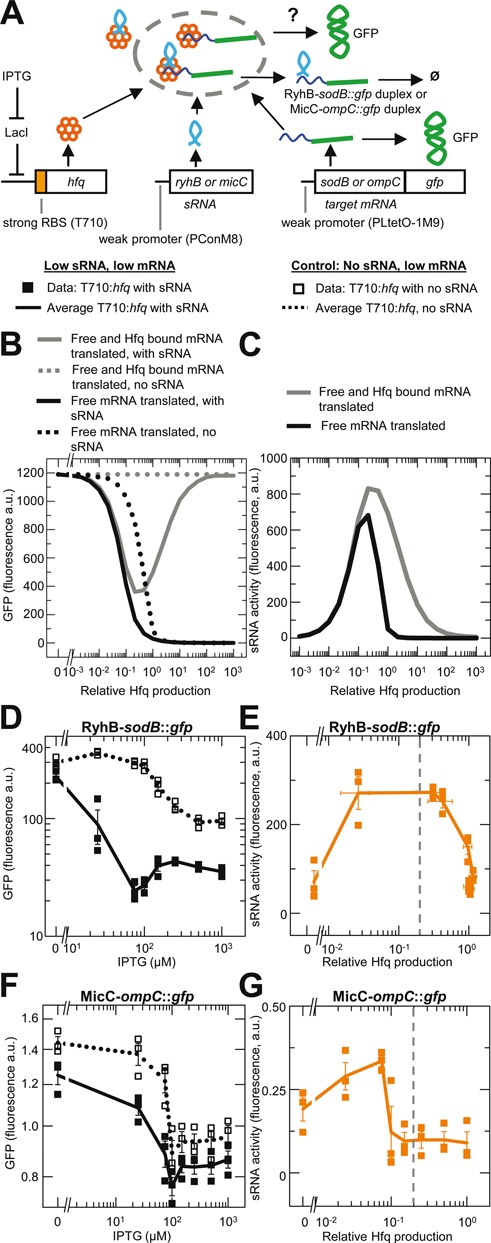
RyhB and MicC silencing sRNAs have an Hfq set-point for maximum activity. Error bars are the SEM. Vertical dash line in panels E and G indicates the approximate level of physiological Hfq (see Figure 4). (A) Reaction scheme and genetic circuit for RyhB and MicC and their cognate target mRNAs, sodB and ompC respectively, fused to gfp. Data and line symbols refer to those used in panels D and F. (B) Simulated GFP expression and duplex formation for silencing sRNAs. In one scenario, only the free mRNA is translated and in the second scenario, both free mRNA and Hfq bound mRNA are translated. Note: arbitrary units (a.u.) in the simulation and experiments are not the same. (C) Simulated sRNA activity as a function of relative Hfq concentration for both scenarios described in panel B. (D) Experimentally measured GFP expression at varying Hfq production for the RyhB–sodB mRNA pair. Total number of samples (n) = 24 for each strain with replicate measurements in three different cultures at each IPTG concentration. Strains: HL6361 (low RyhB, low sodB) and HL6357 (no RyhB, low sodB). Symbols and lines are defined in panel A. (E) Experimentally measured RyhB sRNA activity as a function of relative Hfq production. sRNA activity and relative Hfq concentrations were calculated as described in the main text. (F) Experimentally measured GFP expression at varying Hfq production for the MicC–ompC mRNA pair. Total n = 24 for each strain with replicate measurements in three different cultures at each IPTG concentration. Strains: HL6566 (low MicC, low ompC) and HL6555 (no MicC, low ompC). Symbols and lines are defined in panel A. (G) Experimentally measured MicC sRNA activity as a function of relative Hfq production. sRNA activity and relative Hfq production were calculated as described in the main text.
We simulated the above experimental setup to predict the effect of varying Hfq concentrations on steady state levels of target gene expression and sRNA activity. The simulations were performed using differential equations to describe the kinetics of the system and parameter values obtained from the literature (26,34) (see ‘Materials and Methods’ section and Supplementary Material). Two scenarios were considered: (i) target mRNA is translated only when it is unbound; and (ii) target mRNA is translated in the unbound and Hfq-bound states. If only free target mRNA is translated, then increasing Hfq will cause the free mRNA concentration to decrease as more target mRNA shifts to the Hfq complexes, leading to a decrease in target protein concentrations that is independent of sRNA activity (Figure 2B). To take this effect into account, we measured target gene expression with and without the sRNA, and then subtracted the former from the latter. This difference is the activity of the sRNA in the synthetic circuit, which we refer to as ‘sRNA activity’ (Figure 2C). The control without the sRNA allowed us to take into account the effect that increasing the Hfq concentration may directly have on target mRNA translation as well as the indirect effects that it may have on sRNA and mRNA stability, and the activities of other endogenous sRNAs that may also be acting on the target mRNA.
Following the above simulations we measured target gene expression (GFP) and sRNA activity in our experimental system with the RyhB–sodB::gfp pair. In the absence of RyhB, we observed that increasing Hfq production caused GFP expression from sodB::gfp mRNA to decrease (Figure 2D). With RyhB, increasing Hfq production caused GFP expression to decrease and then to increase (note: small fluctuations in GFP expression between 150 and 1000 μM IPTG were not considered to be significant) (Figure 2D). The values for target gene expression with the sRNA were subtracted from the values without the sRNA to yield the sRNA activity for each level of Hfq induction. The sRNA activity was then plotted as a function of the relative amount of Hfq produced at each IPTG concentration (Figure 2E), which was determined by placing gfp directly under the control of the inducible PLlacO-1 promoter with the T710 RBS (Supplementary Figure S3). This method of quantifying the relative Hfq concentrations was shown in an almost identical system to correlate well with measurements of the Hfq concentration by quantitative western blotting (32). We found the sRNA activity initially increased with increased Hfq production but then decreased with more Hfq production (Figure 2E). That is, maximum sRNA activity occurs at an intermediate Hfq concentration, which is consistent with the predicted Hfq set-point.
The above experiments were repeated with the MicC–ompC::gfp pair. In the absence of MicC, increasing Hfq production primarily decreased GFP expression from ompC::gfp mRNA (Figure 2F). In the presence of MicC, increasing Hfq production caused GFP expression to decrease and then increase. MicC sRNA activity at different levels of Hfq production, which was calculated as described for RyhB, was maximal at an intermediate Hfq production rate (Figure 2G). This result is also consistent with an Hfq set-point.
