Abstract
Endochondral ossification consists of successive steps of chondrocyte differentiation, including mesenchymal condensation, differentiation of chondrocytes, and hypertrophy followed by mineralization and ossification. Loss-of-function studies have revealed that abnormal growth plate cartilage of the Cdc42 mutant contributes to the defects in endochondral bone formation. Here, we have investigated the roles of Cdc42 in osteogenesis and signaling cascades governing Cdc42-mediated chondrogenic differentiation. Though deletion of Cdc42 in limb mesenchymal progenitors led to severe defects in endochondral ossification, either ablation of Cdc42 in limb preosteoblasts or knockdown of Cdc42 in vitro had no obvious effects on bone formation and osteoblast differentiation. However, in Cdc42 mutant limb buds, loss of Cdc42 in mesenchymal progenitors led to marked inactivation of p38 and Smad1/5, and in micromass cultures, Cdc42 lay on the upstream of p38 to activate Smad1/5 in bone morphogenetic protein-2-induced mesenchymal condensation. Finally, Cdc42 also lay on the upstream of protein kinase B to transactivate Sox9 and subsequently induced the expression of chondrocyte differential marker in transforming growth factor-β1-induced chondrogenesis. Taken together, by using biochemical and genetic approaches, we have demonstrated that Cdc42 is involved not in osteogenesis but in chondrogenesis in which the BMP2/Cdc42/Pak/p38/Smad signaling module promotes mesenchymal condensation and the TGF-β/Cdc42/Pak/Akt/Sox9 signaling module facilitates chondrogenic differentiation.
Keywords: Cdc42, condensation, osteoblast, chondrocyte
SEVERAL successive steps, including mesenchymal condensation of undifferentiated cells, chondrocyte differentiation, proliferation of chondrocytes, and differentiation into hypertrophic chondrocytes followed by mineralization and ossification, exist in the process of endochondral ossification (Long and Ornitz 2013). Mesenchymal condensation is a prerequisite for chondrogenesis and is facilitated by cell adhesion molecules. Upregulation of cell adhesion proteins such as N-cadherin is a hallmark of condensing cells, whereas loss of cell adhesion molecules is able to abolish condensation (Delise and Tuan 2002; Bobick et al. 2009). The molecular mechanisms governing condensation are not fully understood, though several genes have been implicated in this process such as bone morphogenic proteins (BMPs) and SRY (sex determining region Y)-box 9 (Sox9) (Fromental-Ramain et al. 1996; Wada et al. 1998; Lu et al. 2008). Conditional inactivationof Sox9 in limb mesenchymal progenitors leads to the absence of condensations, while simultaneous deletion of Bmp2 and Bmp4 in limb mesenchymes leads to the loss of zeugopod elements and to defective joint articulations (Reversade et al. 2005). During endochondral ossification, condensed cells undergo the differentiation into chondrocytes that secrete an abundance of cartilage matrices including types II, IX, and XI collagen. Sox9 is also the earliest known transcription factor that is required for chondrogenesis (Foster et al. 1994). Haploinsufficiency of Sox9 results in murine chondrodysplasia, and complete loss of Sox9 in prechondrogenic limb mesenchymes abolishes chondrogenesis (Bi et al. 1999; Akiyama et al. 2002). After chondrogenic differentiation, cells in the core of limbs undergo progressive maturation and hypertrophy associated with the secretion of type X collagen (Col10α1); eventually, the terminally differentiated chondrocytes undergo apoptosis and the cartilaginous matrix is mineralized and replaced by bone (Kronenberg 2003; Michigami 2013). However, the molecular mechanisms governing chondrogenesis are not fully understood.
The Rho family of small GTPases including RhoA, Rac1, and Cdc42 are molecular switches that have multiple roles in regulation of cell movement, cell cycle, proliferation, differentiation, and apoptosis (Exton 1998; Bishop and Hall 2000). Over the years, a large body of evidence indicates that Rac1 is essential for chondrogenesis, while RhoA antagonizes Rac1’s effects (Wang et al. 2007). Overexpression of Cdc42 in ATDC5 cells results in increases of Sox5, Sox9, and Collagen II expression, suggesting that Cdc42 like Rac1 is a positive regulator of chondrogenesis (Wang and Beier 2005). Recently, Aizawa et al. (2012) have demonstrated the critical roles of Cdc42 in limb development by conditional deletion of Cdc42 in limb mesenchymal progenitors (Prx1Cre;Cdc42fl/fl). Prx1Cre;Cdc42fl/fl embryos demonstrate the delayed mesenchymal condensation at embryonic day 11.5 (E11.5), and Prx1Cre;Cdc42fl/fl mice have short limbs, abnormal calcification of the cranium, cleft palate, disruption of the xiphoid process, and syndactyly. Among the variety of phenotypes in Prx1Cre;Cdc42fl/fl mice, severe defects in long bone growth plate cartilage are notable and related to loss of columnar organization of chondrocytes, as well as thickening and massive accumulation of hypertrophic chondrocytes, resulting in delayed endochondral bone formation associated with reduced bone growth (Aizawa et al. 2012). Moreover, chondrocyte-specific inactivation of Cdc42 (Col2Cre;Cdc42fl/fl) shows shorter limbs and body, and severe defects were found in growth plate chondrocytes of long bone, characterized by a reduced proliferating zone height, wider hypertrophic zone, and loss of columnar organization in proliferating chondrocytes (Suzuki et al. 2015).
Despite data demonstrating the importance of Cdc42 in cartilage development, little is known about the molecular mechanisms underlying Cdc42-mediated chondrogenesis. On the other hand, though activation of FYVE, RhoGEF, and PH domain-containing protein 1 (FGD1), a guanine nucleotide exchange factor (GEF) of Cdc42, promotes the osteogenic differentiation in human mesenchymal stem cells (Gao et al. 2011), whether Cdc42 directly regulates bone formation still lacks genetic evidence. Hence, we have investigated the roles of Cdc42 in osteogenesis and the signaling cascades governing Cdc42-mediated chondrogenic differentiation.
Materials and Methods
Chemicals and antibodies
SB203580, IPA-3, wortmannin, and anisomycin were obtained from Selleckchem (Houston). ML141, Alizarin Red S, Alcian blue, and X-gal were obtained from Sigma (St. Louis). Recombinant BMP-2 and TGF-β1 protein were purchased from R&D Systems (Minneapolis). Antibodies against Col1α1 (sc-59772), Lamin B (sc-6216), and β-actin (sc-8432) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against p-AKT (Ser473, 4060), p-Erk (4370), p-JNK (9255), p-p38 (4511), p-Smad5 (13820), AKT (9272), Erk (4695), JNK (9252), p38 (9212), Smad5 (9517), Pak1 (2612) and N-cadherin (4061) were from Cell Signaling Technology (Danvers, MA). Antibodies against AP (ab95462), BSP (ab33022), Cdc42 (ab64533), OC (ab13420), Osx (ab22552), p-Sox9 (ab59252), Sox9 (ab26414), and Runx2 (ab76956) were purchased from Abcam (Cambridge, UK). The IRDye 680- and 800-sec antibodies were from LI-COR Bioscience (Lincoln, NE).
Mouse strains
Prx1Cre, Col1Cre, and Cdc42fl/fl mice on a C57BL/6J genetic background were provided by Harvard Medical School, McGill University, and Cincinnati Children’s Hospital Medical Center and generated as previously described (Logan et al. 2002; Miao et al. 2005; Yang et al. 2007; Zha et al. 2008). ROSA26 mice were obtained from the Model Animal Research Center of Nanjing University. To conditionally inactivate Cdc42 in mesenchymal progenitors and preosteoblasts, we crossed floxed Cdc42 mice (Cdc42fl/fl) to mice carrying the Cre recombinase transgene under control of Prx1 (Prx1Cre) and Col1α1 promoter (Col1Cre), respectively. Prx1Cre;Cdc42fl/+ or Col1Cre;Cdc42fl/+ mice were then crossed to Cdc42fl/fl mice to generate Prx1Cre;Cdc42fl/fl or Col1Cre;Cdc42fl/fl embryos. To check the activity of Cre recombinase, the Col1Cre transgenic line was bred with ROSA26 mice to generate Col1Cre;R26R mice. All animals were housed at the Zhejiang University Animal Care Facility according to the institutional guidelines for laboratory animals, and the animal protocol was approved by the Zhejiang University Institutional Animal Care and Use Committee.
Whole-mount skeletal staining
Whole-mount skeletal staining with Alizarin-Red S and Alcian blue was performed as previously described (Wu et al. 2008). Briefly, embryos were skinned, eviscerated, and dehydrated in 95% ethanol overnight. The skeletons were then stained overnight with 0.015% Alcian Blue and 10% acetic acid in 75% ethanol, and soft tissues were dissolved in 1% KOH, while the skeletons were additionally stained overnight with 0.0075% Alizarin-Red S in 1% KOH. Finally, the skeletons were cleared in 1% KOH and stored in glycerol/ethanol (1:1). Embryonic tissues were fixed in 10% formalin overnight at room temperature, and then processed and embedded in paraffin prior to sectioning at 4 μm.
LacZ staining
Whole-mount X-gal staining was performed as described previously (Day et al. 2005). Embryos were fixed for 5 hr at room temperature in 0.2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3), then washed three times in PBS, and placed in X-gal staining solution containing 1 mg/ml X-gal in N,N-dimethylformamide, 5 mM K3Fe(CN)6, and 5 mM K4Fe(CN)6 overnight at 37° in the dark. Embryos were embedded in paraffin, sectioned at 5 μm, and counterstained with Nuclear Fast Red.
In situ hybridization and immunostaining
The sections were subjected to in situ hybridization as previously described (Tang et al. 2015). Immunohistochemistry was performed by using the SP Rabbit HRP kit (Kangwei Reagents, Beijing) as previously described (Tang et al. 2015). Briefly, after deparaffinized and rehydrated in xylene and a graded series of ethanol, the sections were subjected to antigen retrieval in 10 mM sodium citrate and 10 mM citric acid. Tissue sections were incubated with 3% H2O2 in methanol to quench endogenous peroxidase followed by sequential incubation with normal serum, control IgG, and primary antibodies overnight at 4°. After sections were incubated with HRP-labeled secondary antibody, the diaminobenzidine (DAB) solution was used for development of brown color. The sections were counterstained with hematoxylin, and the primary antibodies used were as follows: anti-p-p38 (1:100, Cell Signaling Technology), anti-p-Smad1/5 (1:100, Cell Signaling Technology).