An activating sRNA (GlmZ) has an Hfq set-point
In the next set of experiments we examined whether an activating sRNA (GlmZ) also has an Hfq set-point. GlmZ binds the intergenic region of the glmUS target mRNA after the mRNA has been cleaved by RNase E resulting in a GlmZ–glmS duplex that increases translation and decreases glmS degradation (39,40).
An experimental system was constructed for the GlmZ–glmS pair that was very similar to that used for the silencing sRNAs. The system was modified to test the prediction that the Hfq set-point depends not on the absolute Hfq concentration but on the Hfq concentration relative to that of the sRNA and mRNA concentrations. The sRNA and mRNA concentrations were altered by transcribing them with strong or weak promoters (Figure 3A). We removed 6 nt from the 3’ end of the PCon and PConM8 promoter sequences because they prevented GlmZ activity (probably because they altered GlmZ secondary structure) to create strong and weak promoters (PConshort and PConM8short respectively). The glmS::gfp fusion target mRNA was transcribed from the strong PLtetO-1 promoter or weak PLtetO-1M9 promoter (Figure 3A and Supplementary Figure S2B). GlmZ and glmS were transcribed in four combinations to produce: (i) low sRNA, low mRNA; (ii) low sRNA, high mRNA; (iii) high sRNA, low mRNA; and (iv) high sRNA, high mRNA. The measurements were performed in a strain without chromosomal glmZ and hfq. As with the silencing sRNAs, Hfq production was varied by transcribing hfq from the PLlacO-1 promoter with the T710 RBS and adding different IPTG concentrations to the media (Figure 3A).
Figure 3.
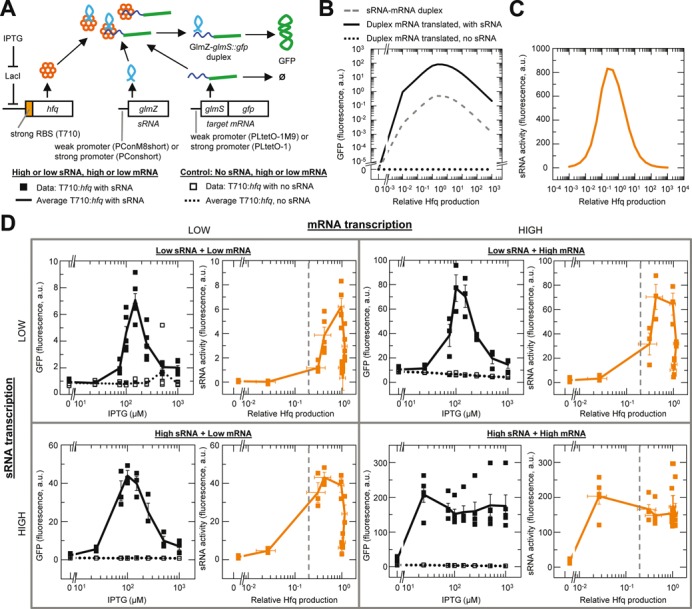
GlmZ activating sRNA has an Hfq set-point for maximum activity. (A). Reaction scheme and genetic circuit for GlmZ and glmS target mRNA. GFP expression only occurs when the GlmZ–glmS::gfp duplex is translated. Data and line symbols refer to panel D. (B) Simulated GFP expression and duplex formation for an activating sRNA. (C) Simulated GlmZ activity as a function of relative Hfq production (see main text). (D) Experimentally measured GFP expression and sRNA activity at varying Hfq induction levels and different combinations of low and high transcription rates for the sRNA and mRNA. sRNA activity was calculated by subtracting the average GFP fluorescence value without GlmZ from the average value with GlmZ at each level of Hfq induction. Relative Hfq production was determined as described in the main text. Vertical dash lines indicate the approximate level of physiological Hfq (see Figure 4 and main text). Error bars are the SEM for GFP expression or the root mean square error calculated from the SEM for sRNA activity. In each GFP expression plot n = 24 for each strain with replicate measurements in three different cultures at each IPTG concentration, except at high sRNA and high mRNA and at low sRNA and low mRNA where n = 40 for each strain with measurements in quintuplicate cultures at each IPTG concentration. Strains: HL6526 (low GlmZ, low glmS), HL6527 (low GlmZ, high glmS), HL6476 (high GlmZ, low glmS), HL6477 (high GlmZ, high glmS), HL6379 (no GlmZ, low glmS) and HL6380 (no GlmZ, high glmS).
Simulations of our model of the GlmZ–glmS system show the level of GFP expression should be proportional to the sRNA–mRNA duplex concentration (Figure 3B) and that an Hfq set-point should also occur for this activating sRNA (Figure 3C). Furthermore the simulations predict the Hfq set-point will be most evident at low sRNA and low mRNA production rates.
Measurements in our experimental system confirmed the predictions of the model. Specifically, our experimental measurements show a clear Hfq set-point at low glmZ and/or low glmS::gfp transcription; that is, increasing Hfq production up to a certain level increases sRNA activity and then further increases in Hfq production decrease sRNA activity (left upper, left lower and right upper panels, Figure 3D). At high glmZ and high glmS::gfp transcription rates, increasing Hfq production up to a certain level also increases sRNA activity but then further increases in Hfq production result in comparatively little decrease in sRNA activity (lower right panel, Figure 3D). These experiments demonstrate an Hfq set-point for an activating sRNA, and this set-point depends on the relative (rather than the absolute) concentration of Hfq to the sRNA and mRNA concentrations.
Comparison of physiological and synthetic Hfq concentrations
In E. coli the Hfq hexamer concentration is in the range of 4000–10 000 per cell (41–43), which should be sufficient to create a set-point for many endogenous sRNAs and mRNAs that are at relatively low concentrations (44–48). To explore this point we compared the physiological Hfq concentration to that in our synthetic system. This was performed by measuring target protein expression (GFP) at fixed production rates of sRNA and target mRNA and either with varying levels of induced Hfq or with physiological Hfq. GFP expression should be the same when the amount of induced Hfq equals the physiological amount of Hfq. In our system, RyhB and sodB::gfp were constitutively transcribed from weak promoters and hfq was transcribed either from the inducible PLlacO-1 promoter (in a strain with chromosomal hfq deleted) or from the endogenous chromosomal hfq (Figure 4A). The circuit with the inducible hfq was the same as in Figure 2D, E except that we chose a moderately strong st7 RBS for hfq because we expected physiological Hfq levels to be much less than that generated with the combination of a very strong T710 RBS, a strong promoter and many copies of hfq because it is on a plasmid.
Figure 4.
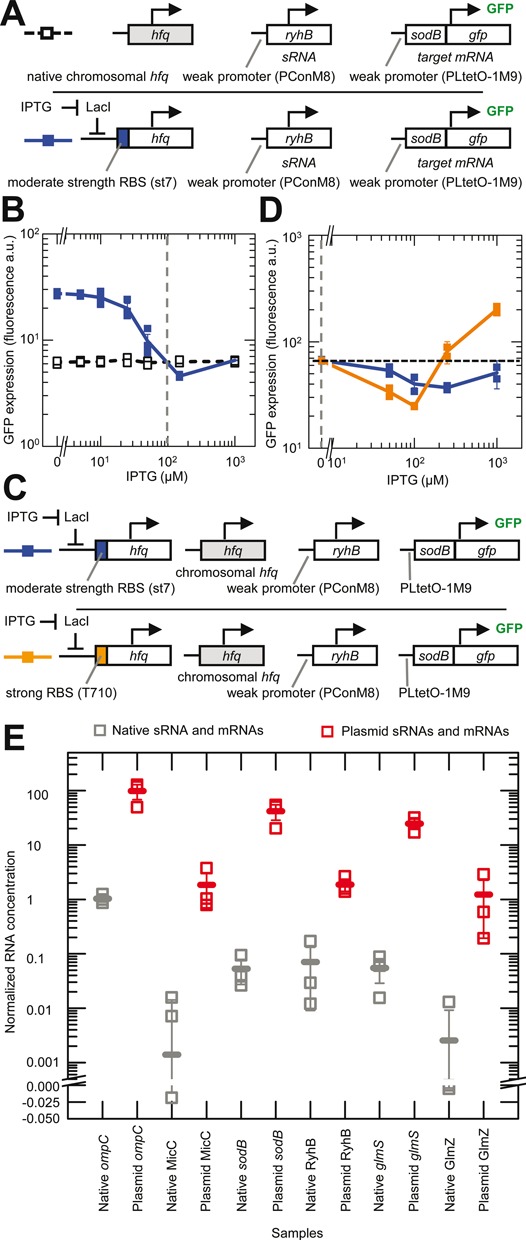
Comparison of physiological and induced concentrations of Hfq, sRNAs and mRNAs. Error bars are the SEM. (A) Schematic of the experimental system. sRNA activity with chromosomal (chr.) hfq was measured by its effect on the expression of the target mRNA (sodB::gfp). For comparison, Hfq production was increased from a plasmid copy of hfq under the control of PLlacO-1 in an hfq deletion strain until the same expression levels were achieved as with chromosomal hfq. The color of the boxes to the left indicate the corresponding data and line colors in panel B. Strains: HL6370 (plasmid hfq with st7 RBS) and HL6581 (chr. hfq). (B) Experimentally measured GFP expression (with constant transcription of target mRNA and sRNA) as a function of IPTG concentration for the systems shown in panel A. Connecting lines are guides to the eye that connect the mean GFP expression at each IPTG concentration. Vertical dash line indicates the IPTG concentration predicted to produce an induced level of Hfq that is equivalent to the level from chr. hfq (see main text). n = 21 for each strain with measurements in three replicate cultures at each concentration. (C) Schematic of the experimental system, which is the same as that shown in the lower row of panel A except that chr. hfq is present and the induced hfq either had a moderately strong RBS (st7) or a very strong RBS (T710). The color of the boxes to the left indicate the corresponding data and line colors in panel D. (D) Experimentally measured GFP expression from the target mRNA (with constant transcription of target mRNA and sRNA) as a function of IPTG concentration for the systems shown in panel C. Lines are guides to the eye that connect the mean GFP expression at each IPTG concentration. Horizontal dash line shows the starting level of GFP expression with only chromosomal hfq expressed and no induction from PLlacO-1. Note: GFP expression was measured on a different FACS machine to the experiments in panel B and therefore the absolute values cannot be directly compared. Vertical dash line indicates the concentration of Hfq from chr. hfq alone. n = 10 for each strain with measurements in duplicate at each concentration. Strains: HL6199 (chr. hfq + plasmid hfq with st7 RBS) and HL6200 (chr. hfq + plasmid hfq with T710 RBS). (E) Experimentally measured relative sRNA and mRNA concentrations in cells with endogenous genes on the chromosome (in HL716) and in cells with constitutive transcription of sRNAs and mRNAs from a plasmid (HL1178, HL2839, HL3373, HL3395, HL5963 and HL6257). The sRNA and mRNA concentrations are normalized to the 16S loading control. Each sRNA and mRNA was measured in three replicate cultures (each measurement is shown by an unfilled square). The mean value in each set of samples is indicated by a horizontal line. In these experiments all the plasmid mRNAs and sRNAs were transcribed from the PLlacO-1 promoter except GlmZ which was transcribed from the pConshort promoter.
With our experimental system we found that increasing Hfq production caused GFP expression to decrease and then increase (Figure 4B). The ‘dip’ in GFP expression is consistent with an Hfq set-point. In contrast, GFP expression was relatively constant in the control strain with chromosomal hfq, which is expected because it had no gene under the control of the PLlacO-1 promoter (Figure 4B). GFP expression with the induced hfq is the same as the chromosomal hfq (i.e. where the two lines intersect in Figure 4B) at two IPTG concentrations: ∼50–100 μM and at 1000 μM. These IPTG concentrations correspond to ∼40 and ∼100% of maximum Hfq induction with the st7 RBS (Supplementary Figure S4).
To determine which level corresponds to the physiological Hfq concentration we varied the induced hfq in the presence of chromosomal hfq in our experimental system (Figure 4C). In other words, induction of Hfq will result in concentrations that are equal to (at 0 μM IPTG) or greater than the physiological Hfq concentration. If the physiological Hfq concentration corresponds to the lower IPTG concentration, then induction of Hfq should cause GFP expression to decrease then increase. If on the other hand, the physiological Hfq concentration corresponds to the higher IPTG concentration, then induction of Hfq should only cause GFP expression to increase. The inducible hfq had the moderately strong st7 RBS or the very strong T710 RBS. We found with the st7 and the T710 RBS that the induction of Hfq initially decreased GFP expression before causing it to increase (Figure 4D). Therefore, the physiological Hfq concentration corresponds to the lower level of induction with the st7 RBS in the previous experiment (which corresponds to ∼40% of maximum induction with st7 or ∼20% of maximum induction with T710). The latter was determined from Supplementary Figures S3 and S4 and this estimated level of physiological Hfq is indicated in Figures 2E, G and 3D by a vertical dash line. In these experiments we noticed again that high Hfq concentrations can almost completely eliminate sRNA silencing.