Cell cultures and treatments
C3H10T1/2 cells were obtained from ATCC and cultured in minimum essential medium medium (MEM) (Life Technologies, Grand Island, NY) containing 10% fetal bovine serum (FBS) (Life Technologies) as previously described (Gong et al. 2014). HEK293T packaging cells for generating the lentiviruses and 293GPG packaging cells for generating retroviruses were cultured as previously described (Ory et al. 1996; Pan et al. 2015). After confluence, C3H10T1/2 cells were starved for 12 hr and then pretreated with various inhibitors (SB203580 at 20 μM, ML141 at 40 μM, IPA-3 at 10 μM, and anisomycin at 5 μM) for 3 hr followed by further treatment with recombinant human BMP2 (rhBMP2) or TGF-β at 10 ng/ml for indicated times. Alternatively, when C3H10T1/2 cells reached 60% confluence, cells were infected with lentiviruses or retroviruses for 24 hr and then treated with rhBMP2 or TGF-β at 10 ng/ml for indicated times. All cell lines were incubated at 37° with 5% CO2.
Generation of Cdc42-shRNA-expressing lentiviruses and da-Cdc42-expressing retroviruses
Lentiviruses expressing Scramble-shRNA, Cdc42-shRNA, Akt1-shRNA, or Smad5-shRNA were generated as previously described (Pan et al. 2015). Briefly, the hairpin shRNA templates of complementary oligonucleotide containing overhangs were digested. The synthesized complementary oligonucleotides were annealed and inserted into a lentiviral shRNA-expressing vector, pSicoR-GFP. The sense and antisense sequences for mCdc42, mAkt1, and mSmad5 are listed in Supporting Information, Table S1. The 293T packaging cells were transfected with 6 μg of each construct by Lipofectamine reagents (Life Technologies); 72 hr after transfection, supernatants were harvested and used for infection. Retroviruses expressing GFP or da-Cdc42 were generated as previously described (Wu et al. 2008). A proper dilution of each virus stock in appropriate growth medium was chosen to achieve >90% infection as per GFP detection. For infections, cells were incubated at ∼60% confluence in virus-containing medium for 24 hr.
Reporter construction, transient transfection, and dual-luciferase assays
The Sox9 reporter construct was generated as previously described. The construct contains four repeats of a 48-bp Collagen2α1 intron 1 enhancer element that is known to be activated by Sox9 (Lefebvre et al. 1996). When C3H10T1/2 cells reached 60% confluence on 24-well plates, the cells were transfected with 2 μl Lipofectamine 2000 reagent, 2 μg Sox9 reporter, or Runx2-dependent 6× OSE reporter constructs, and 0.02 μg Renilla luciferase constructs for 6 hr in the absence of serum. Then, the cells were cultured in the media containing various chemicals in presence or absence of rhBMP2 at 10 ng/ml for 48 hr. In some cases, the cells were infected with lentiviruses or retroviruses for 24 hr, before transfection with reporter constructs and treatment with or without rhBMP2 for indicated times. After transfection and treatments, the cell lysates were prepared in lysis buffer, and the supernatants were used for dual-luciferase assay according to the manufacturer’s instructions (Promega, Madison, WI). The firefly luciferase activities were normalized to Renilla luciferase levels. Two distinct small interfering RNA (siRNA) duplexes each for Scramble, Cdc42, Akt1, and Pak1 were used in experiments; the targeting siRNA sequences are listed in Table S2.
Osteoblast and chondrocyte differentiation assays and quantitative RT-PCR
C3H10T1/2 cells were pretreated with inhibitors for 3 hr as above or infected with viruses for 24 hr; then, cells were stimulated with or without rhBMP2 at 10 ng/ml or TGF-β at 10 ng/ml for 48 hr. Total RNA was extracted using TRIzol reagent (Takara Biotechnology, Dalian, China). Messenger RNA (mRNA) levels of Runx2, OSX, N-cadherin, and Col2α1 were determined by quantitative RT-PCR as described previously (Gong et al. 2014). The relative amounts of each mRNA level were normalized to GAPDH levels, and the differences in mRNA levels were calculated by 2−ΔΔCt method. The primers are listed in Table S3. For mineralization assays, C3H10T1/2 cells were infected with scramble-short hairpin RNA (shRNA)- or Cdc42-shRNA-expressing lentiviruses for 24 hr; then, cells were incubated in the presence or absence of 50 μM ascorbic acid, 100 nM dexamethasone and 10 mM β-glycerophosphate for 21 days. After that, cells were stained with Alizarin-Red S as previously described (Gong et al. 2014), and Alizarin-Red S was further extracted with 10% cetylpyridinium chloride (Sigma) for 2 hr at room temperature and quantified at 570 nm in a microplate reader (BioRad, Hercules, CA). Protein levels were measured using a Pierce Micro BCA kit (Pierce Biotechnology, Rockford, IL).
Cdc42, p38, Akt, and Smad1/5 activation assays and Western blots
C3H10T1/2 cells were starved for 12 hr and then pretreated with or without inhibitors for 3 hr; alternatively, cells were infected with lentiviruses or retroviruses for 24 hr, before cells were further incubated with rhBMP2 or TGF-β at 10 ng/ml for indicated times in the presence or absence of inhibitors. After that, cells were harvested for preparing the cell lysates and centrifugation; the supernatants were subjected to pull-down assays by using Active GTPase Pull-Down and Detection kits (Pierce Biotechnology) as previously described (Yao et al. 2011); alternatively, the supernatants were subjected to Western blot (WB) assays for determining the activation of p38, Akt1, and Smad1/5. Total Cdc42 was used as an internal control for the active form of Cdc42 (GTP binding form).
Micromass cultures and Alcian Blue staining
Micromass cultures were performed as previously described (Denker et al. 1999). Briefly, C3H10T1/2 cells at 2 × 105 were plated as a 20-μl drop at the center of a 24-well plate and allowed to adhere for 1 hr, prior to being flooded with 1 ml of differentiation media in the presence or absence of rhBMP2 at 10 ng/ml. C3H10T1/2 cells were infected with either da-Cdc42-expressing retroviruses or Cdc42-shRNA-expressing lentiviruses for 24 hr; then, the cells were plated for micromass cultures, followed by the addition of differentiation media in the presence or absence of 10 ng/ml rhBMP2 and inhibitors as above for 7 days. After being fixed in ethanol, cells were stained with 0.5% Alcian Blue (Sigma) in 0.1 N HCl (pH 1.0) overnight, then rinsed with distilled water, dissolved by 6 M guanidine-HCl, and quantified at 570 nm in a microplate reader.
Isolation of subcellular fractions and immunoprecipitation
The cytosolic and nuclear fractions of cells were prepared by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoScientific, Waltham, MA) as per the manufacturer’s instructions. β-Actin and lamin-B were used as the internal standards for the cytosolic and nuclear fractions, respectively. For coimmunoprecipitation, cells were treated with rhBMP2 at 10 ng/ml for 1 hr and harvested for preparation of lysates. After centrifugation, the supernatant was subjected to immunoprecipitation by using protein A/G plus agarose beads (Santa Cruz Biotechnology) containing either control IgG or anti-p-p38 antibody. After beads were washed with washing buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1% NP-40, and 2 mM EDTA), the sample was boiled and centrifuged, the supernatant was subjected to SDS-PAGE and Western blots by using anti-p-Smad5 and anti-p-p38 antibodies (1:1000).
Statistical analysis
Numerical data were expressed as means ± SD, and statistical analysis was performed by using the SPSS statistical package (IBM, North Castle, NY). We determined significance by one-way ANOVA and Tukey–Kramer multiple comparisons test. Statistical significance was assessed at levels of P < 0.05 and P < 0.01. Experiments were at least independently triplicated and results were qualitatively identical. Representative experiments are shown.
Results
Expression patterns of Cdc42 in embryonic long bones and deficiency in ossification by genetic ablation of Cdc42
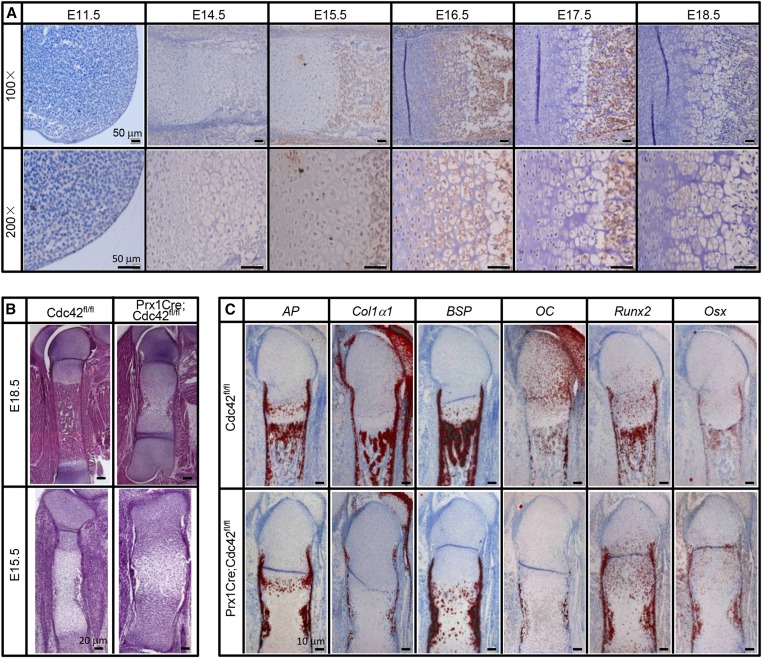
To investigate the expression patterns of Cdc42 in developing long bones, we harvested embryos from E11.5 to E18.5. The expression of Cdc42 weakly detectable in both the zone of polarizing activity (ZPA) and epithelial covering of limb bud at E11.5 was increased gradually from E14.5 and reached a peak at around E16.5 (Figure 1a). Though Cdc42 was predominantly expressed in terminal hypertrophic chondrocytes and trabecular bone areas at E15.5 and E17.5, it was robustly expressed in proliferating chondrocytes, prehypertrophic chondrocytes, hypertrophic chondrocytes, and trabecular bone areas at E16.5 (Figure 1a). At E18.5, Cdc42 expression was exclusively restricted to hypertrophic chondrocytes and trabecular bone areas (Figure 1a). Severe cartilage phenotypes have been reported by several previous studies (Aizawa et al. 2012; Suzuki et al. 2015); however, whether Cdc42 directly regulates osteogenesis still lacks solid evidence. To assess the physiological role of Cdc42 in bone formation, we genetically removed Cdc42 from the skeletal progenitors by using the Cre-loxP technique and harvested the embryos at E18.5 and E15.5. As previously described (Aizawa et al. 2008), Prx1Cre;Cdc42flox/flox (Prx1Cre;Cdc42fl/fl) embryos had normally patterned but shorter limbs as compared with wild-type littermates, and whole-mount skeletal staining revealed that PrxCre;Cdc42fl/fl embryos exhibited thicker and shorter mineralized bones in the forelimbs and hindlimbs as compared with wild-type littermates (Figure S1). At E15.5, when wild-type embryos showed obvious ossification, Cdc42 mutant littermates exhibited a diminution in ossification (Figure S1). To further analyze the phenotypes of the long bones, we performed paraffin-embedded section and hematoxylin and eosin (H&E) staining for femora at E18.5 and E15.5. In gross appearance, the femora of Cdc42 mutants appeared fatter and shorter than those of wild-type littermates. Except for the disorganized growth plates with excessive undifferentiated chondrocytes and nonresorbed hypertrophic chondrocytes, the expansion of hypertrophic zone and consequently diminished and delayed ossification are robust features of the phenotype, though columnar disorganization of the proliferating and hypertrophic chondrocytes and nonresorbed hypertrophic cartilage existed in growth plates (Figure 1b). Consistent with previous findings (Aizawa et al. 2012; Suzuki et al. 2015), our data indicate that genetic ablation of Cdc42 in skeletal progenitors results in severe phenotypes in developing long bones.
Figure 1.
Expression patterns of Cdc42 and deficiency in ossification by genetic ablation of Cdc42. (a) Expression patterns of Cdc42 in embryonic long bones. (b) H&E staining for the sections of femora at E18.5 and E15.5. (c) In situ hybridization analyses for the genes of osteogenic differentiation markers by using paraffin-embedded sections of femur at E18.5.
To examine potential defects in the osteoblast lineage, we assayed the expression of osteoblast differentiation markers on adjacent sections of E18.5 femora. In the wild-type E18.5 femora, robust expression of alkaline phophatase (AP) was detected in the perichondrium surrounding the prehypertrophic and hypertrophic cartilage, in the bone collar flanking the marrow cavity, in the early hypertrophic chondrocytes, as well as in the primary spongiosa (Figure 1c). In Cdc42 mutants, AP was detected in all above mentioned areas except for the primary spongiosa (Figure 1c). Collagen type 1 (Col1a1) was expressed in a similar manner to AP in the wild-type embryo, except that Col1a1 was not expressed by early hypertrophic chondrocytes (Figure 1c). In Cdc42 mutants, only a low level of Col1a1 was detected in the perichondrium, no Col1a1 expression was evident in the primary spongiosa (Figure 1c). Like Col1a1, bone sialoprotein (Bsp) was normally expressed in the perichondrium surrounding the prehypertrophic and hypertrophic cartilage, as well as in the primary spongiosa (Figure 1c). However, Bsp was ectopically and robustly expressed in the nonresorbed hypertrophic chondrocytes, though it was detectable in either the perichondrium or the marrow cavity (Figure 1c). Osteocalcin (OC) was normally expressed by mature osteoblasts present in either bone collar or primary spongiosa, but OC was only detectable at low levels in bone collar of Cdc42 mutants (Figure 1c). Runt-related transcription factor 2 (Runx2) was most robustly expressed in the perichondrium, hypertrophic chondrocytes, and primary spongiosa as well, though it was also expressed at a low level in immature chondrocytes (Figure 1c). In Cdc42 mutants, Runx2 expression was maintained at relatively high levels in the perichondrium, immature chondrocytes, and bone collar, though it was rarely expressed in the hypertrophic chondrocytes (Figure 1c). Ostrix (Osx) was normally expressed at high levels in the perichondrium flanking the prehypertrophic and hypertrophic chondrocytes in addition to a lower level in the prehypertrophic cells (Figure 1c). Like Runx2, Osx was relatively abundant in the perichondrium, immature chondrocytes, and bone collar, though it was also detectable in the diminished primary spongiosa of Cdc42 mutants (Figure 1c). Thus, genetic ablation of Cdc42 leads to no severe deficiency in the osteoblastic lineage; however, the defects in expression patterns of osteoblast differential markers in Cdc42 mutants could be secondary to the delayed cartilage development.
No deficiency in bone formation by osteoblast-specific inactivation of Cdc42
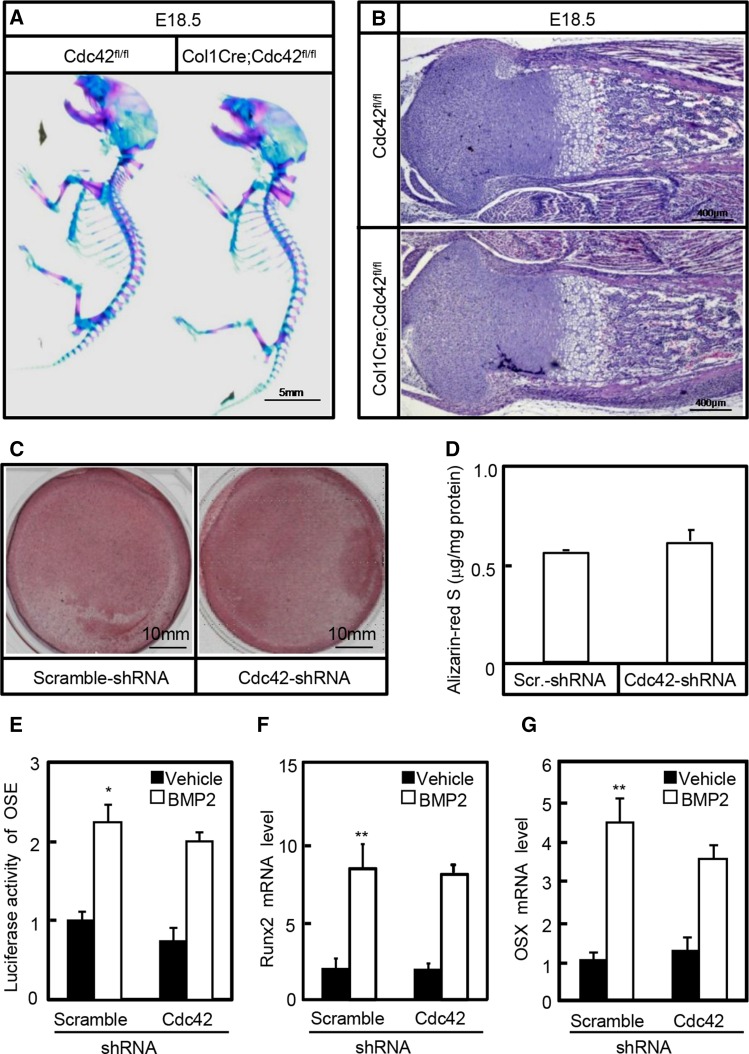
Although severe bone phenotypes have been found in Prx1Cre;Cdc42fl/fl mice, whether Cdc42 has direct effects on bone formation (ossification) remains unknown. To dissect the potential roles of Cdc42 in osteogenesis, we used a well-characterized transgene in which the Cre-recombinase gene is placed under the control of a 2.3-kb proximal fragment of alpha1 (I)-collagen (Col1α1) promoter expressed at high levels in osteoblasts throughout their differentiation. Transgenic mice were bred with ROSA26 reporter (R26R) and Cdc42fl/fl strains to monitor the Cre-recombinase expression and subsequent Cre-recombinase-mediated recombination and delete Cdc42 in osteoblasts, respectively. In R26R mice, LacZ staining was detected in virtually all osteoblasts but not in other cell types of Col1α1Cre;R26R mice (data not shown), indicating that homologous recombination occurred in these cells. Then, we performed whole-mount skeletal preparations of Col1Cre;Cdc42fl/fl and Cdc42fl/fl littermates at E18.5. The gross appearance of Col1Cre;Cdc42fl/fl mutants at E18.5 was as normal as the wild-type littermates, consistent with the gross appearance, H&E staining for the femora showed that Cdc42 mutants exhibited normally patterned cartilage and well-organized bone as compared to their control littermates (Figure 2, a and b). In addition, we examined the effects of Cdc42 on osteogenic differentiation in C3H10T1/2 cells able to differentiate into preosteoblasts in the presence of osteogenic medium. After incubation for 21 days, cells underwent robust mineralization in the presence of osteogenic medium containing 50 μM ascorbic acid, 100 nM dexamethasone and 10 mM β-glycerophosphate; however, knockdown of Cdc42 by Cdc42-shRNA-expressing lentiviruses did not significantly affect the mineralized nodule formation attested by Alizarin-Red S staining and quantitative determination (Figure 2, c and d). Finally, Cdc42 knockdown in C3H10T1/2 cells affected neither the BMP2-induced Runx2-dependent 6× OSE reporter activities nor the mRNA levels of osteogenic marker genes including Runx2 and Osx (Figure 2, e–g). Thus, Cdc42 is not directly involved in osteoblastic lineage differentiation.
Figure 2.
Cdc42 is not involved in osteogenesis. (a) Whole-mount skeletal staining of wild type and Col1Cre;Cdc42fl/fl littermates at E18.5. (b) H&E staining for sections of tibia at E18.5. (c and d) Alizarin-Red S staining and quantitative determination of mineralized nodules. After infection with scramble-shRNA- or Cdc42-shRNA-expressing lentiviruses, C3H10T1/2 cells were cultured in osteogenic differentiation medium for 21 days, and then cells were subjected to Alizarin-Red S staining and quantitative determination. (e) Cdc42 knockdown in C3H10T1/2 cells had no effect on BMP2-induced Runx2 reporter activities. After cotransfection with reporter constructs, C3H10T1/2 cells were infected with scramble-shRNA- (scramble-sh) or Cdc42-shRNA-expressing (Cdc42-sh) lentiviruses. Then, cells were incubated in the presence or absence of rhBMP2 at 10 ng/ml for 48 hr followed by dual-luciferase assays. (f and g) After infection with scramble-sh or Cdc42-sh lentiviruses, cells were incubated in the presence or absence of rhBMP2 at 10 ng/ml for 48 hr followed by quantitative RT-PCR assays for mRNA levels of Runx2 and OSX. *P < 0.05, **P < 0.01 vs. scramble-sh and vehicle treatments.