The above experiments show that the physiological Hfq concentration is just below the Hfq set-point for the levels of RyhB and sodB in our experimental system (i.e. vertical dash line is just to the left of the Hfq concentration where there is maximum sRNA activity and therefore minimum target gene expression; Figure 4B). Even though the levels of RyhB and sodB were generated by transcription from weak promoters they are still likely to be higher than many native sRNAs and mRNAs (44–48). Furthermore even highly transcribed and stable sRNAs and mRNAs will have low concentrations at some periods such as when they are turned on or off and if they are consumed in duplexes. To provide a rough sense of how sRNA and mRNA levels in our system compare to that of endogenous genes in wild-type cells under the same conditions we measured them by quantitative RT-PCR (Figure 4E). As expected, under non-stress, exponential growth conditions most endogenous sRNAs and mRNAs are not highly transcribed and their concentrations are usually <1% of that from the strong promoters in our system (Figure 4E and Supplementary Figure S5). Given that the transcriptional output from our weak promoters is ∼10% of the strong promoters (Supplementary Figure S2A and B) then the endogenous sRNAs and mRNAs are at concentrations that are <10% of that from the weak promoters. That is, during exponential growth under non-stress conditions, the endogenous sRNA and mRNA concentrations are within the regime where there is a relatively high Hfq concentration and suppression of sRNA activity. Care must be taken in further interpreting these relative levels of sRNAs and mRNAs because they do not discriminate between the free, Hfq bound and sRNA–mRNA duplex forms, and because the total levels may not be indicative of the transcription rate of the sRNAs and mRNAs or sRNA activity due to increased degradation or stabilization of sRNAs and mRNAs in Hfq complexes and duplexes.
In summary, our experiments show that suppression of sRNA activity by relatively high endogenous Hfq concentrations can occur under some physiological conditions (i.e. during exponential growth under non-stress conditions).
Random-order binding with a dead-end mRNA–Hfq complex creates an mRNA set-point
We now investigate how sRNA and mRNA concentrations can affect sRNA activity due to random-order binding to Hfq, and how this is affected if the two pathways to duplex formation are unequal. In regard to the latter, there is no reason why the two paths to duplex formation should be the same. For example, an mRNA bound to one face of the Hfq could occlude sRNA binding on the other face thereby making duplex formation via the mRNA–Hfq complex less efficient than via the sRNA–Hfq complex.
A simulation was performed to compare the effect on target gene expression and sRNA activity if the sRNA–Hfq complex cannot bind the mRNA or the mRNA–Hfq complex cannot bind the sRNA (Figure 5A). That is, the sRNA–Hfq complex and mRNA–Hfq complex are dead-ends that only serve to sequester one of the RNAs. The dead-end complexes do not eliminate the Hfq set-point but they do decrease the amount of duplex formation (Figure 5B). In addition, maximum sRNA activity is also dependent on an optimal mRNA production rate if there are dead-end mRNA–Hfq complexes (Figure 5C) or on an optimal sRNA production rate if there are dead-end sRNA–Hfq complexes (Supplementary Figure S6E). We refer to these optimal mRNA and sRNA production rates as the mRNA and sRNA set-points respectively (Figure 5C and Supplementary Figure S6E). The simulated mRNA set-point occurs because increasing the mRNA concentration initially leads to more duplex formation due to more mRNA binding to the sRNA–Hfq complex but then further increasing the mRNA concentration sequesters significant amounts of Hfq causing decreases in the sRNA–Hfq complex concentration and duplex formation. The same logic applies to the sRNA set-point except that in this case high sRNA concentrations sequester Hfq away from the mRNA–Hfq complex to the dead-end sRNA–Hfq complex resulting in less duplex formation. To be clear, sRNA–mRNA pairs that have an mRNA or sRNA set-point will also always have an Hfq set-point but the reverse is not true. That is, the Hfq set-point, mRNA set-point and sRNA set-point all occur due to random-order binding but the latter two set-points also require an mRNA–Hfq dead-end complex or an sRNA–Hfq dead-end complex.
Figure 5.