Involvement of Cdc42 in mesenchymal condensation
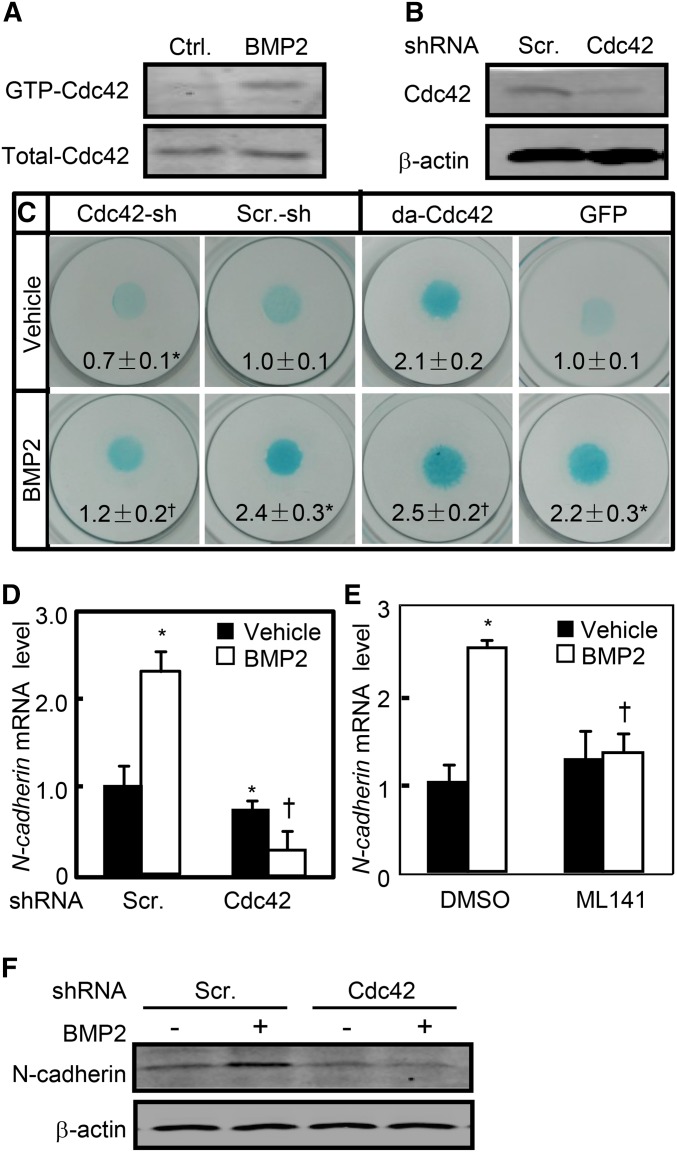
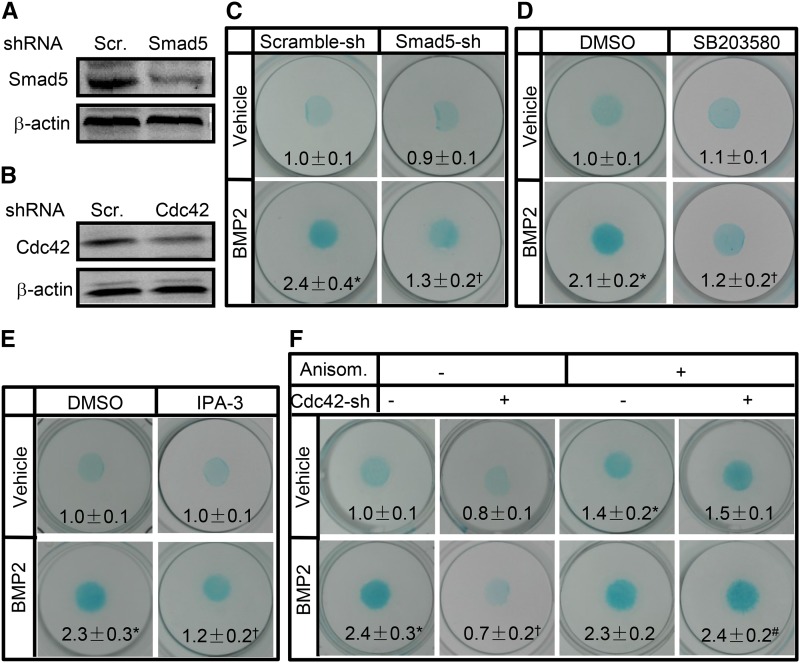
Mesenchymal progenitors in the limb bud initially undergo condensation preceding chondrocyte commitment and BMP signaling plays an essential role in this process (Pizette and Niswander 2000). To determine the potential signaling cascade governing Cdc42-mediated condensation in response to BMP, we performed micromass cultures by using C3H10T1/2 cells that had been shown to form precartilaginous nodules in response to BMP2 (Rosen et al. 1994). Recombinant human BMP2 (rhBMP2) at 10 ng/ml increased the active form of Cdc42 (GTP-Cdc42) up to 2.5-fold, whereas Cdc42-shRNA letiviruses reduced Cdc42 expression by 70% (Figure 3, a and b). In micromass cultures, BMP2 treatment robustly increased the nodule formation as assessed by Alcian Blue staining; however, knockdown of Cdc42 not only reduced the basal levels of nodule formation but also almost completely abolished BMP2-induced nodule formation (Figure 3c). Conversely, constitutive activation of Cdc42 by retroviruses expressing the dominant-active form of Cdc42 (da-Cdc42) enhanced not only the basal but also BMP2-induced nodule formation as compared with the control GFP-expressing (GFP) retroviruses (Figure 3c). In addition, BMP2 treatment induced the mRNA and protein expression of N-cadherin by 1.5- and 1.3-fold, respectively, whereas knockdown of Cdc42 reduced not only the basal levels of N-cadherin mRNA and protein by 30 and 20%, respectively, but also BMP2-induced N-cadherin mRNA and protein levels by 90 and 70%, respectively (Figure 3, d and f). Similarly, inhibition of Cdc42 activity by its specific inhibitor, ML141, at 40 μM almost completely abolished BMP2-induced N-cadherin mRNA expression (Figure 3e). Thus, Cdc42 is required for mesenchymal condensation in response to BMP2.
Figure 3.
Cdc42 is required for mesenchymal condensation. (a) rhBMP2 at 10 ng/ml increased the active form of Cdc42. Cdc42 activation assays in C3H10T1/2 cells treated with or without rhBMP2 for 1 hr. (b) The efficiency of Cdc42 knockdown in C3H10T1/2 cells infected with scramble-sh or Cdc42-sh lentiviruses for 48 hr. (c) Cdc42 affected the nodule formation of C3H10T1/2 cells. After C3H10T1/2 cells were infected with Cdc42-shRNA- or scramble-shRNA-expressing lentiviruses, alternatively, after cells were infected withGFP- or da-Cdc42-expressing retroviruses, cells were subjected to micromass cultures in the presence or absence of rhBMP2 at 10 ng/ml for 7 days followed by Alcian Blue staining. (d and e) Knockdown of Cdc42 reduced the BMP2-induced N-cadherin mRNA expression. C3H10T1/2 cells infected with scramble-sh-RNA- or Cdc42-shRNA-expressing lentiviruses or pretreated with vehicle or ML141 were incubated in the presence or absence of rhBMP2 for 48 hr followed by quantitative RT-PCR assays for N-cadherin mRNA levels. (f) Knockdown of Cdc42 reduced the BMP2-induced N-cadherin protein expression. C3H10T1/2 cells infected with lentiviruses were incubated in the presence or absence of rhBMP2 for 48 hr followed by WB. *P < 0.05 vs. scramble-sh and vehicle treatments. †P < 0.05 vs. scramble-sh and BMP2 treatments.
Involvement of Cdc42 in BMP2/p38/Smad signaling cascade
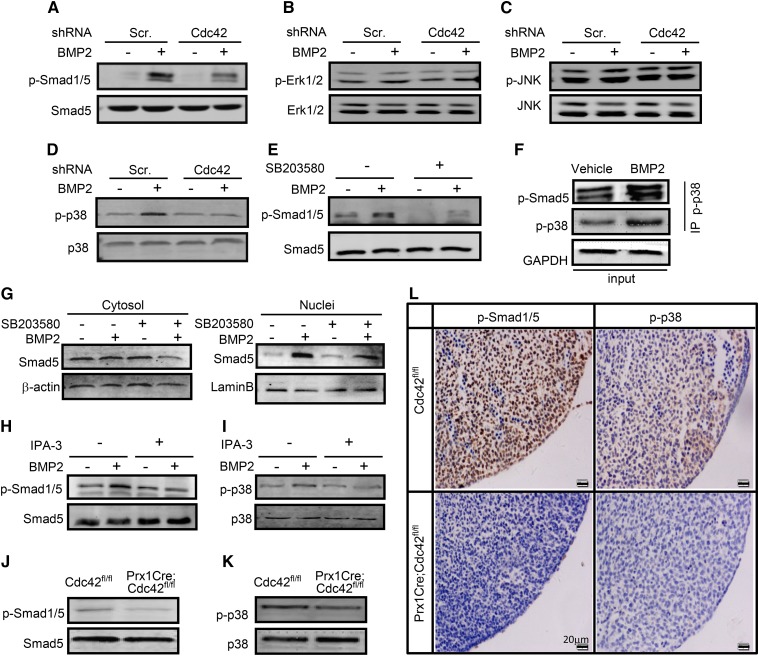
Canonical BMP-Smad as well as noncanonical BMP-mitogen-activated protein kinase (MAPK) signaling pathways play critical roles in mesenchymal condensation and chondrocyte differentiation as well (Denker et al. 1999; Schmitt et al. 2003). To explore the potential mechanism governing Cdc42-mediated mesenchymal condensation, we examined the roles of Cdc42 in BMP2/Smad signaling in C3H10T1/2 cells. BMP2 at 10 ng/ml increased the phosphorylated Smad1/5 (p-Smad1/5) levels by 5.0-fold, whereas knockdown of Cdc42 reduced the basal and BMP2-induced p-Sma1/5 levels by 20 and 50%, respectively (Figure 4a). Expectedly, BMP2 induced the phosphorylation of p38 (p-p38) by 2.0-fold but had no significant effects on phosphorylation of both extracellular regulated kinase (ERK1/2) and c-Jun N-terminal kinase (JNK), and knockdown of Cdc42 almost completely abolished BMP2-induced increases in phosphorylation of p38, whereas it had no effect on phosphorylation of both ERK and JNK (Figure 4, b–d). Likewise, inhibition of p38 activity by its specific inhibitor, SB203580, at 20 μM reduced the basal and BMP2-induced p-Sma1/5 by 40 and 74%, respectively (Figure 4e). Furthermore, protein complexes precipitated with a p-p38 antibody contained an abundance of p-Smad5 as well as p-p38 as expected, even though the protein complex from BMP2-treated cells contained more p-p38 and p-smad5 than that from vehicle-treated cells (Figure 4f), suggesting that physical interactions exist between endogenous active p38 and Smad1/5. Then, the subcellular localization of Smad5 was examined in C3H10T1/2 cells followed by treatment with SB203580. In the absence of BMP2, Smad5 was predominantly distributed in the cytoplasmic fractions, whereas in the presence of BMP2, Smad5 was predominantly localized in the nuclear fractions; inhibition of p38 by SB203580 at 20 μM reduced the cytoplasmic and nuclear fractions of Smad5 by 33 and 40%, respectively (Figure 4g). Among the downstream effectors of Cdc42, P21-activated kinases (Pak) mediate a variety of Cdc42-controlling cell functions (Morreale et al. 2000; Roig et al. 2000; Royal et al. 2000). To determine the roles of Pak in Cdc42-mediated cell signaling, we treated C3H10T1/2 cells with Pak inhibitor, IPA-3. IPA-3 at 10 μM negated BMP-induced increases of p-Smad1/5 and p-p38 by 65 and 73%, respectively (Figure 4, h and i). Thus, these data suggest that Cdc42 lies on the upstream of Pak and p38 to activate Smad1/5 in response to BMP2.
Figure 4.
Cdc42 is involved in BMP2/Pak/p38/Smad signaling. (a–d) Cdc42 regulated BMP2 signaling in C3H10T1/2 cells. C3H10T1/2 cells were infected with scramble-shRNA- or Cdc42-shRNA-expressing lentiviruses for 48 hr and then were stimulated with or without rhBMP2 at 10 ng/ml for an additional 30 min. p-Smad1/5, total Smad1/5, p-JNK, total JNK, p-Erk, total Erk, p-p38, and total p38 were measured by Western blots. p-Smad1/5, p-Erk1/2, p-p38, and p-JNK were normalized to total Smad1/5, Erk1/2, p38, and JNK, respectively. (e) SB203580 inhibited the activation of Smad1/5. C3H10T1/2 cells were pretreated with vehicle (−) or SB203580 (+) at 20 μM for 3 hr and then were stimulated with rhBMP2 at 10 ng/ml for an additional 30 min. (f) Coimmunoprecipitation of endogenous p-Smad5 and p-p38 in cells treated with or without BMP2. After rhBMP2 treatments for 30 min, C3H10T1/2 cells were subjected to immunoprecipitation and Western blot analyses by using the indicated antibodies. (g) SB203580 reduced Smad5 protein levels. Western blot analyses of Smad5 levels in cytosolic and nuclear fractions of C3H10T1/2 cells treated with vehicle (−) or SB203580 (+) at 20 μM in the presence or absence of rhBMP2 for 12 hr. (h and i) IPA-3 inhibited the activation of Smad1/5 and p38. C3H10T1/2 cells were pretreated with vehicle (−) or IPA-3 (+) at 10 μM for 3 hr and then were stimulated with rhBMP2 for an additional 30 min. (j and k) Cdc42fl/fl and PrxCre;Cdc42fl/fl limb buds at E11.5 were harvested for Western blot analyses of p-Smad1/5 and p-p38. (l) The abovementioned limb buds were fixed in 10% formalin overnight and then processed and embedded in paraffin prior to sectioning at 4 μm, followed by immunohistochemistry analyses of p-Smad1/5 and p-p38.
To further confirm the signaling cascade involved in Cdc42-mediated Smad1/5 activation, we examined the expression of p-Smad1/5 and p-p38 in Cdc42 mutant limbs at E11.5 by immunohistochemistry and Western blots. The expression of p-Smad1/5 and p-p38 in Cdc42 mutant limbs was reduced by 60 and 50%, respectively, as compared with that in wild-type limbs (Figure 4, j and k). Moreover, p-Smad1/5 was robustly and evenly expressed in ZPA and epithelial surroundings, whereas p-p38 was predominantly expressed in ZPA but not epithelial surroundings of limb bud at E11.5. Genetic ablation of Cdc42 resulted in considerable decreases in the expression of both p-Smad1/5 and p-p38 in ZPA (Figure 4l). Taken together, BMP2 signals through Cdc42/Pak/p38 to activate Smad1/5.
Roles of Cdc42/Pak/p38/Smads signaling in mesenchymal condensation
To confirm the roles of p38 and Smad5 in mesenchymal condensation, we performed micromass cultures of C3H10T1/2 cells followed by various treatments. Smad5-shRNA expressing lentiviruses (Smad5-sh) knocked down the expression of Smad5 by 50% as compared with the scramble (Scr)-shRNA expressing lentiviruses (Figure 5a). In the absence of BMP2, Smad5-sh had no significant effect on the nodule formation, however, in the presence of BMP2, Smad5-sh reduced nodule formation by 50% (Figure 5c). Likewise, inhibition of p38 or Pak1 activity by their specific inhibitors, SB203580 and IPA-3 attenuated BMP2-induced nodule formation by 43 and 48%, respectively (Figure 5, d and e). Furthermore, Cdc42-shRNA expressing lentiviruses (Cdc42-sh) knocked down the expression of Cdc42 by 50% over Scr-shRNA expressing lentiviruses (Figure 5b). Though knockdown of Cdc42 attenuated BMP2-induced nodule formation by 71%, in the presence of anisomycin (anisom), an activator of p-38 and JNK, the inhibitory effect of Cdc42-shRNA on nodule formation in response to BMP2 was almost completely diminished (Figure 5f). Thus, Pak/Cdc42/p38/Smad5 signaling module is involved in mesenchymal condensation in response to BMP2.
Figure 5.
p38 and Smad activation were required for condensation. (a and b) The efficiency of Smad5 and Cdc42 knockdown in C3H10T1/2 cells infected with lentiviruses for 48 hr. (b) Knockdown of Smad5 blocked the condensation of C3H10T1/2 cells. C3H10T1/2 cells infected with scramble-shRNA- or Smad5-shRNA-expressing lentiviruses were placed in micromass culture in the presence or absence of rhBMP2 at 10 ng/ml for 7 days and stained with Alcian Blue. (d and e) SB203580 at 20 μM and IPA-3 at 10 μM inhibited nodule formation. Micromass cultures of C3H10T1/2 cells treated with DMSO, SB203580, or IPA-3 were stimulated with rhBMP2 for 7 days and stained with Alcian Blue. (f) Anisomycin at 5 μM attenuated Cdc42-shRNA-reduced nodule formation. Micromass cultures of C3H10T1/2 cells infected with scramble-shRNA- or Cdc42-shRNA-expressing lentiviruses were treated with vehicle (−) or anisomycin (+) in the presence or absence of rhBMP2 at 10 ng/ml for 7 days and stained with Alcian Blue. *P < 0.05 vs. scramble-sh and vehicle treatments. †P < 0.05 vs. scramble-sh and BMP2 treatments.