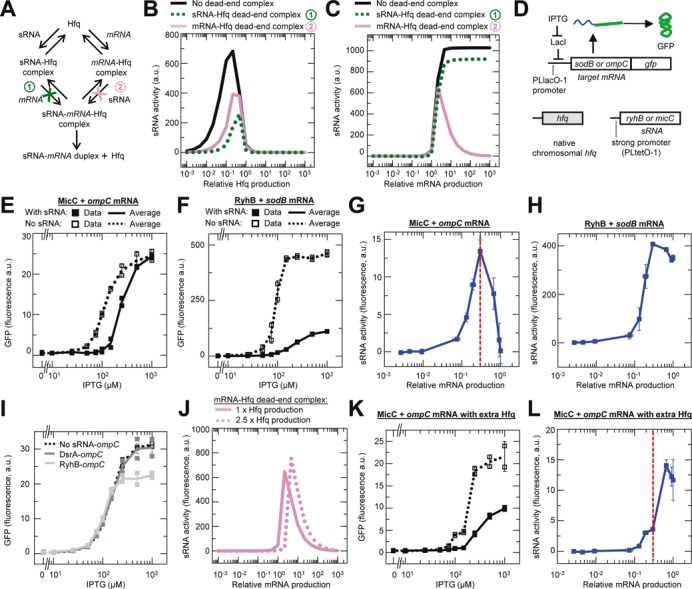
Effect of random-order binding and a dead-end mRNA–Hfq complex on sRNA activity. Error bars are the SEM of duplicate measurements (panels E, F,I and K) or root mean square error calculated from the SEM (panels G,H and L). (A–C) Simulated sRNA activity for a target mRNA as a function of Hfq production or mRNA production. Only the free mRNA is translated. Three different reactions schemes are simulated: (1) a dead-end sRNA–Hfq complex that cannot bind the mRNA; (2) a dead-end mRNA–Hfq complex that cannot bind the sRNA; and (3) no dead-end complex and both the sRNA–Hfq and mRNA–Hfq complexes can bind the mRNA and sRNA respectively (panel A). Panel B shows an Hfq set-point and panel C shows an mRNA set-point. The simulated sRNA set-point is shown in Supplementary Figure S6E. (D) Genetic circuit used to determine if an mRNA set-point occurs in vivo at varying target mRNA concentrations in the presence of chromosomal (chr.) hfq. (E and F) Experimentally measured GFP expression as a function of ompC and sodB mRNA transcription. Total n = 20 for each strain with replicate measurements in two cultures. (G and H) Experimentally measured MicC and RyhB sRNA activity as a function of relative mRNA production. sRNA activity was calculated by subtracting the average GFP fluorescence value with the sRNA from the average value without the sRNA at each level of mRNA induction. Relative mRNA production was determined using previously reported data (32). Strains: HL2304 (MicC, ompC), HL3186 (no MicC, ompC), HL2300 (RyhB, sodB) and HL2817 (no RyhB, sodB). Vertical dash line indicates the relative mRNA production that results in maximum sRNA activity (i.e. the mRNA set-point). (I) Experimentally measured GFP expression as a function of ompC mRNA transcription in the presence of chr. hfq and DsrA or RyhB. The genetic circuit was identical to that in panel D except that DsrA or RyhB substituted for MicC. Strains: HL3619 (DsrA, ompC), HL3186 (No DsrA or RyhB, ompC) and HL3612 (RyhB, ompC). (J) Simulated effect of two different Hfq production rates (1× and 2.5×) on the mRNA set-point for a target mRNA that is only translated in the free form and with a dead-end mRNA–Hfq complex. (K and L) Experimentally measured GFP expression and sRNA activity as a function of mRNA production in the presence of additional Hfq. Vertical dash line from panel G is shown to highlight the shift in the mRNA set-point (see main text). The genetic circuit used to investigate the effect of additional Hfq on the mRNA set-point is the same as that shown in panel D except there are additional copies of hfq that are on a multicopy plasmid and transcribed from the PLtetO-1 promoter with the st7 RBS. Strains: HL2325 (MicC, ompC) and HL3611 (no MicC, ompC). n = 20 for each strain with measurements in duplicate cultures.
We created an experimental system to investigate the above predictions for the mRNA set-point (Figure 5D). In the system, production of the target mRNA (sodB or ompC fused to gfp) was varied using the inducible PLlacO-1 promoter. In addition, transcription of the cognate sRNAs was constitutive and chromosomal hfq was present. We measured GFP expression at different levels of target mRNA transcription with and without the cognate sRNA (Figure 5E and F). The amount of sRNA activity is the difference in GFP expression with and without the sRNAs. We found that MicC activity increased with increasing ompC production up to a point but then clearly decreased until there was no activity (Figure 5G). This pattern for MicC-ompC is consistent with a dead-end mRNA–Hfq complex generating an mRNA set-point under the conditions measured. RyhB activity was also increased with increasing sodB transcription until very high levels of sodB mRNA transcription were achieved and then it decreased by a small amount (Figure 5H). The small decrease in RyhB activity was associated with a lower growth rate in the control strain without RyhB and therefore is probably a non-specific physiological effect of high sodB mRNA levels (due to the absence of RyhB silencing) rather than evidence for an mRNA set-point.
According to our model the mRNA set-point occurs due to ompC mRNA sequestering Hfq away from the MicC–Hfq complex which is the only viable pathway to duplex formation. Therefore the mRNA set-point should not occur if ompC transcription is increased without its cognate sRNA. We experimentally tested this prediction of the model by repeating the above experiments with chromosomal hfq and either RyhB or DsrA, which are sRNAs that are not known to interact directly with ompC. We observed that gene expression with RyhB or DsrA is almost identical to the control without an sRNA, except at high ompC production rates with RyhB, which reaches a plateau for reasons that are unclear (Figure 5I). The curves were in stark contrast to that seen with MicC (Figure 5E). Therefore as predicted a cognate sRNA–mRNA pair is required for the mRNA set-point, and this experiment excludes the possibility that high production rates were somehow causing ompC to directly or indirectly inhibit its own translation.
Because the mRNA set-point is due to sRNAs and mRNAs competing for Hfq, additional Hfq should alter the mRNA set-point. To predict precisely how additional Hfq will affect the mRNA set-point, we simulated an increase in Hfq production in our model of 2.5-fold (the approximate increase in Hfq with an additional copy of hfq under the control of the PLtetO-1 promoter and st7 RBS). The simulation predicted that additional Hfq should: (i) shift the mRNA set-point to a higher mRNA production rate, (ii) minimally increase the maximum amount of sRNA activity, (iii) substantially decrease the amount of sRNA activity at the mRNA concentration that was the set-point without the additional Hfq and (iv) decrease the drop in sRNA activity to the right of the peak (if there is an upper limit to mRNA production) (Figure 5J). Points (ii) and (iii) above are particularly important predictions because they would not be expected to be observed if the additional Hfq simply allows more duplexes to form due to it having been a limiting factor for duplex formation. Specifically, if the mRNA set-point is due to Hfq being a limiting factor for duplex formation, then we would expect the additional Hfq to substantially increase the maximum amount of sRNA activity (assuming sRNA was not also limiting) and result in the same or more sRNA activity at the mRNA concentration that was the set-point without the additional Hfq.
We experimentally tested the predicted effects of additional Hfq on the mRNA set-point by repeating the previous set of MicC-ompC experiments with Hfq expressed from a plasmid in addition to the endogenous chromosomal hfq (Figure 5K and L). Measurements of target gene expression and sRNA activity confirmed all four predictions of the simulations: (i) a shift in the mRNA set-point to a higher mRNA production rate, (ii) a very small to no increase in the maximum amount of sRNA activity, (iii) a substantial decrease in sRNA activity at the mRNA concentration that was the set-point without the additional Hfq (vertical dash line in Figure 5L) and (iv) a reduction in the amount of drop in sRNA activity at maximum mRNA production. Therefore our experiments provide strong support for the mRNA set-point and the proposed mechanism that generates it.