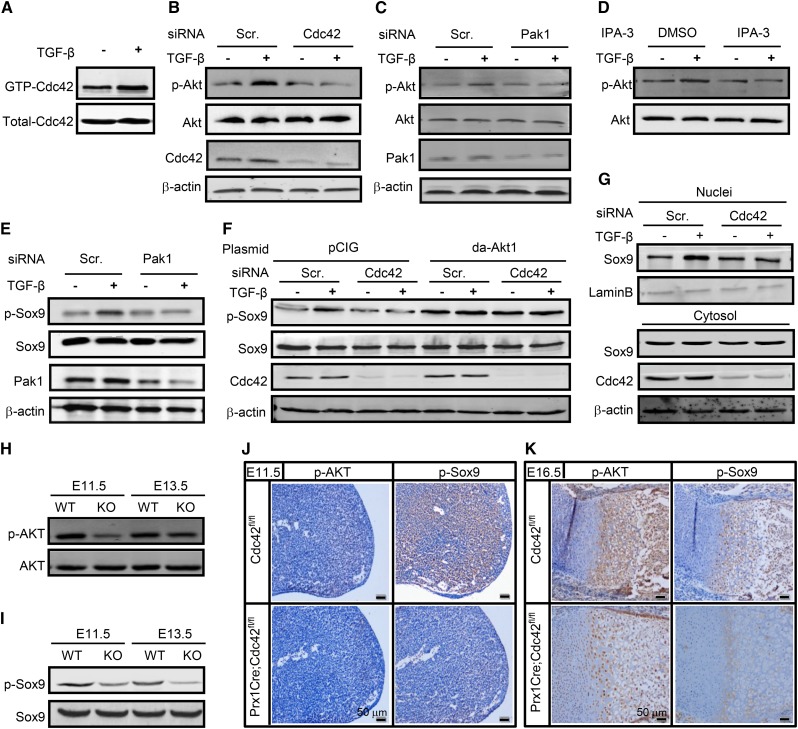
Involvement of Cdc42 in TGF-β-induced Sox9 activation
Chondrocyte-specific inactivation of Cdc42 (Col2Cre;Cdc42fl/fl) has demonstrated that Cdc42 is essential for cartilage development during endochondral ossification (Suzuki et al. 2015). To explore the underlying mechanism governing Cdc42-mediated chondrogenic differentiation, we stimulated C3H10T1/2 cells with transforming growth factor-β (TGF-β), a critical regulator in the process of chondrocyte differentiation and hypertrophy (Zhang et al. 2004). TGF-β at 10 ng/ml increased the active form of Cdc42 (GTP-Cdc42) by 1.8-fold and induced the phosphorylation of Akt (p-Akt) by 2.0-fold, whereas knockdown of Cdc42 almost completely diminished TGF-β-induced activation of Akt (Figure 6, a and b). Moreover, either inhibition of Pak by IPA-3 at 10 μM or knockdown of Pak1 by its specific siRNA almost completely abolished TGF-β-induced activation of Akt (Figure 6, c and d). Sox9 is a critical transcription factor responsible for the chondroprogenitor differentiation except for hypertrophy, and the DNA binding activity and cellular localization of Sox9 are regulated by post-translational modifications including phosphorylation at Ser181 (Huang et al. 2000; Song et al. 2013). To evaluate the potential effects of Cdc42 on Sox9 activation, we performed Western blot assays. TGF-β at 10 ng/ml increased the phosphorylation of Sox9 at Ser181 (p-Sox9) by ∼1.5-fold, whereas knockdown of Pak1 reduced TGF-β-induced p-Sox9 by 61% and knockdown of Cdc42 reduced TGF-β-induced p-Sox9 by 68% (Figure 6, e and f). Constitutive activation of Akt1 by the dominant-active form of Akt1 (da-Akt1) increased the p-Sox9 activities by ∼2.0-fold in both the absence and presence of TGF-β, whereas da-Akt1 almost completely reversed Cdc42-shRNA-negated Sox9 phosphorylation (Figure 6f), suggesting that Akt1 lies on the downstream of Cdc42 to activate Sox9 in response to TGF-β. Then, the subcellular localization of Sox9 was examined in C3H10T1/2 cells treated withTGF-β at 10 ng/ml. TGF-β led to the increases in nuclear Sox9 levels by 1.9-fold and cytoplasmic Sox9 levels by 20%, respectively (Figure 6g). Knockdown of Cdc42 decreased nuclear fraction of Sox9 by 52% in the presence of TGF-β, and had no significant effect on the levels of cytoplasmic Sox9 in both the presence and absence of TGF-β (Figure 6g). Finally, to confirm the activation of AKT and Sox9 by Cdc42 in vivo, we performed Western blots and immunohistochemistry staining by using limbs from Prx1Cre;Cdc42fl/fl and Cdc42fl/fl embryos at E11.5, E13.5, and E16.5. Knockout (KO) of Cdc42 led to decreases in p-AKT and p-Sox9 by 60 and 50%, respectively, at E11.5, and 40 and 60%, respectively, at E13.5 (Figure 6, h and i). Moreover, genetic ablation of Cdc42 resulted in considerable decreases in the expression of both p-AKT and p-Sox9 in limb mesenchymal progenitors at E11.5 (Figure 6j). In Cdc42fl/fl long bone, p-AKT was predominantly expressed in prehypertrophic and hypertrophic chondrocytes as well as in trabecular bone areas at E16.5, whereas deletion of Cdc42 resulted in marked attenuation of p-AKT in prehypertrophic and hypertrophic chondrocytes and trabecular bone areas but slight increases in proliferating chondrocytes (Figure 6k). In Cdc42fl/fl long bone, p-Sox9 was predominantly expressed in prehypertrophic and hypertrophic chondrocytes as well as in trabecular bone areas at E16.5, whereas deletion of Cdc42 led to a significant diminishment of p-Sox9 in above-mentioned areas (Figure 6k). Thus, TGF-β/Cdc42 signals through Pak/Akt to activate Sox9.
Figure 6.
Cdc42 is involved in TGF-β/Pak1/Akt1/Sox9 signaling. (a) TGF-β increased the active form of Cdc42. Cdc42 activation assays in C3H10T1/2 cells treated with or without TGF-β at 10 ng/ml for 1 hr. (b) Knockdown of Cdc42 inhibited the activation of Akt. C3H10T1/2 cells were transfected with Cdc42 siRNAfor 48 hr and then were stimulated with or without TGF-β at 10 ng/ml for an additional hour. p-Akt at S473 and total Akt were measured by Western blot. (c and d) Pak1 affected the activation of Akt. C3H10T1/2 cells pretreated with IPA-3 at 10 μM or transfected with Pak1 siRNA were stimulated with or without TGF-β for 1 hr, followed by Western blot analyses. (e) Pak1 affected the activation of Sox9. (f) Cdc42 and Akt affected the activation of Sox9. C3H10T1/2 cells were transfected with Cdc42 siRNA or da-Akt1 plasmids for 48 hr and then were stimulated with or without TGF-β for an additional hour, followed by Western blot analyses of p-Sox9. (g) Knockdown of Cdc42 reduced Sox9 protein levels. Western blot analyses of cytosolic and nuclear fractions of Sox9 in cells tranfected with Cdc42 siRNA in the presence or absence of TGF-β for 24 hr. (h and i) Cdc42fl/fl and PrxCre;Cdc42fl/fl limb buds at E11.5 or E13.5 were harvested for Western blot analyses of p-AKT and p-Sox9. (j and k) The abovementioned limb buds were fixed in 10% formalin and embedded in paraffin prior to sectioning at 4 μm, followed by immunohistochemistry analyses of p-AKT and p-Sox9 (1:100).
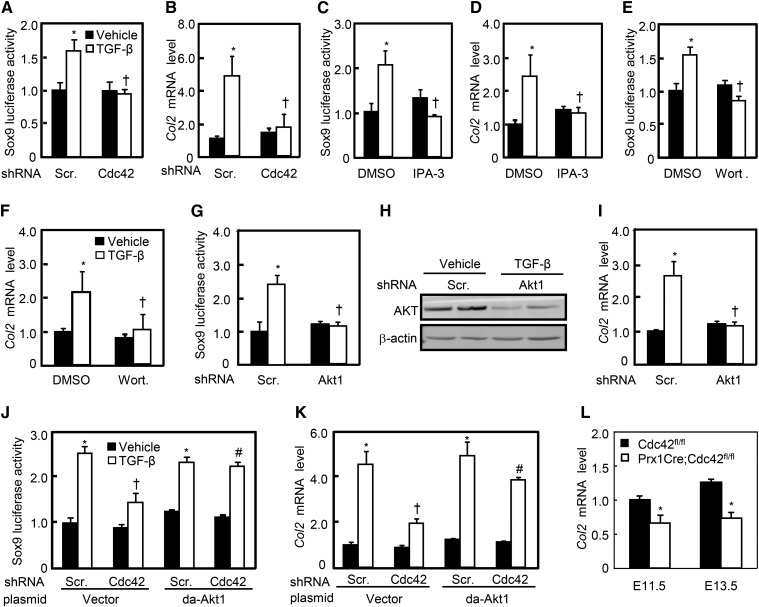
Cdc42/Pak/Akt signaling cascade in TGF-β-induced chondrogenic differentiation
To determine the roles of Cdc42/Pak/Akt/Sox9 signaling module in TGF-β-induced chondrocyte differentiation, we performed the Sox9-luciferase assays and quantitative RT-PCR in C3H10T1/2 cells. TGF-β significantly induced Sox9-luciferase activities as well as the mRNA levels of collagen type 2α1 (Col2), a chondrocyte marker, whereas knockdown of Cdc42 almost completely abolished the effects of TGF-beta induced (Figure 7, a and b). Likewise, inhibition of either Pak by IPA-3 at 10 μM or Akt by wortmannin at 10 μM almost completely diminished TGF-β-induced Sox9-luciferase activities as well as Col2 mRNA expression (Figure 7, c–f). Moreover, Akt1-shRNA expressing lentiviruses reduced the Akt1 expression by ∼50–60% in either the presence or absence of TGF-β, and completely abolished TGF-β-induced Sox9-luciferase activity as well as Col2 mRNA expression (Figure 7, g–i). Finally, knockdown of Cdc42 robustly attenuated TGF-β-induced Sox9 luciferase activities and Col2 mRNA expression as well, and overexpression of da-Akt1 was as potent as TGF-β treatment in induction of Sox9-luciferase activity and Col2 mRNA expression (Figure 7, j and k). However, da-Akt1 almost completely reversed Cdc42-shRNA-negated Sox9-luciferase activity and Col2 mRNA expression in response to TGF-β (Figure 7, j and k). In addition, knockout of Cdc42 in mesenchymal progenitors resulted in decreases in Col2 mRNA levels by 30 and 46% at E11.5 and E13.5, respectively (Figure 7l). Thus, TGF-β through Cdc42/Pak/Akt/Sox9 signaling cascade induces chondrogenesis.
Figure 7.
Cdc42 is involved in TGF-β-induced Sox9 transactivities and Col2 mRNA expression. (a, c, e, and g) Cdc42, Pak, and Akt affected the Sox9 luciferase activity. After cotransfection with the Sox9 and Renilla luciferase plasmids, C3H10T1/2 cells were infected with Cdc42-sh, Akt1-sh lentiviruses, or treated with indicated concentrations of IPA-3 or wortmannin for 24 hr in the presence or absence of TGF-β. (b, d, f, and i) Cdc42, Pak, and Akt affected the mRNA levels of Col2. C3H10T1/2 cells infected with Cdc42-sh, Akt1-sh lentiviruses, or pretreated with 10 μM IPA-3 or wortmannin were stimulated with or without TGF-β for an additional 48 hr. (h) The efficiency of AKT knockdown in C3H10T1/2 cells infected with scramble-sh or Akt1-sh lentiviruses for 48 hr. (j and k) da-Akt1 reversed the effects of Cdc42-sh on Sox9-luciferase activity and Col2 mRNA expression. C3H10T1/2 cells infected with Cdc42-sh lentiviruses and/or transfected with da-Akt1 plasmids were stimulated with or without TGF-β for 48 hr. (l) Cdc42fl/fl and PrxCre;Cdc42fl/fl limb buds at E11.5 and E13.5 were harvested for RT-PCR analyses of Col2 mRNA expression. *P < 0.05 vs. scramble-sh or DMSO and vehicle treatments; †P < 0.05 vs. scramble-sh or DMSO and TGF-β treatments; #P < 0.05 vs. cells infected with Cdc42-sh and transfected with empty vector.
Discussion
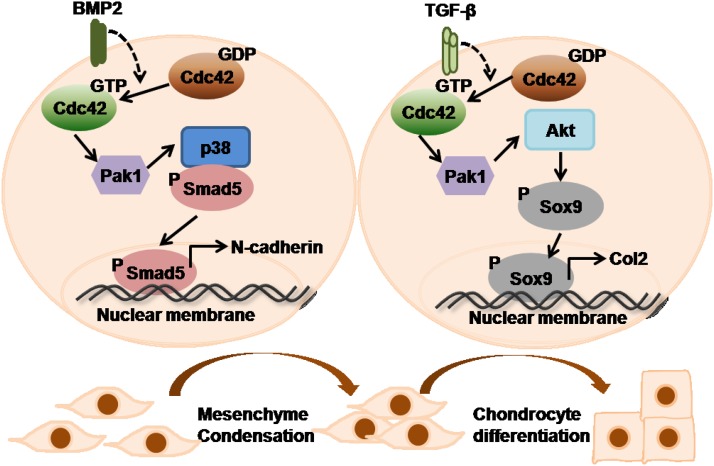
By using biochemical and genetic approaches, we have demonstrated that Cdc42 is involved not in osteogenesis but chondrogenesis in which the BMP2/Cdc42/Pak/p38/Smad signaling cascade promotes mesenchymal condensation and the TGF-β/Cdc42/Pak/Akt/Sox9 signaling cascade facilitates chondrogenic differentiation (Figure 8). In combination with previous studies that have demonstrated the essential roles of Cdc42 in terminal hypertrophy and apoptosis of chondrocytes (Wang and Beier 2005; Aizawa et al. 2012), eventually, we suggest that Cdc42 is required in successive steps of the chondrocyte differentiation pathway.
Figure 8.
A model for the role of Cdc42 in endochondral ossification. In mesenchymal condensation, Cdc42 lies on the upstream of Pak and p38 to activate Smad1/5 in response to BMP2. In chondrocyte differentiation, Cdc42 lies on the upstream of Pak and Akt to activate Sox9 in response to TGF-β.
Though severe bone phenotypes, including disappearance of trabecular bone and diminished primary ossification center, have been observed in the Cdc42 mutant long bone, mesenchymal progenitor-specific ablation of Cdc42 leads to no significant changes in expression of osteoblastic differential markers in Prx1Cre;Cdc42fl/fl mutants; preosteoblast-specific ablation of Cdc42 results in no obvious bone phenotypes in Col1Cre;Cdc42fl/fl mutants; and knockdown of Cdc42 affects neither the mineralization nor the expression of osteoblast differential markers in vitro. Such findings prompt us to speculate that ossification deficiency in Cdc42 mutants could be the consequence of improper cartilage degradation caused by overexpansion and nonresorption of terminal hypertrophic chondrocytes.This notion has been supported by previous studies showing that accumulation of hypertrophic chondrocytes may contribute to delayed terminal maturation of chondrocytes and apoptosis in the chondro–osseous junction in Prx1Cre;Cdc42fl/fl and Col2Cre;Cdc42fl/fl long bones (Aizawa et al. 2012; Suzuki et al. 2015). Though a positive correlation between the decreased number of osteoclasts adjacent to chondrocytes and delayed mineralization formation has been found in Col2Cre;Cdc42fl/fl long bones and Cdc42 indeed regulates osteoclast formation and function (Ito et al. 2010; Suzuki et al. 2015), whether osteoclasts are directly or indirectly involved in the malformation of cartilage and bone in Cdc42 mutants needs to be further studied.
In BMP signaling pathways, p38 is generally recognized to be involved in noncanonical BMP signaling but not canonical BMP/Smad signaling (Guicheux et al. 2003; Jin et al. 2006; Ulsamer et al. 2008). However, in the current study, BMP2 activates Smad1/5 through the Cdc42/Pak/p38 signaling module, and inhibition of p38 attenuates BMP2-induced C-terminal phosphorylation of Smad1/5 that is functionally redundant with Smad4 (Retting et al. 2009). Moreover, the interaction between p-p38 and p-samd1/5 in immunocomplex is potentiated by BMP2 stimulation, though it is still unknown whether p-p38 and p-smad1/5 interacts directly or indirectly. These findings support the idea that Cdc42/Pak lies on the upstream of p38 to activate Smad1/5 in regulating the mesenchymal condensation. Consistent with our findings showing the critical roles of BMP2/Cdc42/p38 signaling in mesenchymal condensation, a previous study in osteoblastic cells demonstrated that p38 was activated in response to BMP2 treatment and that inhibition of p38 activity suppressed BMP2-induced Smad1 phosphorylation as well as its translocation to the nucleus (Noth et al. 2003). Similarly, another previous study indicated that overexpression of Sma7 in mesenchymal progenitors (Prx1Cre) led to disturbed mesenchymal condensation associated with decreased Sox9 expression and poor cartilage formation by down-regulating the BMP-activated p38 pathway (Iwai et al. 2008). TGF-β signals through Smad2/3 to interact with Sox9 and induces Sox9 transactivity; alternatively, TGF-β recruits the transcriptional coactivators CBP/p300 to increase Sox9 transactivity (Furumatsu et al. 2005; Kozhemyakina et al. 2015). Regardless of the little known facts about the post-translational regulation of Sox9, phosphorylation of Sox9 by protein kinase A (PKA) at Ser181 is associated with increase in transactivity (Huang et al. 2001; Malki et al. 2005). In the present study, we have demonstrated that Cdc42 through Pak activates Akt-mediated Sox9 transcription in chondrogenic differentiation in response to TGF-β. Though we have not shown evidence that Akt directly phosphorylates Sox9 at Ser181, it is plausible to suggest that other players lie on the downstream of Akt to phosphorylate Sox9 at Ser181 and subsequently induce chondrogenic differentiation. However, Ikegami et al. (2011) have indicated that Sox9 binds to the promoter and enhances the transcription of PI3Kca (P110α), resulting in an increase of phosphorylated Akt, which promotes chondrocyte survival and hypertrophy. Cheng et al. (2009) have demonstrated that PI3K/Akt signaling controls aggrecan gene expression, in part by modulating Sox9 expression and transactivity in nucleus pulposus cells. The discrepancy in our findings could be explained by the different cell types and treatments among these studies. Consistent with our findings indicating that Sox9 is involved in TGF-β/Cdc42-mediated chondrogenesis, transgenic mice misexpressing Sox9 in hypertrophic chondrocytes reveal the features of growth plates that are similar to those of Prx1Cre;Cdc42fl/fl mice (Hattori et al. 2010). In addition, a study of TGF-β type II receptor (Tgfbr2) knockout mice (Prx1Cre;Tgfbr2fl/f) also reveals that Col2-expressing cells exhibited a less-organized columnar distribution than the controls, whereas hypertrophic chondrocytes are larger but decreased in expression of Col10 characterized in both Prx1Cre;Cdc42fl/fl and Col2Cre;Cdc42fl/fl long bones (Spagnoli et al. 2007; Aizawa et al. 2012; Suzuki et al. 2015).
Mutations in human FYVE, RhoGEF, and PH domain-containing 1 (FGD1) cause faciogenital dysplasia (FGDY) (also known as Aarskog syndrome, AAS), an X-linked disorder that affects multiple skeletal structures (Aarskog 1970), and FGD1 is the only known causative gene of AAS (Pasteris et al. 1994). FGD1 encodes a guanine nucleotide exchange factor (GEF) that specifically activates the Cdc42 (Whitehead et al. 1998; Egorov et al. 2009). Cdc42 is predicted to play important roles in skeletal development. Consistent with our findings showing that Cdc42 is involved in chondrogenesis but not in osteogenesis, a previous study has demonstrated that MAP3K mixed-lineage kinase 3 (MLK3) functions as downstream of FGD1/Cdc42 to regulate ERK and p38, which in turn phosphorylate and activate Runx2 in mineralization, and that loss-of-function mutation in MLK3 displays multiple defects in bone formation (Zou et al. 2011). Another previous study has suggested that defects in bone remodeling in FGDY show an important role for FGD1/Cdc42 signaling in osteogenesis (Gao et al. 2011). The inconsistency could be explained by the multifaceted cellular functions of FGD1 and MLK3. In summary, our current study and the body of work in the literature have substantiated the critical roles of Cdc42 in cartilage development; therefore, the present study elucidating that the role of Cdc42 signaling in cartilage development is fundamental for a mechanistic understanding of the AAS and for potential future therapeutic interventions.
Acknowledgments
We thank Yi Zheng (Department of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center) for Cdc42fl/fl mice, Andrew C. Karaplis (Department of Medicine, McGill University) for Col1Cre mice, and Cliff Tabin (Department of Genetics, Harvard Medical School) for Prx1Cre mice. This work was supported by the National Natural Science Foundation of China (31571493, 31271561, 31071292, and 81171748), the 973 Program (2011CB944403), and the Natural Science Foundation of Zhejiang Province, China (R2110269). The authors declare that they have no conflict of interest.
Footnotes
Communicating editor: T. Magnuson
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180109/-/DC1.