Additional control experiments were performed to support the above findings (Supplementary Figure S6). In brief, we repeated the above experiments for the RyhB–sodB and MicC–ompC pairs with varying mRNA production rates in strains without hfq (Supplementary Figure S6A–D). As expected, we found that without hfq there was much less sRNA activity (a small amount of sRNA activity can occur without Hfq as previously reported (32)). In addition, we examined the effect of varying sRNA transcription for the RyhB–sodB and MicC–ompC pairs and did not find any evidence of an sRNA set-point (Supplementary Figure S6E–H). We did not expect an sRNA set-point for MicC-ompC because it has an mRNA set-point, and the mRNA and sRNA set-points are mutually exclusive for a given sRNA–mRNA pair. For the RyhB–sodB pair, the absence of evidence for an sRNA set-point may be because it does not form an sRNA–Hfq dead-end complex or because we did not achieve sufficiently high sRNA levels to sequester enough Hfq to reduce duplex formation via the mRNA–Hfq complex.
In summary, our model predicted that the combination of random-order binding of sRNAs and mRNAs to Hfq and a dead-end mRNA–Hfq complex can create an mRNA set-point where maximum sRNA activity requires not only an optimal Hfq concentration but also an optimal mRNA concentration. The requirement of an optimal mRNA concentration was predicted to occur because if there is too little mRNA then this limits duplex formation and if there is too much mRNA then it sequesters Hfq causing a lack of free Hfq and sRNA–Hfq complexes which limits duplex formation. We experimentally identified an mRNA set-point for the MicC–ompC pair. In addition, we experimentally confirmed for the MicC–ompC pair that: (i) additional Hfq alters the properties of the mRNA set-point in four very specific ways that were predicted by the model; and (ii) the ompC mRNA set-point is due to competition for Hfq between the mRNA and its cognate sRNA (by showing that the mRNA set-point does not occur between the ompC mRNA and the non-cognate sRNAs RyhB and DsrA). One of the consequences of an sRNA–mRNA pair having an mRNA set-point is that it reduces maximum sRNA activity at the Hfq set-point (compare the simulated peak in sRNA activity for the Hfq set-point with and without the dead-end mRNA–Hfq complex in Figure 5B); this may be a contributing factor in why MicC caused a lower fold change in gene expression compared to RyhB in an earlier experiment (Figure 2D and F).
DISCUSSION
In this study we investigated the consequences of random-order binding of sRNAs and their target mRNAs to Hfq. We have now experimentally confirmed the prediction of an Hfq set-point (26) for three sRNA–mRNA pairs. Earlier experimental studies probably did not identify the Hfq set-point because the Hfq concentration was not sufficiently high relative to the high sRNA and mRNA concentrations, even in studies that expressed additional Hfq (32,43). Therefore previous studies measured sRNA activity under conditions that were to the left of the Hfq set-point (Figure 1B) where maximum sRNA activity is threshold-limited; that is, the amount of duplex formation is determined by the amount of whatever factor is limiting.
The Hfq set-point is likely to be physiologically important. We and others have already experimentally demonstrated that if the physiological Hfq concentration is too low relative to the sRNA and mRNA concentrations then maximum sRNA activity will be reduced (32,43,45,49). Here we experimentally show the opposite scenario may occur when physiological Hfq concentrations are too high relative to the sRNA and mRNA concentrations. This finding is predicted by the model to occur because of an increase in singly-bound Hfq complexes causing a decrease in duplex formation. There is also supporting evidence from in vitro studies that high Hfq concentrations can sequester sRNAs and mRNAs (24,25). Conditions where the physiological Hfq concentration is relatively high are likely to be common given that Hfq is one of the most abundant proteins in the cell (41–43) and that many native sRNAs and mRNAs are present at low concentrations (44–48). Furthermore, all sRNAs and mRNAs will have transiently low concentrations when transcription is first turned on or after a period following transcription being turned off. We have shown in our model (Figure 1B) and in our experiments with chromosomal hfq that negative feedback regulation of Hfq (50) does not affect the Hfq set-point (it only makes it harder to alter the Hfq concentration).
The Hfq set-point is unlikely to be due to aggregation of Hfq hexamers. The aggregation of Hfq (if it occurs) could cause sRNA activity to plateau at high Hfq concentrations but it should not by itself cause sRNA activity to decrease. For sRNA activity to decrease there would also need to be some sort of threshold-limited positive feedback mechanism associated with aggregate formation, and we are not aware of any evidence for this. Furthermore, if aggregation occurs at high Hfq production rates causing less free Hfq hexamer than at lower Hfq productions rates, then our controls without the cognate sRNA should have shown less direct and indirect effects of Hfq on target mRNA expression and therefore higher target gene expression at high Hfq production rates; but this was not observed (Figure 2D and F).
The Hfq set-point appears to constrain sRNA activity because maximum duplex formation will only occur at specific sRNA, mRNA and Hfq concentrations. In contrast, duplex formation without Hfq or compulsory-order binding of Hfq only requires that the sRNA, mRNA and/or Hfq reach a threshold level to achieve a specified level of maximum activity (32,34,37,51–53). The properties of the Hfq set-point showed differences among the MicC–ompC, RyhB–sodB and GlmZ–glmS pairs. These differences include the maximum amount of sRNA activity and the minimum Hfq concentration needed to achieve maximum sRNA activity (Figures 2E, G and 3D), which we have previously shown in simulations (26) to depend on the rates of: (i) association of sRNAs and mRNAs with Hfq; (ii) dissociation of sRNAs and mRNAs from Hfq complexes; (iii) duplex formation and release from the Hfq ternary complexes; and (iv) formation and dissociation of non-cognate ternary Hfq complexes (26). We also observed differences in the range over which the Hfq concentration was able to mediate efficient sRNA activity (previously referred to as ‘Hfq robustness’) among the MicC–ompC, RyhB–sodB and GlmZ–glmS pairs (Figures 2E, G and 3D). Simulations have shown that robustness depends on all four of the above factors as well as whether there is preferential association of sRNAs over mRNAs (or vice versa) for free or singly bound Hfq complexes, and whether there is cooperativity in the association or dissociation of sRNAs and mRNAs with Hfq (26).
One factor that simulations show does not have much impact on the properties of the Hfq set-point is the relative magnitude of the degradation rates for free and Hfq bound sRNAs (26). Many sRNAs including MicC, RyhB and GlmZ have a greater degradation rate when they are not bound to Hfq (32,54). This difference in the degradation rates of sRNAs in the free and Hfq bound states has minimal effect on sRNA activity because at low Hfq concentrations free sRNAs are in relative excess and therefore greater degradation in the free state has minimal impact on sRNA–Hfq concentrations and consequently on duplex formation. At intermediate or high Hfq concentrations, sRNAs efficiently form duplexes or sRNA–Hfq complexes resulting in low free sRNA concentrations, therefore greater degradation of free sRNAs under these conditions also has minimal impact on duplex formation.
So why has E. coli evolved an Hfq set-point (or alternatively evolved not to be constrained by it)? One explanation might be that Hfq is essential for one or more reaction steps in duplex formation (e.g. chaperone activity) but there is plenty of evidence that sRNAs in E. coli and other bacteria can evolve to form duplexes without Hfq (see ‘Introduction’ section). A second explanation is that Hfq is not essential for duplex formation but it does make it more efficient. However, others have shown that more efficient Hfq-independent duplex formation can occur when an Hfq-dependent sRNA is mutated (21). Furthermore, if the primary role of Hfq was to make duplex formation more efficient then we would have expected compulsory-order binding of sRNAs and mRNAs to Hfq to always occur (26).
A third explanation for random-order binding is that it can produce properties with advantages that outweigh its disadvantages. These properties may include: (i) further minimizing the effect that low sRNA levels, which may arise from leaky or transient transcription, have on target gene expression by sequestering them; and (ii) enabling the Hfq concentration to select which sRNA–mRNA pairs can function based on their kinetics. To elaborate on the latter, because the kinetics of duplex formation for each sRNA–mRNA pair are different, the Hfq set-point allows the Hfq concentration to selectively limit the activity of some sRNA–mRNA pairs but not others (compare sRNA activity at a relative Hfq concentration of 2 × 10−1 in the experiments shown in Figure 2E and G). The above properties increase the potential for greater control of sRNA regulation by making it conditional on specific requirements that do not occur with Hfq-independent duplex formation or compulsory-order binding. It should be noted that random-order binding should not impact other properties of sRNA regulation including generating threshold-linear responses (37,52), filtering low amplitude and transient signals in gene regulation (51,52) and propagating signals with less delay (34,53).
The question of why random-order binding occurs directly relates to: (i) why mRNAs can directly bind Hfq (rather sRNAs bound to Hfq always serving as a guide to find their cognate mRNAs, which would prevent Hfq being sequestered by mRNAs that do not have any cognate sRNA present); and (ii) why Hfq is able to directly repress translation without an sRNA (as we observed in experiments with ompC and sodB). Both of these features have also been shown in vitro so it is clear that the ability of Hfq to bind directly to mRNAs and inhibit their translation (55,56) is not simply because of some missed in vivo interaction of Hfq with another unknown sRNA. One explanation is that Hfq may be binding and silencing some mRNAs to dampen the effect of fluctuations in their concentration due to transcription, which is typically one of the ‘noisiest’ processes in gene expression (57). That is, Hfq may act like a pH buffer to prevent changes in the free mRNA concentration. There are two appealing features to this explanation. The first is that it may explain why Hfq binds mRNAs in bacteria that do not need Hfq for duplex formation (e.g. S. aureus and Bacillus subtilis) (58,59). The second is that it shifts the perspective from thinking about how the binding of mRNAs directly to Hfq makes sense in the context of duplex formation to how duplex formation fits with Hfq's role in providing robust control of target gene expression. From this latter perspective, the constraints on sRNA regulation due to the Hfq set-point can be seen as advantageous because they minimize the impact of transient signals and stochastic fluctuations in sRNA production (the latter being more likely to occur at low concentrations) thereby making target gene expression more robust.
We have shown in our models and experiments that an additional consequence of Hfq being able to bind directly to many mRNAs without first binding to their cognate sRNAs is that it can create an mRNA set-point if the mRNA–Hfq complex is a dead-end and the sRNA is also able to bind directly to Hfq. As stated earlier, in the presence of a dead-end mRNA–Hfq complex the maximum sRNA activity depends not only on having an optimal Hfq concentration but also on having an optimal mRNA concentration. The latter occurs because at low mRNA concentrations there is insufficient mRNA, which limits the amount of duplex formed, and at high mRNA concentrations the mRNA can sequester the Hfq resulting in decreased concentrations of sRNA–Hfq complexes and consequently less duplex formation. To be clear, an mRNA set-point does not occur simply because the mRNA concentration exceeds the sRNA concentration. There must be an mRNA–Hfq dead-end complex; without the dead-end complex, once the mRNA concentration exceeds the sRNA concentration then the amount of sRNA activity simply reaches a maximum level at a particular mRNA concentration and further increases in mRNA concentration have no effect (rather than decreasing sRNA activity as we observed; Figure 5G). While it remains unclear whether mRNA set-points occur with endogenous sRNA–mRNA pairs, our demonstration that it can be observed in vivo with an engineered sRNA regulatory system shows that it needs to be considered in future studies and factored into the design of synthetic circuits. Furthermore, the accurate prediction of the mRNA set-point and the effects of additional Hfq on its properties provide strong additional supporting evidence for the model and for the Hfq set-point since the random-order binding that is necessary for mRNA set-point to occur (note: without random-order binding a dead-end complex could not occur and decrease sRNA activity) is also required for the Hfq set-point.
In conclusion, this study advances our understanding of the kinetics and properties of sRNA regulation in several ways. First, it highlights the necessity of explicitly including Hfq and its complexes in models to obtain accurate quantitative predictions of the properties and regulation of target gene expression and sRNA activity. As we have demonstrated by multiple experiments, the model was able to provide useful, counterintuitive and specific predictions. This includes the Hfq and mRNA set-points, which we would not have investigated and discovered if the models had not explicitly included the kinetics of Hfq complex formation and Hfq mediated duplex formation. Second, it experimentally demonstrates that random-order binding of sRNAs and mRNAs to Hfq results in distinctly different gene regulatory properties compared to those expected to occur with Hfq-independent duplex formation and with non-coding RNA regulatory mechanisms that have compulsory-order reaction schemes of duplex formation. For instance random-order binding is able to produce non-monotonic responses where increasing the concentration of Hfq, sRNA and/or mRNA leads to increasing sRNA activity up to a point followed by decreasing sRNA activity. Third, the simulations and experiments demonstrate that there is an Hfq set-point, and this finding should be applicable to almost every sRNA–mRNA pair that requires Hfq for duplex formation. All sRNAs and mRNAs will likely have low concentrations relative to the Hfq concentration at some point (e.g. when transcription is first turned on, after transcription has been turned off, if most of the sRNAs and mRNAs are consumed in duplexes, or if the sRNAs and mRNAs have weak promoters and short lifetimes) leading to inhibition of sRNA activity. Fourth, an mRNA set-point or sRNA set-point may occur due to the formation of dead-end complexes and this can impact the level of sRNA activity. Fifth and lastly, the experiments in this study reveal that sRNA activity is very dependent on the specific conditions within a cell; this presents a challenge to quantitative analysis of sRNA activity in physiological and synthetic systems but it also presents an opportunity for fine-tuning and robust control of gene regulation.