Literature Cited
- Aarskog D., 1970. A familial syndrome of short stature associated with facial dysplasia and genital anomalies. J. Pediatr. 77: 856–861. [DOI] [PubMed] [Google Scholar]
- Aizawa R., Yamada A., Suzuki D., Iimura T., Kassai H., et al. , 2008. Cdc42 is required for chondrogenesis and interdigital programmed cell death during limb development. Mech. Dev. 129: 38–50. [DOI] [PubMed] [Google Scholar]
- Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B., 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16: 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W. M., Deng J. M., Zhang Z. P., Behringer R. R., de Crombrugghe B., 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85–89. [DOI] [PubMed] [Google Scholar]
- Bishop A. L., Hall A., 2000. Rho GTPases and their effector proteins. Biochem. J. 348(Pt 2): 241–255. [PMC free article] [PubMed] [Google Scholar]
- Bobick B. E., Chen F. H., Le A. M., Tuan R. S., 2009. Regulation of the chondrogenic phenotype in culture. Birth Defects Res. C Embryo Today 87: 351–371. [DOI] [PubMed] [Google Scholar]
- Cheng C. C., Uchiyama Y., Hiyama A., Gajghate S., Shapiro I. M., et al. , 2009. PI3K/AKT regulates aggrecan gene expression by modulating Sox9 expression and activity in nucleus pulposus cells of the intervertebral disc. J. Cell. Physiol. 221: 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T. F., Guo X., Garrett-Beal L., Yang Y., 2005. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 8: 739–750. [DOI] [PubMed] [Google Scholar]
- Delise A. M., Tuan R. S., 2002. Analysis of N-cadherin function in limb mesenchymal chondrogenesis in vitro. Dev. Dyn. 225: 195–204. [DOI] [PubMed] [Google Scholar]
- Denker A. E., Haas A. R., Nicoll S. B., Tuan R. S., 1999. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation 64: 67–76. [DOI] [PubMed] [Google Scholar]
- Egorov M. V., Capestrano M., Vorontsova O. A., Di Pentima A., Egorova A. V., et al. , 2009. Faciogenital dysplasia protein (FGD1) regulates export of cargo proteins from the golgi complex via Cdc42 activation. Mol. Biol. Cell 20: 2413–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exton J. H., 1998. Small GTPases minireview series. J. Biol. Chem. 273: 19923. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Dominguezsteglich M. A., Guioli S., Kwok C., Weller P. A., et al. , 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an Sry-related gene. Nature 372: 525–530. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain, C., X. Warot, N. Messadecq, M. LeMeur, P. Dolle et al, 1996. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122: 2997–3011. [DOI] [PubMed] [Google Scholar]
- Furumatsu T., Tsuda M., Yoshida K., Taniguchi N., Ito T., et al. , 2005. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J. Biol. Chem. 280: 35203–35208. [DOI] [PubMed] [Google Scholar]
- Gao L., Gorski J. L., Chen C. S., 2011. The Cdc42 guanine nucleotide exchange factor FGD1 regulates osteogenesis in human mesenchymal stem cells. Am. J. Pathol. 178: 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y., C. Y. Xu, J. R. Wang, X. H. Hu, D. Hong et al., 2014 Inhibition of phosphodiesterase 5 reduces bone mass by suppression of canonical Wnt signaling. Cell Death Dis. 5: e1544. [DOI] [PMC free article] [PubMed]
- Guicheux J., Lemonnier J., Ghayor C., Suzuki A., Palmer G., et al. , 2003. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J. Bone Miner. Res. 18: 2060–2068. [DOI] [PubMed] [Google Scholar]
- Hattori T., Muller C., Gebhard S., Bauer E., Pausch F., et al. , 2010. SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification. Development 137: 901–911. [DOI] [PubMed] [Google Scholar]
- Huang W., Chung U. I., Kronenberg H. M., de Crombrugghe B., 2001. The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc. Natl. Acad. Sci. USA 98: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhou X., Lefebvre V., de Crombrugghe B., 2000. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol. Cell. Biol. 20: 4149–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami D., Akiyama H., Suzuki A., Nakamura T., Nakano T., et al. , 2011. Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138: 1507–1519. [DOI] [PubMed] [Google Scholar]
- Ito Y., Teitelbaum S. L., Zou W., Zheng Y., Johnson J. F., et al. , 2010. Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J. Clin. Invest. 120: 1981–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai T., Murai J., Yoshikawa H., Tsumaki N., 2008. Smad7 Inhibits chondrocyte differentiation at multiple steps during endochondral bone formation and down-regulates p38 MAPK pathways. J. Biol. Chem. 283: 27154–27164. [DOI] [PubMed] [Google Scholar]
- Jin E. J., Lee S. Y., Choi Y. A., Jung J. C., Bang O. S., et al. , 2006. BMP-2-enhanced chondrogenesis involves p38 MAPK-mediated down-regulation of Wnt-7a pathway. Mol. Cells 22: 353–359. [PubMed] [Google Scholar]
- Kozhemyakina E., Lassar A. B., Zelzer E., 2015. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 142: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H. M., 2003. Developmental regulation of the growth plate. Nature 423: 332–336. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Zhou G., Mukhopadhyay K., Smith C. N., Zhang Z., et al. , 1996. An 18-base-pair sequence in the mouse proalpha1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell. Biol. 16: 4512–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., et al. , 2002. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33: 77–80. [DOI] [PubMed] [Google Scholar]
- Long F., Ornitz D. M., 2013. Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5: a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P. F., Yu Y., Perdue Y., Werb Z., 2008. The apical ectodermal ridge is a timer for generating distal limb progenitors. Development 135: 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S., Nef S., Notarnicola C., Thevenet L., Gasca S., et al. , 2005. Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phosphorylation. EMBO J. 24: 1798–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao D., He B., Jiang Y., Kobayashi T., Soroceanu M. A., et al. , 2005. Osteoblast-derived PTHrP is a potent endogenous bone anabolic agent that modifies the therapeutic efficacy of administered PTH 1–34. J. Clin. Invest. 115: 2402–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michigami T., 2013. Regulatory mechanisms for the development of growth plate cartilage. Cell. Mol. Life Sci. 70: 4213–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., et al. , 2000. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 7: 384–388. [DOI] [PubMed] [Google Scholar]
- Noth U., Tuli R., Seghatoleslami R., Howard M., Shah A., et al. , 2003. Activation of p38 and Smads mediates BMP-2 effects on human trabecular bone-derived osteoblasts. Exp. Cell Res. 291: 201–211. [DOI] [PubMed] [Google Scholar]
- Ory D. S., Neugeboren B. A., Mulligan R. C., 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93: 11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. B., Y. Gong, H. F. Ruan, L. Y. Pan, X. K. Wu et al., 2015 Sonic hedgehog through Gli2 and Gli3 is required for the proper development of placental labyrinth. Cell Death Dis. 6: e1653. [DOI] [PMC free article] [PubMed]
- Pasteris N. G., Cadle A., Logie L. J., Porteous M. E., Schwartz C. E., et al. , 1994. Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 79: 669–678. [DOI] [PubMed] [Google Scholar]
- Pizette S., Niswander L., 2000. BMPs are required at two steps of limb chondrogenesis: formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev. Biol. 219: 237–249. [DOI] [PubMed] [Google Scholar]
- Retting K. N., Song B., Yoon B. S., Lyons K. M., 2009. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136: 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversade B., Kuroda H., Lee H., Mays A., De Robertis E. M., 2005. Depletion of Bmp2, Bmp4, Bmp7 and Spemann organizer signals induces massive brain formation in Xenopus embryos. Development 132: 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J., Huang Z., Lytle C., Traugh J. A., 2000. p21-activated protein kinase gamma-PAK is translocated and activated in response to hyperosmolarity. Implication of Cdc42 and phosphoinositide 3-kinase in a two-step mechanism for gamma-PAK activation. J. Biol. Chem. 275: 16933–16940. [DOI] [PubMed] [Google Scholar]
- Rosen V., Nove J., Song J. J., Thies R. S., Cox K., et al. , 1994. Responsiveness of clonal limb bud cell lines to bone morphogenetic protein 2 reveals a sequential relationship between cartilage and bone cell phenotypes. J. Bone Miner. Res. 9: 1759–1768. [DOI] [PubMed] [Google Scholar]
- Royal I., Lamarche-Vane N., Lamorte L., Kaibuchi K., Park M., 2000. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol. Biol. Cell 11: 1709–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt B., Ringe J., Haupl T., Notter M., Manz R., et al. , 2003. BMP2 initiates chondrogenic lineage development of adult human mesenchymal stem cells in high-density culture. Differentiation 71: 567–577. [DOI] [PubMed] [Google Scholar]
- Song S., Maru D. M., Ajani J. A., Chan C. H., Honjo S., et al. , 2013. Loss of TGF-beta adaptor beta2SP activates notch signaling and SOX9 expression in esophageal adenocarcinoma. Cancer Res. 73: 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnoli A., O’Rear L., Chandler R. L., Granero-Molto F., Mortlock D. P., et al. , 2007. TGF-beta signaling is essential for joint morphogenesis. J. Cell Biol. 177: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki W., Yamada A., Aizawa R., Suzuki D., Kassai H., et al. , 2015. Cdc42 is critical for cartilage development during endochondral ossification. Endocrinology 156: 314–322. [DOI] [PubMed] [Google Scholar]
- Tang C., Pan Y., Luo H., Xiong W., Zhu H., et al. , 2015. Hedgehog signaling stimulates the conversion of cholesterol to steroids. Cell. Signal. 27: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsamer A., Ortuno M. J., Ruiz S., Susperregui A. R., Osses N., et al. , 2008. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J. Biol. Chem. 283: 3816–3826. [DOI] [PubMed] [Google Scholar]
- Wada N., Kimura I., Tanaka H., Ide H., Nohno T., 1998. Glycosylphosphatidylinositol-anchored cell surface proteins regulate position-specific cell affinity in the limb bud. Dev. Biol. 202: 244–252. [DOI] [PubMed] [Google Scholar]
- Wang G., Beier F., 2005. Rac1/Cdc42 and RhoA GTPases antagonistically regulate chondrocyte proliferation, hypertrophy, and apoptosis. J. Bone Miner. Res. 20: 1022–1031. [DOI] [PubMed] [Google Scholar]
- Wang G., Woods A., Agoston H., Ulici V., Glogauer M., et al. , 2007. Genetic ablation of Rac1 in cartilage results in chondrodysplasia. Dev. Biol. 306: 612–623. [DOI] [PubMed] [Google Scholar]
- Whitehead I. P., Abe K., Gorski J. L., Der C. J., 1998. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol. Cell. Biol. 18: 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Tu X., Joeng K. S., Hilton M. J., Williams D. A., et al. , 2008. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell 133: 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang L., Geiger H., Cancelas J. A., Mo J., et al. , 2007. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc. Natl. Acad. Sci. USA 104: 5091–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H. Y., Chen L., Xu C., Wang J., Chen J., et al. , 2011. Inhibition of Rac activity alleviates lipopolysaccharide-induced acute pulmonary injury in mice. Biochim. Biophys. Acta 1810: 666–674. [DOI] [PubMed] [Google Scholar]
- Zha L., Hou N., Wang J., Yang G., Gao Y., et al. , 2008. Collagen1alpha1 promoter drives the expression of Cre recombinase in osteoblasts of transgenic mice. J. Genet. Genomics 35: 525–530. [DOI] [PubMed] [Google Scholar]
- Zhang X., Ziran N., Goater J. J., Schwarz E. M., Puzas J. E., et al. , 2004. Primary murine limb bud mesenchymal cells in long-term culture complete chondrocyte differentiation: TGF-beta delays hypertrophy and PGE2 inhibits terminal differentiation. Bone 34: 809–817. [DOI] [PubMed] [Google Scholar]
- Zou W., Greenblatt M. B., Shim J. H., Kant S., Zhai B., et al. , 2011. MLK3 regulates bone development downstream of the faciogenital dysplasia protein FGD1 in mice. J. Clin. Invest. 121: 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]