Supplementary Material
Acknowledgments
We thank Chris Lin, Katya Frazier and Justin Choe for constructing some of the strains.
Author Contributions: S.S., J.-E.S. and R.H. performed the experiments and analyzed the data. H.N.L. designed the project, analyzed data, performed simulations and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of California, Berkeley; Hellman Fellows Fund (to H.N.L.). Funding for open access charge: Berkeley Research Impact Initiative (BRII).
Conflict of interest statement. None declared.
REFERENCES
- 1.Michaux C., Verneuil N., Hartke A., Giard J.C. Physiological roles of small RNA molecules. Microbiology. 2014;160:1007–1019. doi: 10.1099/mic.0.076208-0. [DOI] [PubMed] [Google Scholar]
- 2.Chao Y., Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Desnoyers G., Bouchard M.P., Masse E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2012;29:92–98. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Frohlich K.S., Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Papenfort K., Vanderpool C.K. Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 2015;39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sledjeski D.D., Gupta A., Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 7.Lease R.A., Cusick M.E., Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soper T., Mandin P., Majdalani N., Gottesman S., Woodson S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon K., Six D.A., Lee H.J., Raetz C.R., Gottesman S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013;89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masse E., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prevost K., Salvail H., Desnoyers G., Jacques J.F., Phaneuf E., Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 12.Vanderpool C.K., Balasubramanian D., Lloyd C.R. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–1949. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moll I., Leitsch D., Steinhauser T., Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Lay N., Schu D.J., Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasini A., Francois P., Howden B.P., Fechter P., Romby P., Caldelari I. The importance of regulatory RNAs in Staphylococcus aureus. Infect. Genet. Evol. 2014;21:616–626. doi: 10.1016/j.meegid.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Brantl S., Bruckner R. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol. 2014;11:443–456. doi: 10.4161/rna.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey S.P., Winkler J.A., Li H., Camacho D.M., Collins J.J., Walker G.C. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics. 2014;15 doi: 10.1186/1471-2164-15-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castillo-Keller M., Vuong P., Misra R. Novel mechanism of Escherichia coli porin regulation. J. Bacteriol. 2006;188:576–586. doi: 10.1128/JB.188.2.576-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidtke C., Abendroth U., Brock J., Serrania J., Becker A., Bonas U. Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas. PLoS Pathog. 2013;9:e1003626. doi: 10.1371/journal.ppat.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y., Zhang Z.J., Erickson D.W., Huang M., Huang Y., Li J., Hwa T., Shi H. Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12473–12478. doi: 10.1073/pnas.1100432108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Updegrove T.B., Shabalina S.A., Storz G. How do base-pairing small RNAs evolve. FEMS Microbiol. Rev. 2015;39:379–391. doi: 10.1093/femsre/fuv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissmann T.A., Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lease R.A., Woodson S.A. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Fender A., Elf J., Hampel K., Zimmermann B., Wagner E.G. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamson D.N., Lim H.N. Essential requirements for robust signaling in Hfq dependent small RNA networks. PLoS Comput. Biol. 2011;7:e1002138. doi: 10.1371/journal.pcbi.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ipsaro J.J., Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015;22:20–28. doi: 10.1038/nsmb.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luteijn M.J., Ketting R.F. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 29.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 30.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussein R., Lim H.N. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamson D.N., Lim H.N. Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13120–13125. doi: 10.1073/pnas.1308476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussein R., Lim H.N. Direct comparison of small RNA and transcription factor signaling. Nucleic Acids Res. 2012;40:7269–7279. doi: 10.1093/nar/gks439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner E.G. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., Zhang A., Blyn L.B., Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine E., Zhang Z., Kuhlman T., Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban J.H., Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban J.H., Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalamorz F., Reichenbach B., Marz W., Rak B., Gorke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 41.Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajitani M., Kato A., Wada A., Inokuchi Y., Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J. Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon K., Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghavan R., Groisman E.A., Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papenfort K., Said N., Welsink T., Lucchini S., Hinton J.C., Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 46.Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J. An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfeiffer V., Sittka A., Tomer R., Tedin K., Brinkmann V., Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 48.Guantes R., Cayrol B., Busi F., Arluison V. Positive regulatory dynamics by a small noncoding RNA: speeding up responses under temperature stress. Mol. Biosyst. 2012;8:1707–1715. doi: 10.1039/c2mb05479e. [DOI] [PubMed] [Google Scholar]
- 49.Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vecerek B., Moll I., Blasi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine E., Hwa T. Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 2008;11:574–579. doi: 10.1016/j.mib.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehta P., Goyal S., Wingreen N.S. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol. Syst. Biol. 2008;4 doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimoni Y., Friedlander G., Hetzroni G., Niv G., Altuvia S., Biham O., Margalit H. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gopel Y., Papenfort K., Reichenbach B., Vogel J., Gorke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013;27:552–564. doi: 10.1101/gad.210112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vecerek B., Moll I., Afonyushkin T., Kaberdin V., Blasi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 56.Vytvytska O., Moll I., Kaberdin V.R., von Gabain A., Blasi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 57.Maheshri N., O'Shea E.K. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu. Rev. Biophys. Biomol. Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 58.Dambach M., Irnov I., Winkler W.C. Association of RNAs with Bacillus subtilis Hfq. PLoS One. 2013;8:e55156. doi: 10.1371/journal.pone.0055156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y., Wu N., Dong J., Gao Y., Zhang X., Mu C., Shao N., Yang G. Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS One. 2010;5:e13069. doi: 10.1371/journal.pone.0013069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


